Abstract
The identification of the estrogen-related receptors (ERRs) as the first orphan nuclear receptors ignited a new era in molecular endocrinology, which led to the discovery of new ligand-dependent response systems. Although ERR subfamily members have yet to be associated with a natural ligand, the characterization of these orphan receptors has demonstrated that they occupy a strategic node in the transcriptional control of cellular energy metabolism. In particular, ERRs are required for the response to various environmental challenges that require high energy levels by the organism. As central regulators of energy homeostasis, ERRs may also be implicated in the etiology of metabolic disorders, such as type 2 diabetes and metabolic syndrome. Here, we review the recent evidence that further highlights the role of ERRs in metabolic control, particularly in liver and skeletal muscle, and their likely involvement in metabolic diseases. Consequently, we also explore the promises and pitfalls of ERRs as potential therapeutic targets.
Keywords: diabetes, energy metabolism, fatty liver, glucose, metabolic syndrome, mitochondria, nuclear receptors, oxidative phosphorylation, skeletal muscle
Introduction
The term “Big Bang” sometimes refers to the beginning of the universe; however, the “Bang” could perhaps be better referred to as a rapid period of inflation of the universe. What preceded the “Big Bang”, the true beginning of the universe, if indeed there was such a moment, remains a matter of intense debates, hypotheses, experimentations and soul searching. In the modern era of molecular investigations of nuclear hormone receptor actions and biological functions, of which the 30th anniversary is celebrated in this issue, the identification of retinoid X receptor (RXR) and its ability to heterodimerize with a multitude of nuclear receptors has been referred to as the ''RXR Big Bang'' in a recent review1. This is unquestionably a suitable assessment of the importance of this moment in nuclear receptor research because of its substantial impact on the subsequent discovery of entirely new physiological signaling pathways, and the corresponding inflation of the nuclear receptor universe. However, not only do we not know what preceded the “Big Bang” but the inflationary period that the universe went through immediately after the “Big Bang” may represent only one of many inflationary periods, which have given rise to the concept of multiple parallel universes. Although the molecular cloning of the classic steroid hormone receptors could be viewed as the pre-“Big Bang” era2,3,4, the subsequent identification of distinct classes of nuclear receptors, both RXR-dependent and -independent, could also be perceived as multiple universes; furthermore, each universe could significantly contribute to the inflation of nuclear receptor-regulated developmental, physiological and pathological processes. In this review, we will describe the parallel universes of the estrogen-related receptors (ERRs), which were identified as the first orphan nuclear receptors and therefore suggested the existence of previously unknown hormone response systems5. We will focus on the extensive role of ERRs in the transcriptional control of metabolic genes involved in the generation and utilization of cellular energy, with a particular emphasis on the impact of ERRs on gene regulation in skeletal muscles and the liver in relation to type 2 diabetes, metabolic syndrome and insulin resistance. Finally, the prospect and potential pitfalls of ERRs as therapeutic targets will be discussed.
Estrogen-related receptors: not estrogen receptors
The identification and mode of action of the ERR subfamily of nuclear receptors, which comprises three members referred to as ERRα (NR3B1), ERRβ (NR3B2), and ERRγ (NR3B3), has been extensively reviewed in the past6,7,8,9,10; thus, only a few key points will be discussed here. Although their name accurately reflects their molecular origin, the ERRs do not bind natural estrogens and they do not directly participate in classic estrogen signaling pathways or biological processes. In addition, recent functional genomic experiments have clearly demonstrated that ERRs and the estrogen receptor (ERα) display strict binding site specificities11; in contrast to ERα, the addition of a hormone/ligand is not required to observe strong and widespread ERR binding to the genome. The ERRs also have a marked preference for the recognition of the sequence TCAAGGTCA, which is referred to as an estrogen-related response element (ERRE). Furthermore, although the ERRE is similar to the half-sites contained within the estrogen response element (ERE), a relatively small overlap exists between the gene set bound by ERα and ERRα11. When direct overlap in binding to DNA does occur, it occurs through an ERRE embedded within an ERE11, and when in place, this functional relationship can be used to fine tune the expression of a small subset of specific genes11,12,13,14. Another functional distinction between the ERRs and ERα is that, in general, the ERRs exhibit a preference for binding in close proximity to the promoters of genes, whereas ERα typically recognizes enhancers that are often located very distant from transcriptional start sites8,15,16,17,18. This functional characteristic of the ERRs provides a vast advantage to the ERRs that facilitates the identification and characterization of their target genes. Finally, the ERRs display significant constitutive transcriptional activity that is dependent on the presence of coactivator proteins, most prominently peroxisome-proliferator activated receptor γ coactivator-1α and peroxisome-proliferator activated receptor γ coactivator-1β (PGC-1α and PGC-1β), which regulate metabolic pathways and exhibit high specificity for the ERRS19,20,21. As such, these coactivators are often considered to function as protein ligands for these receptors. The presence of functional ERRs is required for maximal mitochondrial biogenesis and activity induction by PGC-1α and PGC-1β22,23,24.
The identification of ERR binding sites in the genome has demonstrated that they are preferentially associated with metabolic genes involved in processes such as glucose and glutamine metabolism, mitochondrial activity, lipid handling, and energy sensing8,25. Consequently, the ERRs are now considered major transcriptional regulators of energy metabolism, particularly in the context of high-energy demands by cells, tissues or the whole organism in response to physiological and/or environmental challenges7,10. In the next sections, ERR gene regulation will be dissected to understand their roles in energy metabolism pathways. The discussion will be limited herein to ERRα and ERRγ because the expression of ERRβ is higher in embryonic tissues; therefore, less is known regarding its role in adult physiology.
ERRα, ERRγ, and cellular energy metabolism
Carbohydrate and energy metabolism
Glycolysis is a process that requires several enzymes to convert glucose into pyruvate, the latter of which is used to feed the mitochondria or is anaerobically converted to lactate. This process produces ATP, but it is also required for the synthesis of several building blocks of the cell, thereby comprising nucleotides, lipids, and amino acids. Most genes involved in glycolysis have been identified as bound and regulated by ERRα and ERRγ, including genes that encode key enzymes, such as phosphofructokinase, hexokinase 2 (HK2), glyceraldehyde dehydrogenase (GAPDH), and enolase 1 (ENO1)16,26,27. Moreover, ERRα also binds to glucose transporter family members (Glut1, Glut2, Glut4, and Glut12)18,28, and shRNA-mediated knock down of both ERRα and ERRγ decreases glucose uptake in breast cancer cells26. Based on their regulation of most genes in the glycolysis pathway, the ERRs are key components of cellular glucose metabolism.
Another crucial step during glucose utilization is the conversion of pyruvate to acetyl coenzyme A (CoA), which is catalyzed by the pyruvate dehydrogenase complex (PDC). The pyruvate dehydrogenase kinase (PDK) can phosphorylate PDC to inhibit its activity, which results in a switch from glucose oxidation to fatty acid oxidation (FAO). The key role of PDK enzymes, including PDK isoenzyme 4 (PDK4), in energy homeostasis has been demonstrated in PDK4-null mice: these mice exhibit lower blood glucose levels as a result of elevated glucose utilization29. Importantly, ERRα and ERRγ have been demonstrated to positively control PDK4 transcription in muscle and hepatic cells30,31,32,33. Moreover, PDK4 upregulation by the PGC-1α/ERRα axis downregulated glucose oxidation rates in C2C12 myotubes30. In addition, ERRα also binds to Pdk1 and Pdk2 in mouse livers18. Therefore, both ERRs can regulate the switch between glucose and lipid usage by mitochondria.
When oxygen is lacking or when cells are rapidly proliferating, glucose can be converted in lactate (anaerobic) instead of processed by aerobic respiration in the mitochondria. This reversible process is catalyzed by lactate dehydrogenase (LDH), and several genes that encode LDHs are targets of and regulated by ERRα and ERRγ in mouse and human cells, including LDHB and LDHD16,27,34,35. Importantly, ERRγ has been shown to be a negative regulator of anaerobic glycolysis in breast cancer cells, and the inhibition of ERRγ by miR-378* leads to an increase in lactate production and a decrease in aerobic respiration27. In addition, inhibition using an siRNA against ERRα and ERRγ leads to a decrease and increase in LDHA expression, respectively27. The biological impact of LDH regulation by the ERRs can also be supported by the observation that siRNA-mediated knock down of ERRα indicates this nuclear receptor is indispensable for the switch from oxidative to glycolytic metabolism16. The recent demonstration that all three ERRs could interact with hypoxia-inducible factor (HIF) and positively coordinate transcriptional activation in hypoxic conditions via the modulation of angiogenic and glycolytic genes is another demonstration of their crucial roles in glucose handling36,37,38. One of the hallmarks of energy metabolism reprogramming following HIF pathway activation is the induction of anaerobic respiration39. ERRγ has been shown to be upregulated and to increase several key hypoxic-response genes, such PDK4, LDHA, and VEGF, under hypoxic conditions36. These data demonstrate that ERRs are involved in anaerobic glycolysis and in the response to hypoxic conditions. However, the exact role of each isoform in the differential regulation of genes involved in this process remains to be investigated.
Together, these results demonstrate ERRs can regulate glucose metabolism at several key steps, including glucose uptake, glucose to pyruvate conversion, pyruvate entry in the TCA cycle and anaerobic respiration (Figure 1). For some genes, such as LDHA, an opposing effect on transcriptional regulation has been observed, and the global portrait of how each ERR isoform orchestrates glucose metabolism in the cell will require further investigation.
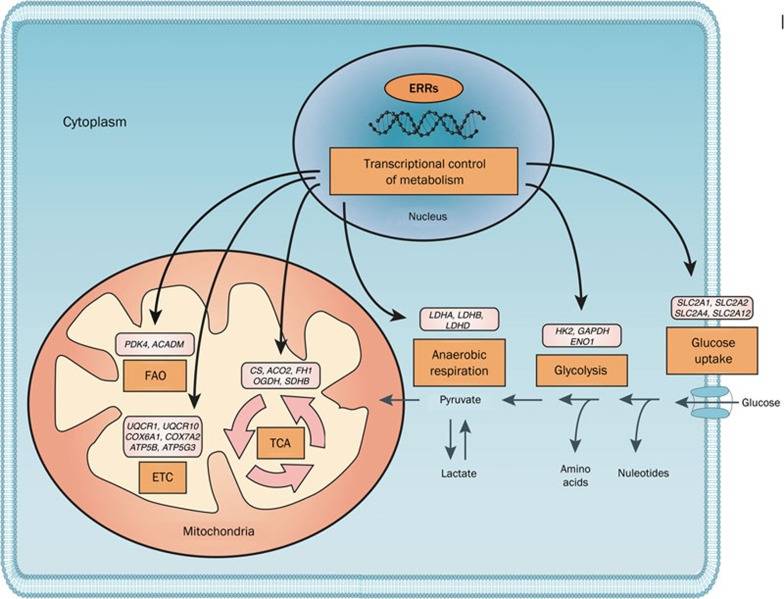
Figure 1.
ERRs are key transcriptional factors that regulate cellular energy metabolism. Through the control of vast transcriptional networks, the ERRs are master regulators of energy metabolism in the cell. Their target genes include several glucose transporters (SLC2A1, SLC2A2, SLC2A4, and SLC2A12, also known as GLUT1, GLUT2, GLUT4, and GLUT12, respectively), most genes in the glycolysis pathway, and several genes that encode different LDH isoforms, which indicate the ERRs are key components in cellular glucose metabolism. By regulating the levels of key gatekeepers (such as PDK4), they also control the switch between glucose and fatty acid oxidation (FAO) by the mitochondria. ERRs are also essential for mitochondrial activity by targeting most genes in the TCA cycle and the electron transport chain (ETC). Recent results indicate that they also play a role in other metabolic functions of the cell, including amino acid and nucleotide synthesis and metabolism, which remain to be fully elucidated. Several examples of the regulated genes are provided for each function, which are based on microarray and ChIP-sequencing analysis in human cell lines and mouse models.
Lipid synthesis and fatty acid oxidation
Acadm encodes the medium-chain acyl-coenzyme A dehydrogenase (MCAD), which is the initial step in mitochondrial fatty acid β-oxidation (FAO), and it is considered one of the gatekeepers that controls the FAO rate in the cell. Therefore, the identification of Acadm as the first target gene of ERRα was a good hint to support the role of this nuclear receptor in the modulation of energy metabolism40,41. ERRα is highly expressed in tissues involved in lipid metabolism and energy homeostasis, such as white and brown adipose tissues, and tissues that require high oxidative capacity, such as heart and skeletal muscles5,42.
Genetic ablation of ERRα leads to viable and fertile mice with no apparent defects compared with wild-type mice, with the exception of reduced body weight and peripheral fat deposits, as well as a lower capacity for lipid absorption by the intestine of ERRα-null pups43,44. Unexpectedly, these mice were resistant to high-fat diet-induced obesity as a consequence of the altered expression of genes involved in lipid and energy metabolism in white adipose tissue, such as fatty acid synthase (FAS, encoded by Fasn), fatty acid coenzyme A ligase long-chain 5 (Facl5), Acadm, and several other genes associated with lipid synthesis and mitochondrial activity. These changes in gene expression lead to a decrease in triglyceride synthesis capacity in ERRα-null mice adipocytes. Furthermore, these mice exhibit an important mass decrease in fat deposits, with up to a 50%–60% decrease in inguinal, epididymal and peritoneal fat pads. These observations are consistent with the results obtained in cell culture in which ERRα was up-regulated in adipocytes during differentiation and promoted lipid accumulation under an adipogenic condition40,41,45,46,47. Notably, Acadm and Fasn are directly regulated by ERRα in adipocytes and during differentiation. In differentiated adipocytes, ERRα is a positive regulator of mitochondrial biogenesis and triglyceride production46,47.
Less is known regarding ERRγ in lipid handling; however, because it binds to most ERRα target genes, it most likely also regulates the transcription of genes involved in lipid synthesis and metabolism. Using an siRNA mediated knock-down of ERRγ in vitro and studying mice treated with a high fat diet, ERRγ was shown to be positively regulated in adipogenic conditions and to act positively for the induction of adipogenic genes and lipid accumulation48. Moreover, ERRγ is essential to activate the FAO switch in the neonatal heart, which is discussed in more detail later in this review49. Therefore, both ERR isoforms positively regulate lipid synthesis, which can be further used to produce energy through FAO in the mitochondria.
Mitochondrial activity and oxidative phosphorylation
The ERRα and ERRγ targets include several genes related to mitochondrial activity and biogenesis16,17,18,27,28,49,50,51,52,53,54,55. The two receptors bind in the vicinity of the transcriptional start site of most genes that encode the enzymes involved in the tricarboxylic acid (TCA) cycle, including: Aco2, Idh3a, Idh3b, Sdha, Sdhb, Sdhc, Sdhd, Ogdh, Cs, and Fh1. Recent chromatin-immunoprecipitation coupled to a deep sequencing (ChIP-seq) analysis of ERRα binding in mouse liver17 demonstrated that this nuclear receptor binds the regulatory region of more than one hundred genes involved in the mitochondrial electron transport chain. Some of these genes include members of the NADH dehydrogenase complex (over 30 genes, including several Ndufa and Ndufb genes), ubiquinol-cytochrome c reductases (Uqcr genes) and several cytochrome c oxidase subunits, and more than 100 peaks were detected in the proximity of 56 genes of the ATPase superfamily. The biological consequences of ERR isoforms binding to these mitochondrial genes are evident because when their activity is impaired, decreases in mitochondrial integrity, activity and ATP production have been demonstrated16,17,26,27,50,53,54,55,56. Conversely, enhanced ERR expression leads to an increase in mitochondrial biogenesis and activity, which, in turn, stimulates respiration capacity52,56.
In addition to ATP synthesis and aerobic respiration, the target genes of ERRs include genes involved in all aspects of mitochondrial functions, such as biosynthetic functions25. ERR transcriptional control of these pathways correlates with the finding that ERRα is a positive regulator of glutamine metabolism in breast cancer cells24. Overall, ERRα and ERRγ are master regulators of mitochondrial activity, including the positive regulation of aerobic respiration and ATP synthesis. Their roles in other mitochondrial functions, such as amino acid and nucleotide synthesis, remain to be investigated.
ERRs as key players in adaptive energy metabolism
The phenotypes of the three ERR-null mice are markedly different and range from embryonic lethality for ERRβ-null mice, death shortly after birth for ERRγ-null mice, and viability for ERRα-null mice43,49,57. The complete knockdown of an individual ERR isoform does not abolish mitochondrial biogenesis or activity. Therefore, each ERR isoform is not per se essential for mitochondria functions, which could be explained, in part, by a compensatory mechanism through increased expression of other ERR subfamily members. For example, ERRγ is increased in ERRα-null tissues, such as the liver17. The results, primarily obtained from genetically engineered mice, have shown that they are required in a tissue-dependent manner for adaptive energy metabolism. Thus, in response to various environmental insults or stimuli, tissue-specific high-demanding energy processes are activated in an ERR-dependent manner to address a specific physiological challenge.
In various conditions where high energy levels are required, in general, ERRα-null mice display a deficient biological response compared with their wild-type littermates. The complete ablation of ERRα leads to defects in ATP production and transport in the mitochondria of the heart, which develops signatures of heart failure after ventricular pressure overload compared with wild-type mice28,58. In rodents, adaptive thermogenesis is a process that relies on a high mitochondrial activity in brown adipose tissues to produce heat after cold exposure, and this process is defective in ERRα-null mice. These mice are unable to adapt to cold because of dysfunctional mitochondria and decreased mitochondrial mass, which thereby compromises survival54. ERRα has also been implicated in muscle cell energy metabolism and is essential in skeletal muscle cells for recovery during regeneration through sustaining mitochondrial size and activity35,59,60. Interferon-γ-induced activation of ROS production by mitochondria in macrophages for the clearance of Listeria monocytogenes was also decreased in ERRα-null mice; this effect was again because of decreased expression of mitochondrial genes in KO mice53. ERRα has also been shown to be a metabolic regulator in T-cell activation and differentiation because the inhibition of this nuclear receptor led to decreased T-cell activation and proliferation51. Finally, following a xenobiotic insult, ERRα-null hepatocytes have been shown to have a defective apoptotic program that favored necrosis and inflammation, which thereby promoted hepatocarcinogenesis in diethylnitrosamine-induced hepatocellular carcinoma55. ATP production was dependent on the presence of ERRα, and genetic disruption of Esrra decreased mitochondrial activity, which blocked the regular stress response-induced apoptotic program. In summary, ERRα is required in a tissue-specific manner to promote the acute energetic response following various metabolic cues and environmental changes, particularly to control mitochondrial biogenesis and activity.
In contrast to ERRα, ERRγ is crucial for mice to survive after birth, which complicates the study of its role in energy metabolism. However, in a manner similar to ERRα, ERRγ is required to reprogram energy producing pathways in response to specific challenges. For example, the transition from cardiac carbohydrate-based fetal metabolism to lipid-predominant adult metabolism is blunted in ERRγ-null mice, which reduces mitochondrial activity and induces lactatemia, thereby contributing to the poor health of newborns and death shortly after birth 49. ERRγ overexpression in mouse skeletal muscles results in a subsequent coordinate increase in mitochondrial function, in part, through increased TCA cycle activity, larger mitochondria and improved oxidative capacity52,56. In these experiments, a gene set analysis identified an increase in the genes associated with the mitochondrial electron transport chain, TCA cycle, and FAO56. What is even more impressive is that ERRγ expression in skeletal muscle was sufficient to increase exercise capacity, activate mitochondrial activity, and force muscle fibers to switch from type II glycolytic to type I oxidative fibers. Conversely, ERRγ+/− mice exhibited lower exercise capacity compared with their wild-type littermates. ERRγ has also been associated with several roles during pregnancy, particularly through trophoblast-related gene regulation. In particular, ERRγ was necessary for the transcription of several voltage-gated K+ channels (Kcnq1, Kcne1, and Kcne3), as well as for the induction of hormone secretion and angiogenesis61 and the positive regulation of mitochondrial biogenesis62.
ERRα, ERRγ, and metabolic disorders
The major metabolic disorders include type 2 diabetes and metabolic syndrome, which are closely linked by their clinical features. Type 2 diabetes is characterized by insulin resistance, hyperglycemia and hyperinsulinemia. Metabolic syndrome includes central obesity, dyslipidemia, hypertension, hyperglycemia and/or type II diabetes63,64. Obesity can also lead to insulin resistance and is recognized as a major risk factor for type 2 diabetes. The transition to insulin-resistance is, in part, a result of an insensitivity to insulin in the skeletal muscle and liver; these two distinct metabolic tissues have been associated with several metabolic alterations identified in type 2 diabetic patients65. Skeletal muscle exhibits a defect in mitochondrial activity, whereas the liver exhibits an increase in the gluconeogenesis rate and glucose production, which increases blood glucose levels. As previously discussed, ERRα and ERRγ are key transcriptional regulators of these processes in these tissues. The next sections will discuss their roles in global energy homeostasis with a special emphasis on the contributions of ERRα and ERRγ in skeletal muscle and liver.
ERRα, ERRγ, and skeletal muscle
A decrease in the expression of genes linked to oxidative phosphorylation (OXPHOS) occurs early in the development of insulin resistance in human diabetic muscle66. ERRα and ERRγ are both expressed in skeletal muscles and positively associated with mitochondrial biogenesis and activity. In addition, ERR activity is associated with increased respiration and mitochondrial activity in skeletal muscles, which therefore suggests the potential for ERR-based interventions to reverse this molecular hallmark of insulin-resistant muscles.
ERRα cooperates with PGC-1α in muscle cells to activate the transcription of mitochondrial genes, such as ATP-synthase 5O (Atp5o) and cytochrome c oxidase 5b (Cox5b), which are necessary for maximal respiration induction by PGC-1α67. The inhibition of ERRα activity induced a type 2 diabetes-like phenotype in skeletal muscle cells, which indicates pharmacological activation of ERRα might have a beneficial effect in humans in this tissue. Interestingly, the expression of transcription factors and coactivators known to positively regulate these genes, such as the coactivators PGC-1α and PGC-1β, is also decreased in insulin-resistant skeletal muscles66,68. In addition to a positive regulation of mitochondrial genes, ERRα binds to several genes associated with the insulin pathway, including several glucose transporters and the insulin receptor (Insr)18,28. Moreover, ERRα-null mice exhibit time-dependent hypoglycemia and hypoinsulinemia that appears to be the result of increased glucose uptake because of improved insulin sensitivity18. Another way that ERRα may affect the insulin pathway is through the transcriptional regulation of several genes in the AMP-kinase (AMPK) pathway, including some of its subunits (Prkag2), its upstream regulator Lkb1, and some if its downstream targets (Acacb)28. AMPK is a master regulator of energy homeostasis in the cell; it is activated in response to a decrease in ATP/AMP ratios and is a major downstream target of the most prescribed drug for patients with type 2 diabetes, metformin69. Of note, AMPK pathway activity was decreased in the hearts of ERRα-null compared with wild-type mice based on the Acetyl-Coa-Carboxylase 2 (ACC2, encoded by Acacb) phosphorylation status18. ACC2 is the predominant ACC form in skeletal muscle and regulates both fatty acid synthesis and oxidation. Acacb-null mice exhibit enhanced FAO and enhanced insulin sensitivity70,71. Therefore, ERRα regulation of key genes in the insulin and AMPK pathways and its influence on the activities of these pathways are likely factors that affect glucose handling and insulin sensitivity by ERRα.
In the skeletal muscle, ERRγ is highly expressed in oxidative type 1 fibers, and forced expression of ERRγ in glycolytic type 2 fibers triggers aerobic transformation, vascularization and mitochondrial biogenesis in the absence of exercise52. These effects were not dependent on PGC-1α induction, but were linked to AMPK. In these transgenic mice, the AMPK pathway was constitutively activated, which suggests a convergence between ERRγ and AMPK to direct oxidative metabolism in type 1 fibers in skeletal muscle. Moreover, treatment with the dual ERRβ/γ agonist GSK4716 led to an increase in the expression of genes involved in mitochondrial activity56. This finding indicates that pharmacological molecules that target ERRs may help restore insulin sensitivity through the restoration of mitochondrial mass and activity. Although the full relationship between ERRs, the transcriptional modulation of insulin pathway target genes, and the AMPK pathway remain to be defined, it appears clear that there is a convergence between these major pathways of energy metabolism. In summary, both ERRα and ERRγ positively regulate genes associated with mitochondrial functions in skeletal muscle. Moreover, the transcriptional controls of other key pathways that mediate energy homeostasis, including the insulin and AMPK pathways, may also have a beneficial impact on insulin response in this tissue.
ERRα, ERRγ, and glucose metabolism in liver
Type 2 diabetes is also characterized by the impairment in insulin's ability to inhibit gluconeogenesis in the liver, the main organ in which this pathway is active in vertebrates. The end product of gluconeogenesis is the production of glucose to maintain constant blood levels, particularly during a fasting period. The inability of insulin to shut down this pathway will result in increased glucose production by the liver and higher blood glucose levels. Understanding how gluconeogenesis is controlled in the liver is therefore critical to treat high blood glucose levels and related metabolic disorders.
In contrast to its positive role on OXPHOS, ERRα acts as a repressor of gluconeogenesis72. Through the repression of Pepck expression, a key enzyme in gluconeogenesis, ERRα activity could contribute to the control of high glucose levels in the fed state. ERRα represses the gluconeogenic program through direct binding to a Pepck enhancer and, in contrast to its activity on most of its target genes, antagonizes the stimulatory effect of PGC-1α. Paradoxically, a negative regulation of gluconeogenesis should lead to an increase in blood glucose ERRα-null mice; however, these mice do not exhibit increased plasma glucose levels in the fed state, even though several key gluconeogenic genes are up-regulated in the livers of these mice18,72. In fact, ERRα-null mice exhibit significantly lower glucose levels at Zeitgeber times (ZT) 12 and 0/24, which represent the start and end of the dark cycle, respectively18. This observation parallels the observation that hepatic glucose production relies on the metabolic state of an organism in a circadian manner because nutrient blood levels display diurnal variation. Consistent with the results obtained using ERRα-null mice as a model, pharmacological inhibition of ERRα in a diet-induced model of obesity improved insulin sensitivity and glucose tolerance and reduced circulating glucose, free fatty acids and triglyceride levels73. Consistent with the circadian cycle role in energy homeostasis, ERRα levels are modulated in a circadian manner in the liver and are also induced by fasting18,74.
Unlike ERRα, ERRγ has been shown to activate key gluconeogenic genes, such as Pepck and G6pc, in response to fasting conditions75,76. ERRγ overexpression in the liver also induced gluconeogenic gene expression, increased blood glucose levels and caused a delayed response to glucose tolerance tests, without significantly changing insulin, triglyceride or fatty acid blood levels76. These results indicate that in patients with diabetes, increased hepatic ERRγ activity could play a role in increasing glucose serum levels. Consistent with these observations, the knock down of endogenous ERRγ in the liver decreased hepatic gluconeogenic gene expression and fasting blood glucose levels in db/db mice76. Moreover, db/db diabetic mice treated with GSK5182, a specific inhibitor of ERRγ, exhibited lower hyperglycemia, hepatic fat accumulation, food intake and body weight75,76. Therefore, the regulation of the ERRγ-dependent gluconeogenic program in the liver could be considered a target to control hepatic insulin resistance.
Although ERRα and ERRγ are positive regulators of mitochondrial activity, they appear to antagonize each other in the regulation of gluconeogenesis. Nonetheless, ERRα inhibition or down-regulation is associated with an improvement in glucose handling most likely through the decreased expression of gluconeogenesis, decreased PDK4 expression, transcriptional regulation of genes in the insulin signaling pathway, and control of mitochondrial activity. Moreover, ERRα-null mice exhibit lower blood glucose and increased insulin sensitivity. In a similar manner, ERRγ inhibition in the liver leads to an improvement in diabetic symptoms and a decrease in gluconeogenic gene expression. It is therefore surprising to observe similar beneficial effects on the diabetic condition via the inhibition of ERRα or ERRγ because they act in an antagonistic manner on gluconeogenic genes in the liver. However, their positive regulatory activity on other pathways, including mitochondrial biogenesis, FAO and glucose handling, may be dominant in vivo, which could explain why the inhibition of either ERRα or ERRγ has similar global effects. ERR inhibition would act in a manner similar to metformin and reduce mitochondrial activity, which would lead to increased AMPK pathway activity, with all its beneficial effects on metabolic disorders.
ERRα, mTOR, and lipid metabolism in liver
Non-alcoholic fatty liver disease (NAFLD) is the most common liver disorder in Western countries and is now regarded as the liver manifestation of the metabolic syndrome64,77. NAFLD is associated with impaired insulin response and type 2 diabetes, which leads to increased gluconeogenesis, and more than 90% of obese patients with type 2 diabetes have NAFLD77,78. As previously discussed, both ERRα and ERRγ are opposite regulators of hepatic gluconeogenesis. However, NAFLD is also a disease associated with increased lipogenesis in the liver and impaired hepatic FAO. The mechanistic target of rapamycin (mTOR), a phosphatidylinositol 3 kinase (PI3K)-related serine/threonine kinase, has been demonstrated to play an important role in the control of energy metabolism, particularly in lipid biosynthesis79. mTOR inhibition by pharmacological tools, such as rapamycin, has been reported to induce side effects, such as hyperlipidemia, hypercholesterolemia, and the induction of hepatic gluconeogenesis, as well as NAFLD in murine models17,80,81,82. Remarkably, mTOR inhibition in ERRα-null mice or mice exposed to an ERRα inhibitor led to an exacerbated rapamycin-induced NAFDL17. This finding might be considered surprising because these mice are resistant to high-fat diet-induced obesity43. However, ERRα-null mice have a blunted TCA cycle after rapamycin treatment, with a down-regulation of several genes in this pathway (Ogdh, Aco2, Idh3a, and Sdhc), which leads to an accumulation of citrate and other TCA cycle intermediates. The observed rapamycin-induced change in ERRα-dependent activity has been related to a rapid increase in ERRα degradation following mTOR inhibition. The impaired TCA cycle activity in mice with reduced or null ERRα activity while treated with rapamycin suggests that these mice not only have a decreased capacity to oxidize lipids but the accumulation of citrate is reoriented toward lipid biosynthesis. Rapamycin induces a diabetic-like state in which excess energy is converted to fat. A significant number of patients treated with rapamycin and its analogs have suffered from a diabetes-like syndrome, which is associated with insulin resistance and hepatic hyperlipidemia83. Therefore, considering the molecular and genetic interactions observed between the ERRα and mTOR regulatory pathways in the liver, this functional crosstalk might play a more global role in the etiology of other metabolic disorders, such as diabetes and metabolic syndrome.
ERRs as pharmacological targets in metabolic disorders
ERRα is an interesting target for the treatment of type 2 diabetes because it can increase muscle mitochondrial oxidative capacity and suppress hepatic glucose production50,72. Paradoxically, one could have assumed that increased ERRα activity would be associated with the improvement of metabolic disorders; however, the results obtained in murine models indicate the opposite effect. In a polygenic mouse model of diet-induced obesity characterized by hypertriglyceridemia and hyperinsulinemia, treatment with the ERRα inverse agonist C29 decreased body fat without changing lean mass73. Similar results were also observed in ERRα-null mice43. Treatment with C29 also decreased insulin levels by approximately 66% without changing fed glucose levels, which indicates an improvement in peripheral insulin sensitivity. Circulating triglyceride levels were also decreased by half. In microarrays performed in the skeletal muscles and hepatic tissues of mice treated with C29, the liver was most responsive to ERRα pharmacological inhibition73. Despite the phenotypes that affect the physiology of multiple tissues observed ERRα-null mice11,18,28,51,53,54,55,58,84, no apparent toxicities or severe adverse effects were reported in mice treated with C29. In various murine models of diet-induced obesity and type 2 diabetes, C29 induced significant improvements in insulin sensitivity, glucose tolerance, and reduced triglyceride levels73. Therefore, the pharmacological inhibition of ERRα had a similar beneficial impact on glucose handling, diabetes and obesity to that of the genetic loss of ERRα18,43,73. Furthermore, ERRα inhibition may also facilitate the fight against obesity because ERRα-null mice are protected against high-fat diet-induced obesity43. Moreover, the observation that the liver was the main target for ERRα inhibition by pharmacological activation indicates that pharmacological compounds may have a tissue-specific modulation of this nuclear receptor's activity. These molecules, if developed, would be similar to the well-known selective estrogen-receptor modulators (SERMs), which display antagonist activity against ERα in the breast, but agonist activity in peripheral tissues, such as the bones and uterus85.
Inactivation of ERRγ by GSK5182 improves insulin signaling via the inhibition of LIPIN1-induced protein kinase C activation86. Moreover, the same inhibitor decreased hyperglycemia in db/db mice and gluconeogenic genes, such as Pck1, G6pc and Pdk4, in the liver75. GSK5182 completely restored blood glucose levels and gluconeogenic gene expression and also improved liver toxicity caused by diabetic progression. Similar to the effects of C29 on ERRα in a tissue-specific manner, the pharmacological inhibition of ERRγ only decreased the expression of its target genes in the liver without altering gene expression in skeletal muscle75.
Overall, the hepatic inhibition of both ERRs by pharmacological tools has beneficial effects on insulin sensitivity and glucose handling in mouse models. Moreover, at least in these models, the effects of these therapeutic tools appear to primarily target the liver and not skeletal muscles. This is a crucial requirement for targeted ERR disruption. Thus, considering the beneficial effects of the ERRs in skeletal muscle on glucose uptake and handling, hepatic inhibition of ERRs combined with their activation in skeletal muscle would be an interesting avenue for the design of therapeutic tools (Figure 2).
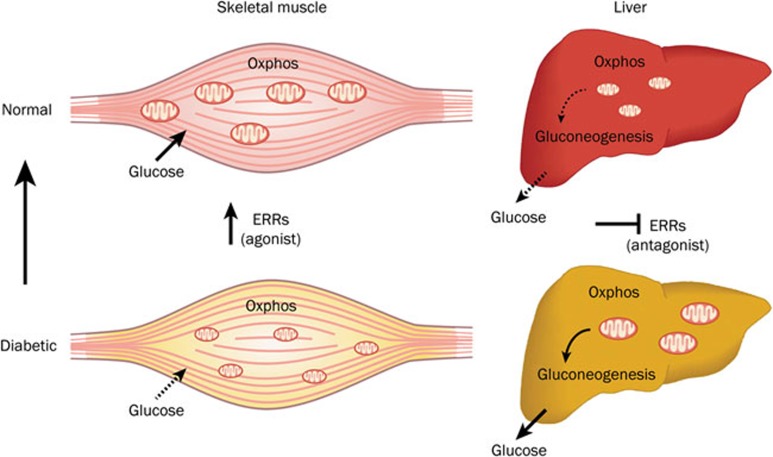
Figure 2.
ERRs as pharmacological targets to manage type 2 diabetes. In the skeletal muscles of patients with type 2 diabetes, mitochondrial activity is deficient and cells are less responsive to insulin regarding glucose uptake and usage. Increasing ERR activity using a compound with agonist-like activity would result in increased glucose uptake and usage, as well as increased mitochondrial activity, which would therefore ameliorate insulin signaling in skeletal muscle. In the livers of patients with type 2 diabetes, elevated energy production sustains an increased gluconeogenesis that results in higher blood glucose levels, which leads to hyperglycemia. ERR inhibition would have the benefit of blocking mitochondrial activity, which would therefore reduce the ATP production that feeds gluconeogenesis and restore normal blood glucose levels. This mechanism is similar to metformin, the most prescribed drug for the treatment of type 2 diabetes, which inhibits hepatic mitochondrial activity and gluconeogenesis. In addition, the inhibition of ERRγ activity would also have the advantage of directly blocking gluconeogenesis.
Hepatic insulin resistance is associated with increased mitochondrial respiration and performance87,88. Patients with type 2 diabetes have an up-regulation of OXPHOS genes in the liver, including a significant positive correlation between ERRα mRNA and OXPHOS89. This finding is in contrast to the decreased mitochondrial gene expression in diabetic skeletal muscles65,66,67,90. It was hypothesized that the up-regulation of OXPHOS genes in type 2 diabetic livers would result in increased gluconeogenesis and therefore contribute to hyperglycemia89. This finding is consistent with the reduction in glucose production by the liver following metformin treatment in diabetic patients, a drug that targets mitochondrial activity69. It therefore appears that to ameliorate insulin sensitivity and metabolic disorders in the liver, decreased mitochondrial activity is desired as opposed to increased activity in the skeletal muscles. In these conditions, hepatic inhibition of ERR activity would also be beneficial to inhibit hepatic mitochondrial activity. This concept is consistent with the observation that the liver is also the main target for metformin action91.
The modulation of ERRα and ERRγ activities demonstrates promise for the control and recovery of metabolic disorders, such as type 2 diabetes, obesity and metabolic syndrome. However, the use of pharmacological modulators must be carefully performed because it could also lead to adverse side effects. Despite the positive results of C29 and GSK5182 in mouse models, ERR inhibition in vivo must still be approached with caution. As previously discussed, several potential side effects of ERR downregulation can be thought of, including the induction of NAFLD17, the sensitivity to hepatic carcinogenesis by xenobiotics55, and heart failure58 as a result of FAO blockade in cardiac mitochondria.
Perspectives and conclusions
ERRα and ERRγ primarily bind to the same target genes as homo- or heterodimers28,92. The functional consequence of this interaction is not yet understood because ERRα is not required for ERRγ activity and vice-versa; however, heterodimers have been reported to have lower activity in vitro. It also appears that they are able to counterbalance for the absence of the other because in ERRα-null cells, ERRγ can be up-regulated and vice versa60,93. Nevertheless, it appears that in specific contexts and for specific genes, the two ERR isoforms do not have the same role, such as for the transcriptional control of the key gluconeogenic gene Pepck. This duality of the ERRs has also been studied in the context of breast cancer, where ERRα and ERRγ expression is inversely correlated, and the former is associated with a more aggressive disease, whereas the latter is considered a tumor suppressor14,94,95,96. In the cellular environment, both nuclear receptors appear to act in an opposite manner on cancer cell metabolism because ERRα contributes to the suppression of oxidative metabolism, which is stimulated by ERRγ27. Additional work must be completed in this area of research, which will certainly highlight their precise molecular mechanisms of action, as well as the interesting and diverging biological functions of ERRα and ERRγ.
It is clear that the ERRs control vast metabolic gene networks and are key regulators of energy metabolism, particularly in response to various environmental challenges or biological stresses. However, data from multiple genome-wide binding site location analyses, which have yet to be completely investigated, have indicated that the ERRs might also be major orchestrators of other biosynthetic pathways and biological processes; furthermore, this regulation is likely to be cell- and tissue-specific. Therefore, research will continue to explore the multiple parallel ERR universes to fully understand the role of these nuclear receptors in physiology and disease and evaluate their true potential as therapeutic targets.
Acknowledgments
The authors thank all members of the laboratory, particularly Drs R Butler and M Vernier for critical reading of the manuscript, C Flageole for fruitful discussions and S Labrecque for figure design. EAW is recipient of postdoctoral fellowships from the Canadian Institutes of Health Research (CIHR), the Fonds de recherche du Québec-Santé (FRQS) and the McGill Integrated Cancer Research Training Program (MICRTP). This work was supported by grants to VG by the CIHR (MOP-111144 and 125885) and the Terry Fox Research Institute (TFF-116128).
References
- Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the Big Bang. Cell 2014; 157: 255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornet JM, Argos P, et al. Human oestrogen receptor cDNA: sequence, expression and homology to v-erbA. Nature (London) 1986; 320: 134–39. [DOI] [PubMed] [Google Scholar]
- Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. Sequence and expression of human estrogen receptor complementary DNA. Science 1986; 231: 1150–54. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, et al. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature (London) 1985; 318: 635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature 1988; 331: 91–94. [DOI] [PubMed] [Google Scholar]
- Giguère V. To ERR in the estrogen pathway. Trends Endocrinol Metab 2002; 13: 220–25. [DOI] [PubMed] [Google Scholar]
- Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev 2008; 29: 677–96. [DOI] [PubMed] [Google Scholar]
- Deblois G, Giguère V. Functional and physiological genomics of estrogen-related receptors (ERRs) in health and disease. Biochim Biophys Acta 2011; 1812: 1032–40. [DOI] [PubMed] [Google Scholar]
- Tremblay AM, Giguère V. The NR3B subgroup: an ovERRview. Nucl Recept Signal 2007; 5: e009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA, Kralli A. ERRα: a metabolic function for the oldest orphan. Trends Endocrinol Metab 2008; 19: 269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblois G, Hall JA, Perry MC, Laganière J, Ghahremani M, Park M, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor α as a determinant of breast cancer heterogeneity. Cancer Res 2009; 69: 6149–57. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol 2006; 20: 1707–14. [DOI] [PubMed] [Google Scholar]
- Deblois G, Chahrour G, Perry MC, Sylvain-Drolet G, Muller WJ, Giguère V. Transcriptional control of the ERBB2 amplicon by ERRα and PGC-1β promotes mammary gland tumorigenesis. Cancer Res 2010; 70: 10277–87. [DOI] [PubMed] [Google Scholar]
- Deblois G, Giguere V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer 2013; 13: 27–36. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009; 462: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, et al. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev 2010; 24: 537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, et al. Molecular and genetic crosstalks between mTOR and ERRα are key determinants of rapamycin-induced non-alcoholic fatty liver. Cell Metab 2013; 17: 586–98. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Levasseur MP, Pham NHH, Eichner LJ, Wilson BJ, Charest-Marcotte A, et al. Genomic convergence among ERRα, Prox1, and Bmal1 in the control of metabolic clock outputs. PLoS Genet 2011; 7: e1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N, et al. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A 2003; 100: 12378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S, Dwyer MA, McDonnell DP. Definition of the molecular basis for estrogen receptor-related receptor-α-cofactor interactions. Mol Endocrinol 2007; 21: 62–76. [DOI] [PubMed] [Google Scholar]
- Gaillard S, Grasfeder LL, Haeffele CL, Lobenhofer EK, Chu TM, Wolfinger R, et al. Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol Cell 2006; 24: 797–803. [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, et al. The estrogen-related receptor alpha (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A 2004; 101: 6472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor ERRα. J Biol Chem 2003; 278: 9013–8. [DOI] [PubMed] [Google Scholar]
- McGuirk S, Gravel SP, Deblois G, Papadopoli DJ, Faubert B, Wegner A, et al. PGC-1α supports glutamine metabolism in brreast cancer cells. Cancer Metab 2013; 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner LJ, Giguère V. Estrogen-related receptors (ERRs): a new dawn in the control of mitochondrial gene networks. Mitochondrion 2011; 11: 544–52. [DOI] [PubMed] [Google Scholar]
- Cai Q, Lin T, Kamarajugadda S, Lu J. Regulation of glycolysis and the Warburg effect by estrogen-related receptors. Oncogene 2013; 32: 2079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner LJ, Perry M-C, Dufour CR, Bertos N, Park M, St-Pierre J, et al. mir-378* mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab 2010; 12: 352–61. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, et al. Genome-wide orchestration of cardiac functions by orphan nucler receptors ERRα and γ. Cell Metab 2007; 5: 345–56. [DOI] [PubMed] [Google Scholar]
- Jeoung NH, Wu P, Joshi MA, Jaskiewicz J, Bock CB, Depaoli-Roach AA, et al. Role of pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) in glucose homoeostasis during starvation. Biochem J 2006; 397: 417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1α coactivates PDK4 gene expression via the orphan nuclear receptor ERRα: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol 2005; 25: 10684–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard S, Wang F, et al. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J Biol Chem 2006; 281: 39897–906. [DOI] [PubMed] [Google Scholar]
- Connaughton S, Chowdhury F, Attia RR, Song S, Zhang Y, Elam MB, et al. Regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) gene expression by glucocorticoids and insulin. Mol Cell Endocrinol 2010; 315: 159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Hazen BC, Russell AP, Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J Biol Chem 2013; 288: 25207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirebeau-Prunier D, Le Pennec S, Jacques C, Fontaine JF, Gueguen N, Boutet-Bouzamondo N, et al. Estrogen-related receptor alpha modulates lactate dehydrogenase activity in thyroid tumors. PLoS One 2013; 8: e58683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summermatter S, Santos G, Perez-Schindler J, Handschin C. Skeletal muscle PGC-1alpha controls whole-body lactate homeostasis through estrogen-related receptor alpha-dependent activation of LDH B and repression of LDH A. Proc Natl Acad Sci U S A 2013; 110: 8738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim EJ, Kim DK, Lee JM, Park SB, Lee IK, et al. Hypoxia induces PDK4 gene expression through induction of the orphan nuclear receptor ERRgamma. PLoS One 2012; 7: e46324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci U S A 2008; 105: 7821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Yu S, Xu Z, Wu D, Ng CF, Yao X, et al. ERRalpha augments HIF-1 signalling by directly interacting with HIF-1alpha in normoxic and hypoxic prostate cancer cells. J Pathol 2014; 233: 61–73. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 2013; 123: 3664–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Bader JA, Giguère V. The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol 1997; 17: 5400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega RB, Kelly DP. A role for estrogen-related receptor α in the control of mitochondrial fatty acid β-oxidation during brown adipocyte differentiation. J Biol Chem 1997; 272: 31693–9. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006; 126: 789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguère V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol Cell Biol 2003; 23: 7947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier JC, Deblois G, Champigny C, Levy E, Giguère V. Estrogen related-receptor α (ERRα) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J Biol Chem 2004; 279: 52052–8. [DOI] [PubMed] [Google Scholar]
- Ijichi N, Ikeda K, Horie-Inoue K, Yagi K, Okazaki Y, Inoue S. Estrogen-related receptor alpha modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem Biophys Res Commun 2007; 358: 813–8. [DOI] [PubMed] [Google Scholar]
- Nie Y, Wong C. Suppressing the activity of ERRalpha in 3T3-L1 adipocytes reduces mitochondrial biogenesis but enhances glycolysis and basal glucose uptake. J Cell Mol Med 2009; 13: 3051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju D, He J, Zhao L, Zheng X, Yang G. Estrogen related receptor alpha-induced adipogenesis is PGC-1beta-dependent. Mol Biol Rep 2012; 39: 3343–54. [DOI] [PubMed] [Google Scholar]
- Kubo M, Ijichi N, Ikeda K, Horie-Inoue K, Takeda S, Inoue S. Modulation of adipogenesis-related gene expression by estrogen-related receptor γ during adipocytic differentiation. Biochim Biophys Acta 2009; 1789: 71–7. [DOI] [PubMed] [Google Scholar]
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRγ directs and maintains the transition to oxidative metabolism in the post-natal heart. Cell Metab 2007; 6: 16–24. [DOI] [PubMed] [Google Scholar]
- Huss JM, Pineda Torra I, Staels B, Giguère V, Kelly DP. Estrogen-related receptor α directs peroxisome proliferator-activated receptor α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol 2004; 24: 9079–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Nichols AG, Inoue M, Kazmin D, Chang CY, et al. Estrogen-related receptor-α is a metabolic regulator of effector T-cell activation and differentiation. Proc Natl Acad Sci U S A 2011; 108: 18348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab 2011; 13: 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Laganière J, Mehl IR, Barish GD, Chong LW, Li X, et al. Nuclear receptor ERRα and coactivator PGC-1β are effectors of IFN-γ induced host defense. Genes Dev 2007; 21: 1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA, Hock MB, Giguère V, Kralli A. Orphan nuclear receptor ERRα is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A 2007; 104: 1418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E-J, Levasseur M-P, Dufour CR, Perry M-C, Giguère V. Loss of estrogen-related receptor α promotes hepatocellular carcinogenesis development via metabolic and inflammatory disturbances. Proc Natl Acad Sci U S A 2013; 110: 17975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem 2010; 285: 22619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sladek R, Bader J-A, Rossant J, Giguère V. Placental abnormalities in mouse embryos lacking orphan nuclear receptor ERRβ. Nature 1997; 388: 778–82. [DOI] [PubMed] [Google Scholar]
- Huss JM, Imahashi K-I, Dufour C, Weinheimer CJ, Courtois M, Kovacs A, et al. The nuclear receptor ERRα is required for the bioenergetic and functional adaption to cardiac pressure overload. Cell Metab 2007; 6: 25–37. [DOI] [PubMed] [Google Scholar]
- Foletta VC, Brown EL, Cho Y, Snow RJ, Kralli A, Russell AP. Ndrg2 is a PGC-1alpha/ERRalpha target gene that controls protein synthesis and expression of contractile-type genes in C2C12 myotubes. Biochim Biophys Acta 2013; 1833: 3112–23. [DOI] [PubMed] [Google Scholar]
- LaBarge S, McDonald M, Smith-Powell L, Auwerx J, Huss JM. Estrogen-related receptor-alpha (ERRalpha) deficiency in skeletal muscle impairs regeneration in response to injury. FASEB J 2014; 28: 1082–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Kumar P, Mendelson CR. Estrogen-related receptor gamma (ERRgamma) regulates oxygen-dependent expression of voltage-gated potassium (K+) channels and tissue kallikrein during human trophoblast differentiation. Mol Endocrinol 2013; 27: 940–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poidatz D, Dos Santos E, Brule A, De Mazancourt P, Dieudonne MN. Estrogen-related receptor gamma modulates energy metabolism target genes in human trophoblast. Placenta 2012; 33: 688–95. [DOI] [PubMed] [Google Scholar]
- Cauchy F, Fuks D, Zarzavadjian Le Bian A, Belghiti J, Costi R. Metabolic syndrome and non-alcoholic fatty liver disease in liver surgery: The new scourges? World J Hepatol 2014; 6: 306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel RH, Alberti KG, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet 2010; 375: 181–3. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science 2005; 307: 384–7. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1a-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–73. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, et al. ERRα and GABPAα/β specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A 2004; 101: 6570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 2003; 100: 8466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M. Potential applications for biguanides in oncology. J Clin Invest 2013; 123: 3693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, Savage DB, Abu-Elheiga L, Liu ZX, Kim S, Kulkarni A, et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci U S A 2007; 104: 16480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elheiga L, Oh W, Kordari P, Wakil SJ. Acetyl-CoA carboxylase 2 mutant mice are protected against obesity and diabetes induced by high-fat/high-carbohydrate diets. Proc Natl Acad Sci U S A 2003; 100: 10207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog B, Cardenas J, Hall RK, Villena JA, Budge PJ, Giguère V, et al. Estrogen-related receptor α is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem 2006; 281: 99–106. [DOI] [PubMed] [Google Scholar]
- Patch RJ, Searle LL, Kim AJ, De D, Zhu X, Askari HB, et al. Identification of diaryl ether-based ligands for estrogen-related receptor α as potential antidiabetic agents. J Med Chem 2011; 54: 788–808. [DOI] [PubMed] [Google Scholar]
- Ranhotra HS. Up-regulation of orphan nuclear estrogen-related receptor α expression during long-term caloric restriction in mice. Mol Cell Biochem 2009: 59–65. [DOI] [PubMed]
- Kim DK, Ryu D, Koh M, Lee MW, Lim D, Kim MJ, et al. Orphan nuclear receptor estrogen-related receptor gamma (ERRgamma) is key regulator of hepatic gluconeogenesis. J Biol Chem 2012; 287: 21628–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Gang GT, Ryu D, Koh M, Kim YN, Kim SS, et al. Inverse agonist of nuclear receptor ERRgamma mediates antidiabetic effect through inhibition of hepatic gluconeogenesis. Diabetes 2013; 62: 3093–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Samuel VT, Petersen KF, Shulman GI. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 2014; 510: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameer F, Scandiuzzi L, Hasnain S, Kalbacher H, Zaidi N. De novo lipogenesis in health and disease. Metabolism 2014; 63: 895–902. [DOI] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 2012; 52: 381–400. [DOI] [PubMed] [Google Scholar]
- Houde VP, Brule S, Festuccia WT, Blanchard PG, Bellmann K, Deshaies Y, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes 2010; 59: 1338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy G, Schmidli H, Punch J, Tuttle-Newhall E, Mayer D, Neuhaus P, et al. Safety, tolerability, and efficacy of everolimus in de novo liver transplant recipients: 12- and 36-month results. Liver Transpl 2006; 12: 1640–8. [DOI] [PubMed] [Google Scholar]
- Patsenker E, Schneider V, Ledermann M, Saegesser H, Dorn C, Hellerbrand C, et al. Potent antifibrotic activity of mTOR inhibitors sirolimus and everolimus but not of cyclosporine A and tacrolimus in experimental liver fibrosis. J Hepatol 2011; 55: 388–98. [DOI] [PubMed] [Google Scholar]
- Watt KD. Metabolic syndrome: is immunosuppression to blame? Liver Transpl 2011; 17 Suppl 3: S38–42. [DOI] [PubMed] [Google Scholar]
- Tremblay AM, Dufour CR, Ghahremani M, Reudelhuber TL, Giguère V. Physiological genomics identifies estrogen-related receptor α as a regulator of renal sodium and potassium homeostasis and the renin-angiotensin pathway. Mol Endocrinol 2010; 24: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komm BS, Mirkin S. An overview of current and emerging SERMs. J Steroid Biochem Mol Biol 2014; 143C: 207–22. [DOI] [PubMed] [Google Scholar]
- Kim DK, Kim JR, Koh M, Kim YD, Lee JM, Chanda D, et al. Estrogen-related receptor gamma (ERRgamma) is a novel transcriptional regulator of phosphatidic acid phosphatase, LIPIN1, and inhibits hepatic insulin signaling. J Biol Chem 2011; 286: 38035–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko A, von Kleist-Retzow JC, Neschen S, Wu M, Schommers P, Bose M, et al. Liver adapts mitochondrial function to insulin resistant and diabetic states in mice. J Hepatol 2014; 60: 816–23. [DOI] [PubMed] [Google Scholar]
- Jelenik T, Sequaris G, Kaul K, Ouwens DM, Phielix E, Kotzka J, et al. Tissue-specific differences in the development of insulin resistance in a mouse model for type 1 diabetes. Diabetes 2014; 63: 3856–67. [DOI] [PubMed] [Google Scholar]
- Misu H, Takamura T, Matsuzawa N, Shimizu A, Ota T, Sakurai M, et al. Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia 2007; 50: 268–77. [DOI] [PubMed] [Google Scholar]
- Razak F, Anand SS. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. Vasc Med 2004; 9: 223–4. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Giacomini KM, McKeon C, Shuldiner AR, Florez JC. Metformin pharmacogenomics: current status and future directions. Diabetes 2014; 63: 2590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppunen J, Wohlfahrt G, Aarnisalo P. Requirements for transcriptional regulation by the orphan nuclear receptor ERRgamma. Mol Cell Endocrinol 2004; 219: 151–60. [DOI] [PubMed] [Google Scholar]
- Murray J, Auwerx J, Huss JM. Impaired myogenesis in estrogen-related receptor gamma (ERRgamma)-deficient skeletal myocytes due to oxidative stress. FASEB J 2013; 27: 135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Kazmin D, Jasper JS, Kunder R, Zuercher WJ, McDonnell DP. The metabolic regulator ERRα, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell 2011; 20: 500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor α and estrogen-related receptor γ associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res 2002; 62: 6510–8. [PubMed] [Google Scholar]
- Suzuki T, Miki Y, Moriya T, Shimada N, Ishida T, Hirakawa H, et al. Estrogen-related receptor α in human breast carcinoma as a potent prognostic factor. Cancer Res 2004; 64: 4670–6. [DOI] [PubMed] [Google Scholar]