HJURP exhibits a centromere expansion activity that is separable from its CENP-A–binding activity. It is proposed that this centromere expansion activity reflects the functional properties of HJURP, which uses this activity to contribute to the plastic establishment of a centromeric chromatin structure.
Abstract
The CENP-A–specific chaperone HJURP mediates CENP-A deposition at centromeres. The N-terminal region of HJURP is responsible for binding to soluble CENP-A. However, it is unclear whether other regions of HJURP have additional functions for centromere formation and maintenance. In this study, we generated chicken DT40 knockout cell lines and gene replacement constructs for HJURP to assess the additional functions of HJURP in vivo. Our analysis revealed that the middle region of HJURP associates with the Mis18 complex protein M18BP1/KNL2 and that the HJURP-M18BP1 association is required for HJURP function. In addition, on the basis of the analysis of artificial centromeres induced by ectopic HJURP localization, we demonstrate that HJURP exhibits a centromere expansion activity that is separable from its CENP-A–binding activity. We also observed centromere expansion surrounding natural centromeres after HJURP overexpression. We propose that this centromere expansion activity reflects the functional properties of HJURP, which uses this activity to contribute to the plastic establishment of a centromeric chromatin structure.
INTRODUCTION
The kinetochore is an essential structure required for chromosome segregation and is assembled at the centromeric region of each chromosome. In most organisms, the centromere is specified by sequence-independent epigenetic mechanisms that involve the centromere-specific histone H3 variant centromere protein-A (CENP-A; Fukagawa and Earnshaw, 2014). To understand the basis for centromere specification, many studies have analyzed the molecular mechanisms by which CENP-A is incorporated specifically into centromeric chromatin (Black and Cleveland, 2011; Westhorpe and Straight, 2013).
Although CENP-A is a histone H3 variant, unlike canonical histone H3, its chromatin incorporation is not coupled with DNA replication in vertebrate cells (Shelby et al., 1997; Jansen et al., 2007). Newly synthesized CENP-A is incorporated into centromeres specifically during early G1 in human cells (Jansen et al., 2007). The deposition of CENP-A at centromeres requires the action of several proteins. The Mis18 protein was originally identified from fission yeast mutants that displayed defects in CENP-A incorporation (Hayashi et al., 2004). There are two vertebrate homologues of Mis18, Mis18α and Mis18β, which function in CENP-A deposition. In addition, the Mis18α/β-binding protein M18BP1 (Fujita et al., 2007), a homologue of the Caenorhabditis elegans centromere protein KNL2 (Maddox et al., 2007), forms a complex with Mis18α/β that functions in CENP-A incorporation. Human Mis18α/β begin to localize to centromeres during anaphase/telophase, just before CENP-A incorporation, but are restricted from centromeres from late G1 until the next anaphase (Fujita et al., 2007). This temporal centromere localization of Mis18α/β is regulated by two major kinases, CDK and Plk1 (Silva et al., 2012; McKinley and Cheeseman, 2014). CDK activity is high during mitosis and drops upon anaphase onset. As Mis18 proteins are phosphorylated by CDK during mitosis, the kinase negatively regulates the centromere localization (Silva et al., 2012) and assembly (McKinley and Cheeseman, 2014) of the Mis18 complex. In contrast, Plk1 phosphorylation positively regulates the centromere localization of the Mis18 complex to promote CENP-A deposition (McKinley and Cheeseman, 2014). Based on its localization profile, the Mis18 complex appears to be responsible for licensing centromere chromatin for CENP-A incorporation.
Although the Mis18 complex is functionally required for CENP-A incorporation, it does not bind directly to CENP-A. Biochemical purifications identified the CENP-A specific chaperone Holliday junction recognition protein (HJURP; Dunleavy et al., 2009; Foltz et al., 2009), which binds to CENP-A and mediates its deposition at centromeres in human cells (Barnhart et al., 2011). HJURP also displays structural and functional conservation with the fungal protein Scm3 (Mizuguchi et al., 2007; Pidoux et al., 2009; Sanchez-Pulido et al., 2009; Williams et al., 2009). Both HJURP and Scm3 act as CENP-A chaperones, and the N-terminal region of HJURP binds directly to the CENP-A–H4 complex (Shuaib et al., 2010; Hu et al., 2011). In addition, ectopic targeting of HJURP to noncentromeric regions results in CENP-A deposition at those sites (Barnhart et al., 2011; Hori et al., 2013). A recent study suggested that dimer formation through a C-terminal region of human HJURP is crucial for CENP-A targeting (Zasadzinska et al., 2013). In addition, phosphorylation of HJURP may control its centromeric recruitment (Muller et al., 2014).
Although it is clear that HJURP functions as a CENP-A–specific chaperone and that its N-terminal domain is responsible for CENP-A binding, it is unknown how other regions of HJURP contribute to centromere specification, formation, and maintenance. In this study, we generated a conditional knockout chicken DT40 cell line for HJURP and selected HJURP mutant proteins to assess the functions for each region of HJURP. In addition, we generated artificial kinetochores at a noncentromeric LacO site (Hori et al., 2013) using LacI fusion proteins with different HJURP mutant proteins. By analyzing these ectopic kinetochores, we found that HJURP possesses a unique activity for controlling CENP-A distribution and expansion into neighboring DNA.
RESULTS
HJURP is essential for mitotic progression
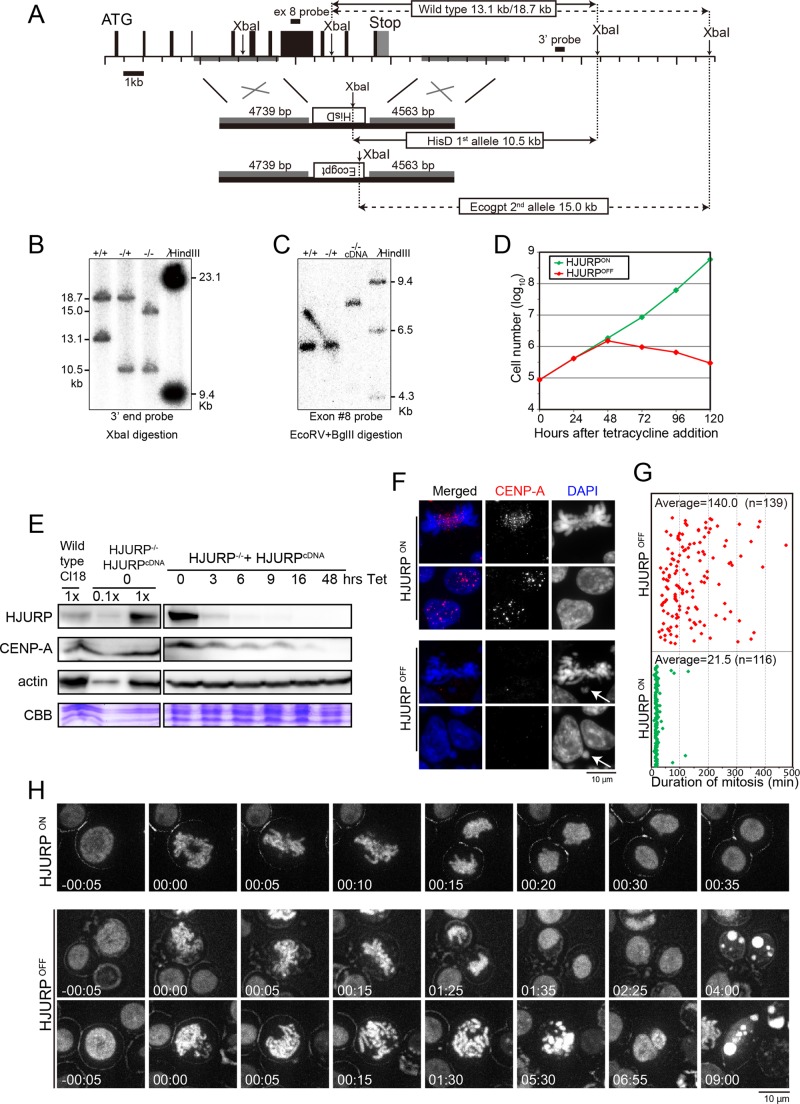
To analyze the functional domains of HJURP in vivo, we first created HJURP-deficient chicken DT40 cell lines (Figure 1, A–C). Human HJURP localizes to centromeres during G1 for CENP-A deposition and dissociates from centromeres after CENP-A deposition (Dunleavy et al., 2009; Foltz et al., 2009). Similar to human HJURP, the centromere localization of chicken HJURP was only detected in G1 phase (Supplemental Figure S1A). Because we predicted that HJURP would be essential for cell viability, we generated a conditional knockout cell line by expression of the HJURP cDNA under the control of a tetracycline-responsive promoter after disruption of two HJURP alleles (Figure 1A). We confirmed the disruption of the two HJURP alleles by Southern hybridization (Figure 1, B and C). Next we analyzed the growth of the conditional cell line in the presence (HJURPOFF) or absence of tetracycline (HJURPON). HJURP expression was undetectable within 9 h after tetracycline addition (Figure 1E). As expected, growth of these cells stopped by 48 h after addition of tetracycline and displayed cell death (Figure 1D and Supplemental Figure S1B), indicating that HJURP is essential for cell viability.
FIGURE 1:
HJURP is essential for mitotic progression in chicken DT40 cells. (A) Genome map around the chicken HJURP locus and targeting constructs. Restriction sites for XbaI are shown. The left arm of the knockout construct contains the genomic region 19090–23829 of chromosome 1 and the right arm, 30513–34990. There is a polymorphism for XbaI sites. Solid and dashed lines show the respective alleles. After the first targeting, the 13.1-kb fragment was reduced to 10.5 kb (solid line). After the second targeting, the 18.7-kb fragment was reduced to 15.0 kb (dashed line). (B) Southern blot analysis for confirmation of gene disruption. The probe is shown in A. (C) Additional confirmation of gene disruption based on Southern blot analysis using a probe for exon 8 of the HJURP gene. Exon 8 hybridized with the HJURP gene locus before the second allele knockout, but it hybridized with a rescue construct (HJURP cDNA) integrated at a different locus after the second allele knockout. (D) Growth curve of HJURP conditional knockout cells in the presence (HJURPOFF) or absence (HJURPON) of tetracycline. Numbers indicate hours (h) after addition of tetracycline. (E) Western blot analysis of HJURP conditional knockout cell lines with anti-HJURP or anti–CENP-A antibodies. Numbers indicate hours (h) after addition of tetracycline. Lysate from Cl18 (clone 18: wild-type DT40 cells) and conditional knockout cells (HJURP−/−, HJURPcDNA) were prepared. CBB, protein samples stained by Coomassie brilliant blue. (F) Typical images of HJURP-deficient cells (HJURPOFF). Cells were stained with anti–CENP-A antibody (red). Chromosomes and nuclei were counterstained with DAPI (blue). Arrows shows micronuclei (HJURPOFF bottom) and misaligned chromosomes (HJURPOFF top). Scale bar, 10 μm. (G) Average time span necessary for HJURPON (green diamonds) or HJURPOFF (red diamonds) cells to complete mitosis, starting from nuclear envelope breakdown (NEBD) to anaphase onset. Each diamond represents one cell. This measurement was performed by live-cell imaging observation. n, number of analyzed cells. (H) Typical images from live-cell imaging of HJURP conditional knockout cells stably expressing H2B-RFP in the presence (HJURPOFF) or absence (HJURPON) of tetracycline. Time 0 indicates entry of mitosis, recognized as nuclear envelope breakdown and start of nuclear condensation. The live-cell observation was started at 48 h after tetracycline addition, and time-lapse images were taken every 5 min for the next 16 h. Scale bar, 10 μm.
Because HJURP is a CENP-A–specific chaperone, we next assessed CENP-A localization and abundance in the HJURP-deficient cells (Figure 1, E and F). Immunofluorescence analysis at 48 h after tetracycline addition revealed that CENP-A levels were reduced in HJURP-deficient cells, consistent with the dilution of CENP-A during DNA replication after four to five cell cycles (Figure 1F). In addition, Western blot analysis revealed that CENP-A abundance gradually decreased after tetracycline addition, indicating that CENP-A degradation correlates with HJURP disruption (Figure 1E). This suggests that CENP-A is unstable when it is not associated with HJURP, consistent with previous observation in human cells (Dunleavy et al., 2009). The reduction in CENP-A localization and levels would be predicted to prevent the formation of functional kinetochore structures in HJURP-deficient cells. Indeed, we frequently observed abnormal mitotic cells in the presence tetracycline (HJURPOFF) based on live-cell imaging. In addition, although it takes ∼21 min to complete mitosis in wild-type DT40 cells, HJURP-deficient cells remained in mitosis for ∼140 min (Figure 1, G and H, and Supplemental Movies S1–S3). During the prolonged mitosis, cells exhibited abnormal behavior. We observed cells with disorganized chromosomes, as well as cells with a metaphase plate but a subset of unaligned chromosomes (Figure 1, F and H). Some cells were able to segregate their chromosomes and progress to telophase/G1 despite the presence of lagging chromosomes or unequal chromosome segregation (Figure 1H, HJURPOFF top). Other cells struggled to progress into anaphase and chromosome segregation, and cytokinesis failed to occur (Figure 1H, HJURPOFF bottom). However, after the prolonged mitotic arrest of these cells, the chromosomes decondensed and the cells entered G1 and subsequently died in interphase (Figure 1H and Supplemental Movies S2 and S3). Consistent with these observations, we detected an accumulation of sub-G1 DNA content cells after the addition of tetracycline, which indicates an accumulation of dead cells (Supplemental Figure S1). This suggests that, due to incomplete kinetochore assembly, the spindle assembly checkpoint signal is weakened and cells progress into G1 after an abnormal mitosis. In summary, on the basis of the observation of HJURP-knockout cells, we confirmed that HJURP is essential for CENP-A deposition at centromeres and is crucial for mitotic progression in chicken DT40 cells.
The centromere localization of HJURP occurs upstream of most centromere proteins
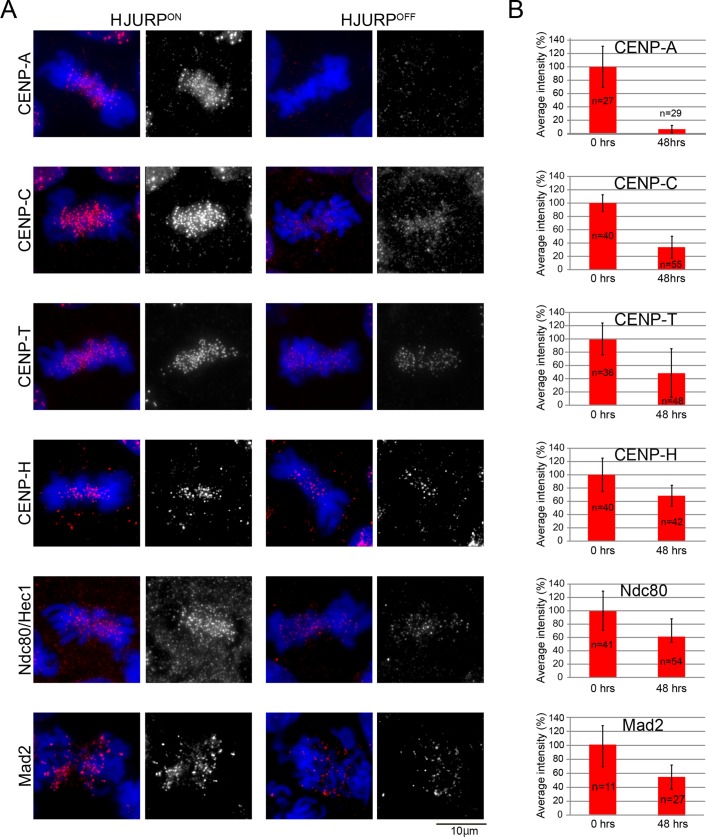
Because live-cell imaging suggested the presence of impaired kinetochore structures in HJURP-deficient cells (Figure 1, G and H), we quantitatively examined the immunofluorescence signals for representative centromere proteins in these cells (Figure 2A). The centromeric localization of CENP-A was dramatically decreased in the HJURP-deficient cells, with an average intensity of ∼7.3% relative to those observed in wild-type cells. Because the doubling time of DT40 cells is 10–11 h, at the time point of our analysis (48 h), preexisting centromeric CENP-A would have been diluted at each cell division. Although we could detect centromere signals for other kinetochore proteins, the localization of most of these proteins (CENP-C, CENP-H, CENP-T, Ndc80, Mad2) was significantly decreased (Figure 2, A and B). Of importance, although HJURP-deficient cells exhibit a dramatic reduction of the centromeric CENP-A and an abnormal mitotic phenotype (Figure 1H and Supplemental Movies S1 and S2), these cells retain some localization of downstream centromere proteins, including CENP-C and CENP-T, consistent with previous observations of CENP-A–knockout chicken and human cells (Regnier et al., 2005; Fachinetti et al., 2013).
FIGURE 2:
Most centromere proteins are gradually lost in HJURP-deficient cells. (A) Representative images of immunofluorescence staining for indicated centromere proteins in HJURP conditional knockout cells in the absence (HJURPON) or presence (48hrs, HJURPOFF) of tetracycline. Antibodies against CENP-A, CENP-C, CENP-T, CENP-H, and Ndc80 were used. (B) Quantification of signal intensities for each centromere protein in HJURP conditional knockout cells in the presence (48 h, HJURPOFF) or absence (HJURPON) of tetracycline.
The middle region of HJURP is essential for cell viability
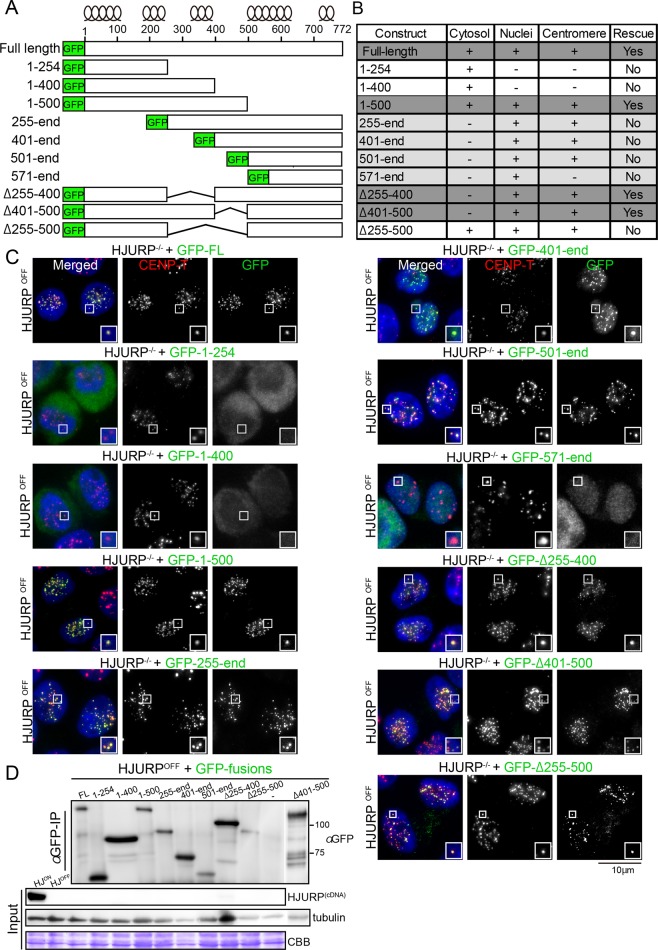
Although HJURP is a CENP-A–specific chaperone (Dunleavy et al., 2009; Foltz et al., 2009), it may also have additional functional domains. To investigate the functional contribution of each HJURP domain in vivo, we generated replacement constructs in the HJURP-conditional knockout cell line. We introduced selected green fluorescent protein (GFP)–tagged mutant proteins of chicken HJURP (Figure 3A and Supplemental Figure S2) into the conditional knockout cell line and assessed their ability to complement the function of HJURP in the presence of tetracycline (Figure 3, B and C, and Supplemental Figure S2).
FIGURE 3:
Middle region of HJURP is essential for cell viability. (A) Scheme of different HJURP GFP-tagged expression constructs. The predicted position of α-helixes obtained with PSIPRED version 3.3 software. (B) Summary of results for localization and complementation assay of indicated GFP-tagged HJURP mutant proteins. Supplemental Figure S2 shows the results of cell viability assay. (C) Representative images showing cellular localization of different GFP-HJURP mutant proteins (green). CENP-T was used as a centromere marker (red). DAPI was used for DNA staining (blue). HJURPON refers to cells grown in the absence of tetracycline and HJURPOFF to those grown for 48 h in the presence of tetracycline. Because cells expressing C-terminal truncation of HJURPs (HJURP1-254 and HJURP1-400) restrictively localize to the cytoplasm, it is possible that the middle region may be related to the nuclear localization of HJURP. (D) Western blot analysis with anti-GFP antibody showing the expression levels for each of the GFP-mutant proteins. Although GFP-tagged HJURP1-500 shows unexpectedly slower migration, similar to GFP-tagged HJURPFL, expression of GFP-tagged HJURP1-500 was confirmed by genomic PCR.
On its own, the conserved N-terminal region of HJURP (chicken HJURP1-254, Figure 3, A and D, and Supplemental Figure S2) was unable to localize to the nucleus and did not rescue cell viability (Figure 3, B and C, and Supplemental Figure S2). In contrast, HJURP constructs that contained the remaining regions of the protein (HJURP255-end, HJURP401-end, and HJURP501-end) localized to centromeres (Figure 3, A, C, and D), consistent with previous observations (Zasadzinska et al., 2013). However, even though centromere localization was observed in these mutant proteins, they failed to rescue cell viability due to the lack of the CENP-A–binding region (Figure 3D and Supplemental Figure S2). We tested further truncation mutants and found that HJURP571-end does not localize to centromeres (Figure 3, B and C), indicating that the 500– to 570–amino acid (aa) region is crucial for centromere targeting of HJURP.
Because both the CENP-A–binding and centromere-targeting domains are essential for HJURP function, we next sought to identify a minimal HJURP domain that was sufficient to rescue cell viability. A C-terminal deletion mutant (HJURP1-500) showed centromere localization, despite the absence of the 500- to 570-aa region, and rescued cell viability (Figure 3, B–D, and Supplemental Figure S2). In contrast to HJURP1-500, a further C-terminal deletion (HJURP1-400) did not rescue cell viability or localize to centromeres (Figure 3, A–D, and Supplemental Figure S2). We also prepared mutant proteins lacking this middle region (HJURPΔ255-400, HJURPΔ255-500, and HJURPΔ401-500) and introduced these mutant proteins into HJURP-deficient cells. HJURP1-500, HJURP501-end, and HJURPΔ255-500 each localized to centromeres (Figure 3C). However, we found that HJURPΔ255-400 and HJURPΔ401-500 rescued the HJURP deficiency, whereas HJURPΔ255-500 did not (Figure 3B and Supplemental Figure S2; unpublished data). Because HJURPΔ255-500 does not rescue the HJURP deficiency, we concluded that the 255- to 500-aa region is essential for HJURP function. However, HJURPΔ255-500 localized to centromeres due to the presence of the C-terminal centromere-targeting domain (aa 500–570). This suggests that there are multiple regions within HJURP that contribute to its centromere targeting and function.
The middle region of HJURP associates with M18BP1/KNL2
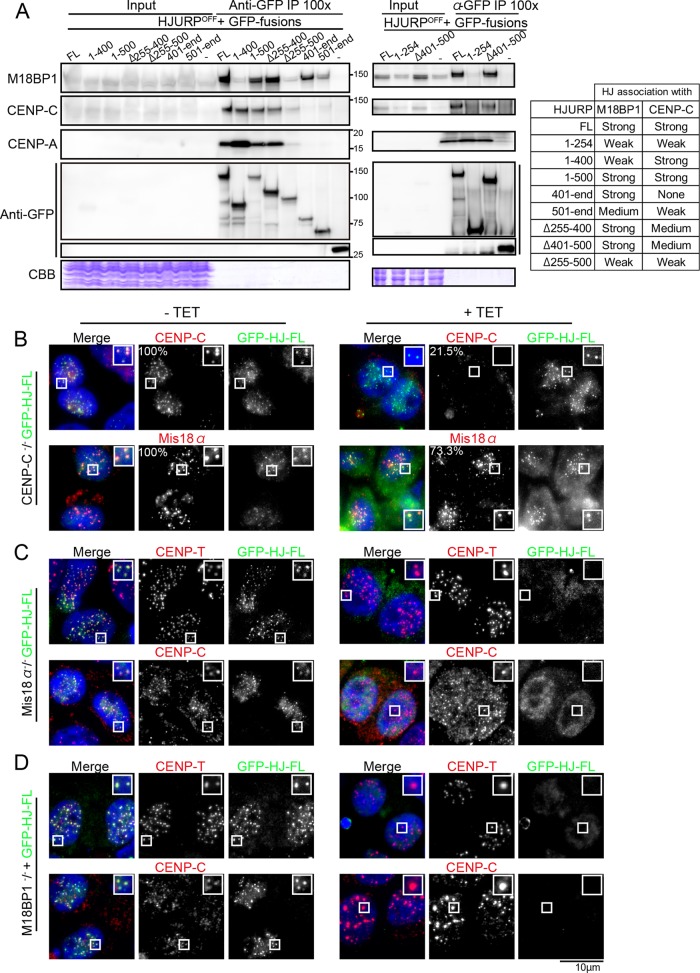
To test the functional significance of each HJURP domain, we sought to identify interacting molecules. On the basis of immunoprecipitation (IP)/mass spectrometry analyses with cells expressing Mis18α-FLAG, we found potential interactions between HJURP and the Mis18 complex (Supplemental Figure S3), although we did not detect HJURP by Western blot analysis from IPs with endogenous Mis18α antibodies (Supplemental Figure S3B). It is possible that the endogenous antibody might interfere with this interaction. Consistent with our data, Wang et al. (2014) reported that Mis18β binds to HJURP in human cells. On the basis of IP/mass spectrometry analyses, we also detected an interaction between CENP-C and HJURP in the soluble fraction from cells expressing GFP-CENP-C644-864 (Supplemental Figure S3A).
Several recent studies indicated that CENP-C binds directly to the Mis18 complex in human cells (Moree et al., 2011; Dambacher et al., 2012) and that the Mis18 localization occurs downstream of CENP-C (McKinley and Cheeseman, 2014). However, we observed Mis18α localization in CENP-C–deficient DT40 cells (Figure 4B and Supplemental Figure S3), although its intensity was slightly reduced (73% compared with intensities of control cells). This suggests that the localization of Mis18α depends only in part upon CENP-C. Consistent with this, we also detected GFP-HJURPfull length (FL) localization in CENP-C–deficient cells, again with a slight reduction in signal intensity (Figure 4B). We speculate that the Mis18 complex might recognize not only CENP-C, but also an unknown structure in centromeric chromatin in chicken cells. In contrast, centromere GFP-HJURPFL signals were strongly reduced in both Mis18α- and M18BP1/KNL2-deficient cells (Figure 4, C and D). These results suggest that the centromere localization of HJURP depends on the Mis18 complex in chicken cells. We also analyzed the localization of GFP-HJURP401-end (Supplemental Figure S3, F–H) and GFP-HJURP501-end (unpublished data) and observed a similar dependence on the Mis18 complex for their localization.
FIGURE 4:
The Mis18 complex is associated with HJURP. (A) Immunoprecipitation/Western blot analysis showing preferential associations of HJURP with M18BP1/KNL2, CENP-C, and CENP-A. Coprecipitation was also confirmed by mass spectrometry analysis. Whole-cell lysates of HJURP conditional knockout cell lines in which expression of HJURP was replaced with GFP-tagged-HJURPFL, -HJURP1-400, -HJURP1-500, -HJURPΔ255-400, -HJURPΔ255-400, and -HJURP401-end (left gel) and -HJURP1-254 and -HJURPΔ401-500 (right gel) were immunoprecipitated with anti-GFP antibody, and the eluates were applied to SDS–PAGE and subjected to Western blot analysis with the indicated antibodies. Because the majority of M18BP1/KNL2 localizes in nuclei and HJURP mainly exists in the cytoplasm (Supplemental Figure S3B), cytoplasmic and nuclear soluble fractions were prepared, and IP experiments were also done using these two mixed fractions (Supplemental Figure S3C). (B) Immunofluorescence with anti-Mis18α and anti–CENP-C antibodies in CENP-C–deficient cells (+TET). CENP-C was expressed in the absence of tetracycline (–TET). GFP-HJURP-FL was stably expressed in these cells. Mis18α and CENP-C are visible in CENP-C–deficient cells (+TET). (C) Immunofluorescence with anti–CENP-T and –CENP-C antibodies in Mis18α-deficient cells (+TET). Mis18α was expressed in the absence of tetracycline (–TET). GFP–HJURP-FL was stably expressed in these cells. HJURP signals were not detected in Mis18α-deficient cells (+TET). (D) Immunofluorescence with anti–CENP-T and –CENP-C antibodies in M18BP1/KNL2-deficient cells (+TET). M18BP1/KNL2 was expressed in the absence of tetracycline (–TET). GFP–HJURP-FL was stably expressed in these cells. HJURP signals were not detected in M18BP1/KNL2-deficient cells (+TET).
Because HJURP associates with the Mis18 complex, it is possible that CENP-C may coimmunoprecipitate with HJURP through its association with the Mis18 complex. However, we found that although HJURP401-end strongly associated with M18BP1/KNL2, this mutant protein did not copurify with CENP-C from DT40 cells (Figure 4A), suggesting that HJURP may associate with CENP-C independently of the Mis18 complex. Although we cannot rule out the possibility of indirect interaction between CENP-C and HJURP, they may interact directly before centromere deposition, as this interaction was detected in the nuclear soluble fraction (Supplemental Figure S3, A and C). Recently, Tachiwana et al. (2015) demonstrated that human CENP-C directly binds to HJURP. Whereas the HJURP N-terminal domains (HJURP1-400 and HJURP1-500) associated with CENP-C based on IP/immunoblotting experiments, the C-terminal domain (HJURP401-end) did not (Figure 4A). This indicates that the N-terminal domain of HJURP, which is distinct from its centromere-targeting region, is responsible for its coprecipitation with CENP-C.
Whereas the N-terminal domain of HJURP is specific for its association with CENP-A and CENP-C, we hypothesized that the middle region of HJURP (aa 255–500) might be responsible for its coprecipitation with M18BP1/KNL2. Indeed, as shown in Figure 4A, HJURPΔ255-500 did not efficiently associate with M18BP1/KNL2, but the HJURP1-500 domain strongly associated with M18BP1/KNL2. Because the HJURPΔ255-400 deletion mutant protein, but not HJURPΔ255-500, associated with M18BP1/KNL2 (Figure 4A), the 401- to 500-aa region contributes to the association with M18BP1/KNL2. However, HJURPΔ401-500 rescued the HJURP deficiency (Figure 3B) and associated with M18BP1/KNL2 (Figure 4A), suggesting that HJURP associates with M18BP1/KNL2 at multiple sites in the middle region and the entire 255- to 500-aa region is crucial for this association. Of interest, the CENP-C–HJURP association was reduced in cells expressing HJURPΔ255-500. This suggests that, whereas the N-terminal portion of HJURP is crucial for its association with CENP-C, the middle region might facilitate the stable association of HJURP with CENP-C and CENP-A. Therefore we conclude that the 255- to 500-aa region is essential for HJURP function, acting as a mediator between HJURP–CENP-A, the Mis18 complex, and CENP-C.
The middle region of HJURP is involved in centromere expansion
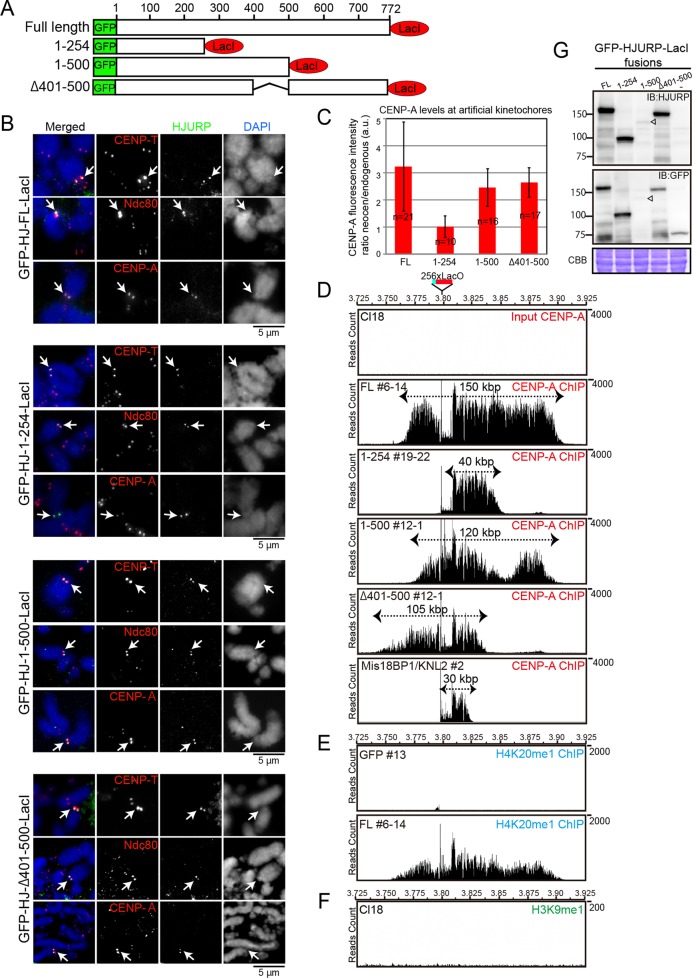
We next analyzed the role of HJURP in the process of de novo establishment of centromeric chromatin. Using the combination of the LacO-LacI system coupled with the excision of an endogenous centromere, we previously generated artificial kinetochores at the LacO site by tethering full-length HJURP, CENP-C, or CENP-I (Hori et al., 2013). Here we induced artificial kinetochores by tethering different HJURP mutant proteins to the noncentromeric LacO locus (Figure 5A) after excision of the endogenous centromere of chromosome Z. Full-length HJURP-derived artificial kinetochores exhibited brighter immunofluorescence signals for all kinetochore proteins analyzed than did native kinetochores (Figure 5, B and C). To understand this larger appearance of the artificial kinetochores, we performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis using anti–CENP-A antibodies (Figure 5D). The experimental scheme for this analysis is described in Supplemental Figure S4. Although the LacO site is ∼10 kb in length (256 copies of single LacO sequence; see Figure 5D), after HJURP-LacI expression, CENP-A incorporation expanded to reside over an ∼150-kb region surrounding the LacO site (Figure 5D). Given that we performed the ChIP experiments with anti–CENP-A under non–cross-linked condition, we conclude that precipitated DNA was from CENP-A nucleosomes. This suggests that when HJURP is targeted continuously to a particular site, the size of the assembled CENP-A chromatin expands. We also detected an extended distribution for H4K20me1 (Hori et al., 2014), suggesting that functional centromere chromatin also expanded (Figure 5E). This expansion was maintained even after cells were treated with isopropyl-β-d-thiogalactoside (IPTG; Supplemental Figure S4), which disrupts the interaction of LacI with LacO, for >1 mo (unpublished data). Because LacI-GFP signals were not detected after IPTG addition (Supplemental Figure S4B), we conclude that once expanded centromeres are established, a seeding protein is no longer needed and the chromatin information is preserved.
FIGURE 5:
Middle region of HJURP contributes to centromere expansion. (A) Schematic representation of different HJURP fusion constructs with GFP and LacI used for induction of artificial kinetochores at the LacO site on chromosome Z. (B) Immunofluorescence images with anti–CENP-T, -Ndc80, and -CENP-A antibodies on artificial kinetochores induced by HJURPFL, HJURP1-254, HJURP1-500, and -HJURPΔ401-500 tethering. Mitotic spreads were used for immunofluorescence experiments. Arrows shows artificial kinetochores. (C) Quantification of signal intensity of CENP-A from B for each type of HJURP-induced artificial kinetochore. The intensity at induced kinetochores was calculated relative to that of centromeres of chromosome 1 or 2. Error bars are SDs. Cell numbers (n) are shown. (D) ChIP-seq profiles with anti–CENP-A antibody around LacO sites in indicated artificial centromeres induced by HJURPFL (#6-14), HJURP1-254 (#19-22), HJURP1-500 (#12-1), HJURPΔ401-500 (#2-1), and M18BP1 (#2). DNA purified from wild-type DT40 cells (Cl18) before ChIP was used as input DNA sample. Raw reads of sequence were mapped, as CENP-A peaks around centromere regions were highly enriched and normalization was not necessary. The 256 copies of LacO sequences were inserted into the chicken galGAL4 genome database (UCSC Genome Browser), which was further used as a reference for mapping. Note that CENP-A incorporation at the LacO site is not high and is expanded in adjacent regions, but some clones shows still high incorporation of CENP-A at the LacO site. (E) ChIP-seq profile with anti-H4K20me1 antibody around LacO sites in the artificial centromeres induced by HJURPFL (#6-14). Cells in which LacI-GFP was tethered at the LacO site were used as control. (F) ChIP-seq profile with anti-H3K9me3 antibody around LacO sites in DT40 cells (Cl18). (G) Western blot analysis showing expression level of each LacI-fused HJURP protein. Anti-HJURP and anti-GFP antibodies were used.
Next we created artificial kinetochores using the tested domains of HJURP (Figure 5, A and B). The C-terminal region of HJURP (aa 254–end) was unable to induce formation of an ectopic kinetochore (unpublished data). Although HJURP1-254 did not rescue the HJURP deficiency due to its lack of centromere localization (Figure 3), this domain was sufficient to induce artificial kinetochores upon tethering at the LacO site as a LacI fusion. This suggests that CENP-A incorporation is sufficient for the creation of an artificial kinetochore. However, when we examined the distribution of CENP-A using ChIP-seq, we found that the size of the CENP-A domain in cells expressing HJURP1-254-LacI was ∼40 kb surrounding the LacO site, one-third of the size of the domain formed by expression of full-length HJURP-LacI (Figure 5D). This suggests that the middle or C-terminal regions of HJURP are responsible for the observed centromere expansion activity.
Finally, we tested HJURP1-500 and HJURPΔ401-500, which associate with M18BP1/KNL2, for their ability to induce artificial kinetochore formation (Figure 5A). HJURP1-500 and HJURPΔ401-500 induced artificial kinetochores, and the size of CENP-A domain was ∼120 and ∼105 kb, respectively, reflecting a centromere expansion similar to full-length HJURP-derived kinetochores. We confirmed that the expansion was maintained in the presence of IPTG. Although the regions of CENP-A occupancy in HJURP1-500- and HJURPΔ401-500-derived kinetochores are slightly shorter than those induced by full-length HJURP, they are still longer than that of HJURP1-254-derived kinetochores. This expansion ratio between full-length and mutant HJURPs also correlated with the CENP-A intensity observed by immunofluorescence (Figure 5, B and C).
Because the M18BP1/KNL2-HJURP association appears to be important for centromere expansion, we directly tethered M18BP1/KNL2 at the LacO site and examined centromere expansion. As shown in Figure 5D, the centromere expansion was not observed around the M18BP1/KNL2-induced centromere, suggesting that, although the M18BP1-binding middle region of HJURP is crucial for centromere expansion, M18BP1 is not sufficient for the centromere expansion.
Finally, we examined whether expression levels of truncated proteins were related to the expansion activity. Although expression levels varied (Figure 5G), expression of HJURP1-254 was similar to that of HJURPFL. However, HJURPFL, but not HJURP1-254, displays the centromere expansion activity, suggesting that total expression levels are not responsible for the observed differences. In addition, we confirmed that centromere expansion occurred similarly in independent two clones of each construct (unpublished data). Thus we propose that HJURP contributes to centromere expansion, and the middle region is important for this activity.
CENP-A expansion occurs at endogenous centromeres without heterochromatin after HJURP overexpression
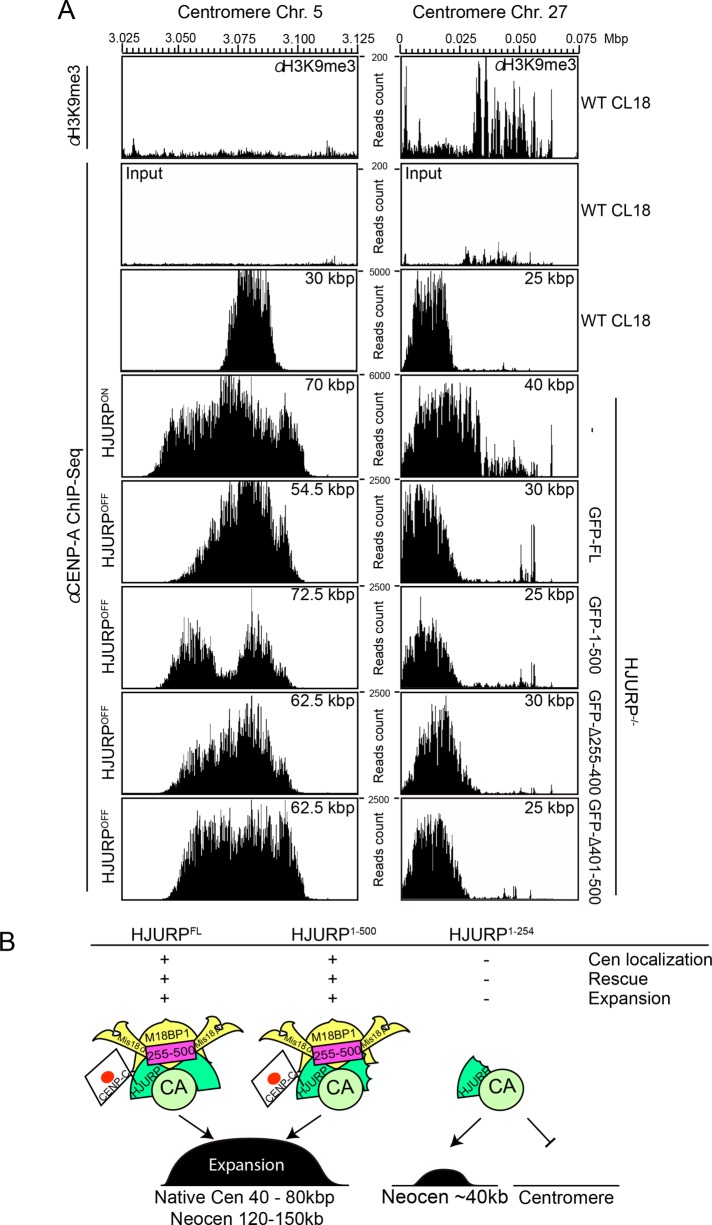
To understand the centromere expansion activity of HJURP, we also investigated CENP-A distribution at native centromeres in wild-type DT40 cells and in HJURP conditional knockout cells expressing full-length HJURP, HJURP1-500, HJURPΔ255-400, or HJURPΔ401-500 (Figure 6A). In HJURP conditional knockout cells, full-length or truncated HJURPs were expressed under the control of the cytomegalovirus promoter at high levels compared with endogenous HJURP expression in wild-type DT40 cells. ChIP-seq analysis using anti–CENP-A antibodies revealed that the CENP-A domain at the nonrepetitive centromeres on chromosomes 5 and 27 is ∼30–40 kb in wild-type DT40 cells. However, overexpression of the full-length and mutant HJURPs that rescue the HJURP deficiency resulted in expansion of the CENP-A domain to 50–70 kb at centromere 5. In contrast, we did not observe centromere expansion at centromere 27. Although there are no heterochromatin markers such as H3K9me3 surrounding centromere 5 based on ChIP-seq analysis, H3K9me3 exists near centromere 27 (Figure 6A; Shang et al., 2013). Note that this centromere is located near the telomere on chicken chromosome 27 and that the genome sequence information at the telomere region was omitted from the galGal4 chicken genome database. Owing to this, we detected H3K9me3 enrichment at only the right flank of centromere 27. In addition, we confirmed that H3K9me3 does not accumulate around the LacO region (Figure 5F) where we observed centromere expansion after HJURP tethering. On the basis of these data, we propose that high amounts of HJURP result in centromere expansion even at native centromeres but that the presence of surrounding heterochromatin might suppress this expansion.
FIGURE 6:
Centromere expansion around native centromeres upon HJURP overexpression. (A) Distribution of CENP-A and heterochromatic marker H3K9me3 around native centromere of chromosomes 5 and 27 in wild-type DT40 cells (Cl18, top). CENP-A distribution was examined around native centromere of chromosomes 5 and 27 in cells overexpressing HJURPFL, HJURP1-500, and HJURPΔ255-400 and HJURPΔ401-500, which could rescue HJURP deficiency. Centromere expansion was observed around centromere 5 upon overexpression of HJURPs but was not around centromere 27, where heterochromatin was enriched around centromeres. (B) Schematic summary emphasizing the importance of the middle region for HJURP function in centromere formation. HJURPFL or HJURP1-500 containing the middle region (aa 255–500), which efficiently binds the Mis18 complex, shows chromatin expansion activity, whereas HJURP1-254 cannot establish a native centromere or form an expanded centromere even if HJURP1-254 is tethered to the LacO site.
DISCUSSION
The middle region of HJURP is essential for its function
HJURP is the CENP-A–specific chaperone required for CENP-A deposition at centromeres (Dunleavy et al., 2009; Foltz et al., 2009). Previous studies highlighted the importance of the N-terminal (for CENP-A binding: corresponding to aa 1–254 for chicken HJURP) and C-terminal (for centromere targeting: corresponding to 500 aa–end for chicken HJURP) domains (Shuaib et al., 2010; Hu et al., 2011; Zasadzinska et al., 2013). In this study, on the basis of the combination of our genetic and biochemical analyses, we conclude that the middle region of chicken HJURP (aa 255–500) is also crucial for its function, possibly through its association with the Mis18 complex protein M18BP1/KNL2. The Mis18 complex appears to function as the HJURP receptor to target it to centromeres. In addition, we propose that the HJURP-M18BP1 association plays an additional important role in the process of establishment of centromere-specific chromatin because CENP-A and CENP-C levels were weak in cells expressing HJURPΔ255-500 based on immunofluorescence analysis (unpublished data).
HJURP is involved in centromere expansion
We hypothesized that the middle region (255–500 aa) of HJURP has an additional role in HJURP function, and it is possible that this region may be involved in establishment of centromere-specific chromatin. We obtained insights into this potential role based on our analysis of CENP-A distribution at artificial kinetochores induced by HJURP mutant proteins. Tethering of full-length HJURP, or the Δ401-500 and 1-500 mutants that associate with M18BP1/KNL2, induced the expansion of kinetochores along the chromosome. Thus we propose that the HJURP-M18BP1 association facilitates this expansion activity. However, we could not directly test whether this expansion depends on the Mis18 complex because an artificial kinetochore could not be established after depletion of the Mis18 complex due to cell death. In addition, because CENP-C binds to HJURP, it is possible that the HJURP–CENP-C interaction may contribute to this expansion activity. However, given that CENP-C preferentially binds to N-terminal region of HJURP at cytoplasm (Supplemental Figure S3), CENP-C–HJURP interaction may not be related to the centromere expansion.
Human HJURP contains a dimerization domain in its C-terminal region (human HJURP554-614), and dimer formation appears to be crucial for HJURP function in human cells (Zasadzinska et al., 2013). It is difficult to detect a homologous dimerization domain in chicken HJURP by sequence comparison due to high sequence divergence. Given that we created the artificial kinetochore in the presence of endogenous HJURP, it is possible that dimer formation of HJURP may facilitate centromere expansion or the association of HJURP with M18BP1/KNL2.
Although we believe that the expansion activity of HJURP is essential for its function, we note that the size of natural kinetochores is 30–40 kb in chicken cells (Shang et al., 2013), and we observed a threefold to fivefold expansion of CENP-A under the tested artificial conditions. Although this expansion appears abnormal compared with natural kinetochores, we propose that this reflects the functional properties of HJURP. In fact, we observed centromere expansion at natural centromeres when HJURP was overexpressed. Because we found that CENP-A overexpression did not simply induce centromere expansion (Shang et al., 2013), our finding is specific for HJURP. Taking the results together, we propose that HJURP contributes to the establishment of a centromere-specific chromatin structure through this chromatin expansion activity.
Because the size of normal centromeres in chicken cells is maintained at 30–40 kb (Shang et al., 2013), there must be mechanisms to suppress centromere expansion. We found that centromere 27, which contains heterochromatin adjacent to its CENP-A domain, did not expand after HJURP overexpression. Therefore it is possible that heterochromatin acts as a barrier for the centromere expansion to maintain centromere size. Indeed, heterochromatin is detected surrounding most native centromeres (Shang et al., 2013). Alternatively or coordinately, there may be a mechanism by which the amount of HJURP is tightly regulated. The mechanism responsible for centromere chromatin expansion through HJURP is an important question for future work. Chromatin remodeling factors such RSF1 or FACT, which localize to kinetochores (Okada et al., 2009; Perpelescu et al., 2009), may be also involved in this process. Identifying these factors is an important next challenge.
In summary, on the basis of the combination of our genetic, biochemical, and chromosome-engineering analyses, we conclude that HJURP functions for CENP-A deposition, as well as contributes to the plastic establishment of a centromeric chromatin structure through its CENP-A expansion activity (Figure 6F).
MATERIALS AND METHODS
Cell culture and transfection
DT40 cells were grown on DMEM supplemented with 10% fetal bovine serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin at 38.5°C. HJURP conditional knockout cells were cultured in the presence of tetracycline (2 μg/ml). Stable cell lines were generated using a plasmid of interest by electroporation (Gene Pulser II; Bio-Rad, Tokyo, Japan). The stable HJURP−/−/GFP-HJURP fusions and HJURP−/−/H2B-RFP clones were positively selected in 2 mg/ml G418 (Sigma-Aldrich, St. Louis, MO). Mis18α−/−/GFP-HJURP and M18BP1−/−/GFP-HJURP clones were obtained through selection with Ecogpt. Artificial kinetochore induction was performed according to a previously described protocol (Hori et al., 2013).
Indirect immunofluorescence and live-cell imaging
Cells probed for anti–CENP-A, -Mis18, and -M18BP/1 were fixed in methanol at −20°C for 20 min and blocked for 30 min in 0.5% bovine serum albumin (BSA)/phosphate-buffered saline (PBS) before the primary antibody exposure. For the rest of the antibodies, cells were fixed in 3% PFA/PBS for 10 min at room temperature, permeabilized with 0.5% NP-40/PBS, and blocked in 0.5% BSA/PBS. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI), and mounting was performed in VectaShield antifading agent (Vector Lab, Burlingame, CA)
Quantification of centromere signals was performed exclusively on mitotic cells for 10 kinetochores on different chromosomes per cell as described previously (Johnston et al., 2010). Images were acquired with a 100× objective lens on an Olympus IX71 inverted microscope (Olympus, Tokyo, Japan) equipped with a CoolSNAP HQ camera (Roper Scientific, Tokyo, Japan). Z-stacks of 14 slices at 0.2-μm steps were collected at exposures of 100 ms for DAPI, 2000–2500 ms for fluorescein isothiocyanate, and 200-500 ms for Cy3. The image analysis was performed using MetaMorph (Molecular Devices, Sunnyvale, CA). The maximum Z-stacked images were adjusted at the same intensity for controls and tetracycline-treated cells.
Living cell analysis was performed as described (Hori et al., 2013). Cells were cultured in the absence or for 48 h in the presence of tetracycline and then transferred to Iwaki dishes and for live-cell imaging with Cell-Voyager CV1000 (Yokogawa, Tokyo, Japan).
Immunoprecipitation
Immunoprecipitation experiments in Figures 3 and 4 and Supplemental Figure S3A were performed in phosphate buffer as previously described (Hori et al., 2008). Anti-GFP antibody (MBL) preincubated with Protein G-Dyna Beads (Novex, Life Technology, Tokyo, Japan) was used. Immunoprecipitations were performed at 4°C overnight. Beads were washed in IP buffer and eluted through incubation for 20 min at 37°C in sample buffer. The anti-FLAG immunoprecipitation in Supplemental Figure S3C was performed as previously described (Dignam et al., 1983) with slight modifications.
ChIP-seq
The chromatin immunoprecipitations and subsequent deep sequencing in Figures 5 and 6 were also performed according to previous reports (Shang et al., 2010, 2013). The MNase (Takara, Kyoto, Japan)-digested chromatin of 1.5 × 109 cells was extracted with 0.5 M NaCl–containing buffer, and the extract was exposed for 4 h at 4°C to Sepharose-Protein G beads preincubated with rabbit anti–CENP-A or -H3K9me3 antibodies (MBL, Nagoya, Japan). Beads were washed, and the bound DNA was purified and applied to deep sequencing.
ChIP-seq libraries were constructed as described in the Illumina TruSeq DNA LT Sample Prep Kit protocols. Briefly, ∼50 ng of purified DNAs were end repaired, followed by the 3′ addition of a single adenosine nucleotide and ligation to universal library adapters. DNA libraries were prepared by eight cycles of PCR amplification. ChIP DNA libraries were sequenced using the Illumina HiSeq 2500. Sequencing of libraries was performed up to 2 × 151 cycles. Image analysis and base calling were performed with the standard Illumina pipeline version RTA1.17.21.3.
Sequencing data were mapped to chicken genome database galGAL4 (UCSC Genome Browser) with a BWA 0.6.2 mapping tool (Li and Durbin, 2009). A 4% mismatch was allowed for the mapping. Figures 5 and 6 represent raw sequence reads. Input data are also shown.
Supplementary Material
Acknowledgments
We are very grateful to Mayumi Takahashi, Kaeko Nakaguchi, and Michiko Arii for technical assistance and Iain Cheeseman for useful comments and critical reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.F. M.P. dedicates this work to the memory of her advisor Kinya Yoda, who performed leading biochemical research on human centromeres.
Abbreviations used:
- CENP
centromere protein
- ChIP
chromatin immunoprecipitation
- HJURP
Holliday junction recognition protein
- KNL
kinetochore null
- M18BP1
Mis18-binding protein 1
- Mis
minichromosome instability.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-02-0094) on June 10, 2015.
REFERENCES
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, Cleveland DW. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev Cell. 2014;29:740–749. doi: 10.1016/j.devcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 2014;8:190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, Trouche D, Earnshaw W, Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, Kimura H, et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell. 2013;24:635–648. doi: 10.1016/j.devcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Muller S, Blumer J, Klare K, Musacchio A, Almouzni G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015;11:22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone Holliday junction recognition protein (HJURP) J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadzinska E, Barnhart-Dailey MC, Kuich PH, Foltz DR. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.