CCTε, a subunit of the oligomeric CCT molecular chaperone, interacts with the cotranscription factor MRTF-A, thus influencing MRTF-A nuclear accumulation upon serum stimulation. This establishes a novel function for CCTε when monomeric and reveals an extended role for CCT in the stringent quality control of actin.
Abstract
Correct protein folding is fundamental for maintaining protein homeostasis and avoiding the formation of potentially cytotoxic protein aggregates. Although some proteins appear to fold unaided, actin requires assistance from the oligomeric molecular chaperone CCT. Here we report an additional connection between CCT and actin by identifying one of the CCT subunits, CCTε, as a component of the myocardin-related cotranscription factor-A (MRTF-A)/serum response factor (SRF) pathway. The SRF pathway registers changes in G-actin levels, leading to the transcriptional up-regulation of a large number of genes after actin polymerization. These genes encode numerous actin-binding proteins as well as actin. We show that depletion of the CCTε subunit by siRNA enhances SRF signaling in cultured mammalian cells by an actin assembly-independent mechanism. Overexpression of CCTε in its monomeric form revealed that CCTε binds via its substrate-binding domain to the C-terminal region of MRTF-A and that CCTε is able to alter the nuclear accumulation of MRTF-A after stimulation by serum addition. Given that the levels of monomeric CCTε conversely reflect the levels of CCT oligomer, our results suggest that CCTε provides a connection between the actin-folding capacity of the cell and actin expression.
INTRODUCTION
The actin filament network is essential for many cellular events, such as receptor signaling, cytokinesis, and cell migration, and enables the cell to respond to changes in the extracellular environment (reviewed by Pollard and Borisy, 2003; Grantham et al., 2012). Actin is a highly abundant protein that is dependent upon interactions with the molecular chaperone CCT in order to reach its native structure (e.g., Llorca et al., 1999), and thus CCT has a major role in maintaining actin homeostasis. The CCT oligomer is comprised of two rings of eight protein subunits (named α to θ in mammalian cells and CCT1–8 in yeast), each encoded by an essential gene (Stoldt et al., 1996; Kalisman et al., 2013). Although there has been much focus on the role of CCT as an oligomeric folding machine, a pool of free, monomeric subunits exists in the cell (Liou and Willison, 1997). This led to the discovery that CCT subunits have additional roles outside of the CCT oligomer, some of which involve actin function. For example, in mammalian cells, small interfering RNA (siRNA) depletion of individual CCT subunits disrupts the oligomer, increasing the levels of nontargeted subunits as monomers, an approach that has revealed a novel role for CCTε in determining cell shape (Brackley and Grantham, 2010). In yeast, mutational analysis of individual CCT subunits reveals specific changes in cell size and sensitivity to the actin-depolymerizing drug latrunculin, confirming the likelihood of CCT subunits functioning outside of the assembled oligomer (Amit et al., 2010). These observations are consistent with monomeric CCT functions being conserved from yeast to mammals. The concept of subunits of multimeric assemblies having additional functions is emphasized by Matalon et al. (2014), who show that not all components of oligomeric assemblies are present in cells in equal amounts. Additional CCT subunit functions, together with the actin-filament severing and capping protein gelsolin being identified as a CCT oligomer–binding protein (Brackley and Grantham, 2011), provide evidence that the role of CCT extends beyond the folding of newly synthesized actin to include modulation of assembled actin structures.
The levels of assembled actin are connected to the transcription of actin via the myocardin-related cotranscription factor-A (MRTF-A)/serum response factor (SRF) pathway. MRTF-A, a cotranscription factor, is retained in the cytosol by interactions with nonassembled actin molecules (G-actin; Mouilleron et al., 2008, 2011). When cell surface receptor–mediated stimulation of actin polymerization leads to a depletion in levels of G-actin, the MRTF-A:actin interaction is disrupted, allowing MRTF-A to accumulate in the nucleus, where it coactivates the transcription factor SRF, leading to increased actin transcription (Vartiainen et al., 2007). SRF also controls the expression of several actin-binding proteins, such as gelsolin, cofilin, and profilin (Sun et al., 2006).
To extend our knowledge of the activity of free, monomeric CCT subunits in mammalian cells, we investigated whether modulation of CCT subunit levels using siRNA depletion affected the SRF signaling pathway. This approach has established that the levels of one CCT subunit, CCTε, are closely coupled to SRF activation. The use of a novel strategy to overexpress CCTε exclusively in its monomeric form, together with protein:protein interaction assays, has revealed an interaction between the apical domain of CCTε and the C-terminal half of MRTF-A. Overexpression of monomeric CCTε changes the rate of MRTF-A accumulation in the nucleus upon serum stimulation, consistent with monomeric CCTε being a component of the SRF pathway. This study emphasizes that the role of CCT extends beyond the folding of actin and shows that the protein quality control mechanisms ensuring the correct folding of actin extend to influencing actin transcription.
RESULTS
Depletion of CCTε by siRNA results in a dramatic increase in SRF activity
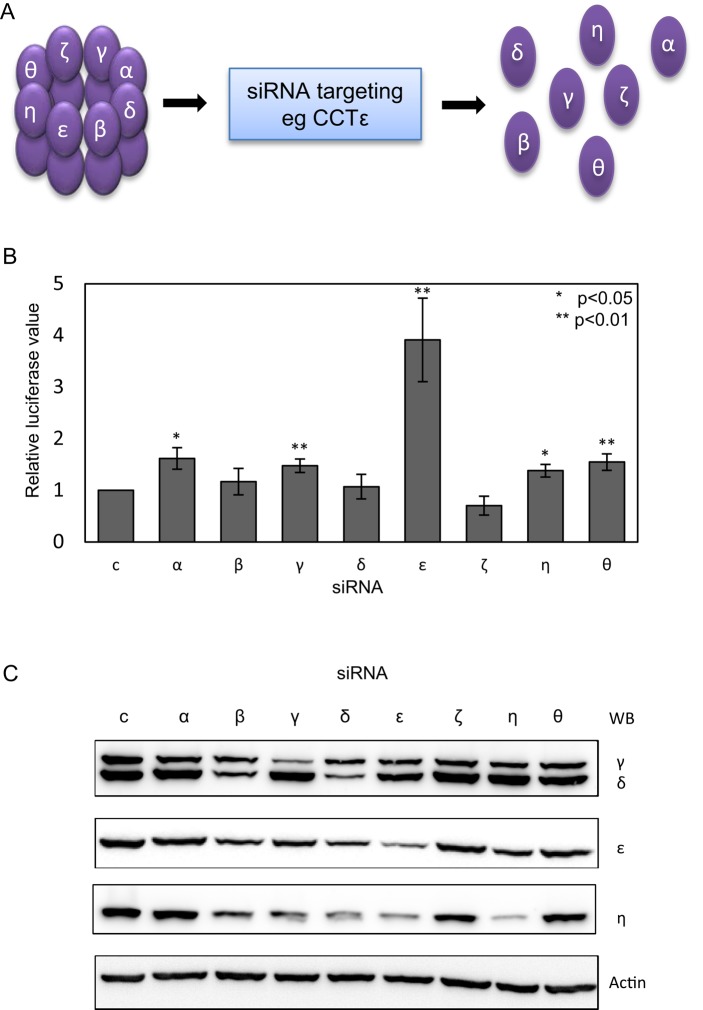
We used an established luciferase-based SRF reporter gene assay previously described by Vartiainen et al. (2007) to determine whether altered levels of CCT subunits have an effect on the MRTF-A/SRF pathway. After depletion of all eight CCT subunits individually by siRNA, we measured SRF-dependent reporter gene expression. The removal of any CCT subunit disrupts the CCT oligomer, resulting in an excess of the nontargeted CCT subunits as free monomers (Grantham et al., 2006; Brackley and Grantham, 2010), as depicted in Figure 1A. The depletion of CCTε resulted in a fourfold increase in luciferase activity (Figure 1B).
FIGURE 1:
Depletion of CCTε by siRNA increases serum response factor activity. (A) SiRNA targeting of a single CCT subunit leads to disassembly of the CCT oligomer and an excess of the nontargeted subunits as free monomers. For simplicity, only the CCT subunits in one ring of the oligomer are labeled. (B) MDA-MB-436 cells were treated with siRNAs targeting each CCT subunit or an Oligofectamine control and an SRF-luciferase reporter gene system used to assess SRF activation after 16 h in 10% serum. Results shown are the mean ± SEM from three experiments. Student's t test was performed for each siRNA compared with control. (C) Western blot analysis of CCT subunit levels after siRNA depletion in cell lysates standardized by cell counting. Antibodies are affinity-purified rabbit polyclonal antibodies to CCTγ and CCTε and rat monoclonal antibodies η81a against CCTη and δ8g against CCTδ.
This effect is specific and not due to differences in the efficiencies of CCT subunit depletion, which were determined for CCTγ (65%), CCTδ (68%), CCTε (73%), and CCTη (87%) by Western blot analysis of equally loaded cell lysate samples standardized by cell counting (Figure 1C). That SRF-activated transcription is substantially increased only upon CCTε depletion is remarkable, since disruption of actin folding will occur due to loss of the assembled CCT oligomer when depleting any of the CCT subunits (Grantham et al., 2006; Brackley and Grantham, 2010). This provides strong evidence for monomeric CCTε being coupled to SRF-dependent gene expression.
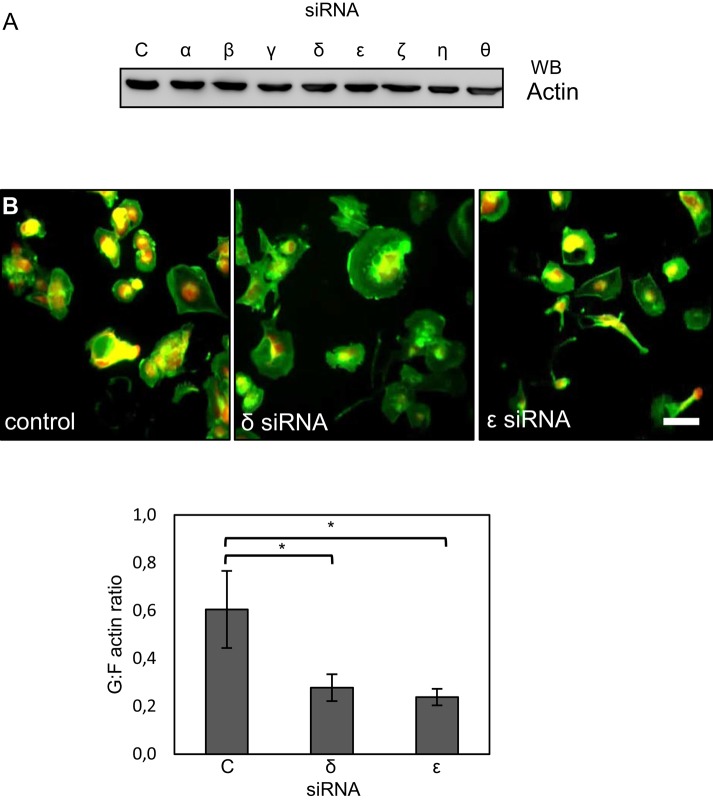
Depletion of CCT subunits reduces the ratio of G- to F-actin
Because the SRF pathway responds to changes in actin polymerization, we decided to investigate whether CCTε depletion enhanced SRF activity via changes in the assembly state of actin. Initially, we confirmed that none of the eight CCT subunit depletions changed the total levels of actin in MDA-MB-436 cells by analyzing actin levels in cell lysates that were equally loaded based on total protein levels (Figure 2A). This is consistent with previous observations in BE cells, in which total actin levels remain unchanged after CCT subunit depletions (Grantham et al., 2006; Brackley and Grantham, 2010). In contrast, assessment of the ratio of G- to F-actin, using an established cell staining approach (Irving et al., 2012), after depletion of either CCTδ or CCTε revealed a decrease in nonfilamentous actin (Figure 2B) similar to the effect of depleting CCTζ (Grantham et al., 2006). Therefore the increase in the MRTF-A/SRF activity specifically upon depletion of CCTε cannot be attributed to a reduction in the G-actin pool due to loss of the CCT oligomer.
FIGURE 2:
Changes in SRF signaling after depletion of CCTε are not a result of CCTε depletion-specific changes in the actin assembly state. (A) Western blot analysis of MDA-MB-436 cell lysates standardized by total protein content show that total actin levels are not changed after CCT siRNA treatment. (B) siRNA targeting of both CCTδ and CCTε reduces the relative levels of nonfilamentous actin, as shown by visualizing filamentous actin with FITC-phalloidin (green) and G-actin with tetramethylrhodamine isothiocyanate–labeled DNaseI (red). Fluorescence intensities in the green and red channels for individual cells were then measured using ImageJ, and the G:F actin ratio was calculated. The mean ± SEM from 10 fields of view in three experiments is shown using the Student's t test for comparison to control cells (*p < 0.05). Scale bar, 20 μm.
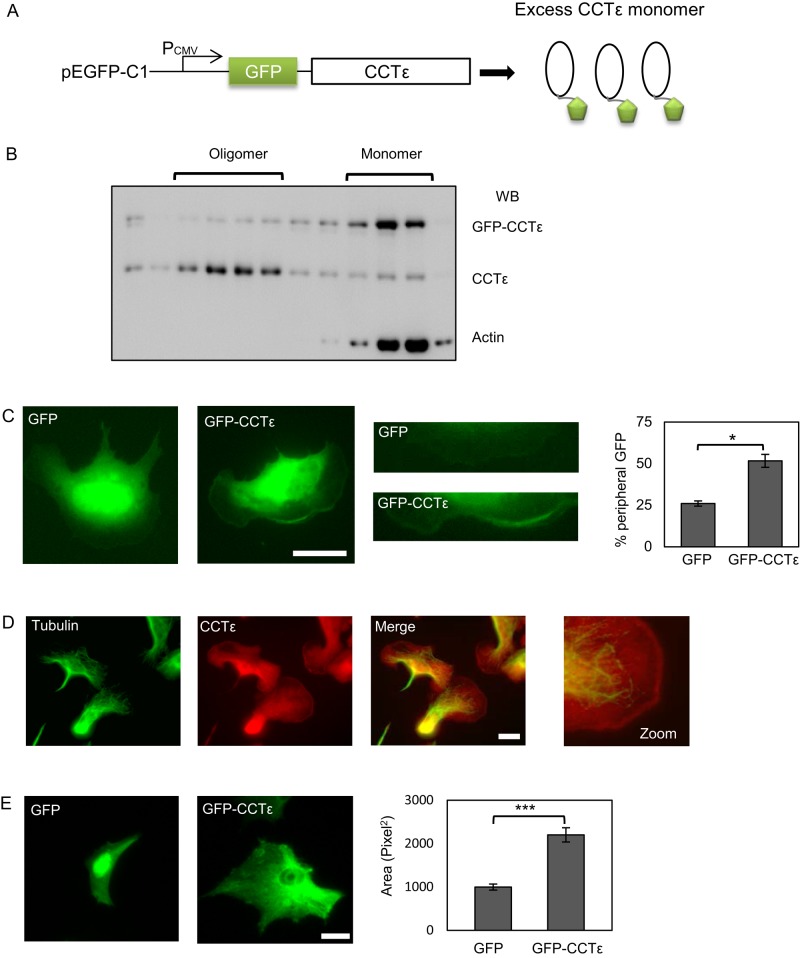
Green fluorescent protein–CCTε as a probe for assessing monomeric CCTε function
Because the effects on SRF signaling after the depletion of CCTε could not be explained by changes to the G-actin pool, we sought to assess directly the function of the monomeric form of CCTε. To this end, we generated an N-terminal green fluorescent protein (GFP)–CCTε fusion protein, which, due to the positioning of the GFP, should be incompatible with incorporation into the CCT oligomer (Figure 3A). Sucrose density gradient fractionation of a transfected BALB 3T3 lysate determined that GFP-CCTε is predominantly monomeric, thereby confirming the validity of this approach to express only the monomeric form of CCTε (Figure 3B). When GFP-CCTε or GFP alone was expressed in B16F1 cells for 24 h, no difference in cell size was observed (unpublished data). However, the GFP-CCTε displayed an increased lamellipodial localization compared with GFP alone (Figure 3C). We then enhanced actin dynamics in B16F1 cells by treatment with AlF4 (Hahne et al., 2001), which activates RhoGTPases and thereby causes increased lamellipodia formation and migration. We then stained cells for CCTε and used an antibody to α-tubulin for costaining, as the microtubules do not extend to the peripheral edges of the lamellipodia, thus eliminating concerns about bleedthrough from the green to the red channel during imaging. This approach clearly visualizes endogenous CCTε at the cell periphery, indicating that the endogenous CCTε and the GFP-CCTε are both able to accumulate at the cell periphery (Figure 3D). We went on to determine the effect of GFP-CCTε expression for 96 h. In comparison to GFP-expressing control cells, expression of GFP-CCTε dramatically increased cell spreading (Figure 3E). These observations of monomeric CCTε enhancing cell spreading are consistent with the depletion of CCTε inducing a narrow phenotype in BE cells after 96 h, as described previously (Brackley and Grantham, 2010), and confirm the suitability of a GFP-CCTε fusion as a probe for CCTε monomer function.
FIGURE 3:
Expression of GFP-CCTε as a probe for CCTε monomer function. (A) Construct design for expression of a GFP-CCTε fusion. (B) Lysate from BALB 3T3 cells expressing GFP-CCTε was fractionated on a 40–10% sucrose density gradient and then analyzed by SDS–PAGE and Western blotting to confirm that placement of the GFP tag renders the subunit monomeric. (C) GFP vector alone and GFP-CCTε localization in B16F1 cells 24 h posttransfection were compared. GFP-CCTε is found specifically to accumulate at the cell periphery and is quantified by counting the percentage of cells with peripheral staining from separate transfections (mean ± SEM, n = 3; *p < 0.05). (D) Staining of B16F1 cells with antibodies to tubulin and CCTε after stimulation with AlF4 shows an accumulation of endogenous CCTε at the cell periphery, using the εAD1 monoclonal antibody. (E) Expression of GFP-CCTε but not GFP alone for 96 h results in increased cell spreading (***p < 0.001). Scale bars, 20 μm.
CCTε interacts with MRTF-A
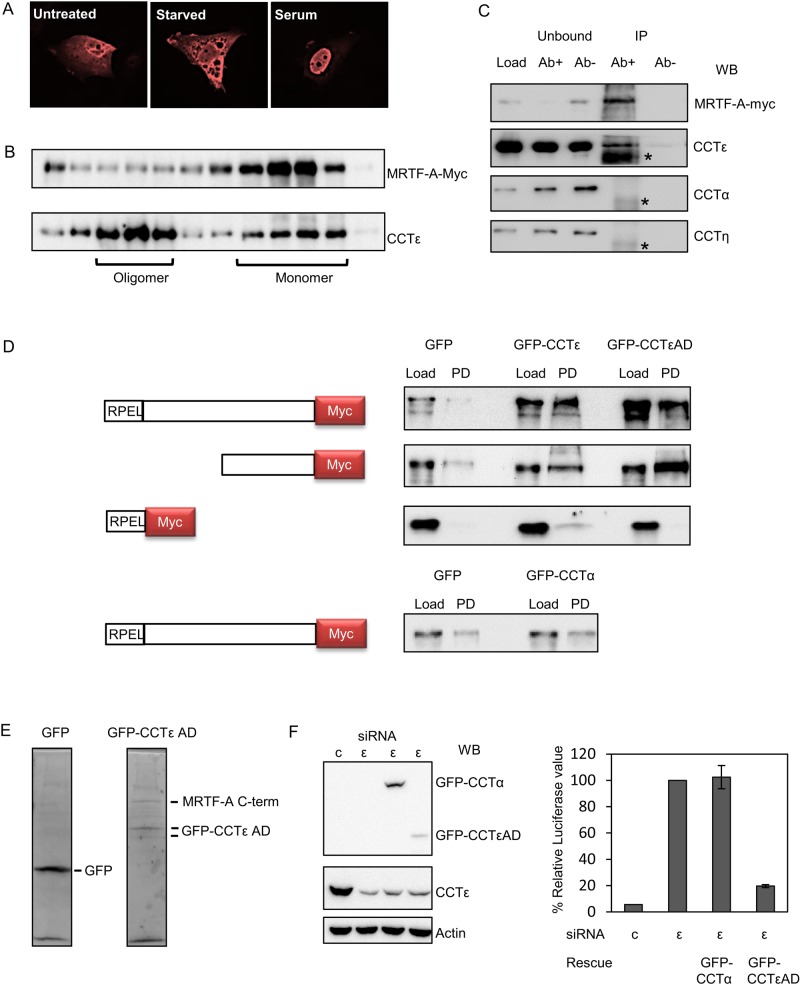
We addressed whether CCTε interacted with MRTF-A by using a C-terminally myc-tagged MRTF-A. The MRTF-A–myc was found to be functional with respect to serum-stimulated translocation to the nucleus (Figure 4A). The profile of MRTF-A–myc was analyzed by sucrose gradient density fractionation of lysates from transfected cells and compared with that of CCTε as a marker for both the CCT oligomer and the pool of free, monomeric CCT subunits. This revealed that the MRTF-A–myc–containing fractions overlapped with those of the CCT monomer pool, whereas cosedimentation with the CCT oligomer was not observed (Figure 4B). An interaction between MRTF-A and endogenous CCTε was then established by coimmunoprecipitation of the endogenous CCTε subunit with MRTF-A–myc, whereas no interaction between MRTF-A–myc and CCTα or CCTη was observed (Figure 4C).
FIGURE 4:
CCTε binds to the cotranscription factor MRTF-A. (A) Nuclear localization of MRTF-A–myc upon serum stimulation by immunofluorescence staining. (B) BALB 3T3 cells transfected with MRTF-A–myc were lysed and soluble proteins analyzed by sucrose density gradient fractionation. Western blotting revealed that the elution profile of MRTF-A overlaps with the free CCT subunit pool but not the CCT oligomer, as defined using antibodies to CCTε. (C) BALB 3T3 cells were transfected with MRTF-A–myc and immunoprecipitation performed with anti–myc antibody–coupled beads (Ab+) or beads alone (Ab–) to show that MRTF-A binds to endogenous CCTε. Samples of the remaining supernatants after incubation with beads (unbound) and the immunoprecipitated material (IP) were resolved by SDS–PAGE and proteins transferred to nitrocellulose followed by Western blotting. Asterisk indicates immunoglobulin G heavy chain. Antibodies used for Western blotting were rat monoclonals α91a, εAD1, and η81a. (D) BALB 3T3 cotransfected with GFP vector, GFP-CCTε, GFP-CCTα, or GFP-CCTε apical domain (AD) and full-length MRTF-A-myc, the C-terminal half of MRTF-A-myc, or the RPEL domain of MRTF-A with a myc tag as indicated. Lysates were incubated with GFP-nanobody beads to pull down GFP proteins (PD), and the collected proteins were analyzed by Western blot using an anti-myc antibody, confirming that the CCTε apical domain recognizes the C-terminal region of MRTF-A. (E) Ponceau staining of the pull-down lanes of GFP and GFP-CCTε apical domain cotransfected with the C-terminal half of MRTF-A. (F) Transfection of GFP-CCTε apical domain, but not GFP-CCTα, rescues the increased SRF signal induced after depletion of CCTε by siRNA.
To determine which region of MRTF-A might interact with CCTε, we prepared two additional constructs encoding either the C-terminal portion of MRTF-A or the actin-binding N-terminal RPEL domain (Mouilleron et al., 2008). A GFP-CCTε apical domain (GFP-CCTεAD) construct was included, as this region is the least conserved between CCT subunits (Kim et al., 1994) and is responsible for subunit-specific interactions (Pappenberger et al., 2002; Spiess et al., 2006). Although the apical domains are divergent between subunits, they are conserved between orthologues (Archibald et al., 2000), consistent with subunit-specific binding properties being conserved. Binding to CCTε was determined by double transfections of BALB 3T3 cells with each of the MRTF-A constructs and GFP, GFP-CCTε, or GFP-CCTεAD, followed by isolation of protein complexes with GFP-nanobody beads. This revealed that full-length GFP-CCTε interacts with full-length MRTF-A. Moreover, using the truncated versions of MRTF-A and CCTε demonstrated that it is the CCTε apical substrate-binding domain that binds to the C-terminal half of MRTF-A (Figure 4D). To address the CCT subunit specificity of the MRTF-A interaction, we assessed MRTF-A binding to GFP-CCTα and found it to be comparable to that of GFP alone, which was used as a negative control throughout (Figure 4D). Bands corresponding to GFP, GFP-CCTεAD, and the C-terminal half of MRTF-A from the nanobody pull down were identified by the alignment of the Ponceau-stained membrane with Western blot images. This did not reveal any significant additional protein bands, consistent with the interaction between CCTε and MRTF-A being direct (Figure 4E). We then asked whether addition of CCTε could rescue the enhanced SRF signaling observed when depleting CCTε by siRNA (Figure 1B). To this end, we used the GFP-CCTεAD construct, as this is not targeted by the CCTε siRNA duplex, and observed a substantial reduction in SRF signaling (Figure 4F). GFP-CCTα, however, did not reduce the SRF signaling, indicating that GFP-CCTα cannot rescue the loss of CCTε (Figure 4F). The binding of MRTF-A to CCTε provides an explanation of how levels of CCTε can modulate SRF signaling.
The CCTε subunit specifically affects accumulation of MRTF-A in the nucleus
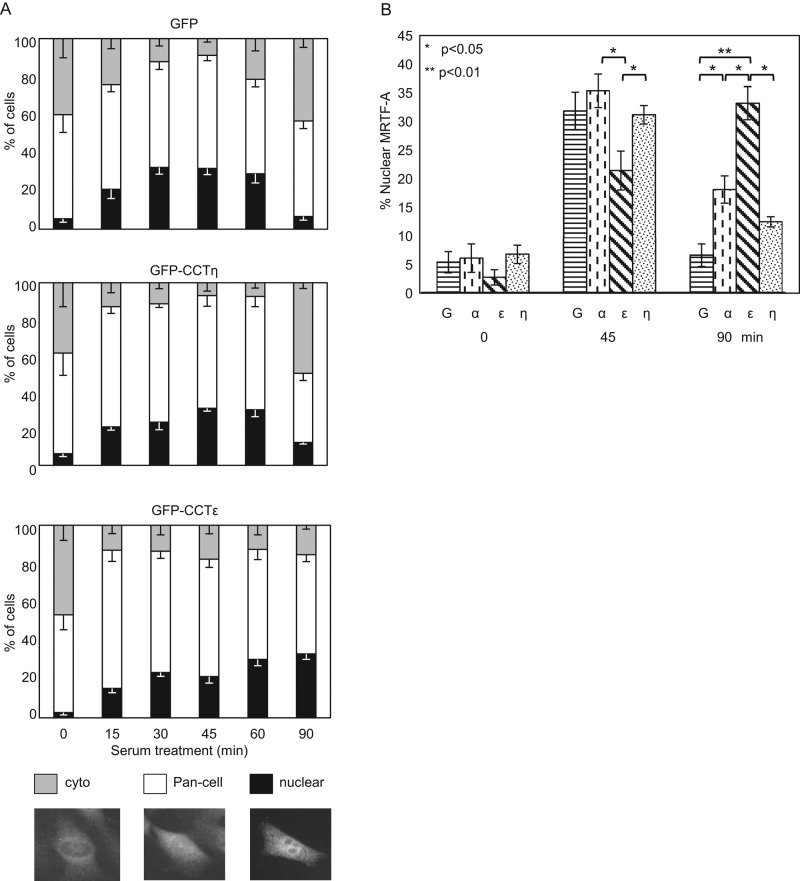
We then addressed the in vivo significance of the CCTε:MRTF-A interaction by assessing MRTF-A nucleocytoplasmic shuttling after overexpression of monomeric CCTε. Cells were transfected with GFP vector, GFP-CCTε, or GFP-CCTη before serum stimulation and fixation. The localization of endogenous MRTF-A was determined by immunostaining and cells classified as having cytosolic, pan-cell, or nuclear MRTF-A (Figure 5A). Under all conditions, the nuclear localization of MRTF-A increased upon serum stimulation. However, cells expressing GFP-CCTε displayed a distinctly different time-dependent profile of MRTF-A accumulation in the nucleus than that of cells expressing GFP-CCTη or GFP alone. Figure 5B shows the quantification of the nuclear accumulation of MRTF-A at 45 min after serum addition, when cells expressing GFP-CCTε have significantly fewer MRTF-A–positive nuclei than cells expressing CCT-GFP-α, GFP-CCTη, or GFP alone. For cells expressing GFP-CCTε, it is not until 90 min after serum addition that cells reach the maximum levels of positive nuclei seen for the other transfections. However, in cells expressing GFP-CCTα, GFP-CCTη, and GFP, at 90 min after serum addition, the MRTF-A distribution was returning to that seen before serum stimulation. Therefore overexpression of CCTε delays the rate of MRTF-A accumulation in the nucleus after addition of serum.
FIGURE 5:
CCTε retains MRTF-A in the cytosol. (A) After transfection of GFP or GFP-CCTε or η, BALB 3T3 cells were starved for 16 h and then stimulated with serum for the times indicated, and the localization of endogenous MRTF-A was analyzed by immunofluorescence. MRTF-A localization was categorized as cytosolic (cyto), pan-cell, and nuclear (mean ± SEM, n = 3). Of note, shifts in cytoplasmic and nuclear MRTF-A between different time points were congruent with each other, whereas pan-cell levels remained relatively constant throughout. (B) Comparison of nuclear localization of MRTF-A upon serum stimulation after transfection with GFP or GFP-CCTα, ε, or η (mean ± SEM, n = 3).
DISCUSSION
In this study, we show that two major pathways in actin modulation, namely its folding and its transcriptional control, are linked, and we identify the CCTε subunit of the CCT molecular chaperone as being central to this interplay. We describe a mechanism by which an interaction between CCTε and the cotranscriptional activator MRTF-A modulates SRF signaling. Our model, summarized in Figure 6, is based on the well-established dependence of actin on the CCT oligomer for its folding (e.g., Sternlicht et al., 1993). The actin-folding capacity of the cell is reduced if levels of assembled CCT oligomer are low, but the resulting increase in the pool of monomeric CCT subunits would provide sufficient monomeric CCTε to delay MRTF-A entering the nucleus. Conversely, high levels of CCT oligomer will result in low levels of monomeric CCTε, releasing the inhibition on the SRF pathway. Because CCT subunit levels are roughly equimolar (with the exception of CCT4 and CCT8, which are higher in yeast; Matalon et al., 2014), it would be necessary to couple the levels of only one of the CCT subunits to such a sensing mechanism. It is probable that either changes between the monomeric and oligomeric conformations of CCTε or the close proximity of neighboring CCT subunits prevents the binding of MRTF-A to CCTε when CCTε is present in the oligomer.
FIGURE 6:
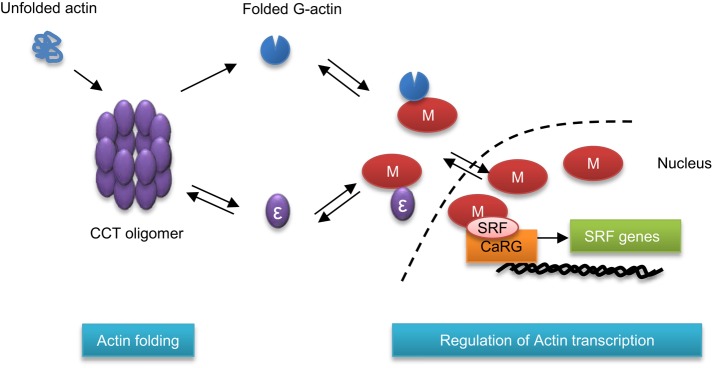
Linking the actin-folding capacity to MRTF-A/SRF signaling. Summary of the interplay between the folding of actin by the CCT oligomer and the SRF pathway. The CCT oligomer is essential for the folding of newly synthesized actin (blue). If less CCT oligomer is assembled, then levels of monomeric CCTε will increase and prevent MRTF-A (shown here as M) from entering the nucleus.
In the context of the siRNA experiments presented in Figure 1, the extremely low levels of CCTε after its depletion mimic the situation in which most CCTε is assembled within oligomers, thus depleting the monomeric CCTε pool. Therefore depletion of CCTε triggers the same cellular response as high levels of CCT oligomer. According to our model, despite there being a decrease in G-actin levels when targeting the other seven CCT subunits (Figure 2; see also Grantham et al., 2006), the higher levels of monomeric CCTε would act as a counterbalance to an increase in SRF signaling activity by ensuring that MRTF-A is retained in the cytosol. This explains why there is no substantial change in SRF signaling after siRNA depletion of the other seven CCT subunits, which leads to CCT oligomer disassembly/reduced G-actin (Figure 1).
Because there are no structural data for full-length MRTF-A, it is impossible to predict whether CCTε would bind via the C-terminal binding site on MRTF-A while actin is bound simultaneously to the N-terminal RPEL domain. Nevertheless, regardless of whether MRTF-A would need to be released by actin in order to allow CCTε binding, CCTε would still exert a secondary level of control on SRF signaling should there be a drop in G-actin levels when assembled CCT is sparse. This ensures that an increase in actin synthesis does not occur when there is insufficient CCT oligomer. The changes to the nuclear accumulation of MRTF-A seen when overexpressing GFP-CCTε are consistent with a mechanism by which the affinity of the MRTF-A:CCTε interaction is sufficient to delay MRTF-A entering the nucleus but does not result in a complete block in translocation after stimulation by serum (Figure 5). Such a mechanism would reduce actin transcription until additional factors are able to stimulate CCT assembly to increase the folding capacity of the cell. As a consequence, the CCTε pool would be depleted, thereby releasing the block on SRF signaling. This is consistent with the observations that the CCT oligomer is highly dynamic with regard to stability in cell lysates but not when purified, suggesting that additional factors influence its assembly (Roobol et al., 1999).
In summary, the work described here establishes a novel function for the CCTε subunit when in its monomeric form and provides evidence of a complex interplay between CCT and actin that extends beyond folding. This emphasizes the stringency of actin modulation and provides a new perspective on SRF pathway signaling.
MATERIALS AND METHODS
Plasmids
Mouse sequences encoding CCTε, CCTε apical domain, CCTη, and CCTα were cloned into pEGFP-C1 to give GFP-CCT subunit fusion proteins. The MRTF-A sequence was amplified from MRTF-A–GFP and cloned into pcDNA3.1 with a C-terminal myc tag.
Cells
MDA-MB-436 and BALB 3T3 cells were grown in RPMI or DMEM, respectively, containing 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin sulfate (PEST), and 5 μg/ml plasmocin and, for MDA-MB-436, 5 μg/ml insulin. B16F1 cells were grown in DMEM containing FCS and PEST. For serum stimulation, cells were starved in medium containing 0.5% FCS for 16 h, then stimulated with medium containing 10% FCS. siRNA depletions targeting each of the human CCT subunits are described by Brackley and Grantham (2010). For plasmid transfections, Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions.
Antibodies
Commercial antibodies were anti–α-tubulin B512 and anti-actin AC15 (Sigma-Aldrich, St. Louis, MO), anti-GFP (Roche, Basel, Switzerland), anti-Mkl1 (MRTF-A; ab49311; Abcam, Cambridge, United Kingdom). Rat monoclonals to CCT subunits were η81a (Llorca et al., 2000), δ8g (Llorca et al., 1999), and α91a and εAD1 (Llorca et al., 2001). Rabbit affinity-purified polyclonals were anti-CCTγ and ε. The anti-myc Jac6 antibody was from the Institute of Cancer Research (London, United Kingdom).
Cell staining and microscopy
Cells were stained as in Brackley and Grantham (2010), except for MRTF-A staining, for which permeabilization was 30 min and the primary antibody was incubated overnight at 4°C. Cells were imaged using a Zeiss Axioplan 2 Imaging microscope with an AxioCamHR camera and AxioVision software.
Sucrose gradient fractionation
Sucrose gradient fractionation and immunoblot analysis were performed as in Brackley and Grantham (2010).
Luciferase assay
MDA-MB-436 cells (100,000 cells/well) were plated in a six-well dish (day 0) and transfected with siRNA targeting CCT subunits on day 1. On day 4, cells were transfected with SRF reporter p3DA.luc, and reference reporter ptk-RL plasmids were then maintained in 10% FCS for 15–20 h. For rescue experiments, the rescue plasmid was transfected together with the SRF reporter and reference reporter plasmids. Luciferase activity was measured and normalized for Renilla luciferase activity using the Dual Luciferase System (Promega, Madison, WI).
Estimation of the ratio of filamentous to nonfilamentous actin
After siRNA treatment, MDA-MB-436 cells were stained with Alexa 594 DNase I (red) and fluorescein isothiocyanate (FITC)–phalloidin (green) for 20 min as described by Irving et al. (2012). The intensity of staining in the green and red channels for individual cells was quantified using ImageJ (National Institutes of Health, Bethesda, MD).
Total actin determination
After siRNA treatment for all CCT subunits, MDA-MB-436 cells were lysed directly into SDS sample buffer and total proteins resolved by SDS–PAGE. Quantification of Coomassie-stained gels using ImageJ was then performed to standardize loadings for quantitative immunoblotting.
Immunoprecipitation of MRTF-A–myc
MRTF-A–myc was immunoprecipitated with Jac6-coupled protein G beads as described by Brackley and Grantham (2010) with an additional incubation for 1 h with nontransfected lysate to enrich for coprecipitating proteins.
GFP nanobody pull-down assay
Transfected cells were lysed on ice by addition of 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.6, 90 mM KCl, and 0.5% Igepal for 5 min. Extracted proteins were clarified by centrifugation at 13,000 rpm for 5 min at 4°C and then incubated with GFP-nanobody beads for 1 h on a rotating wheel in the cold. Beads were washed three times in ice-cold lysis buffer and recovered proteins analyzed by SDS–PAGE and Western blotting.
Acknowledgments
SRF luciferase plasmids (P3DA.luc, pRL.TK, SRF-VP16) and MRTF-A–GFP in pEF N3 were kind gifts from Maria Vartiainen (University of Helsinki, Helsinki, Finland). Rabbit polyclonals to CCT subunits were a gift from Martin Carden (University of Kent, Canterbury, United Kingdom) and α91a a gift from Keith Willison (Imperial College, London, United Kingdom). We thank Peter Carlsson (University of Gothenburg, Gothenburg, Sweden) for comments on the manuscript, as well as the Imaging Facility at Stockholm University. J.G. acknowledges funding from Vinnova, Carl Tryggers Stiftelse, the W and M Lundgrens Fond, and the Royal Society of Arts and Sciences in Gothenburg. J.G., R.K., and K.E. acknowledge funding from the Stiftelsen Olle Engkvist Byggmästare.
Abbreviations used:
- CCT
chaperonin containing tailless complex polypeptide 1
- F-actin
filamentous-actin
- G-actin
globular-actin
- MRTF-A
myocardin-related cotranscription factor-A
- SRF
serum response factor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0048) on June 10, 2015.
REFERENCES
- Amit M, Weisberg SJ, Nadler-Holly M, McCormack EA, Feldmesser E, Kaganovich D, Willison KR, Horovitz A. Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J Mol Biol. 2010;401:532–543. doi: 10.1016/j.jmb.2010.06.037. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Logsdon JM, Jr, Doolittle WF. Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol Biol Evol. 2000;17:1456–1466. doi: 10.1093/oxfordjournals.molbev.a026246. [DOI] [PubMed] [Google Scholar]
- Brackley KI, Grantham J. Subunits of the chaperonin CCT interact with F-actin and influence cell shape and cytoskeletal assembly. Exp Cell Res. 2010;316:543–553. doi: 10.1016/j.yexcr.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Brackley KI, Grantham J. Interactions between the actin filament capping and severing protein gelsolin and the molecular chaperone CCT: evidence for nonclassical substrate interactions. Cell Stress Chaperones. 2011;16:173–179. doi: 10.1007/s12192-010-0230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham J, Brackley KI, Willison KR. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp Cell Res. 2006;312:2309–2324. doi: 10.1016/j.yexcr.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Grantham J, Lassing I, Karlsson R. Controlling the cortical actin motor. Protoplasma. 2012;249:1001–1015. doi: 10.1007/s00709-012-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne P, Sechi A, Benesch S, Small JV. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 2001;492:215–220. doi: 10.1016/s0014-5793(01)02239-6. [DOI] [PubMed] [Google Scholar]
- Irving AT, Wang D, Vasilevski O, Latchoumanin O, Kozer N, Clayton AH, Szczepny A, Morimoto H, Xu D, Williams BR, Sadler AJ. Regulation of actin dynamics by protein kinase R control of gelsolin enforces basal innate immune defense. Immunity. 2012;36:795–806. doi: 10.1016/j.immuni.2012.02.020. [DOI] [PubMed] [Google Scholar]
- Kalisman N, Schroder GF, Levitt M. The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure. 2013;21:540–549. doi: 10.1016/j.str.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Willison KR, Horwich AL. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Liou AK, Willison KR. Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J. 1997;16:4311–4316. doi: 10.1093/emboj/16.14.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Grantham J, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. The “sequential allosteric ring” mechanism in the eukaryotic chaperonin-assisted folding of actin and tubulin. EMBO J. 2001;20:4065–4075. doi: 10.1093/emboj/20.15.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000;19:5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- Matalon O, Horovitz A, Levy ED. Different subunits belonging to the same protein complex often exhibit discordant expression levels and evolutionary properties. Curr Opin Struct Biol. 2014;26:113–120. doi: 10.1016/j.sbi.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 2008;27:3198–3208. doi: 10.1038/emboj.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal. 2011;4:ra40. doi: 10.1126/scisignal.2001750. [DOI] [PubMed] [Google Scholar]
- Pappenberger G, Wilsher JA, Roe SM, Counsell DJ, Willison KR, Pearl LH. Crystal structure of the CCTgamma apical domain: implications for substrate binding to the eukaryotic cytosolic chaperonin. J Mol Biol. 2002;318:1367–1379. doi: 10.1016/s0022-2836(02)00190-0. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Roobol A, Grantham J, Whitaker HC, Carden MJ. Disassembly of the cytosolic chaperonin in mammalian cell extracts at intracellular levels of K+ and ATP. J Biol Chem. 1999;274:19220–19227. doi: 10.1074/jbc.274.27.19220. [DOI] [PubMed] [Google Scholar]
- Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison K, Yaffe MB. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt V, Rademacher F, Kehren V, Ernst JF, Pearce DA, Sherman F. Review: the Cct eukaryotic chaperonin subunits of Saccharomyces cerevisiae and other yeasts. Yeast. 1996;12:523–529. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C523::AID-YEA962%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Sun Q, Chen G, Streb JW, Long X, Yang Y, Stoeckert CJ, Jr, Miano JM. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]