Extracellular vesicle (EV) shedding from ciliated cells is an evolutionarily conserved phenomenon. Caenorhabditis elegans is an excellent in vivo model with which to address fundamental questions regarding the biology of ciliary EVs. The myristoylated protein CIL-7 regulates EV biogenesis, and the myristoylation motif is essential for EV cargo association and function.
Abstract
The cilium both releases and binds to extracellular vesicles (EVs). EVs may be used by cells as a form of intercellular communication and mediate a broad range of physiological and pathological processes. The mammalian polycystins (PCs) localize to cilia, as well as to urinary EVs released from renal epithelial cells. PC ciliary trafficking defects may be an underlying cause of autosomal dominant polycystic kidney disease (PKD), and ciliary–EV interactions have been proposed to play a central role in the biology of PKD. In Caenorhabditis elegans and mammals, PC1 and PC2 act in the same genetic pathway, act in a sensory capacity, localize to cilia, and are contained in secreted EVs, suggesting ancient conservation. However, the relationship between cilia and EVs and the mechanisms generating PC-containing EVs remain an enigma. In a forward genetic screen for regulators of C. elegans PKD-2 ciliary localization, we identified CIL-7, a myristoylated protein that regulates EV biogenesis. Loss of CIL-7 results in male mating behavioral defects, excessive accumulation of EVs in the lumen of the cephalic sensory organ, and failure to release PKD-2::GFP-containing EVs to the environment. Fatty acylation, such as myristoylation and palmitoylation, targets proteins to cilia and flagella. The CIL-7 myristoylation motif is essential for CIL-7 function and for targeting CIL-7 to EVs. C. elegans is a powerful model with which to study ciliary EV biogenesis in vivo and identify cis-targeting motifs such as myristoylation that are necessary for EV–cargo association and function.
INTRODUCTION
Cilia and extracellular vesicles (EVs) are signaling organelles. Cilia act as a cellular antennae and function in sensation (Seeger-Nukpezah and Golemis, 2012), with defects resulting in human ciliopathies. EVs act as intercellular signaling parcels that contain and deliver donor cell cargo to recipient cells without requiring direct contact. In Chlamydomonas, Caenorhabditis elegans, and mammals, EVs are closely associated with cilia, suggesting that cilia may be essential in EV-mediated communication as both senders and receivers (Tanaka et al., 2005; Hogan et al., 2009; Masyuk et al., 2010; Bakeberg et al., 2011; Pampliega et al., 2013; Wood et al., 2013; Wood and Rosenbaum, 2015; Wang et al., 2014).
In humans, the PKD1 and PKD2 genes are needed for kidney function; loss of PKD gene function leads to autosomal dominant polycystic kidney disease (ADPKD; frequency 1/400 to 1/1000), one of the most common monogenic diseases (Harris and Torres, 2014). The PKD gene products, polycystin-1 (PC1) and polycystin-2 (PC2), localize to cilia, as well as to urinary EVs released from renal epithelial cells (Pazour et al., 2002; Yoder et al., 2002; Pisitkun et al., 2004; Hogan et al., 2009). PC ciliary trafficking defects may be an underlying cause of ADPKD (Cai et al., 2014).
The nematode C. elegans is a powerful model system in which to study mechanisms regulating polycystin ciliary receptor localization (Peden and Barr, 2005; Qin et al., 2005; Bae et al., 2006, 2008, 2009; Hu et al., 2006, 2007; Knobel et al., 2008; Wang et al., 2010; Morsci and Barr, 2011; O'Hagan et al., 2011). The C. elegans cilium is a source of bioactive polycystin-containing EVs (Wang et al., 2014). In C. elegans and mammals, PC1 (C. elegans LOV-1) and PC2 (C. elegans PKD-2) act in the same genetic pathway, act in a sensory capacity, localize to cilia, and are contained in secreted EVs (Bae and Barr, 2008; Hogan et al., 2009; Semmo et al., 2014; Wang et al., 2014), indicating ancient conservation.
In mammals, EVs mediate a broad range of physiological processes (Cocucci et al., 2009; Gyorgy et al., 2011; Robbins and Morelli, 2014). Urinary EVs are enriched in PC1, PC2, and the autosomal recessive PKD protein fibrocystin (Hogan et al., 2009) and are a source of biomarkers for renal diseases that include ADPKD (Pisitkun et al., 2004, 2006; Hogan et al., 2014). In autosomal recessive PKD patients and mice, EVs are associated with renal primary cilia (Hogan et al., 2009). In vitro, PC-containing EVs interact with primary cilia of cultured renal cells (Hogan et al., 2009). Consistent with a possible role for EVs in PKD and other ciliopathies, EVs play diabolical roles in the spread of toxic cargo in cancer, infectious diseases, and neurodegenerative disorders (El Andaloussi et al., 2013; Vader et al., 2014). Despite their profound importance, a fundamental understanding of EV biology and the relationship between cilia and EVs is lacking, including mechanisms regulating EV biogenesis, cargo selection, release, interaction with cilia, and in vivo functions.
Here we use a forward genetic screen to identify cil-7, a regulator of LOV-1 and PKD-2 ciliary localization in C. elegans. cil-7 encodes a conserved myristoylated coil-coil protein that is required for the environmental release of PC-containing EVs. cil-7 mutants accumulate EVs in the luminal space surrounding the intact EV-releasing cilium and are defective in PC-mediated male sensory behaviors, suggesting that EVs may be important for the integrity of the sensory organ. The use of C. elegans to identify in vivo ciliary EV regulators such as CIL-7 provides a way to study mechanisms controlling EV biogenesis and signaling and the relationship between cilia, EVs, and disease.
RESULTS
Myristoylated CIL-7 is required for polycystin localization
The C. elegans PCs LOV-1 and PKD-2 localize to the cilia of 21 male-specific ciliated sensory neurons (Figure 1, A and G), which are the cephalic male neurons (CEMs), the hook B type (HOB) neuron, and the ray B type (RnB, where n = 1–9, excluding 6) neurons (Barr and Sternberg, 1999; Barr et al., 2001). lov-1 and pkd-2 are required for several male-specific mating behaviors, including sex drive, response to hermaphrodite contact, and location of the hermaphrodite's vulva (Barr and Sternberg, 1999; Barr et al., 2001; Barrios et al., 2008). C. elegans is a powerful system for identifying genes that are important for PC localization and function (O'Hagan et al., 2014). One such regulator of PKD-2 localization is the kinesin-like protein 6 (KLP-6) of the kinesin-3 family (Peden and Barr, 2005). klp-6 regulates intraflagellar transport (IFT), EV release, and PKD-2::green fluorescent protein (GFP) targeting to EVs (Morsci and Barr, 2011). klp-6 mutant males accumulate PKD-2::GFP at ciliary bases and are response (Rsp) and location of vulva (Lov) defective (Peden and Barr, 2005). klp-6 is coexpressed with lov-1 and pkd-2 in the 21 male-specific B-type ciliated sensory neurons and also in six shared IL2 neurons (found in both males and hermaphrodites). Each of these 27 klp-6–expressing sensory neurons has an environmentally exposed cilium that penetrates the cuticle and possesses the unique ability to shed and release EVs to the environment. Here we refer to these as the EV-releasing ciliated sensory neurons.
FIGURE 1:
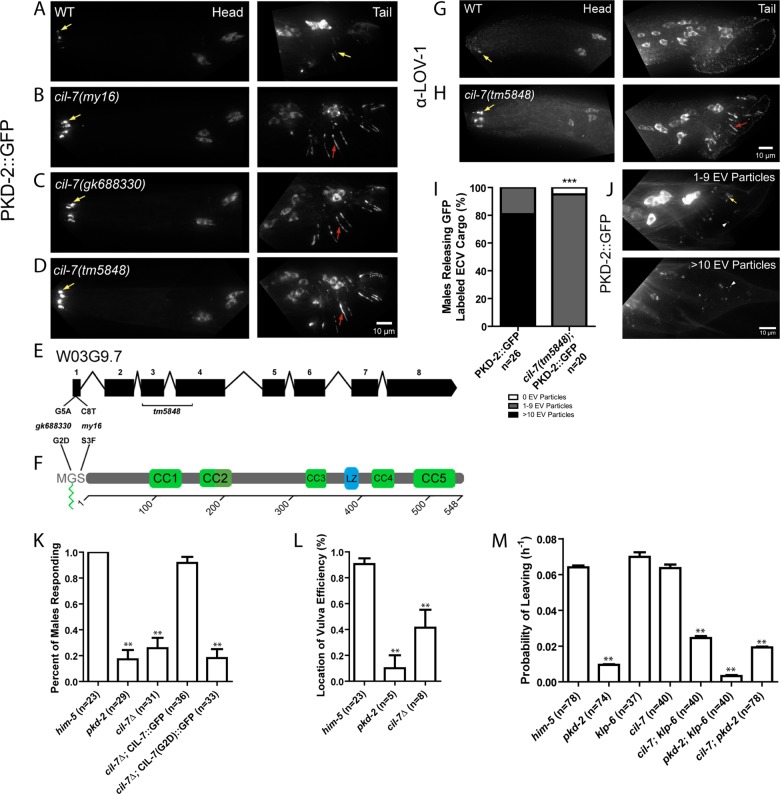
cil-7 is required for the localization of the PCs and for male mating behaviors. (A) PKD-2::GFP localized to the cilia and cell bodies of the CEM, RnB, and HOB neurons of males. (B–D) In cil-7(my16), cil-7(gk688330) and cil-7(tm5848) males, head CEM neurons and tail HOB and RnB neurons accumulated PKD-2::GFP along the dendrites and cilia. (E) cil-7 genomic structure. my16 (C8 → T) and gk688330 (G5 → A) are missense SNPs. tm5848 is an out-of-frame deletion. (F) cil-7 encodes a predicted protein with an N-terminal myristoylation motif, five coiled-coil domains, and a leucine zipper. (G) The anti–LOV-1 antibody showed that endogenous LOV-1 localized to the cilia and cell bodies of the CEM, HOB, and RnB neurons. (H) In cil-7(tm5848) males, endogenous LOV-1 accumulated at the base and along the cilia of the CEM neurons. Excess LOV-1 appeared along the cilia and the dendrites of the RnB neurons. (I) In the cuticle of L4 molting wild-type males, >10 PKD-2::GFP–containing EV particles were observed in 80% of animals scored. In cil-7(tm5848) L4 molting males, much fewer PKD-2::GFP–containing EVs were observed. (J) Representative images of PKD-2::GFP EV particle ranges (1–9 or >10) observed to be trapped in the molted tail cuticle of late L4 males. (K) cil-7(tm5848) males did not respond to hermaphrodite contact. The translational reporter Pcil-7::CIL-7::GFP rescued the response defect of cil-7(tm5848) males. By contrast, the myristoylation-defective reporter Pcil-7::CIL-7(G2D)::GFP failed to rescue response defects of cil-7(tm5848) males. (L) cil-7(tm5848) males were location-of-vulva (Lov) defective. (M) cil-7(tm5848) males were not-leaving-assay defective (nonLas). The cil-7(tm5848); klp-6(my8) double mutant was Las, unlike the cil-7(tm5848) or klp-6(my8) single mutants. In I, ***p < 0.0001 by the Mann–Whitney test. In K, data were analyzed with Fisher's exact test between all groups, followed by Holm–Bonferroni multiple comparison adjustment with a α of 0.01 (**). In L, data were analyzed with the pairwise Mann–Whitney U-test between all groups, followed by Holm–Bonferroni multiple comparison adjustment with a total α of 0.01 (**). In M, PL values were compared by Holm–Bonferroni multiple comparison adjustment with a total α of 0.01 (**). Yellow arrow, cilia; red arrow, dendrite; white arrowhead, EV particle.
The cil-7(my16) mutant was isolated in a forward genetic screen for regulators of PKD-2::GFP ciliary localization (Bae et al., 2008). my16 males accumulated PKD-2::GFP at ciliary bases and displayed a ciliary localization (Cil) phenotype (Figure 1B). We used a combination of single nucleotide polymorphism (SNP) mapping, deficiency mapping, and whole-genome sequencing and determined that my16 was a mutation in the open reading frame of W03G9.7. A fosmid or a single-gene construct of W03G9.7 rescued the my16 Cil phenotype (Supplemental Figure S1A). Two other alleles, gk688330 and tm5848, phenocopy the Cil phenotype of my16 (Figure 1, C and D), fail to complement my16 (Supplemental Figure S1B), and so affect W03G9.7 (Figure 1E). We conclude that my16 is a missense mutation in W03G9.7, which we refer to as cil-7.
cil-7 encodes a predicted protein with a myristoylation motif followed by five coiled-coil domains and a leucine zipper (Figure 1F). CIL-7 contains a 17–amino acid (aa) sequence predicted to be recognized by N-myristoyltransferase (NMT), which cotranslationally adds a 14-carbon saturated fatty acid to the N-terminal glycine (Eisenhaber et al., 2003). The myristoyl group is usually accompanied by a polybasic region or a palmitoyl addition to enable stable membrane association (Resh, 2013). Homology searches using the complete National Center for Biotechnology Information (NCBI) nr database reveal that CIL-7 has homologues in many invertebrate genomes but not in vertebrate lineages (Supplemental Figure S1C). Further, the 17-aa myristoylation sequence is completely conserved in Caenorhabditis species, and the CIL-7 Gly-2 is conserved in most species identified. The third amino acid (CIL-7 Ser-3) is a Cys in more highly diverged species (Supplemental Figure S1D).
The CIL-7 myristoylation motif is disrupted by the my16 and gk688330 alleles (Figure 1, E and F). Proteins that are covalently myristoylated generally contain the sequence Met-Gly-X-X-X-Ser/Thr at the amino terminus. In the NMT recognition sequence, the glycine residue is where the myristoyl moiety is added. In the cil-7(gk688330) mutant, this Gly is changed to Asp. The binding pocket of NMT is narrow, requiring the residue following Gly to be small, such as Ser in CIL-7 (Maurer-Stroh et al., 2002). If this residue is Phe, Lys, Tyr, Trp, or Arg, myristoylation is inhibited (Utsumi et al., 2001). In the cil-7(my16) mutant, this serine residue is changed to Phe (S3F). The tm5848 deletion allele of cil-7 removes the third and part of the fourth exon, producing an out-of-frame deletion (Figure 1F). cil-7 deletion, myristoylation (G2D and S3F), and deficiency/my16 (unpublished data) display similar phenotypes, indicating that each allele is likely a reduction or loss of function. We conclude that CIL-7 myristoylation is essential for its function in localizing PKD-2::GFP to cilia. Here we further characterize the tm5848 deletion allele.
We determined whether cil-7 regulated localization of endogenous LOV-1 and endogenous PKD-2. In wild-type males, anti–LOV-1 and anti–PKD-2 monoclonal antibodies detected endogenous LOV-1 and PKD-2 at the cilia and the cell bodies of the CEM, HOB, and RnB neurons (Wang et al., 2014; Figure 1G and Supplemental Figure S2). In wild-type CEM cilia in the nose, PKD-2 localized to the cilium and ciliary base and was excluded from the transition zone (Supplemental Figure S2, A–C). In cil-7 males, PKD-2 localized to the cilium, was excluded from the transition zone, and abnormally accumulated at the ciliary base (Supplemental Figure S2, D–F). In cil-7 males, LOV-1 also accumulated at ciliary bases of CEM, HOB, and RnB neurons (Figure 1H). PKD-2::GFP and α-LOV-1 staining colocalized in both wild-type and cil-7(tm5848) males (Supplemental Figure S3, A–F). We conclude that CIL-7 is required for the localization of both PCs LOV-1 and PKD-2.
cil-7 is required for release of polycystin-containing EVs
We previously reported that the EV-releasing ciliated neurons shed and release PC-containing EVs into the environment and that these EVs function in animal–animal communication (Wang et al., 2014). To determine whether CIL-7 plays a role in EV shedding or release, we scored the number of GFP-tagged PKD-2 EVs released by the B-type male tail neurons. We counted the number of PKD-2::GFP–containing EVs trapped by the cuticle of late L4 molting males. The majority of wild-type males exhibited an abundance of PKD-2::GFP EVs trapped in the molted cuticle (Figure 1, I and J). In cil-7 males, fewer PKD-2::GFP EVs were released by male tail neurons (Figure 1, I and J). We conclude that the abnormal ciliary base accumulation of the polycystins in cil-7 mutants may reflect a defect in EV shedding or release.
Myristoylation is essential for CIL-7 function
The EV-releasing RnB and HOB neurons are required by the male for response to hermaphrodite contact and location of the hermaphrodite's vulva. cil-7(tm5848) males were Rsp and Lov defective, similar to lov-1, pkd-2, and klp-6 mutants (Figure 1, K and L). A full-length CIL-7::GFP translation fusion reporter rescued the mating defects of cil-7(tm5848) males, demonstrating that this reporter is functional (Figure 1K). By contrast, the myristoylated mutant CIL-7(G2D)::GFP failed to rescue the mating defects of cil-7(tm5848) males (Figure 1K), which is consistent with myristoylation being essential for CIL-7 function.
C. elegans males will leave a food source in search of a mate if no hermaphrodite is present. If both food and a mate are present, males are retained in the food source (Barrios et al., 2008). Mate searching behavior is a form of sex drive and requires the RnB neurons and functional LOV-1 and PKD-2 (Barrios et al., 2008). To our surprise, neither the klp-6 nor the cil-7 single mutant displays a defect in mate searching behavior: their probability of leaving (PL) food in search of a mate was not significantly different from that of wild type (Figure 1M). However, the cil-7; klp-6 double mutant was defective in mate searching (Figure 1M). The cil-7; pkd-2 and pkd-2; klp-6 double mutants resembled the pkd-2 single mutant, indicating that pkd-2 is epistatic to cil-7 and klp-6. Combined, these results suggest that, with respect to male mate searching, klp-6 and cil-7 act in genetically redundant pathways that act upstream of pkd-2.
cil-7 is expressed in the 27 EV-releasing ciliated sensory neurons
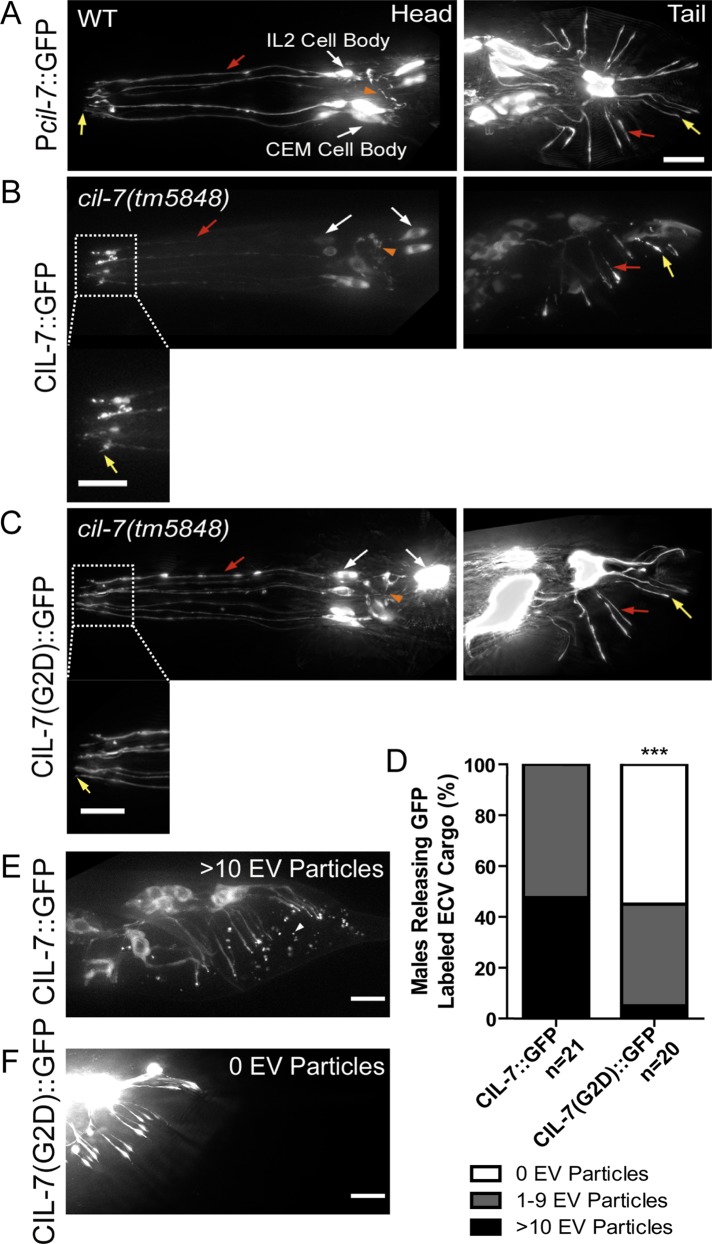
A cil-7 transcriptional GFP reporter (cil-7 promoter [Pcil-7::gfp]) was expressed in the 27 EV-releasing neurons in males and in the 6 EV-releasing IL2 neurons in hermaphrodites (Figure 2A). The translational and functional CIL-7::GFP reporter localized to cell bodies (excluding nuclei), dendrites, axons, and the cilia and ciliary bases of the EV-releasing sensory neurons (Figure 2B). Of interest, CIL-7::GFP was visible in EVs in 100% of males scored. While the myristoylation mutant CIL-7(G2D)::GFP did not perturb the overall distribution of CIL-7 within neurons (Figure 2C), CIL-7(G2D)::GFP was not enriched at ciliary bases and CIL-7(G2D)::GFP–containing EVs were observed in <50% of animals scored (Figure 2, D–F). We conclude that CIL-7 is EV cargo and that the CIL-7 myristoylation motif is required for efficient targeting or tethering of CIL-7 to EVs.
FIGURE 2:
cil-7 is expressed in the 27 EV-releasing ciliated sensory neurons and is targeted to EVs in a myristoylation motif–dependent manner. (A) Pcil-7::GFP was expressed in CEM, RnB, HOB, and IL2 neurons. (B) Pcil-7::CIL-7::GFP in cil-7(tm5848) males localized throughout neurons, including cilia, but was excluded from the nucleus. Pcil-7::CIL-7::GFP rescues cil-7(tm5848) defects (Figure 1K), indicating that this reporter is functional. (C) Pcil-7::CIL-7(G2D)::GFP in cil-7(tm5848) males localized throughout neurons, including cilia, but was excluded from the nucleus. Pcil-7::CIL-7(G2D)::GFP does not rescue cil-7(tm5848) defects (Figure 1K), indicating that this reporter is not functional. (D) CIL-7::GFP as EV cargo was trapped in the molted tail cuticle of the late L4 male in a range from either 1–9 EV particles or >10 EV particles per male tail. The CIL-7(G2D)::GFP myristoylation mutant is not efficiently targeted to EVs. (E) In 100% of animals, CIL-7::GFP was observed in the molted tail cuticle in varying ranges; depicted here is an example of >10 EV particles. (F) In <50% of animals, CIL-7 (G2D)::GFP was observed as EV cargo. In E, ***p < 0.0001 by the Mann–Whitney test. Yellow arrow, cilia; red arrow, dendrite; white arrow, cell body; orange arrowhead, axon; white arrowhead, EV particle.
CIL-7 EVs are released in a klp-6 dependent manner
Similar to cil-7, the kinesin-3 KLP-6 is expressed in EV-releasing neurons, is required for response and vulva location behavior (Peden and Barr, 2005) but not sex drive (Figure 1M), and regulates release of PC-containing EVs (Wang et al., 2014). To determine whether klp-6 controls CIL-7 EV release, we examined CIL-7::GFP in klp-6 mutant animals. In klp-6(my8) mutants, CIL-7::GFP accumulated at ciliary bases of CEM and IL2 sensory neurons (Supplemental Figure S4, A and B), which is consistent with CIL-7 being EV cargo and klp-6 mutants being defective in EV release. In the cil-7 mutant, KLP-6::GFP was not altered (Supplemental Figure S4, C and D). This last result is not surprising, given that, unlike CIL-7, KLP-6 functions within the cilium (Morsci and Barr, 2011) and is not a cargo of ciliary EVs (Wang et al., 2014).
EVs accumulate in the cephalic lumen of cil-7 males
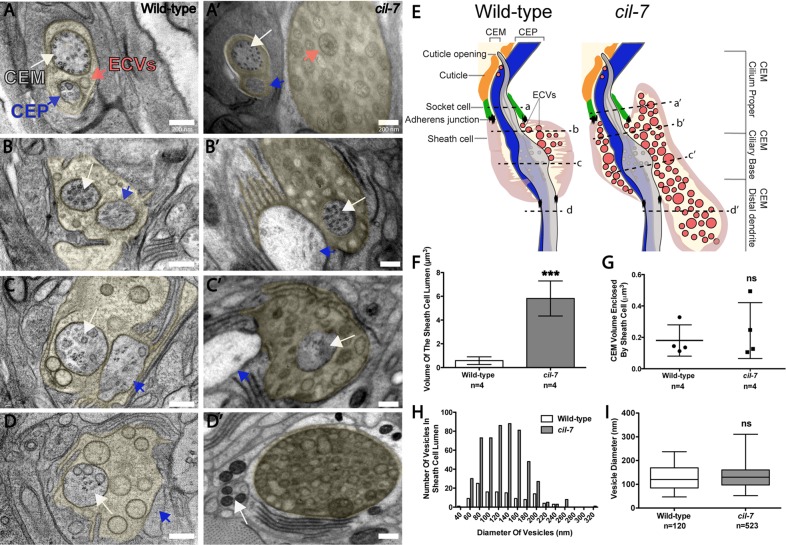
We used transmission electron microscopy (TEM) to analyze the ultrastructure of EV-releasing neurons in cil-7(tm5848) animals. In the wild-type male cephalic sensillum, EVs are found in the cephalic lumenal space that is created by the encapsulating cuticle, socket, and sheath cells and surrounds the CEM and CEP neurons (Wang et al., 2014). In hermaphrodites, EVs are not found in the cephalic lumen that lacks a CEM neuron (Wang et al., 2014). In klp-6 mutant males, a large number of EVs accumulate in the cephalic lumen (Wang et al., 2014). In cil-7 mutant males, an abundance of EVs was also observed in the lumen, spanning approximately the space between the level of the adherens junction connecting the sheath and socket cell and to the level of the distal dendrite below the transition zone (Figures 3, A′– D′). This lumenal space was also distended and increased in volume compared with wild type (Figure 3, E and F). The cil-7 lumenal space distension may be the result of increased quantity of EVs. Sheath cell spanning volumes of the CEM and CEP were not significantly different between wild-type and cil-7 males, suggesting that EV shedding into the sheath cell lumen is not perturbed (Figure 3G and unpublished data). In wild-type males, diameters of the EVs range from 46 to 237 nm, with a mean ± SD of 129.1 ± 49.4 nm, consistent with published results (Wang et al., 2014). cil-7 animals have a similar EV diameter distribution, with a mean ± SD of 131.8 ± 43.3 nm. However, cil-7 animals showed a slightly positive skew (skewness: wild type, 0.46; cil-7, 0.57), suggesting that cil-7 mutants possess a higher count of larger-diameter vesicles (Figure 3, H and I). CEM ciliary ultrastructure appeared normal in cil-7 mutant males. We conclude that CIL-7 is both a cargo of EVs (Figure 2, D and E) and a regulator of EV biogenesis.
FIGURE 3:
cil-7 mutants accumulate EVs in the cephalic lumen but are not defective in ciliogenesis. (A–D) Series of TEM images in a wild-type animal displaying EVs in the sheath cell lumen that surrounds the CEM and CEP distal dendritic region (lumen is pseudocolored yellow). (A′–D′) TEM images revealed that cil-7(tm5848) males had excessive amounts of EVs in the sheath cell lumen. Both CEM and CEP axonemes appeared normal. (E) Cartoon depiction of the cil-7(tm5848) mutant phenotype compared with wild type, including increased quantity of EVs and distended sheath cell lumen. Dashed lines indicate the location of the A–D and A′–D′ TEM images with respect to the cephalic sensillum. (F) Compared to wild type (0.6 ± 0.3 μm3), the cil-7(tm5848) sheath cell lumen is 10 times larger (5.8 ± 1.5 μm3 [average volume ± SD]; n = number of cephalic sensilla). (G) The average CEM volume enclosed within the sheath cell that surrounds the CEM distal dendrite and axoneme in wild type was 0.2 ± 0.1 μm3, and the corresponding volume in cil-7(tm5848) was 0.24 ± 0.2 μm3 (average volume ± SD; n = number of cephalic sensilla), indicating that the integrity of CEM neurons is normal in cil-7 mutants. (H) Compared to wild type, cil-7(tm5848) animals displayed an increased number of EVs in the sheath cell lumen. The frequency distribution of EV sizes in cil-7(tm5848) was slightly favored toward EVs above the average mean (skewness of the diameter distribution as measured in Excel: wild type, 0.46; cil-7, 0.57). (I) The average EV diameter of wild-type and cil-7(tm5848) males was not significantly different. The average EV diameter for wild type was 129.1 ± 49.4 nm and the average vesicle diameter for cil-7(tm5848) was 131.8 ± 43.3 nm (average diameter ± SD). In F, ***p < 0.001 by the unpaired t test. In G, a nonsignificant difference was determined by the unpaired t test. In I, a nonsignificant difference was determined by Welch's t Test. Box-and-whisker plots are included in I, where the plot is divided into limits that are each 25% of the population.
DISCUSSION
Here we identify a novel ciliary protein CIL-7 that is required for PC-mediated sensory signaling and regulates EV biogenesis, particularly the release of PKD-2::GFP–containing EVs into the environment. Myristoylation is essential for CIL-7 function, including its association with EVs (Figure 2, D–F) and its role in PC-dependent mating behaviors (Figure 1K). N-myristoylation is used by proteins for membrane anchoring and for ciliary localization of proteins in Trypanosome flagella, C. elegans sensory neurons, mammalian photoreceptors, and retinal pigment epithelial cells (Ramulu and Nathans, 2001; Evans et al., 2010; Maric et al., 2010; Wright et al., 2011). In Jurkat T-cells, myristoylation signals target proteins to EVs (Shen et al., 2011). In the cpk mouse model of PKD, the cpk mutation lies in the cystin gene, which encodes a myristoylated cilia- and EV-associated protein (Hogan et al., 2009; Tao et al., 2009). The cystin myristoylation signal is necessary for ciliary targeting in inner medullary collecting duct cells (Tao et al., 2009). Ours is the first identification of a cis-acting motif that is essential for EV targeting in vivo and demonstrates that our C. elegans system has the power to identify EV biogenesis regulators, EV cargo, and EV targeting sequences.
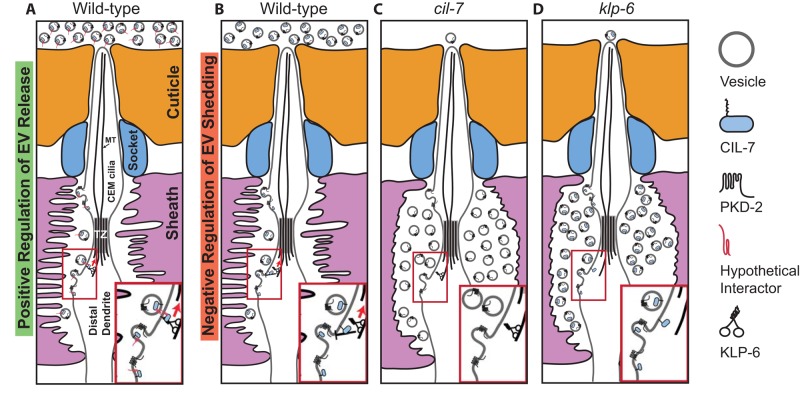
Ultrastructural analysis indicates that C. elegans EVs may bud from the base of the cilium and that, in living animals, GFP-tagged EVs are released from the cuticular pore to the environment (Figures 1J and 2E; Wang et al., 2014). How are EVs shed, transported from the lumen, and released to the environment? klp-6 and cil-7 may control EV biogenesis as positive regulators of EV release (Figure 4A) or negative regulators of shedding (Figure 4B). In Chlamydomonas, polystyrene microspheres adhere to and are moved bidirectionally along the flagellar surface (Bloodgood, 1988, 1995). IFT drives flagellar gliding motility and the transport of the major flagellar surface glycoprotein protein FMG1-B (Shih et al., 2013). When an anti–FMG1-B antibody is attached to beads, beads and IFT trains move at similar speeds. In ctenophores, or comb jellies, individual cells are transported distally up the surface of the cilia, independent of ciliary beating, to build the statolith, a gravity-sensing organ (Noda and Tamm, 2014). In a similar scenario, IFT and KLP-6 may propel EVs along the external ciliary surface (Figure 4A). In this model, EVs express an unidentified surface protein that may act between CIL-7 and a motor. EVs would move in a KLP-6–dependent and IFT-dependent manner, consistent with PKD-2::GFP EV release being blocked in klp-6 and IFT mutant backgrounds (Wang et al., 2014). Alternatively, CIL-7 and KLP-6 may be negative regulators of EV shedding (Figure 4B). In either scenario, the absence of cil-7 or klp-6 results in the accumulation of EVs in the cephalic lumen (Figure 4, C and D).
FIGURE 4:
Models for CIL-7– and KLP-6–mediated EV biogenesis. (A) KLP-6 and CIL-7 may be positive regulators of EV environmental release. In this model, EVs bud from the ciliary base and are shed into the cephalic lumen. From here, KLP-6 and IFT may propel EVs along the ciliary membrane surface to the cuticular opening. Myristoylated, membrane-associated CIL-7 is EV cargo that acts with an unidentified transmembrane tether. The tethered EV binds a ciliary surface protein that is transported by KLP-6 and IFT. Inset, the EV tethered to an unidentified protein such as a flagellar membrane glycoprotein. (B) CIL-7 and KLP-6 may act as dual negative regulators of EV shedding. Inset, a negative feedback loop with CIL-7 and KLP-6 acting in concert to regulate EV shedding. (C, D) In the absence of cil-7 or klp-6 as a positive regulator of EV release or negative regulator of EV shedding, the net result is an accumulation of EVs within the cephalic lumen. In both models, klp-6 acts within the EV-releasing ciliated sensory neurons, whereas cil-7 may act in the EV-releasing cell or in the EV itself. Insets of C and D depict EV shedding that is not regulated in the absence of CIL-7 and KLP-6, respectively. TZ, transition zone.
In Chlamydomonas, C. elegans, and mammals, EVs are closely associated with cilia (Tanaka et al., 2005; Hogan et al., 2009; Pampliega et al., 2013; Wood et al., 2013; Wang et al., 2014), suggesting that the cilium is essential in EV-mediated communication or that EVs are important for the health of the cilium. Alternatively, EVs may not be important for ciliary structure but instead may be for the integrity or function of the sensory organ. Consistent with this possibility, cil-7 and klp-6 EV-release mutants have normal CEM cilia, accumulate EVs in the distended lumen of the cephalic sensory organ (Figure 3), and are defective in the male mating behaviors (Figure 2, K–M).
In addition to their role in intercellular signaling, EVs also signal between animals and between species. In C. elegans and Drosophila, EVs act as animal-to-animal communication devices (Corrigan et al., 2014; Wang et al., 2014). EVs also function as interspecies signaling agents. The gastrointestinal gut nematode Heligmosomoides polygyrus secretes and transfers EVs containing microRNAs (miRNAS) to modulate the innate immune response of mammalian cells (Buck et al., 2014). Therefore the release of EVs has broad implications for communication between cells, between organisms, and between species. Whereas homologues are found in many invertebrate genomes, CIL-7 is absent from vertebrate genomes. In the last decade, evolutionary genetics has shown that genes come and go along lineages with amazing fluidity, even genes that play essential physiological and developmental roles such as miRNAs or transcription factors. Another gene could do the job of CIL-7 in vertebrates; the primary sequence might be diverged but secondary structure conserved. Identifying EV-release regulators such as CIL-7 will provide insight into mechanisms controlling EV biogenesis and signaling and the relationship between cilia, EVs, and disease.
MATERIALS AND METHODS
Strains and maintenance
Transgenic reporters used.
We used myIs1 [PKD-2::GFP + Punc-122::GFP] IV, myEx686 [Pklp-6::GFP::gKLP-6_3′UTR + pBX], myEx815 [Pcil-7::gCIL-7::GFP_3′UTR + ccRFP], myEx816 [Pcil-7::GFP_3′UTR + ccRFP], and myEx847 [Pcil-7::gCIL-7G(2)->D::GFP_3′UTR + ccRFP].
Alleles used.
We used the following alleles:
LGI: cil-7(my16), cil-7(gk688330), cil-7(tm5848)
LGIII: klp-6(my8)
LGIV: pkd-2(sy606)
LGV: him-5(e1490)
Strain list.
We used the following strains:
CB169: unc-31(e169) IV
CB1490: him-5(e1490) V
PT9: pkd-2(sy606) IV; him-5(e1490) V
PT495: myIs1 IV; him-5(e1490) V
PT1194: klp-6(my8) III; him-5(e1490) V
PT1197: klp-6(my8) III; pkd-2(sy606) IV; him-5(e1490) V
PT1319: pha-1(e2123) III; nphp-4(tm925) him-5(e1490)V; myEx514[Ppkd-2::NPHP-4::GFP + pBX]
PT1646: my16; myIs1 pkd-2(sy606) IV; him-5(e1490) V
PT2681: cil-7(tm5848) I; myIs1 IV; him-5(e1490) V
PT2682: cil-7(tm5848) I; him-5(e1490) V
PT2687: him-5(e1490)V; myEx815[Pcil-7::gCIL-7::GFP_3'UTR + ccRFP]
PT2688: him-5(e1490) V; myEx816[Pcil-7::GFP_3'UTR+ccRFP]
PT2763: cil-7(gk688330) I; myIs1 IV; him-5(e1490) V
PT2764: cil-7(tm5848)I; him-5(e1490)V; myEx815[Pcil-7::gCIL-7::GFP_3′UTR+ccRFP]
PT2765: him-5(e1490)V; myEx847[Pcil-7::gCIL-7G(2)->D::GFP_3′UTR + ccRFP]
PT2766: cil-7(tm5848)I; him-5(e1490)V;myEx847[Pcil-7::gCIL-7G(2)->D::GFP_3′UTR + ccRFP]
PT2768: cil-7(tm5848) I; klp-6(my8) III; him-5(e1490) V
PT2770: cil-7(tm5848) I; pkd-2(sy606) IV; him-5(e1490) V
PT2773: klp-6(my8) III; him-5(e1490) V; myEx815[Pcil-7::gCIL-7::GFP_3′UTR + ccRFP]
PT2776: cil-7(tm5848)I; him-5(e1490)V; myEx686[Pklp-6::GFP::gKLP-6_3′UTR + pBX]
PT2983: cil-7(tm5848) I; pha-1(e2123) III; ?nphp-4(tm925) him-5(e1490)V; myEx514[Ppkd-2::NPHP-4::GFP + pBX]
General molecular biology
Mapping.
SNP mapping was performed as described (Davis et al., 2005), followed by deficiency mapping. The following deficiencies complemented the my16 Cil defect: hDf17, qDf7, qDf6, dxDf1, qDf3, and tDf3. However, my16 and sDf4 failed to complement, narrowing the my16 location region to −2.265 and +0.08 cM. Whole-genome sequencing was performed courtesy of Richard Poole of Oliver Hobert's lab (Columbia University, New York, NY) and identified a missense SNP in W03G9.7. From wild-type genomic DNA, W03G9.7 was amplified as a single-gene construct and rescued the my16 Cil phenotype in transgenic animals. gk688330 is a missense SNP, and the nucleotide change involves the G5 → A change. tm5848 is an out-of-frame deletion covering part of the third and fourth exons, resulting in a frameshift.
Construct generation.
The CIL-7 transcriptional and translational reporters were generated by PCR fusion (Hobert, 2002). The cil-7 promoter used 564 base pairs and standard PCR techniques and high-fidelity Phusion DNA Polymerase (New England BioLabs, Ipswich, MA). GFP was amplified from the pPD95.75 plasmid from the Andrew Fire lab (Stanford School of Medicine, Stanford, CA). Both the CIL-7 transcriptional and translational reporters were introduced into the C. elegans germline by microinjection. The transcriptional reporter was injected at a concentration of 35 ng/μl and the translational reporter at a concentration of 1 ng/μl. Injecting the translational reporter at 10 ng/μl caused neuron cell damage. The single-gene fragment of CIL-7 that rescued the cil-7(my16) phenotype also included the CIL-7 promoter and was injected at 0.3 ng/μl.
PCR fusion products were amplified from genomic DNA. Primers sets used for PCR fusion products were as follows and are labeled as published (Hobert, 2002):
CIL-7 transcriptional reporter (myEx816): primer A (P1W03G9.7), 5′GCTGGGAGTCGATACATGGT3′; primer B (P2W03G9.7tr), 5′TTCTTCTCCTTTACTCAGTGAAGAGCCCATAATCAGC3′; primer C (P3W03G9.7tr). 5′GGGCTCTTCACTGAGTAAAGGAGAAGAACTTTTCACT3′; and primer D (P4GFP3′UTR), 5′CAAACCCAAACCTTCTTCCG3′.
CIL-7 translational reporter (myEx815): primer A (P1W03G9.7), 5′GCTGGGAGTCGATACATGGT3′; primer B (P2W03G9.7tl), 5′TTCTTCTCCTTTACTATGATGTGCAGACTTCTTCTTTC3′; primer C (P3W03G9.7tl), 5′AGTCTGCACATCATAGTAAAGGAGAAGAACTTTTCACT3′; and primer D, (P4GFP3′UTR) 5′CAAACCCAAACCTTCTTCCG3′.
The W03G9.7 single-gene rescue product was amplified off of the WRM069cF09 fosmid using the primer set 5′CTCAACAGCAGCGACAACAT3′ and 5′GCTGGGAGTCGATACATGGT3′.
Imaging
All imaging was performed using a Zeiss Axio Imager.D1m microscope using a 100× objective with a Q imaging Regtiga-SRV camera. Images were viewed using MetaMorph version 7.7.7.0 software (Molecular Devices, Sunnyvale, CA). Images were taken at 5-MHz gain and 200-ms exposure. Images were processed using AutoQuant X, Auto Deblur Gold WF Version X2.2.2 software (Media Cybernetics, Rockville, MD) and subsequently with Photoshop CS3 Extended Version 10.0 software (Adobe). Young adult male C. elegans were picked at the L4 stage and imaged 24 h later. C. elegans were imaged using 5% agarose pads and allowed to remain in 5 μl of 50 mM sodium azide for 3 min for anesthetization.
Transmission electron microscopy
cil-7 and wild-type young adult animals were fixed using high-pressure freeze fixation and freeze substitution in 2% OsO4 plus 2% water in acetone as the primary fixative (Weimer, 2006). Samples were slowly freeze-substituted in an RMC freeze substitution device before infiltration with Embed-812 plastic resin.
For TEM, serial sections (75-nm thickness) of fixed animals were collected on copper slot grids coated with Formvar and evaporated carbon and stained with 4% uranyl acetate in 70% methanol, followed by washing and incubating with aqueous lead citrate. Images were captured on a Philips CM10 transmission electron microscope at 80 kV with a Morada 11-megapixel TEM charge-coupled device (CCD) camera driven by iTEM software (Olympus Soft Imaging Solutions).
Extracellular vesicle scoring
Adult males release LOV-1 and PKD-2 EVs from the 21 B-type sensory neurons, and EVs released from the RnB cilia can be trapped in a molted tail cuticle of late L4 males, for which the rays and fan are fully developed (Wang et al., 2014). Late L4 males were chosen for ease of scoring because the molted tail cuticle conveniently traps any EV released from the exposed cilium of the RnB neurons. Late L4 males were picked for imaging in which the shedding cuticle surrounding the tail could be easily visualized. EVs were quantified by generating Z-stacks taken for at least 20 animals for each strain of interest. Imaged Z-stacks were quantified to have 0 PKD-2::GFP–tagged EV particles, 1–9 EV particles, or >10 EV particles.
Cephalic lumen scoring
Both wild-type and cil-7 measurements were derived from serial-section TEM images. Measurements of the inner sheath cell border and the CEM and CEP cell outline were taken in order to estimate the area of the sheath cell lumen. The measurements were taken with the top tight junction made by the sheath cell with the socket cell as the starting point. The lower ending point for taking measurements was the bottom tight junction made by the sheath cell with the CEM-CEP distal dendrites. To obtain volumes, the areas measured were multiplied by the section thickness. To obtain the sheath cell lumen volume, the total lumen volume was subtracted by the corresponding sum of the CEM and CEP volumes. Four CEM sensilla were measured in each wild-type and cil-7 animal.
EV diameter scoring
The TEM images of a wild-type animal were scored for the diameter of EVs in the cephalic lumen of the dorsal right, dorsal left, and ventral left quadrants. The TEM images of a cil-7 animal were scored for the diameter of EVs in the cephalic lumen of the dorsal right, ventral right, and ventral left quadrants. The longest possible diameter for each EV was selected for both the wild-type and cil-7 animals. EVs scored were located approximately between the sheath and socket cell tight junction connection and below the transition zone at the distal dendrite level. Images scored were with a Morada 11-megapixel TEM CCD camera driven by iTEM software. Images were analyzed using MetaMorph version 7.7.7.0 software and ImageJ 1.49k (Wayne Rasband, National Institutes of Health, Bethesda, MD).
Antibody staining
Animals were staged young adults and were washed off plates with M9. Antibody staining against LOV-1 and PKD-2 was performed using a modified Finney–Ruvkun protocol (Bettinger et al., 1996). The monoclonal LOV-1 and PKD-2 primary antibodies were generated by Abmart (Berkeley Heights, NJ; Wang et al., 2014). The monoclonal LOV-1 antibody was created against the extracellular domain that encompasses the first 900 amino acids of LOV-1. The monoclonal PKD-2 antibody was created against amino acids 7–18. The secondary antibody used for both LOV-1 and PKD-2 antibody staining was anti-mouse Alexa Fluor 568 donkey anti-mouse immunoglobulin G (H + L) (2 mg/ml) by Invitrogen. The primary antibody for both LOV-1 and PKD-2 antibody staining was used at a concentration of 1:200 and the secondary antibody for both LOV-1 and PKD-2 antibody staining at a concentration of 1:2000.
Behavioral assays
Male mating assay.
The mating behavioral assay, including the response and location of vulva efficiency of males, was performed as described (Barr and Sternberg, 1999; Bae et al., 2009).
Male leaving assay.
Male leaving behavior was measured as described (Barrios et al., 2008).
Bioinformatics and computer tools
Domain analysis.
The myristoylation motif was identified using NMT—The MYR Predictor (mendel.imp.ac.at/myristate/SUPLpredictor.htm). The leucine zipper was identified using ExPASy PROSITE (prosite.expasy.org/). The coiled-coil domains were identified using the COILS server (ch.embnet.org/software/COILS_form.html).
Phylogenetic analysis.
W03G9.7 protein sequence was used as a query for NCBI p-BLAST, cutoff 1e-5. A multiple sequence alignment was created using full-length sequences from hits identified using Muscle through MEGA6. MEGA6 was used to create a maximum likelihood phylogenetic tree from the sequences sites with a site coverage cut off of 7% and using the JTT substitution model. One hundred bootstrap replications were used. The gene sequence for W03G9.7 was retrieved from Ensembl and was used as an NBCI nBLAST query, using 1e-5 as a cutoff.
Statistical analysis
We used GraphPad Prism 5 version 5.03 software (GraphPad, La Jolla, CA) and Excel version 14.0.7128.5000 (Microsoft).
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center, funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440), the Million Mutation Project, and the National Bioresource Project for the Nematode for providing strains; Richard Poole and Arantza Barrios for help with whole-genome sequencing; Becky Androwski for assistance with confocal microscopy; Deanna De Vore for generating and characterizing the anti–PKD-2 monoclonal antibody; Juan Wang and members of the Barr lab for unpublished data and critical reading of the manuscript; and Christopher Ward, Monica Driscoll, Barth Grant, Chris Rongo, and the entire Rutgers C. elegans community for valuable ideas and advice. This work was funded by National Institutes of Health Grants DK59418 and DK074746 to M.M.B. and OD010943 to D.H.H., National Science Foundation Grant MCB-1161367 to A.D.K., and support by the Rutgers University Human Genetics Institute to M.M.B. and A.D.K.
Abbreviations used:
- ADPKD
autosomal dominant polycystic kidney disease
- EV
extracellular vesicle
- PC1
polycystin-1
- PC2
polycystin-2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0009) on June 3, 2015.
REFERENCES
- Bae YK, Barr MM. Sensory roles of neuronal cilia: cilia development, morphogenesis, and function in C. elegans. Front Biosci. 2008;13:5959–5974. doi: 10.2741/3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, Kim E, L'Hernault SW, Barr MM. The CIL-1 PI 5-phosphatase localizes TRP Polycystins to cilia and activates sperm in C. elegans. Curr Biol. 2009;19:1599–1607. doi: 10.1016/j.cub.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, Lyman-Gingerich J, Barr MM, Knobel KM. Identification of genes involved in the ciliary trafficking of C. elegans PKD-2. Dev Dyn. 2008;237:2021–2029. doi: 10.1002/dvdy.21531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, Qin H, Knobel KM, Hu J, Rosenbaum JL, Barr MM. General and cell-type specific mechanisms target TRPP2/PKD-2 to cilia. Development. 2006;133:3859–3870. doi: 10.1242/dev.02555. [DOI] [PubMed] [Google Scholar]
- Bakeberg JL, Tammachote R, Woollard JR, Hogan MC, Tuan HF, Li M, van Deursen JM, Wu Y, Huang BQ, Torres VE, et al. Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J Am Soc Nephrol. 2011;22:2266–2277. doi: 10.1681/ASN.2010111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- Barrios A, Nurrish S, Emmons SW. Sensory regulation of C. elegans male mate-searching behavior. Curr Biol. 2008;18:1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN- 29 during Caenorhabditis elegans development. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- Bloodgood RA. Gliding motility and the dynamics of flagellar membrane glycoproteins in Chlamydomonas reinhardtii. J Protozool. 1988;35:552–558. doi: 10.1111/j.1550-7408.1988.tb04151.x. [DOI] [PubMed] [Google Scholar]
- Bloodgood RA. Flagellar surface motility: gliding and microsphere movements. Methods Cell Biol. 1995;47:273–279. doi: 10.1016/s0091-679x(08)60820-1. [DOI] [PubMed] [Google Scholar]
- Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Fedeles SV, Dong K, Anyatonwu G, Onoe T, Mitobe M, Gao JD, Okuhara D, Tian X, Gallagher AR, et al. Altered trafficking and stability of polycystins underlie polycystic kidney disease. J Clin Invest. 2014;124:5129–5144. doi: 10.1172/JCI67273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Corrigan L, Redhai S, Leiblich A, Fan SJ, Perera SM, Patel R, Gandy C, Wainwright SM, Morris JF, Hamdy F, et al. BMP-regulated exosomes from Drosophila male reproductive glands reprogram female behavior. J Cell Biol. 2014;206:671–688. doi: 10.1083/jcb.201401072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, Jorgensen EM. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, Neuberger G, Schneider G, Wildpaner M. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res. 2003;31:3631–3634. doi: 10.1093/nar/gkg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Schwarz N, Nagel-Wolfrum K, Wolfrum U, Hardcastle AJ, Cheetham ME. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum Mol Genet. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PC, Torres VE. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J Clin Invest. 2014;124:2315–2324. doi: 10.1172/JCI72272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hogan MC, Bakeberg JL, Gainullin VG, Irazabal MV, Harmon AJ, Lieske JC, Charlesworth MC, Johnson KL, Madden BJ, Zenka RM, et al. Identification of biomarkers for PKD1 using urinary exosomes. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2014040354. ASN.2014040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, Huang BQ, Leontovich AA, Beito TG, Madden BJ, et al. Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol. 2009;20:278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Bae YK, Knobel KM, Barr MM. Casein kinase II and calcineurin modulate TRPP function and ciliary localization. Mol Biol Cell. 2006;17:2200–2211. doi: 10.1091/mbc.E05-10-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wittekind SG, Barr MM. STAM and Hrs down-regulate ciliary TRP receptors. Mol Biol Cell. 2007;18:3277–3289. doi: 10.1091/mbc.E07-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobel KM, Peden EM, Barr MM. Distinct protein domains regulate ciliary targeting and function of C. elegans PKD-2. Exp Cell Res. 2008;314:825–833. doi: 10.1016/j.yexcr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D, Epting CL, Engman DM. Composition and sensory function of the trypanosome flagellar membrane. Curr Opin Microbiol. 2010;13:466–472. doi: 10.1016/j.mib.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D, McGwire BS, Buchanan KT, Olson CL, Emmer BT, Epting CL, Engman DM. Molecular determinants of ciliary membrane localization of Trypanosoma cruzi flagellar calcium-binding protein. J Biol Chem. 2011;286:33109–33117. doi: 10.1074/jbc.M111.240895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, Radtke B, Splinter PL, LaRusso NF. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol. 2010;299:G990–G999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol. 2002;317:523–540. doi: 10.1006/jmbi.2002.5425. [DOI] [PubMed] [Google Scholar]
- Morsci NS, Barr MM. Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of Caenorhabditis elegans. Curr Biol. 2011;21:1239–1244. doi: 10.1016/j.cub.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N, Tamm SL. Lithocytes are transported along the ciliary surface to build the statolith of ctenophores. Curr Biol. 2014;24:R951–R952. doi: 10.1016/j.cub.2014.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan R, Piasecki BP, Silva M, Phirke P, Nguyen KC, Hall DH, Swoboda P, Barr MM. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr Biol. 2011;21:1685–1694. doi: 10.1016/j.cub.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan R, Wang J, Barr MM. Mating behavior, male sensory cilia, and polycystins in Caenorhabditis elegans. Semin Cell Dev Biol. 2014;33:25–33. doi: 10.1016/j.semcdb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol. 2002;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- Peden EM, Barr MM. The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans. Curr Biol. 2005;15:394–404. doi: 10.1016/j.cub.2004.12.073. [DOI] [PubMed] [Google Scholar]
- Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Burnette DT, Bae Y-K, Forscher P, Barr MM, Rosenbaum JL. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Ramulu P, Nathans J. Cellular and subcellular localization, N-terminal acylation, and calcium binding of Caenorhabditis elegans protein phosphatase with EF- hands. J Biol Chem. 2001;276:25127–25135. doi: 10.1074/jbc.M011712200. [DOI] [PubMed] [Google Scholar]
- Resh MD. Covalent lipid modifications of proteins. Curr Biol. 2013;23:R431–R435. doi: 10.1016/j.cub.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger-Nukpezah T, Golemis EA. The extracellular matrix and ciliary signaling. Curr Opin Cell Biol. 2012;24:652–661. doi: 10.1016/j.ceb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmo M, Kottgen M, Hofherr A. The TRPP subfamily and polycystin-1 proteins. Handb Exp Pharmacol. 2014;222:675–711. doi: 10.1007/978-3-642-54215-2_27. [DOI] [PubMed] [Google Scholar]
- Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286:14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SM, Engel BD, Kocabas F, Bilyard T, Gennerich A, Marshall WF, Yildiz A. Intraflagellar transport drives flagellar surface motility. Elife. 2013;2:e00744. doi: 10.7554/eLife.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- Tao B, Bu S, Yang Z, Siroky B, Kappes JC, Kispert A, Guay-Woodford LM. Cystin localizes to primary cilia via membrane microdomains and a targeting motif. J Am Soc Nephrol. 2009;20:2570–2580. doi: 10.1681/ASN.2009020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi T, Sato M, Nakano K, Takemura D, Iwata H, Ishisaka R. Amino acid residue penultimate to the amino-terminal gly residue strongly affects two cotranslational protein modifications, N-myristoylation and N-acetylation. J Biol Chem. 2001;276:10505–10513. doi: 10.1074/jbc.M006134200. [DOI] [PubMed] [Google Scholar]
- Vader P, Breakefield XO, Wood MJ. Extracellular vesicles: emerging targets for cancer therapy. Trends Mol Med. 2014;20:385–393. doi: 10.1016/j.molmed.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Schwartz HT, Barr MM. Functional specialization of sensory cilia by an RFX transcription factor isoform. Genetics. 2010;186:1295–1307. doi: 10.1534/genetics.110.122879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer RM. Preservation of C. elegans tissue via high-pressure freezing and freeze-substitution for ultrastructural analysis and immunocytochemistry. Methods Mol Biol. 2006;351:203–221. doi: 10.1385/1-59745-151-7:203. [DOI] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Rosenbaum JL. Ciliary ectosomes: transmissions from the cell's antenna. Trends Cell Biol. 2015;25:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder BK, Hou X, Guay-Woodford LM. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J Am Soc Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.