The nuclear envelope and endoplasmic reticulum (ER) are part of the same contiguous membrane system and have a common evolutionary history. A combination of genetic, cell biological, and biochemical data is used to identify roles for the cortical ER proteins Lnp1 and Sey1 in nuclear pore complex stability in budding yeast.
Abstract
The nuclear envelope (NE) and endoplasmic reticulum (ER) are components of the same contiguous membrane system and yet have distinct cellular functions. Mounting evidence suggests roles for some ER proteins in the NE for proper nuclear pore complex (NPC) structure and function. In this study, we identify a NE role in Saccharomyces cerevisiae for Lnp1 and Sey1, proteins required for proper cortical ER formation. Both lnp1Δ and sey1Δ mutants exhibit synthetic genetic interactions with mutants in genes encoding key NPC structural components. Both Lnp1 and Sey1 physically associate with other ER components that have established NPC roles, including Rtn1, Yop1, Pom33, and Per33. Of interest, lnp1Δ rtn1Δ mutants but not rtn1Δ sey1Δ mutants exhibit defects in NPC distribution. Furthermore, the essential NPC assembly factor Ndc1 has altered interactions in the absence of Sey1. Lnp1 dimerizes in vitro via its C-terminal zinc finger motif, a property that is required for proper ER structure but not NPC integrity. These findings suggest that Lnp1's role in NPC integrity is separable from functions in the ER and is linked to Ndc1 and Rtn1 interactions.
INTRODUCTION
In eukaryotic cells, the nuclear envelope (NE) and endoplasmic reticulum (ER) are part of the same continuous membrane system and yet have distinct functions. This intrinsic connection is apparent in higher eukaryotes during open mitosis, when the NE is absorbed into the ER and then reformed through the restructuring of cortical ER once mitosis is complete (Hetzer, 2010).
Accordingly, proteins found in the ER are also present in the outer nuclear membrane (ONM) of the NE, whereas the inner nuclear membrane (INM) of the NE has a unique protein composition. However, several of the shared proteins play distinct roles in each membrane domain, with well-defined specific ER functions and roles in the NE at nuclear pore complexes (NPCs), the 60-MDa assemblies embedded in NE pores that allow nucleocytoplasmic exchange (Aitchison and Rout, 2012). In Saccharomyces cerevisiae, these include Sec13, Rtn1, Yop1, Pom33, and Per33 (Hsia et al., 2007; Dawson et al., 2009; Chadrin et al., 2010; Casey et al., 2012; Zhang and Oliferenko, 2014). Of interest, the structures of many NPC proteins (nucleoporins [Nups]) resemble ER coat proteins that bind to and support membrane curvature during vesicle formation (Brohawn et al., 2008). Further study of the connections between ER and NE membrane components is required to understand this focal point of cell physiology.
Studies suggest that the proteins with distinct roles at the ER and NPC are specifically involved in NPC biogenesis and structure (Siniossoglou et al., 2000; Dawson et al., 2009; Chadrin et al., 2010; Casey et al., 2012). In metazoans, NPC biogenesis occurs by way of two processes: postmitotic assembly and interphase de novo assembly. The stepwise progression of postmitotic NPC assembly is well defined. After mitosis, sites of NPC assembly are seeded by the ELYS/Nup107 complex on the chromatin. Integral pore membrane proteins (Poms) of the NPC are recruited as the NE reforms from the cortical ER, stabilizing a pore into which other Nups assemble (Hetzer et al., 2005; Antonin et al., 2008; Doucet and Hetzer, 2010; Doucet et al., 2010). During de novo assembly, the intact double membrane of the NE must fuse to allow the formation of a nascent pore. Ndc1 is the only Pom known to be individually essential for this process (Chial et al., 1998; Mansfeld et al., 2006; Stavru et al., 2006). Owing to functional redundancies among other Poms, the mechanistic steps of this fusion event have been difficult to define further.
S. cerevisiae is a robust model system for analyzing de novo NPC assembly, as Nups are highly conserved and the yeast undergoes closed mitosis, with all NPCs forming de novo (Antonin et al., 2008; Doucet and Hetzer, 2010). It is speculated that Poms, changes in lipid composition, and peripheral membrane–associated Nups all contribute to membrane deformation during NE fusion for pore formation (Antonin et al., 2008; Doucet and Hetzer, 2010; Talamas and Hetzer, 2011; Vollmer et al., 2012). Because the membranes of both nascent pores and fully formed NPCs contain positive and negative curvature, membrane-bound proteins with curvature-stabilizing properties might provide necessary support to nuclear pores. Our previous studies identified Rtn1 and Yop1, membrane-bending proteins required for ER tubule formation, as having a role in S. cerevisiae NPC assembly. Furthermore, in vitro NPC assembly assays using Xenopus extracts found that Rtn1 and Yop1 might promote NPC biogenesis (Dawson et al., 2009; Casey et al., 2012). A model was proposed in which Rtn1 and Yop1 facilitate NE fusion via interactions with NPC membrane proteins and/or stabilize membrane structures during de novo assembly. Once fusion has occurred, structural NPC components further stabilize the highly curved surface of the nascent pore and provide a scaffold onto which other Nups are incorporated (Talamas and Hetzer, 2011).
Environments of high curvature also exist at three-way junctions in reticulated ER. These junctions form by the fusion of two tubules (Chen et al., 2013). In S. cerevisiae, this fusion process is mediated by Sey1 via the formation of a homotypic dimer across opposite membranes in the ER. This dimerization is predicted to induce the GTPase activity of Sey1 and results in a protein conformation change that compels fusion of the two lipid bilayers (Hu et al., 2009; Orso et al., 2009; Bian et al., 2011; Byrnes and Sondermann, 2011; Anwar et al., 2012). Conversely, we previously identified Lnp1 as a regulator of ER tubule structure that seemingly acts as a counterbalance to Sey1 function. In S. cerevisiae, loss of Lnp1 results in regions of collapsed cortical ER, as well as in regions of highly reticulated ER (Chen et al., 2012). Moreover, the presence of mammalian Lnp1 at three-way junctions in the ER stabilizes and decreases the mobility of these structures (Chen et al., 2015). The mechanism by which Lnp1 leads to this stability is unknown. Of interest, Lnp1 coprecipitates and genetically interacts with Rtn1 and Sey1. Furthermore, when Sey1 is inactivated, Lnp1 accumulates on the NE (Chen et al., 2012), which suggests a nuclear role for Lnp1. Thus we tested whether Lnp1 or Sey1 plays a role in NPC assembly.
In this article, we find that in addition to the anticipated ER/NE defects in lnp1Δ rtn1Δ mutants, there are distinct defects in NPC organization and nuclear shape. The lnp1Δ and sey1Δ mutants also exhibit synthetic genetic interactions with mutants in genes encoding key structural components of the NPC. We also find that the C-terminal region of Lnp1 is cytoplasmic and dimerizes in vitro. This dimerization is required for proper ER morphology; however, it does not appear necessary for NPC function. Of interest, we observe functional connections between Lnp1 and Rtn1 at NPCs, as well as both Sey1-dependent and -independent effects of Lnp1 on NPCs, including changes in physical interactions between Ndc1 and Rtn1 in a Sey1-dependent manner. These results provide an important mechanistic context for Lnp1 function. We conclude that Lnp1 plays a key role in NPC integrity independent of ER functions.
RESULTS
lnp1Δ rtn1Δ cells have defects in NPC organization
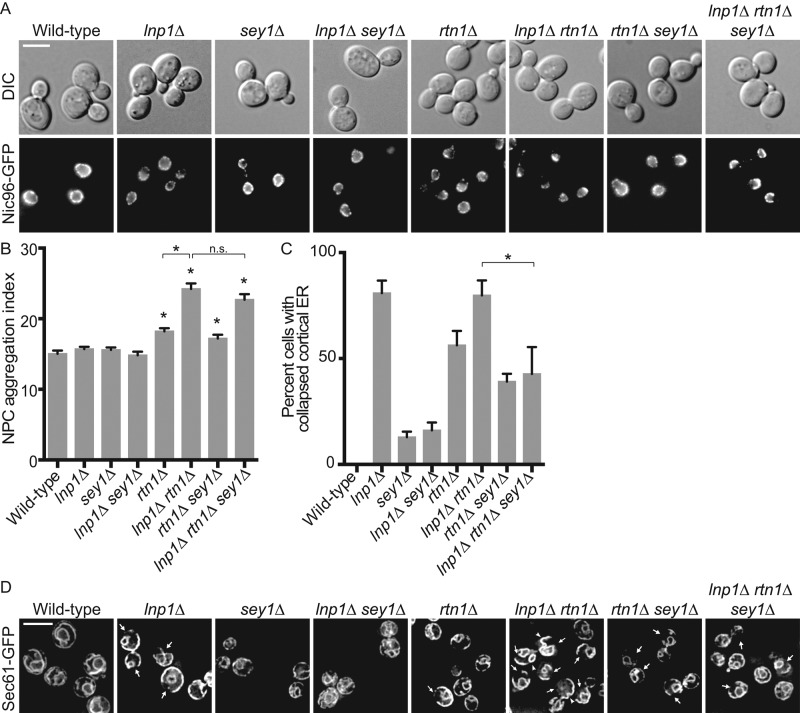
To determine whether Lnp1 or Sey1 has a role in NPC structure or assembly, we tested whether loss of either Lnp1 or Sey1 disrupts NPC organization. Whereas NPCs are distributed throughout the NE in wild-type cells, NPCs with structural and/or assembly defects aggregate in the NE as clusters (Belgareh and Doye, 1997; Bucci and Wente, 1997). To visualize NPCs, we imaged wild-type and mutant cells endogenously expressing Nic96–green fluorescent protein (GFP) by wide-field microscopy (Figure 1A). The distribution of NPCs in the NE was determined by measuring the aggregation index of signal around the perimeter of individual nuclei (Figure 1B), with a higher aggregation index indicating a greater degree of NPC disorder within the NE (Niepel et al., 2013). Whereas a subset of the cell population in rtn1Δ mutants displayed a minor NPC clustering defect, the localization of Nic96-GFP in lnp1Δ and sey1Δ mutants was indistinguishable from that of wild type. Similarly, no increase in NPC clustering was observed in the lnp1Δ sey1Δ or rtn1Δ sey1Δ double mutants compared with rtn1Δ alone. However, an enhanced NPC clustering defect was observed in the lnp1Δ rtn1Δ double mutant. The severity of NPC clustering in lnp1Δ rtn1Δ was not affected by the additional loss of SEY1 in a triple mutant (lnp1Δ rtn1Δ sey1Δ), further indicating that Sey1 function does not alter NPC distribution (Figure 1, A and B). These data indicated a physical or functional link between Lnp1 and Rtn1 function at the NPC.
FIGURE 1:
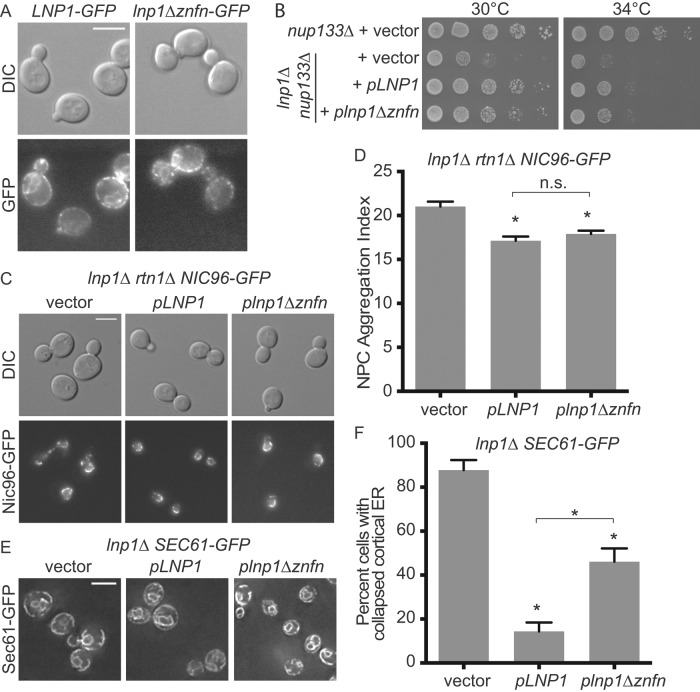
lnp1Δ rtn1Δ cells have defects in NPC organization. (A) Parental or mutant cells expressing Nic96-GFP were grown to early log phase at 25°C and visualized by fluorescence microscopy. Scale bar, 5 μm. (B) The aggregation indexes of Nic96-GFP–expressing cells. Error bars represent SE. Asterisk denotes statistical significance (p < 0.01). n.s., no statistical significance. (C) Percentages of cells with regions of collapsed cortical ER as indicated by lack of peripheral Sec61-GFP staining as quantified from images of Sec61-GFP–expressing cells. Error bars represent SE. Asterisk denotes statistical significance (p < 0.01). (D) Parental or mutant cells expressing Sec61-GFP grown to early log phase at 25°C and visualized by fluorescence microscopy. Scale bar, 5 μm. Arrows mark typical regions of collapsed cortical ER in represented cells. Arrowheads for lnp1Δ rtn1Δ highlight aberrant folds in cortical ER.
To investigate further the NPC aggregation defect in the lnp1Δ rtn1Δ mutant, we examined the nuclei and NPCs of these cells by thin-section transmission electron microscopy (TEM). TEM images of lnp1Δ rtn1Δ cells revealed nuclei with small clusters of NPCs, consistent with live-cell microscopy data (Figure 1A and Supplemental Figure S1, D–F) and a subset of nuclei with abnormal structure (Supplemental Figure S1, H–K). Previously we reported that some of the NPC-like structures in rtn1Δ yop1Δ cells did not fully span the plane of the NE and were associated with only the INM or ONM. Furthermore, spindle pole bodies were also deformed (Dawson et al., 2009; Casey et al., 2012). However, we did not observe these defects in lnp1Δ rtn1Δ cells.
Because rtn1Δ, lnp1Δ, and sey1Δ mutants have ER morphology defects (Hu et al., 2009; Anwar et al., 2012; Chen et al., 2012), we next asked whether the defects in NPC organization correlated with abnormal ER morphology. Wild-type and mutant cells endogenously expressing Sec61-GFP were imaged by wide-field microscopy and visually assessed for defects in ER morphology (Figure 1, C and D). As previously reported (Chen et al., 2012), the ER of lnp1Δ cells was abnormal, with large regions of collapsed cortical ER (indicated by white arrows; Figure 1D). In the lnp1Δ sey1Δ double mutant, this defect in cortical ER structure is rescued. Furthermore, as previously reported (Chen et al., 2012), lnp1Δ rtn1Δ cells had an enhanced ER defect, with regions of collapsed cortical ER and cortical ER folds extending into the cytoplasm (designated by white arrowheads in Figure 1D). This enhanced defect was partially rescued in the lnp1Δ rtn1Δ sey1Δ mutants. Consistent with the reduced ER tubules and increased ER sheets reported for the rtn1Δ sey1Δ mutant (Hu et al., 2009), we observed a modest decrease in collapsed cortical ER defects for this mutant as compared with rtn1Δ; however, this change was not statistically significant. Moreover, the rtn1Δ sey1Δ strain did not display any increase in NPC aggregation (Figure 1, A–D). Taken together, these results revealed that Lnp1 might play a role in NPC and NE organization independent of its role in ER structure.
Lnp1 and Sey1 localize to the NE and physically interact with shared ER and NPC components
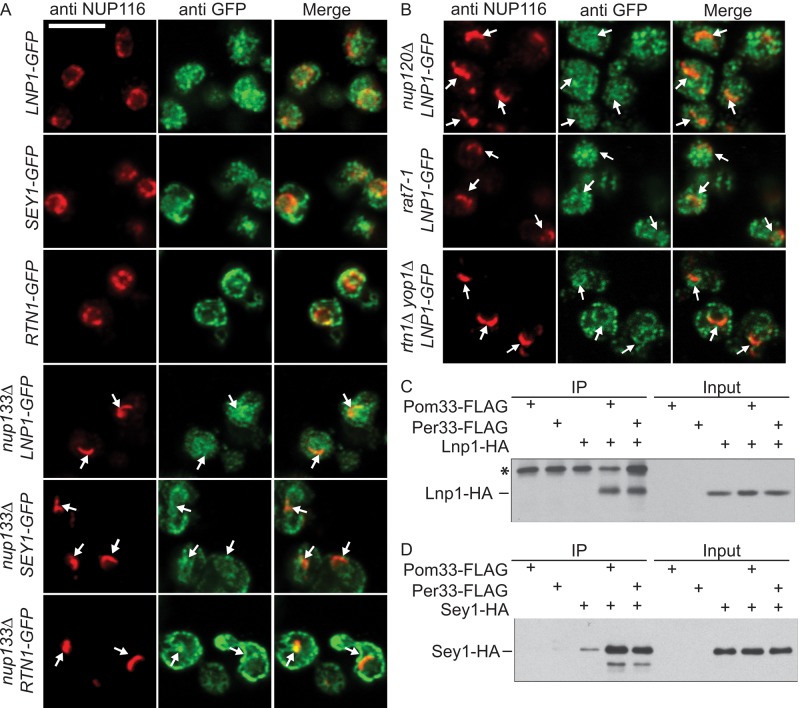
To determine whether Lnp1 and Sey1 are steady-state components of NPCs, we examined Lnp1- GFP and Sey1-GFP localization in NPC clustering mutants. In NPC clustering mutants, localization of NPC structural components shifts from throughout the NE rim to predominantly in the NPC cluster (Figure 2A). Wild-type and nup133Δ mutant cells endogenously expressing Lnp1-GFP, Sey1-GFP, or Rtn1-GFP were grown to log phase, fixed, and labeled by indirect immunofluorescence with anti-GFP and anti-Nup116 antibodies. As previously described (Dawson et al., 2009), Rtn1-GFP localized to both the cortical ER and to the NPC clusters in nup133Δ cells (Figure 2A). In wild-type cells, as reported (Chen et al., 2012), Lnp1-GFP and Sey1-GFP primarily localized as puncta throughout the tubular ER and NE, with some puncta overlapping with NPC signal. In nup133Δ mutants, the distribution of Lnp1-GFP and Sey1-GFP localization in the ER and NE was not noticeably altered (Figure 2A). Whereas foci of Lnp1-GFP and Sey1-GFP signal were visible within nup133Δ NPC clusters, unlike Rtn1-GFP, neither Lnp1-GFP nor Sey1-GFP showed enrichment at these sites. The same Lnp1-GFP localization pattern was observed in nup120Δ and rat7-1 clustering mutants (Figure 2B). Overall both Lnp1-GFP and Sey1-GFP colocalized with NE regions containing NPCs, but it was inconclusive whether Lnp1-GFP and Sey1-GFP are found specifically at NPCs.
FIGURE 2:
Lnp1 and Sey1 localize to the NE and physically interact with shared ER and NPC components. (A, B) Indirect immunofluorescence microscopy was performed with cells using chicken anti-GFP and rabbit anti-Nup116C antibodies. Arrows indicate NPC clusters. (C) Yeast lysates were prepared from cells expressing Pom33-FLAG, Per33-FLAG, Lnp1-HA, Pom33-FLAG and Lnp1-HA, or Per33-FLAG and Lnp1-HA. Lysates were immunoprecipitated with anti-FLAG affinity matrix and blotted using anti-HA antibodies. Asterisk indicates contaminant band. (D) Yeast lysates were prepared from cells expressing Pom33-FLAG, Per33-FLAG, Sey1-HA, Pom33-FLAG and Sey1-HA, or Per33-FLAG and Sey1-HA. Lysates were immunoprecipitated with anti-FLAG affinity matrix and blotted using anti-HA antibodies.
Of interest, in rtn1Δ yop1Δ mutants, Lnp1-GFP localization was markedly perturbed. Without a highly branched ER network, Lnp1-GFP was more evenly distributed throughout the ER and NE in rtn1Δ yop1Δ cells; moreover, the localization of Lnp1-GFP at NPC clusters was diminished (Figure 2B and Supplemental Figure S2). This suggested that the localization of Lnp1 to areas of the NE with NPC clusters is dependent on Rtn1 and Yop1.
We next tested whether the association of Lnp1 with NE-NPC regions was due to protein–protein interactions. Both Lnp1 and Sey1 physically interact with Rtn1 and Yop1 by coimmunoprecipitation (Chen et al., 2012). Here we focused on association of Lnp1 and Sey1 with Pom33 and Per33, other ER components that have roles at the NPC (Chadrin et al., 2010). Pom33 and Per33 have strong connections to NPC organization in both S. cerevisiae and Schizosaccharomyces pombe (Chadrin et al., 2010; Zhang and Oliferenko, 2014). Pom33 and Per33 are also found in the ER, and thus the interactions with Lnp1 could occur in both the ER and the NE. Lysates of yeast cells endogenously expressing Pom33-FLAG or Per33-FLAG and either Lnp1–hemagglutinin (HA) or Sey1-HA were incubated with anti-FLAG affinity matrix. Immunoblots of bound samples revealed that Pom33-FLAG and Per33-FLAG are coisolated with both Lnp1-HA and Sey1-HA (Figure 2, C and D). Taken together, the results indicate that Lnp1 and Sey1 were biochemically and cell biologically linked to the NE and NPC components.
lnp1Δ and sey1Δ mutants genetically interact with mutants in genes of the Nup84 subcomplex
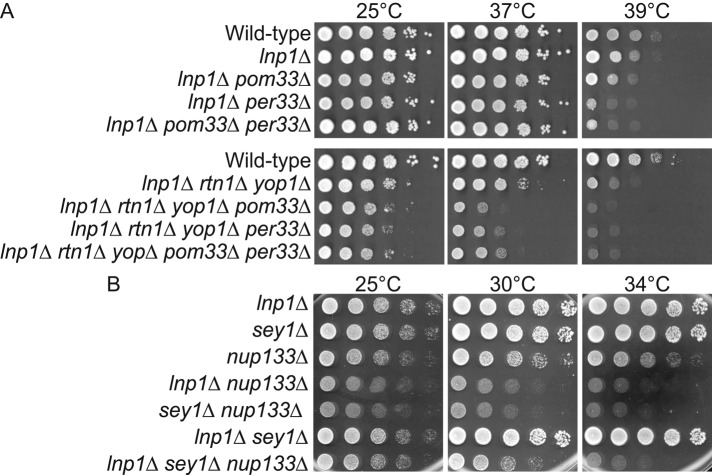
To further evaluate the connections between Lnp1, Sey1, and the NPC, we analyzed growth phenotypes for lnp1Δ and sey1Δ mutants in combination with different nup mutants. The NPC is organized within the NE pore in an apparent eightfold rotational symmetry perpendicular to the membrane plane. Distinct general domains include the nuclear basket, cytoplasmic filaments, and a central core structural scaffold surrounding a central channel. The core NPC scaffold consists of a series of inner, outer, and luminal rings connected by linker complexes (Alber et al., 2007; Aitchison and Rout, 2012). A panel of mutants was tested, including genes encoding Poms, membrane-bound components of the NPC (pom33Δ, per33Δ, pom152Δ, pom34Δ, ndc1-4), structural Nups in the Nup84 subcomplex (nup133Δ, nup120Δ, nup84Δ, nup85Δ, nup145ΔNS, nup145-R4), membrane-binding components of the inner ring (nup53Δ, nup59Δ), and Nups that directly participate in nucleocytoplasmic transport (nup100Δ, rat7-1; Table 1). To determine whether the combinatorial mutants displayed enhanced growth defects, we assayed strains by growth on rich media at a range of temperatures. Whereas per33Δ mutants do not have growth defects alone (Chadrin et al., 2010), the lnp1Δ per33Δ and sey1Δ per33Δ double mutants displayed synthetic fitness defects at higher temperatures (Figure 3A and Table 1). In addition, the growth defects of rtn1Δ yop1Δ per33Δ and rtn1Δ yop1Δ pom33Δ triple mutants were enhanced when combined with lnp1Δ (Figure 3A and Table 1).
TABLE 1:
Genetic interactions with lnp1Δ and sey1Δ.
| lnp1Δ | sey1Δ | |
|---|---|---|
| pom33Δ | No effect | No effect |
| rtn1Δ pom33Δ | No effect | n.d. |
| rtn1Δ yop1Δ pom33Δ | Synthetic sick | n.d. |
| per33Δ | Synthetic sick | Synthetic sick |
| rtn1Δ per33Δ | Synthetic sick | n.d. |
| rtn1Δ yop1Δ per33Δ | Synthetic sick | n.d. |
| pom152Δ | No effect | n.d. |
| pom34Δ | No effect | n.d. |
| pom152Δ pom34Δ | No effect | n.d. |
| ndc1-4 | No effect | Synthetic sick |
| nup53Δ | No effect | No effect |
| nup59Δ | No effect | No effect |
| nup53Δ nup59Δ | No effect | No effect |
| rat7-1 (nup159) | No effect | No effect |
| nup100Δ | No effect | No effect |
| nup133Δ | Synthetic sick | Synthetic sick |
| nup120Δ | Synthetic sick | Synthetic sick |
| nup145ΔNS | Synthetic sick | No effect |
| nup145-R4 | Synthetic sick | No effect |
| nup84Δ | Synthetic sick | Synthetic sick |
| nup85Δ | Synthetic sick | Synthetic sick |
n.d., not determined.
FIGURE 3:
lnp1Δ and sey1Δ mutants genetically interact with mutants in genes of the Nup84 subcomplex. (A) lnp1Δ pom33Δ and lnp1Δ rtn1Δ yop1Δ pom33Δ mutants have enhanced growth defects. Yeast strains were grown at 25°C and fivefold serially diluted onto plates of rich media incubated at the listed temperatures. (B) lnp1Δ nup133Δ and sey1Δ nup133Δ mutants have enhanced growth defects. Yeast strains were grown at 25°C and fivefold serially diluted onto plates of rich media incubated at the temperatures indicated.
On the basis of the genetic and physical interactions for Lnp1 and Sey1 with Rtn1 and Yop1, we predicted that lnp1Δ would genetically interact with mutants in genes encoding NPC components in a manner similar to that found for the rtn1Δ yop1Δ double mutant (Dawson et al., 2009). However, major differences in the genetic interaction profiles of rtn1Δ and lnp1Δ were observed. Of note, all mutants of the Nup84 subcomplex tested (nup133Δ, nup120Δ, nup84Δ, nup85Δ, nup145ΔNS, nup145-R4) had enhanced growth defects in combination with lnp1Δ, whereas other NPC mutants (pom34Δ pom152Δ, nup53Δ nup59Δ, nup100Δ, rat7-1) had no enhanced growth defect with lnp1Δ (Table 1). The sey1Δ mutant also genetically interacted with nup133Δ, nup120Δ, and nup84Δ in a similar manner to lnp1Δ. Of interest, lnp1Δ sey1Δ nup133Δ triple mutants exhibited partial rescue of growth defects compared with either lnp1Δ nup133Δ or sey1Δ nup133Δ double mutants alone (Figure 3B). Of note, both nup145ΔNS and nup145-R4 mutants, with known NPC clustering and RNA export defects, exhibited mildly enhanced synthetic sickness with lnp1Δ but not with sey1Δ (Supplemental Figure S3). Both nup145ΔNS and nup145-R4 mutants result in truncated nup145 proteins, with the first 592 amino acids missing from nup145ΔNS and the C-terminus of nup145-R4 truncated at amino acid 1012 (Emtage et al., 1997). Furthermore, sey1Δ but not lnp1Δ exhibited synthetic sickness with ndc1-4, a mutant with temperature-sensitive defects in NPC assembly. Overall the observed genetic interactions revealed novel and distinct relationships between Lnp1 and Sey1 with NPC components, indicating potential different roles in NPC function and assembly.
The function of Lnp1 and Sey1 at NPCs is coupled with the interaction between Rtn1 and the NPC
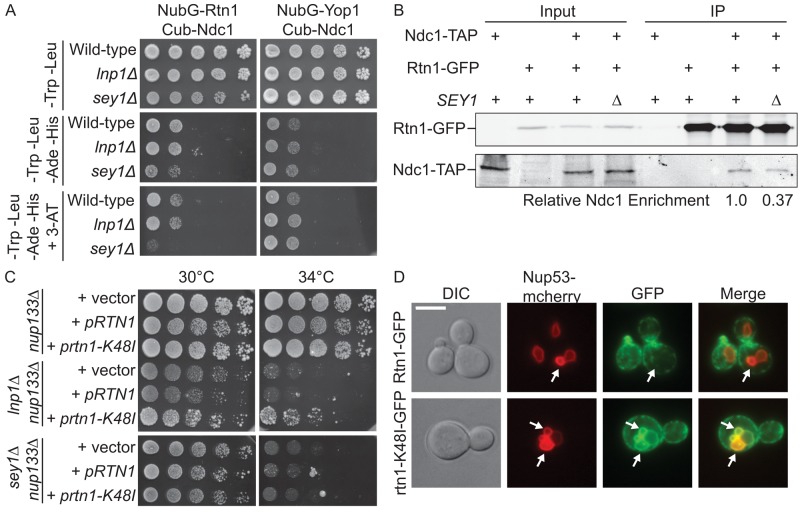
Previously we reported that Rtn1 and Yop1 physically interact with NPC components, including the pore membrane protein Ndc1 (Casey et al., 2012). To test whether lnp1Δ and sey1Δ affect the recruitment of Rtn1 to NPCs, we used a split ubiquitin yeast two-hybrid system to monitor the interaction between Rtn1 and Ndc1. Rtn1 tagged with the N-terminal region of ubiquitin (NubG) was coexpressed with Ndc1 tagged with the C-terminal region of ubiquitin (Cub) and the LexA-VP16 transcription factor. A close physical interaction between the bait (Ndc1-Cub) and prey (NubG-Rtn1) proteins leads to cleavage of the split ubiquitin from the bait and release of the LexA-VP16 transcription factor. Once released, the LexA-VP16 transcription factor can activate reporter genes HIS3 and ADE2 (Snider et al., 2010). Activation of these reporter genes was assayed by growth on synthetic medium lacking histidine and adenine. To increase the stringency of the physical interaction threshold, we added the histidine biosynthesis competitive inhibitor 3-aminotriazole (3-AT) to growth medium to increase the baseline level of HIS3 expression required for cell survival.
We assayed for interaction between Cub-Ndc1 and NubG-Rtn1 in wild-type, lnp1Δ, and sey1Δ reporter strains (Figure 4A). All three strains grew on media lacking histidine and adenine; however, addition of 12 mM 3-AT to the growth medium resulted in loss of growth of the sey1Δ reporter strain. This indicated that sey1Δ mutants exhibit a decreased interaction between NubG-Rtn1 and Cub-Ndc1. We tested whether the interaction between Cub-Ndc1 and Yop1-NubG was similarly affected; however, no changes in the interaction between Ndc1 and Yop1 were observed in lnp1Δ or sey1Δ mutants (Figure 4A). To examine this interaction further, we coimmunoprecipitated Ndc1-TAP and Rtn1-GFP from whole-cell lysates of wild-type and sey1Δ cells (Figure 4B). Compared to wild type, there was a significant and reproducible reduction in the relative levels of Ndc1-TAP coisolated with Rtn1-GFP sey1Δ mutants. The relative enrichment of Ndc1-TAP with Rtn1-GFP from sey1Δ lysates was 31–56% of wild-type levels over three independent experiments.
FIGURE 4:
The function of Lnp1 and Sey1 with NPCs is coupled with the interaction between Rtn1 and the NPC. (A) Split ubiquitin yeast two-hybrid vectors containing genes encoding either NubG-Yop1 or NubG-Rtn1 (preys) were expressed in wild-type or mutant strains and tested for interaction with Ndc1-Cub (Bait). Presence of both bait and prey plasmids was detected on SCM-Leu-Trp. Interaction of bait and prey was assayed by growth on SCM-Leu-Trp-His-Ade at 25°C with and without 3-AT. (B) Yeast lysates were prepared from cells expressing Ndc1-TAP, Rtn1-GFP, and Ndc1-TAP and Rtn1-GFP in wild-type or sey1Δ mutant strains. Lysates were immunoprecipitated with GFP-binding protein resin and blotted using anti-GFP and anti-mouse IgG antibodies. (C) lnp1Δ nup133Δ, nup133Δ lnp1Δ nup133Δ, and sey1Δ nup133Δ mutants were transformed with plasmids encoding RTN1, rtn1-K48I, or empty vector, grown to early log phase at 25°C, and fivefold serially diluted onto SCM-Leu plates at the indicated temperatures. (D) Cells expressing either Rtn1-GFP or rtn1-K48I-GFP were grown to early log phase at 25°C, induced for overexpression of Nup53-mcherry for 8 h, and visualized by fluorescence microscopy. Arrows indicate nuclear karmellae. Scale bar, 5 μm.
Sey1 is required for efficient interaction between Ndc1 and Rtn1
Because lnp1Δ rtn1Δ mutants had defects in NPC organization and loss of Sey1 altered the Rtn1 and Ndc1 interactions, we hypothesized that overexpression of RTN1 might rescue growth defects observed in lnp1Δ nup133Δ and sey1Δ nup133Δ mutants. We also tested overexpression of the prtn1-K48I mutant, which localizes primarily to the NE due to a defect in self-oligomerization. This mutant exhibits severe defects in ER tubule stabilization (Hu et al., 2008; Shibata et al., 2008) but not in NPC function (Dawson et al., 2009). Overexpression of pRTN1 did not alter the growth defect of lnp1Δ nup133Δ or sey1Δ nup133Δ mutant. Intriguingly, overexpression of the prtn1-K48I mutant specifically rescued the growth defect of lnp1Δ nup133Δ mutants but not nup133Δ sey1Δ mutants (Figure 4C). This suggested that the function of Lnp1 might be directly related to Rtn1 function or availability in the NE.
Because the rtn1-K48I protein is deficient in self-oligomerization and more mobile in the ER (Shibata et al., 2008), we hypothesized that the ability of this mutant to rescue lnp1Δ nup133Δ might reflect an increased mobility in the NE. To test this, we compared Rtn1-GFP and rtn1-K48I-GFP for their ability to accumulate in Nup53-induced intranuclear karmellae. On induction of NUP53 overexpression, the nuclei of many cells accumulate flattened intranuclear membranes, and membrane NPC proteins associate within these intranuclear karmellae (Marelli et al., 2001). Whereas Rtn1-GFP did not associate with these structures (n = 30), we found that the rtn1-K48I-GFP was localized to 66% of Nup53 karmellae observed (n = 33) (Figure 4D). Taken together, the rtn1-K48I protein was more mobile in the NE than wild-type Rtn1.
The zinc finger domain of Lnp1 mediates dimerization that is required for ER but not NPC function
The human orthologue of Lnp1 contains two N-terminal transmembrane domains with both the N- and C-termini extending into the cytoplasm. Furthermore, human Lnp1 is N-myristoylated, and the N-myristoylation is necessary for its function in ER morphology (Moriya et al., 2013). However, S. cerevisiae Lnp1 does not contain this N-myristoylation motif (Moriya et al., 2013), suggestive of potential differences in their topology and/or regulation. To determine the orientation of yeast Lnp1's terminal domain, we tagged the C-terminus with Suc2 flanked by myc tags for antibody detection. Suc2 is a target for glycosylation within the ER lumen (Sengstag, 2000). If the C-terminus of a Suc2-tagged domain localizes to the ER lumen, the Suc2 tag will be glycosylated, and treatment with endoglycosidase H (EndoH) will result in a decrease in the molecular mass. Pom152-myc-Suc2-myc and Pom34-myc-Suc2-myc were used as positive and negative controls for glycosylation, respectively (Miao et al., 2006). Lysates were treated with EndoH, precipitated, and analyzed by immunoblotting (Figure 5A). Whereas EndoH digestion of Pom152-myc-Suc2-myc resulted in a reduction of molecular mass, digestion of Lnp1-myc-Suc2-myc and the negative control did not, indicating that the C-terminus of Lnp1 is located in the cytoplasm and not in the ER/NE lumen. This predicted topology is consistent with that reported for human Lnp1 (Moriya et al., 2013; Chen et al., 2015).
FIGURE 5:
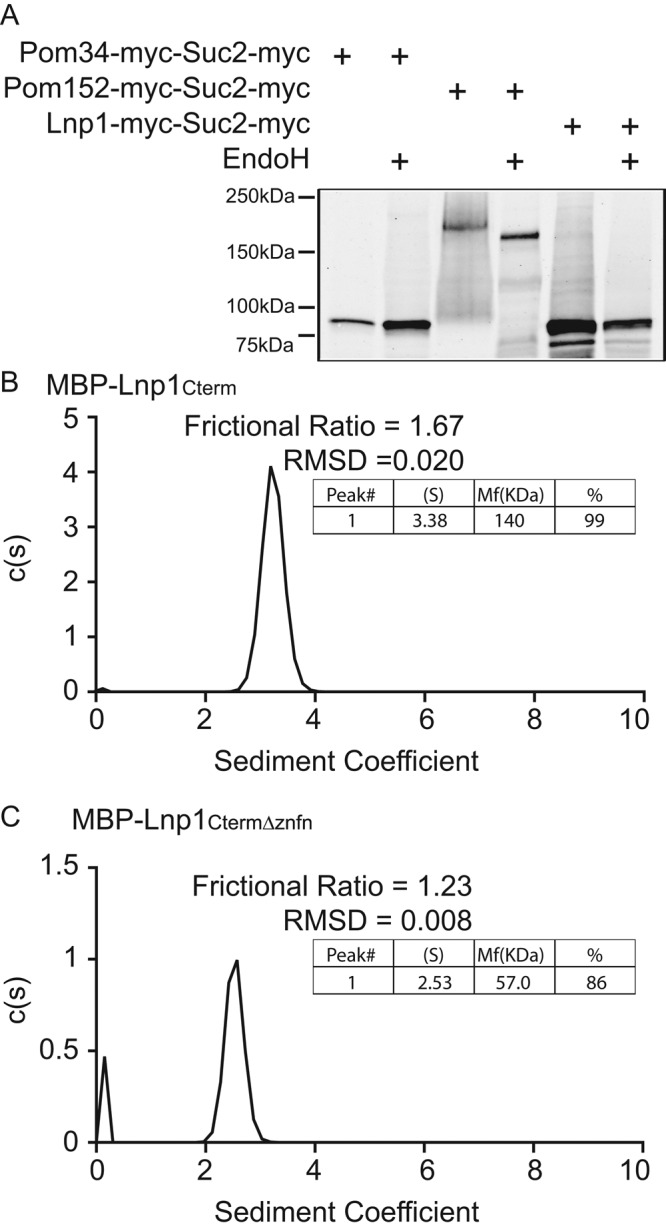
The C-terminal zinc finger domain of Lnp1 is required for dimerization in vitro. (A) Lysates from cells expressing Pom34-myc-Suc2-myc, Pom152-myc-Suc2-myc, or Lnp1-myc-Suc2-myc were either mock digested or treated with EndoH and analyzed by immunoblotting with mouse anti-Myc antibody. (B, C) Sedimentation velocity analytical ultracentrifugation was performed with recombinant MBP-Lnp1Cterm and MBP-Lnp1CtermΔznfn. Determined molecular masses are given for major species.
The C-terminal Lnp1 region contains a zinc finger motif that has a critical yet unknown role in ER function (Chen et al., 2012). Many proteins involved in mediating ER morphology self-interact as a key element in their function (Voeltz et al., 2006; Hu et al., 2008; Shibata et al., 2008; Anwar et al., 2012). We hypothesized that the Lnp1 zinc finger motif mediates dimerization between Lnp1 molecules. To test this hypothesis, we analyzed by analytical ultracentrifugation the oligomeric state of the purified recombinant C-terminal region (amino acids 104–278) of Lnp1 fused to maltose-binding protein (MBP-Lnp1Cterm; Figure 5B). This revealed that MBP-Lnp1Cterm behaved as a dimer in vitro. To determine whether the zinc finger motif of Lnp1 was responsible for the in vitro dimerization of MBP-Lnp1Cterm, we tested purified recombinant MBP-Lnp1CtermΔznfn, which lacks the zinc finger motif (amino acids 221–248). Analytical ultracentrifugation showed that MBP-Lnp1CtermΔznfn migrated as a monomer (Figure 5C). We concluded that the zinc finger motif of Lnp1 is required for the dimerization of Lnp1.
To determine whether the zinc finger motif is required for NE localization, we expressed plasmids encoding full-length Lnp1-GFP and lnp1Δznfn-GFP in wild-type cells. Localization was assessed by epifluorescence wide-field microscopy. Both Lnp1-GFP and lnp1Δznfn-GFP localized similarly in the ER, and thus the zinc finger motif of Lnp1 was not required for its proper localization within the cell (Figure 6A). Because the zinc finger motif is required for the function of Lnp1 in ER morphology (Chen et al., 2012), we next analyzed whether the zinc finger motif dimerization domain is important for the function of Lnp1 at NPCs. Surprisingly, exogenous expression of either pLNP1 or plnp1Δznfn rescued the growth defect of lnp1Δ nup133Δ mutants to the same degree (Figure 6B). However, it is notable that neither pLNP1 nor plnp1Δznfn fully rescued the enhanced growth defect of lnp1Δ nup133Δ mutants. Because plasmid-based expression of genes is highly variable, this might indicate that nup133 mutants are sensitive to the dosage of Lnp1.
FIGURE 6:
The zinc finger of Lnp1 is not required for NPC function. (A) Parental cells exogenously expressing either Lnp1-GFP or lnp1Δznfn-GFP were grown to early log phase at 25°C and visualized by fluorescence microscopy. Scale bar, 5 μm. (B) Expression of lnp1Δznfn results in rescue of lnp1Δ nup133Δ. The lnp1Δ nup133Δ mutants were transformed with pLNP1, plnp1Δznfn, or empty vector and grown to early log phase at 25°C, fivefold serially diluted, and grown at indicated temperatures. (C) Expression of lnp1Δznfn results in rescue of lnp1Δ rtn1Δ NPC aggregation. lnp1Δ rtn1Δ NIC96-GFP mutants were transformed with pLNP1, plnp1Δznfn, or empty vector and grown to early log phase at 25°C and imaged. Scale bar, 5 μm. (D) The aggregation indexes of Nic96-GFP–expressing cells. Error bars represent SE. Asterisk denotes statistical significance (p < 0.01). (E) Expression of lnp1Δznfn is not sufficient to rescue lnp1Δ defects in ER. lnp1Δ SEC61-GFP mutants were transformed with pLNP1, plnp1Δznfn, or empty vector and grown to early log phase at 25°C and imaged. Scale bar, 5 μm. (F) Percentages of cells with regions of collapsed cortical ER as indicated by lack of peripheral Sec61-GFP staining were quantified from images of Sec61-GFP–expressing cells. Error bars represent SE. Asterisk denotes statistical significance (p < 0.01).
Next we tested whether exogenous expression of plnp1Δznfn could rescue the NPC aggregation defects of lnp1Δ rtn1Δ NIC96-GFP mutants. Indeed, both pLNP1 and plnp1Δznfn decreased the NPC aggregation index in lnp1Δ rtn1Δ NIC96-GFP mutants to levels consistent with rtn1Δ NIC96-GFP alone (Figure 6, C and D). As a control for the requirement of the Lnp1 zinc finger motif in ER morphology, we assayed lnp1Δ SEC61-GFP cells expressing pLNP1 or plnp1Δznfn plasmids for ER morphology defects. pLNP1 rescued the ER defects of lnp1Δ SEC61-GFP mutants at 14%, compared with 87% for empty vector. However, plnp1Δznfn did not rescue the ER defects completely, with 46% of cells displaying ER defects. Thus the zinc finger domain of Lnp1 is required for the maintenance of ER structure but is not necessary for Lnp1's role in NPC function and organization.
DISCUSSION
This work identifies a novel role for Lnp1 in NPC organization and structure that is connected with Rtn1 function but is independent of the Lnp1 role in ER structure. This conclusion is based on several lines of evidence. First, loss of Lnp1 and Rtn1 in cells results in aggregation of NPCs. This NPC aggregation defect is not rescued by the further loss of Lnp1's antagonist in ER morphology, Sey1, even though the sey1Δ does rescue the ER defects of lnp1Δ mutants. Moreover, general defects in the ER are not sufficient to cause these NPC aggregation defects, as cells lacking Rtn1 and Sey1 exhibit severe defects in ER morphology but do not display defects in NPC organization.
Second, both Lnp1 and Sey1 physically associate with Pom33 and Per33, NPC components with ties to the ER (Chadrin et al., 2010). Recently S. pombe Tts1, the homologue of S. cerevisiae Pom33 and Per33, was found to also have roles in NE remodeling during mitosis. Loss of Tts1 results in the accumulation of NPCs in ER/NE junctions at the onset of mitosis (Zhang and Oliferenko, 2014). This phenotype parallels our observation that lnp1Δ has genetic interactions with per33Δ. It is also of note that for all the NPC components tested, lnp1Δ and sey1Δ appear to genetically interact only with those that have the most direct ties to the ER. Rtn1, Yop1, Pom33, and Per33 are found within the ER, and the Nup84 subcomplex harbors Sec13 and has evolutionary ties to the COPII coat complex (Siniossoglou et al., 2000; Dawson et al., 2009; Chadrin et al., 2010).
Third, loss of Sey1 and Lnp1 has differential effects on the requirements for Rtn1 at the NPC. Loss of Sey1, but not of Lnp1, results in decreased interaction of Rtn1 with the NPC by yeast two-hybrid and coimmunoprecipitation analysis. This is interesting when considered with previous work showing that loss of Sey1 results in increased Lnp1 at the NE, as well as increased physical interaction between Lnp1 and Rtn1 (Chen et al., 2012). Moreover, overexpression of RTN1 is not sufficient to rescue the synthetic growth defects of sey1Δ nup133Δ mutants. This indicates that loss of Sey1 alters Rtn1's ability to interact with the nuclear pore, but increased levels of Rtn1 are not sufficient to overcome the resulting defect. Intriguingly, this defect is not sufficient to cause obvious defects in NPC organization, as Sey1 loss was not associated with NPC aggregation. Perhaps even more intriguing is the observation that overexpression of the rtn1-K48I mutant that is defective in oligomerization and ER tubule polymerization (Hu et al., 2008, 2009; Shibata et al., 2008) rescues synthetic growth defects of lnp1Δ mutants but not of sey1Δ. The rtn1-K48I altered protein is more mobile in membranes (Shibata et al., 2008) and is localized to Nup53-induced intranuclear karmellae (Figure 4D). Therefore increased mobility of rtn1-K48I might allow it to rescue the loss of Lnp1 function in the NE.
Finally, Lnp1 dimerization is required for maintenance of ER structure but not for NPC function. We find that the zinc finger domain in the C-terminal Lnp1 domain mediates homodimerization in vitro. In concordance with previous studies (Chen et al., 2012), expression of lnp1Δznfn does not fully rescue the ER defects observed in in the ER. However, expression of lnp1Δznfn rescues the synthetic genetic interactions of lnp1Δ and the NPC aggregation defect of lnp1Δ rtn1Δ. Taken together, the results indicate that Lnp1 has distinct and separate roles in ER structure and NPC organization.
There are several possible models for how Lnp1 functions in NPC assembly. The specific nup genetic interactions with lnp1Δ and sey1Δ could be due to a role in stabilizing newly formed pores. Specifically, Lnp1 might mediate Rtn1 function at nascent pores. Nuclear pores contain points of very high membrane curvature in the NE but are surrounded by areas of no curvature. Lnp1 can localize to both flattened and highly curved membranes; however, Rtn1 oligomers are only stably associated with areas of high curvature (Hu et al., 2008; Chen et al., 2012). Through a physical interaction with Lnp1, the oligomerization of Rtn1 could be modulated to increase mobility of Rtn1 in the NE, allowing Rtn1 to be more easily trafficked to sites of new NPC assembly. This is consistent with the ability of rtn1-K48I but not RTN1 to rescue synthetic growth defects of lnp1Δ mutants. Alternatively, changes in the tubular ER network and NE/ER connections could also alter NPC assembly and organization. Decreased connections to the NE could limit the avenues by which membrane proteins are trafficked to the NE. This is consistent with the decreased interaction between Ndc1 and Rtn1 in sey1Δ mutants. The antagonistic relationship between Lnp1 and Sey1 might play a role in its functional link to NPCs, although Lnp1 could function independently of Sey1 as well, as indicated by the NPC aggregation data and the incomplete rescue of growth defects in the lnp1Δ sey1Δ nup133Δ triple mutant. Furthermore, plasmid expression of pLNP1 does not fully rescue the growth defect of lnp1Δ nup133Δ mutants. This result could indicate that increased expression or increased nuclear availability of Lnp1 may be detrimental in nup133Δ mutants, which could also provide an additional explanation for sey1Δ-related growth defects in NPC mutants.
The ER and NE are an interconnected membrane system with a variety of distinct cellular functions (English et al., 2009). Here we build on the paradigm of individual proteins having different functions dependent on different cellular membrane environments: ER versus NE. The roles of Rtn1, Lnp1, and Sey1 in the ER are intimately linked with the fusion of curved membrane tubules. Whereas the mechanism by which Sey1 mediates fusion is well characterized, the mechanism(s) by which Lnp1 functions in the ER are not understood. Both rtn1-K48I and lnp1Δznfn mutants rescue NPC-specific but not ER-specific defects, indicating separate roles at these distinct locations. We predict that determination of Lnp1 and Rtn1 mechanisms in modulation of ER tubules will result in further insights into the function of these proteins in NPC biogenesis.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains and plasmids used in this study are listed in Tables S1 and S2. Unless otherwise noted, yeast genetic techniques were performed by standard procedures described previously (Sherman et al., 1986). All strains were cultured in either rich (YPD: 1% yeast extract, 2% peptone, and 2% dextrose) or complete synthetic minimal (CSM) media lacking appropriate amino acids with 2% dextrose. Kanamycin resistance was selected on medium containing 200 μg/ml G418 (US Biological, Salem, MA). Yeast were serially diluted and spotted onto agar plates to assay fitness and temperature sensitivity as previously described (Tran et al., 2007).
Plasmid pSW3906 was generated by subcloning genomic DNA fragments containing promoter, coding sequence and 3′-untranslated region into the BamHI and PstI sites of pRS425. DNA fragments of LNP1 were isolated by PCR amplification with Phusion (New England BioLabs, Ipswich, MA) using primers 5′-ATGCGGATCCTGCGTGGCTGTGTCGA-3′ and 5′-ATCGCTGCAGCCGCCGCAGAAGGCAG-3′. Plasmid pSW4029 was generated by subcloning genomic DNA fragments containing promoter and coding sequence of LNP1 into the SacI and SacII sites of pRS425. DNA fragments of LNP1-GFP were isolated by PCR amplification of LNP1-GFP:HIS5 from the yeast GFP collection (Huh et al., 2003) with Phusion (New England BioLabs) using primers 5′-ATGCGAGCTCTGCGTGGCTGTGTCGAGATT-3′ and 5′-GGCCGCGCCCGCGGGCCCTATTTGTATAGTTCATCC-3′. Plasmid pSW3975 was generated by subcloning genomic DNA fragments containing the coding sequence of amino acids 104–278 of LNP1 into the EcoRI and SalI sites of pMAL-cRI expression vector. DNA fragments were isolated by PCR amplification using primers 5′-GCTAGAATTCCGCAAGTTGGCAAAACTCCG-3′ and 5′- GCTAGTCGACTCATTTTGTTTTTTCCTTCTCCGAC-3′. Plasmids pSW4032, pSW4071, and pSW4087 were generated by PCR amplification and blunt end ligation of pSW4029, pSW3975, and pSW3906, respectively, using primers 5′-GATTTTTTTGAAGGGAGAG-3′ and 5′-AACCACAAAATAGACGAAGTAAAGG-3′.
Plasmids pSW4000 and pSW4001 were generated using the Gibson Assembly Method (New England BioLabs). pSW4000 was generated with DNA fragments of myc-SUC2-myc coding sequence PCR amplified from pSW3190 using primers 5′-ATATAGAGCTCCTACAGGTCCTCCTCTGAGATCAGCTTCTGCTCGCATTTTACTTCCCTTACTTGG-3′ and 5′-AATTAGAGCTCTGCGAGCAGAAGCTGATCTCAGAGGAGGACCTGATGACAAACGAAACTAGCGATAG-3′ and LNP1 coding sequence using primers 5′- CCTCTGAGATCAGCTTCTGCTCGCAGAGCTCTAATTgTTTTGTTTTTTCCTTCTCCG-3′ and 5′-ctgatctcagaggaggacctgtagGAGCTCTATATCTGATTTTGCGTTAGAATAACTACG-3′ into pRS315. pSW4001 was generated with DNA fragments of Rtn1 coding sequence using primers 5′-AAAAAAAATGAAAAAAAAAAACTGTTAATTTTTTTTTTTACTGATTTACAAATTCCTTG-3′ and 5′-TGTTGTTGGGCTTGGCTATGTTGAGCTGAGGCGGACATATTTGCGTGTGTGAATATGGCCGTAATGGCCACTCTGC-3′ and linearized pR3-N.
Immunoprecipitation
For Pom33-FLAG and Per33-FLAG, yeast cells grown to early log phase were harvested and resuspended in spheroplasting buffer (1.4 M sorbitol, 50 mM NaPi, pH 7.4, 50 mM 2-mercaptoethanol, 10 μg/OD600 Zymolyase-100T). The resuspended cells were incubated at 37°C for 30 min and pelleted through a chilled sorbitol cushion (1.7 M sorbitol, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4), and the pellet was lysed in lysis buffer (25 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protease inhibitor, 1% digitonin, pH 7.4) using a Dounce homogenizer (40 strokes). The lysate was centrifuged at 37,000 × g for 20 min at 4°C, and the protein concentration of the supernatant was measured using the Bradford assay.
The protein concentration of the lysate was adjusted to 2 mg/ml with lysis buffer, and 1.0 ml of the lysate was incubated overnight at 4°C with 20 μl of anti-FLAG antibody (Clone M2, F 1804; Sigma-Aldrich, St. Louis, MO). A 30-μl amount of a 50% slurry of Protein G agarose beads (Thermo) was added to the lysate and incubated at room temperature for 2 h. The beads were pelleted at 5000 rpm for 30 s, washed three times with 1 ml of cold lysis buffer that contained 0.2% digitonin, and heated to 100°C in sample buffer (62.5 mM Tris-HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol, pH 6.8) for 5 min. The eluted protein was subjected to SDS–PAGE and immunoblotted with anti-HA (1:2000 dilution, Clone HA.11, MMS-101R; Covance, Princeton, NJ) antibody. The secondary antibodies used were goat anti-mouse immunoglobulin G (IgG)–horseradish peroxidase (1:10,000 dilution, W402B; Promega, Fitchburg, WI).
For Rtn1-GFP, lysates were prepared from log-phase cultures using a bead beater (BioSpec, Bartlesville, OK) as previously described (Bolger et al., 2008) in lysis buffer (25 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor, 1% Triton X-100, pH 7.4). Soluble fractions were added to 15 μl of GFP-binding protein Sepharose resin (Vanderbilt Antibody and Protein Resource, Nashville, TN) and incubated for 3 h at 4°C. Proteins bound to the GFP-binding beads were washed in buffer (25 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM PMSF, 0.2% Tween-20), eluted by boiling in SDS sample buffer, resolved by SDS–PAGE, and detected with rabbit affinity-purified anti-GFP (1:1000; Life Technologies, Grand Island, NY) and anti-IgG (a gift of M. Linder, Cornell University, Ithaca, NY; 1:1000) and Alexa 700–conjugated anti-rabbit antibodies (1:5000; Life Technologies). Immunoblots were quantified using ImageJ (National Institutes of Health, Bethesda, MD). Relative enrichment of Ndc1-TAP was determined by comparing the ratios of Ndc1-TAP to Rtn1-GFP in input and immunoprecipitation samples.
Membrane yeast two-hybrid system
Bait and prey plasmids were cotransformed into wild-type or mutant reporter strains. Transformants were spotted onto CSM-Leu-Trp, CSM-Leu-Trp-His-Ade, and CSM-Leu-Trp-His-Ade +12 mM 3-AT and analyzed for growth after 4 d at 25°C.
Fluorescence microscopy
To measure the organization of NPCs across the NE, asynchronous cell populations expressing Nic96-GFP were imaged using a microscope (BX50; Olympus, Center Valley, PA) equipped with a motorized stage (Model 999000; Ludl, Hawthorne, NY), a UPlanF1 100×/numerical aperture (NA) 1.30 oil immersion objective, and digital charge-coupled device camera (Orca-R2; Hamamatsu, Middlesex, NJ). Images were collected and scaled using Nikon Elements and processed with ImageJ or Photoshop 12.0 software. The aggregation index of each nucleus was determined as previously described (Niepel et al., 2013) using the Oval Profile Plot plug-in (rsbweb.nih.gov/ij/plugins/oval-profile.html) of ImageJ.
To image the ER, asynchronous cell populations expressing Sec61-GFP were imaged by wide-field microscopy using a Delta Vision OMX (GE Healthcare, Pittsburgh, PA) using a 60×/NA 1.42 oil immersion objective. Images were deconvolved using softWoRx software and scaled using ImageJ or Photoshop 12.0 software.
For immunofluorescence, cells were fixed in 3.7% formaldehyde and 10% methanol for 10 min and processed as previously described (Strawn et al., 2004). Samples were incubated with affinity-purified rabbit anti-Nup116C (1:50; Iovine et al., 1995) and chicken anti-GFP (ASW54; 1:2000) at 4°C overnight. The anti-GFP antibody was generated in chickens against purified hexahistidine-GFP recombinant protein (Covance). IgY was purified from egg yolks using the IgY EggsPress purification system (Gallus Immunotech, Cary, NC). Bound antibodies were detected by incubation with Alexa Fluor 594–conjugated anti-rabbit (1:500) and Alexa Fluor 488–conjugated anti-chicken (1:200). Cells were imaged using a Leica TCS SP5 confocal microscope using a 63×/1.4 NA oil immersion objective.
Electron microscopy
Asynchronous cells were grown in YPD at 25°C to early log phase and processed as previously described (Dawson et al. 2009). Grids were examined on a CM-12 120-keV electron microscope (FEI, Hillsboro, OR). Images were acquired with MegaPlus ES 4.0 camera (Advanced Microscopy Techniques, Woburn, MA) and processed with ImageJ and Photoshop 12.0 software.
Endoglycosidase H treatment
Wild-type cells were transformed with pSW3190, pSW3192, or pSW4000. Transformants were grown in CSM-Leu to early log phase. Cells were harvested, and samples were processed as previously described (Miao et al., 2006). Samples were precipitated with trichloroacetic acid and analyzed by immunoblotting.
Biochemical analysis of recombinant proteins
MBP-Lnp1Cterm and MBP-Lnp1CtermDznfn were expressed in BL21-RIL (DE3) cells (Agilent Technologies, Santa Clara, CA). Bacteria were pelleted and lysed by sonication in buffer (20 mM HEPES, pH7.5, 145 mM NaCl, 5 mM KCl, 10 μM ZnSO4). Affinity purification with amylose resin (New England BioLabs) was performed with the soluble fraction of lysates according to manufacturer recommendations. Proteins were further purified by size exclusion chromatography with a S200 column (GE Healthcare). Sedimentation velocity analytical ultracentrifugation and analysis was performed as previously described (Folkmann et al., 2013).
Supplementary Material
Acknowledgments
We are grateful to the Wente and Ferro-Novick labs for helpful discussions. Preparation and imaging of TEM and Delta Vision images were performed in part through the use of the Vanderbilt University Medical Center Cell Imaging Shared Resource (supported by National Institutes of Health Grants CA68485, DK20593, DK58404, DK59637, and EY08126). The Delta Vision OMX microscope was acquired using funds from a National Institutes of Health High-End Instrumentation Grant (1S10OD012324, principal investigator Matthew Tyska, Vanderbilt University Medical Center). Antibody production and IgY purification were facilitated by the Vanderbilt Antibody Protein Resource, which is supported by the Vanderbilt Institute of Chemical Biology and the Vanderbilt Ingram Cancer Center (P30 CA68485). This work was supported by National Institutes of Health 5R01 GM057438 to S.R.W. and National Science Foundation Graduate Research Fellowship 2011100772 to A.K.C. Salary support for S.C. and S.F.-N. was provided by the Howard Hughes Medical Institute.
Abbreviations used:
- 3-AT
3-aminotriazole
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- INM
inner nuclear membrane
- NE
nuclear envelope
- NPC
nuclear pore complex
- Nup
nucleoporin
- ONM
outer nuclear membrane
- Pom
pore membrane Nup
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0053) on June 3, 2015.
REFERENCES
- Aitchison JD, Rout MP. The yeast nuclear pore complex and transport through it. Genetics. 2012;190:855–883. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol. 2012;197:209–217. doi: 10.1083/jcb.201111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M, Wente SR. In vivo dynamics of nuclear pore complexes in yeast. J Cell Biol. 1997;136:1185–1199. doi: 10.1083/jcb.136.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci USA. 2011;108:2216–2221. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey AK, Dawson TR, Chen J, Friederichs JM, Jaspersen SL, Wente SR. Integrity and function of the Saccharomyces cerevisiae spindle pole body depends on connections between the membrane proteins Ndc1, Rtn1, and Yop1. Genetics. 2012;192:441–455. doi: 10.1534/genetics.112.141465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol. 2010;189:795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Desai T, McNew JA, Gerard P, Novick PJ, Ferro-Novick S. Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2015;112:418–423. doi: 10.1073/pnas.1423026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol. 2012;14:707–716. doi: 10.1038/ncb2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr Opin Cell Biol. 2013;25:428–433. doi: 10.1016/j.ceb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial HJ, Rout MP, Giddings TH, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Hetzer MW. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 2010;119:469–477. doi: 10.1007/s00412-010-0289-2. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage JL, Bucci M, Watkins JL, Wente SR. Defining the essential functional regions of the nucleoporin Nup145p. J Cell Sci. 1997;110:911–925. doi: 10.1242/jcs.110.7.911. [DOI] [PubMed] [Google Scholar]
- English AR, Zurek N, Voeltz GK. Peripheral ER structure to function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Collier SE, Zhan X, Aditi, Ohi MD, Wente SR. Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease. Cell. 2013;155:582–593. doi: 10.1016/j.cell.2013.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;21:347–380. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell . 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Güttinger S, Hawryluk-Gara LA, Panté N, Mall M, Galy V, Haselmann U, Mühlhäusser P, Wozniak RW, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J Cell Biol. 2001;153:709–724. doi: 10.1083/jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Ryan KJ, Wente SR. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 2006;172:1441–1457. doi: 10.1534/genetics.105.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya K, Nagatoshi K, Noriyasu Y, Okamura T, Takamitsu E, Suzuki T, Utsumi T. Protein N-myristoylation pays a critical role in the endoplasmic reticulum morphological change induced by overexpression of protein lunapark, an integral membrane protein of the endoplasmic reticulum. PLoS One. 2013;8:e78235. doi: 10.1371/journal.pone.0078235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell. 2013;24:3920–3938. doi: 10.1091/mbc.E13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- Sengstag C. Using SUC2-HIS4C reporter domain to study topology of membrane proteins in Saccharomyces cerevisiae. Methods Enzymol. 2000;327:175–190. doi: 10.1016/s0076-6879(00)27275-3. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics: Laboratory Course Manual for Methods in Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, Kittanakom S, Damjanovic D, Curak J, Wong V, Stagljar I. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protoc. 2010;5:1281–1293. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]
- Stavru F, Hülsmann BB, Spang A, Hartmann E, Cordes VC, Görlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, Antonin W. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 2012;31:4072–4084. doi: 10.1038/emboj.2012.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Oliferenko S. Tts1, the fission yeast homolog of TMEM33 family, functions in NE remodeling during mitosis. Mol Biol Cell. 2014;25:2970–2983. doi: 10.1091/mbc.E13-12-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.