Abstract
Rap1 is a small GTPase that belongs to Ras superfamily. This ubiquitously expressed GTPase is a key regulator of integrin functions. Rap1 exists in two isoforms: Rap1a and Rap1b. Although Rap1 has been extensively studied, its isoform-specific functions in B cells have not been elucidated. In this study, using gene knockout mice, we show that Rap1b is the dominant isoform in B cells. Lack of Rap1b significantly reduced the absolute number of B220+IgM− pro/pre-B cells and B220+IgM+ immature B cells in bone marrow. In vitro culture of bone marrow-derived Rap1b−/− pro/pre-B cells with IL-7 showed similar proliferation levels but reduced adhesion to stromal cell line compared with wild type. Rap1b−/− mice displayed reduced splenic marginal zone (MZ) B cells, and increased newly forming B cells, whereas the number of follicular B cells was normal. Functionally, Rap1b−/− mice showed reduced T-dependent but normal T-independent humoral responses. B cells from Rap1b−/− mice showed reduced migration to SDF-1, CXCL13 and in vivo homing to lymph nodes. MZ B cells showed reduced sphingosine-1-phosphate-induced migration and adhesion to ICAM-1. However, absence of Rap1b did not affect splenic B cell proliferation, BCR-mediated activation of Erk1/2, p38 MAPKs, and AKT. Thus, Rap1b is crucial for early B cell development, MZ B cell homeostasis and T-dependent humoral immunity.
The small GTPase, Rap1, belongs to the Ras superfamily and cycles between an inactive GDP-bound and an active GTP-bound conformation (1, 2). This ubiquitously expressed molecular switch regulates cell proliferation, differentiation, and adhesion by distinct mechanisms through integrin activation and MAPK cascades (3, 4). Multiple extracellular stimuli can activate Rap1. This activation is mediated by various guanine nucleotide exchange factors (GEFs),3 such as C3G, CalDAG-GEF, and EPAC (5–7). C3G is activated by receptor-associated protein tyrosine kinases (5). CalDAG-GEF responds to calcium and diacylglycerol, downstream of phospholipase C signaling (8). EPAC is a cAMP-dependent GEF (7). Active Rap1-GTPase recruits multiple effectors (e.g., RapL/Nore1B and RIAM) to execute their cellular functions (9, 10). RapL, a Rap1-binding molecule, is a major effector of Rap1-GTPase in immune cells and associates with αLβ2 integrin (9). RIAM contains Ras-associated and pleckstrin homology domains and interacts with profilin and Ena/VASP proteins that regulate actin dynamics (10). Rap1 signaling is terminated by GTPase-activating proteins (GAPs), such as SPA-1 (signal-induced proliferation-associated protein 1) and RapGAPs, which convert active GTP-bound to inactive GDP-bound form (11, 12).
Integrins mediate cellular contacts to extracellular matrix or to their counter receptors, thereby regulating cell motility, polarity, growth, and survival (13). Rap1 is expressed in multiple hematopoietic cells and one of its key functions is to regulate integrin-mediated cell adhesion (4). This amplifies a series of hematopoietic cellular processes including aggregation, migration, extravasation, and homing (4, 14, 15). Mice lacking CalDAG-GEFI show impaired platelet adhesion and aggregation leading to impaired thrombus formation (16). Deficiency of C3G, one of the major Rap-specific GEFs, resulted in severe embryonic lethality, partly due to aberrant cell adhesion and spreading (17). Overexpression of Rap1, RapGEFs, GAPs, or their mutant proteins altered the activation of Rap1 and helped to uncover its signaling functions in different cell types (18–21). Rap1 can be activated through Ag or chemokine receptors (18, 22, 23). Active Rap1 plays an important role in immunological synapse formation between T cells and APCs (22).
Recent studies have well documented that interference in Rap1 functions can result in severe pathological conditions. In human, leukocyte adhesion deficiency syndrome III is associated with defective Rap1 activation and impaired stabilization of integrin bonds (24). Additionally, knockout mice for SPA-1 or RapL have provided critical insights into in vivo functions of Rap1-GTPase (25, 26). SPA-1 is the major Rap1GAP in the hematopoietic progenitors in bone marrow (26). SPA-1−/− mice show an increase in the hematopoietic stem cells and eventually develop leukemia (26). SPA-1 deficiency also results in aberrant BCR repertoire and autoimmune diseases (27). RapL deficiency causes defective chemokine-triggered lymphocyte adhesion and migration (25). However, many of these RapGEFs, GAPs, and effectors are promiscuous and can associate with multiple Rap1 GTPases; therefore the direct evaluation of Rap1 functions is limited in the described knockout mouse models.
Rap1 is closely related to the Rap2 family (Rap2a/2b/2c) and shares 60% homology (1, 2, 28). Rap1 exists in two isoforms, Rap1a and Rap1b, which are encoded by different genes. Rap1 isoforms share more than 95% amino acid homology and differ only in eight amino acids with five located at their C termini. Due to the high homology of Rap1a and Rap1b, their functions have been studied undistinguished in some studies (22, 23, 29). However, the different subcellular localization and tissue expression patterns of these two isoforms have been described and indicate their distinct functions (30–35). Studies have indicated the role of Rap1 isoforms in neuron, T cells, and platelets (19, 34, 35). Rap1a has been shown to positively regulate T cell functions in both transgenic and knockout mouse models (19, 36). Recently, we have generated Rap1b gene knockout mice (34). Approximately 40% of the Rap1b−/− fetuses die during embryogenesis. Surviving Rap1b−/− mice show bleeding disorder due to reduced platelet aggregation that is similar to the phenotype in CalDAG-GEFI−/− mice (16, 34). Irrespective of these findings, the differential expression of Rap1 isoforms and their specific roles in B cell have yet to be determined. In this study, using Rap1b−/− mice, we show that Rap1b is the dominant Rap1 isoform in B cells. Rap1b is crucial for early B cell development, marginal zone (MZ) B cell maturation, and T-dependent humoral responses. Additionally, absence of Rap1b leads to the reduced B cell adhesion and chemotaxis and in vivo homing. However, Rap1b is dispensable for BCR-mediated splenic B cell phosphorylation of Erk1/2, p38, and AKT.
Materials and Methods
Mice and reagents
Rap1b−/− mice were previously described (34). The Rap1b+/− mice were crossed with Rap1b+/− or Rap1b−/− mice to obtain Rap1b+/+ and Rap1b−/− mice. The following Abs and reagents were used in this study: recombinant proteins Rap1a-GST and Rap1b-GST (85–184 aa) (Novus Biologicals); rabbit anti-Rap1 polyclonal Ab, goat anti-rabbit mouse IgG (H+L) HRP (Santa Cruz Biotechnology); rabbit anti-Rap1b mAb (36E1), rabbit anti-Erk1/2 polyclonal Ab, rabbit anti-phospho-Erk1/2 mAb (197G2), rabbit anti-p38 polyclonal Ab, rabbit anti-phospho-p38 mAb (3D7), rabbit anti-AKT polyclonal Ab, rabbit anti-phospho-AKT (Ser473) mAb (193H12), and anti-GST mAb (Cell Signaling Technology); rabbit anti-β-actin Ab (Abcam); rat anti-mouse B220 (RA3-6B2), rat anti-mouse IgM (R6-60.2), rat anti-mouse IgD (11-26c.2a), rat anti-mouse CD19 (1D3), rat anti-mouse CD1d (1B1), rat anti-mouse CD21/CD35 (4E3), rat-anti-mouse CD23 (B3B4), Armenian hamster anti-moue CD3ε (145-2C11), rat anti-mouse CD16/CD32(93), rat anti-mouse CD43, rat anti-mouse CD25 and IL-4 (eBioscience); rat anti-mouse CD4 (H129.19), rat anti-mouse c-Kit, and anti-mouse CD8 (53-6.2) (BD Pharmingen); MOMA-1-FITC (Serotec); goat anti-mouse IgM F(ab′)2 (Jackson ImmunoResearch Laboratories); mouse anti-GAPDH LPS (Escherichia coli O111:B4; Sigma-Aldrich); CFSE (C-1157) and TAMRA (C-300) (Molecular Probes); and stromal cell-derived factor (SDF)-1 and CXCL13 (R&D Systems).
Flow cytometry, cell separation, and cell sorting
Single-cell suspensions were prepared by gently mincing the dissected organs through 70-μm cell strainers. T cells and resting B cells were purified by anti-CD3-PE mAb/anti-PE Microbeads (BD Pharmingen) and B cell isolation kit (Miltenyi Biotec). Separated cells were stained with anti-CD19 and anti-CD3ε mAbs, and their purity was >95% as confirmed by flow cytometry. Flow cytometry analysis was conducted in LSR-II and analyzed with FACSDiva software (BD Biosciences). For isolating newly forming, follicular (FO), and MZ B cell populations, single-cell suspensions from spleens were stained with anti-B220-Pacific Blue, anti-CD21-FITC, and anti-CD23-PE-Cy7 mAbs and sorted using FACSAria (BD Biosciences).
Immunohistochemistry staining
Spleens were embedded in Tissue-Tek (Sakura Finetek), frozen with dry ice-isopropanol mixture, and kept at −80°C until sectioning. The 10-μm cryostat sections were generated, air-dried for 1 h at room temperature, and fixed in acetone for 10 min. The sections were blocked with 0.1% BSA/PBS containing anti-CD16/CD32 mAb for 45 min, incubated for 1 h at room temperature in dark with the rat anti-mouse CD3ε-PE, anti-B220-Cy5, MOMA-1-FITC, anti-mouse IgM-PE, or anti-mouse IgD-FITC mAbs. Sections were mounted in aqueous mounting medium and visualized on inverted Nikon Eclipse TE200 fluorescence microscope. Images are at a magnification of ×40.
Bone marrow transplantation
Bone marrow cells from tibias and femurs of wild-type (WT) and Rap1−/− mice were prepared and resuspended in 2% FBS/RPMI 1640 (Invitrogen Life Technologies). A total of 5 × 106 bone marrow cells (200 μl) were injected i.v. to reconstitute 6- to 8-wk-old, sublethally irradiated (300 rad) JAK3−/− and lethally irradiated (1100 rad) Rap1b−/− mice 4 h after the irradiation. The recipient mice were sacrificed 8 wk later and the spleens were analyzed for reconstitution of B cells by flow cytometry.
Rap1-GTPase pull-down assay and Western blotting
B cells were purified from spleens and resuspended in HEPES buffer (10 mM HEPES/0.5% FCS/RPMI 1640). A total of 15 × 106 B cells were used for each lane. Cells were stimulated with 50 μg/ml goat anti-mouse IgM F(ab′)2 or 100 ng/ml SDF-1 for 2 min at 37°C or left unstimulated. Stimulation was terminated by adding one volume of 2X ice-cold lysis buffer (100 mM Tris (pH 7.4), 150 mM NaCl, 2% Nonidet P-40, 1% deoxycholic acid, 0.2% SDS, 2 mM sodium orthovanadate, with Phosphatase Inhibitor Cocktail (PhosSTOP; Roche Diagnostics)) and Proteinase Inhibitor Cocktail (Sigma-Aldrich). Lysates were incubated for 30 min on ice and centrifuged at 15,000 × g for 15 min at 4°C. An aliquot of cell lysates (1 × 106 cells) was taken out for evaluating the total Rap1 loading. The remaining cell lysates were incubated with glutathione-agarose beads that pre-coupled with GST-RalGDS-Ras binding domain at 4°C (37). This assay is based on the fact that RalGDS binds with high affinity to Rap1-GTPase but not to Rap1-GDP (37). The beads were washed, and 30 μl of 1X Laemmli buffer was added and heated at 95°C to elute the pull-down proteins. The active Rap1 was analyzed by an Ab that recognizes both Rap1a and Rap1b. For Western blotting, cell lysate aliquot (corresponding to 1 × 106 cells) or pull-down protein pellets were separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with primary and secondary Abs conjugated with HRP. The signal was detected by ECL (GE Healthcare Life Sciences).
IL-7-driven bone marrow culture and BrdU labeling
Primary pre-B cell cultures were previously described (38). Briefly, erythrocyte-depleted total bone marrow cells were cultured with 2 ng/ml recombinant murine IL-7 (R&D Systems) at 2 × 106/ml for 5–7 days. The medium was replaced with fresh IL-7 every second day. For BrdU labeling, on day 5, bone marrow cells treated with IL-7 were seeded into 24 wells and cultured for another 3 days with IL-7. These cells were pulsed for 3 h with 10 μM BrdU at the end of day 3. Cells were stained with anti-B220 and anti-IgM Abs, resuspended in cold PBS, fixed with ice-cold ethanol (95%), and incubated on ice for 30 min. Cells were fixed or permeabilized with 1% paraformaldehyde/0.01% Tween 20 at 4°C overnight. Cells were resuspended with 50 Kunitz DNaseI/ml in 0.15 M NaCl, 4.2 mM MgCl2 (pH 5) for 10 min at 37°C, washed, stained with 1/50 anti-BrdU-FITC (BD Biosciences) for 30 min, and analyzed.
[3H]Thymidine incorporation assay
Purified B cells were plated into 96-well plates in triplicates (2 × 104 cells/well) and stimulated with 10 μg/ml goat anti-mouse IgM F(ab′)2, 10 μg/ml goat anti-mouse IgM F(ab′)2 plus 10 ng/ml IL-4, or 1 μg/ml LPS in 10% FBS for 72 h. A total of 1 μCi of [3H]thymidine (GE Healthcare Life Sciences) was added 16 h before harvest. Labeled DNA from cells was collected on GSC filters (Whatman), and the radioactivity was measured in a microplate scintillation counter.
Immunization
For the T-independent immune response, 6- to 8-wk-old mice were i.p. immunized with 100 μg of trinitrophenol (TNP)-Ficoll (Biosearch Technologies) in PBS. A total of 50 μl of blood was collected from the retro-orbital venous plexus with capillary tubes on days 0 and 7 after immunization. For the T-dependent response, mice were i.p. injected with 100 μg of TNP-keyhole limpet hemocyanin (KLH) in 200 μl of aluminum hydroxide adjuvant and Imject Alum (Pierce) and boosted with 10 μg of TNP-KLH without adjuvant on day 21. Sera were collected on days 0, 7, and 28. Sera were analyzed by ELISA for basal and TNP-specific Ig iso-types using Clonotyping System (Southern Biotechnology Associates).
In vitro chemotaxis assay
Chemotaxis assay was performed in triplicates in 5.0-μm Transwell (5.0-μm pore, 6.5 mm; Costar). The lower compartment contained 600 μl of 150 ng/ml SDF-1 or 500 ng/ml CXCL13 in HEPES buffer (RPMI 1640, 10 mM HEPES, 0.5% FBS). Purified B cells were resuspended to 5 × 106/ml in the same buffer. The 100 μl of B cell suspension was added to the upper chamber and allowed to migrate for 3 h. The number of viable cells that migrated into the lower compartment was determined by flow cytometry and expressed as a percentage of the input cells that were directly added to the lower compartment. For sphingosine 1-phosphate (S1P)-induced migration assay, different concentrations of S1P (Biomol) in HEPES buffer were added to the lower compartments and the RBC-depleted splenocytes from WT and Rap1b−/− were added to the upper chambers. At 3 h later, the cells that migrated into the lower compartment were stained with anti-B220, anti-CD21, and anti-CD23 Abs.
In vitro adhesion assay
S1P-induced adhesion of B cells to ICAM-1 was described (39). Briefly, Immulon 4 HBX 96-well flat-bottom plates (ISC BioExpress) were coated with 1% fat-free BSA/PBS, recombinant mouse ICAM-1/Fc 10 μg/ml (R&D Systems), triplicates, keep at 4°C overnight. Splenocytes were incubated in 10% FBS/RPMI 1640 for 30 min at 37°C in a tissue culture flask to eliminate adherent macrophages. Nonadherent floating cells were collected and RBCs were lysed. Cells were resensitized in RPMI 1640 containing 0.5% fatty acid-free BSA and 10 mM HEPES at 37°C for 1 h. The plates were gently washed with PBS twice and blocked with 1% fatty acid-free BSA/PBS at 37°C for 1 h. A total of 3 × 105 cells in 100 μl of buffer with or without 100 nM S1P and incubated at 37°C for 60 min nonadherent cells wells were washed away and adherent cells were released by keeping the plates on ice for 20 min with RPMI 1640/5 mM EDTA. Collected cells were stained for B220, CD23, and CD21 markers. Adhesion was calculated as a percentage of input cells.
To quantify the adhesion of B cell progenitors from bone marrow, IL-7-driven bone marrow cells were labeled on day 7 with 2 μM CFSE and 10 μM TAMRA for 10 min at room temperature, and the reaction was terminated by adding one volume of FBS. Cells were washed and resuspended in 10% FBS medium. A mixture of 0.5 × 106 CFSE- and TAMRA-labeled cells was added onto M2-10B4 confluent stromal cell layer (American Type Culture Collection) and incubated for 5 h. Unbound cells were washed away with warm plain medium and adherent cells were released with 5 mM EDTA/plain medium. Stromal cell layers were removed by filtering before flow cytometry.
In vivo homing assay
RBC-depleted splenocytes from WT and Rap1b−/− were resuspended to 20 × 106 cells/ml in PBS and labeled with 2 μM CFSE and 10 μM TAMRA as bone marrow pro-B cells and resuspended in 2% FBS medium. A mixture of 20 × 106 CFSE- and TAMRA-labeled cells was i.v. injected into recipient WT mice. At 3 h later, single-cell preparations from spleen, peripheral lymph nodes (LNs) (inguinal and auxiliary), and mesenteric LN of recipient mice and a portion of labeled cells were stained with anti-B220-allophycocyanin or anti-CD3-PE-Cy7 mAbs. The homing efficiency of Rap1b−/−/WT B and T cells were calculated compared with the input ratio, which was normalized to 1. A dye swap was also performed.
Statistics
Data were presented as mean ± SE, and statistical significance was calculated using the Student t test. A value of p < 0.05 was considered significant.
Results
Rap1b is the dominant isoform in B cells
Because Rap1 exists in two isoforms, we first analyzed the relative levels of Rap1a and Rap1b in B cells. Expression of total Rap1 protein was assayed using an affinity purified polyclonal IgG that recognizes an identical epitope in the C terminus of both Rap1a and Rap1b isoforms. Fig. 1A shows that the total Rap1 was readily detectable in the WT-derived B cells. However, only a residual level of Rap1 protein was present in B cells from Rap1b−/− mice. With recombinant GST-Rap1A full-length protein and GST-Rap1B (85–184 aa), we show that anti-Rap1 has similar affinities to both Rap1a and Rap1b proteins compared with anti-GST Ab (Fig. 1B). Therefore, the residual protein band in Rap1b−/− B cells represents Rap1a. Similar Rap1 expression pattern was also observed in T cells (Fig. 1A). This finding demonstrates that the Rap1b is the major isoform in B and T cells. Rap1b-specific mAb confirmed the absence of Rap1b in B and T cells from Rap1b−/− mice (Fig. 1A). The dominant expression of Rap1b suggested its potential role in B cell development and function.
FIGURE 1.
Rap1b expression and its role in B cell development. A, Expression of total Rap1 and Rap1b in B and T cells from WT and Rap1b−/− spleens. Total Rap1 was detected with anti-Rap1 polyclonal Ab (upper) and Rap1b was detected with anti-Rap1b mAb (middle). Membranes were stripped and reprobed for actin as loading control (lower). B, Western blot of recombinant GST-Rap1a and GST-Rap1b (85–184 aa). A total of 100 ng of recombinant proteins were detected with anti-Rap1 polyclonal Ab and anti-GST. C and D, Early B cell development in the bone marrow is impaired in Rap1b−/− mice. Bone marrow cells were stained with anti-B220, anti-IgM, CD43, c-Kit, and CD25 mAbs. The absolute cell number was calculated as following: cell number (×106) = cellularity × (percentage of lymphatic cell) × (percentage of lymphocyte subsets). The B cell subset percentage and the absolute cell number are presented. Data presented are the representative of n = 3–5 mice. E, Peripheral blood was stained with anti-B220, IgM, and IgD. The percentage of each subpopulation was shown. Data presented are representative of n = 5 mice. F, IL-7-driven bone marrow culture of WT and Rap1b−/− mice led to the similar purity of B220+IgM− pro/pre-B cells (top) for n = 3 + 3 mice and showed similar proliferation rate of BrdU incorporation (bottom) for n = 3 + 3 mice. G, Reduced Rap1b−/− pro/pre-B cells to M2-10B4 stromal cell line. IL-7-driven bone marrow-derived pro/pre-B cells from WT and Rap1b−/− mice were labeled with CFSE and TAMRA respectively, added to stromal cell line and incubated for 5 h. Adherent cells were analyzed by flow. A dye swap was performed, which showed the same result (data not shown).
Lack of Rap1b impairs B cell development in the bone marrow
As Rap1b is highly expressed in B cells, we analyzed its role in B cell development. The total cellularity of bone marrow in Rap1b−/− mice was significantly less compared with that of WT (WT: 45.4 ± 8.8 × 106 cells vs Rap1b−/−: 31.0 ± 6.4 × 106 cells; n = 6+6 mice; and p = 0.014). The percentage of B220+ B cells were comparable between Rap1b−/− and WT mice. However, because of reduced cellularity, the absolute number of B220+ B cells was significantly reduced in Rap1b−/− mice (Fig. 1C). Based on B220 and IgM expression, B cells in the bone marrow can be divided into different maturation stages: pro/pre-B cells (B220+IgM−), immature B cells (B220+IgM+), and recirculating mature B cells (B220high). The percentages of pro/pre-B cells were reduced in Rap1b−/− mice compared with WT. Likewise, the absolute number of both pro/pre-B cells and immature B cells were significantly reduced in Rap1b−/− mice (Fig. 1C). In contrast, the number of recirculating mature B cell B220high population was comparable between Rap1b−/− and WT mice (Fig. 1C). B cell development in bone marrow can be further characterized through the expression of CD43 (leukosialin), CD117 (c-Kit), and CD25 (IL-2Rα). Early pro-B cells are marked by the expression of B220+CD43+, which develop into B220+CD43− B cells. As pro/pre-B cells mature, c-Kit expression decreases while CD25 level increases. The relative (WT: 2.6 ± 0.7% and Rap1b−/−: 2.8 ± 0.5%) and the absolute (WT: 0.41 ± 0.167 × 106 cells and Rap1b−/−: 0.264 ± 0.076 × 106 cells) B220+CD43+ cell numbers did not differ significantly between Rap1b−/− and WT bone marrow (n = 6 + 6 mice; p = 0.0855) (Fig. 1D). However, B220mediumCD43− B cells were much lower in Rap1b−/− in both relative percentages (WT: 35.2 ± 8.4% and Rap1b−/−: 25.9 ± 10.3%) and absolute number (WT: 6.5 ± 2.09 × 106 cells and Rap1b−/−: 2.58 ± 1.27 × 106 cells, n = 6 + 6 mice; p = 0.003), which is consistent with the reduced B220+IgM− populations (Fig. 1D, upper). The increased B220+c-kit+ population in Rap1b−/− and with relatively decreased B220+CD25+ population (Fig. 1D, middle and lower) indicate a delay in B cell maturation in Rap1b−/− mice. In peripheral blood, there is an increased IgM+IgD− B cell population in Rap1b −/− mice (Fig. 1E). Thus, the lack of Rap1b resulted in a developmental impairment of B cell progenitor maturing into pro/pre-B cell and immature B cell stages.
Adhesion but not proliferation of bone marrow-derived, IL-7-driven pre-B cells is impaired in Rap1b−/− mice
To further gain the insight of B development in Rap1b−/− mice, we conducted in vitro proliferation of pro/pre-B cells derived from IL-7-driven bone marrow culture. The reduced pro/pre-B cells in the bone marrow of Rap1b−/− mice may be due to either reduced proliferation or adhesion to stromal cells. Bone marrow cells cultured with IL-7 contained similar purity of B220+ and B220+IgM− B cells in WT and Rap1b−/− mice (Fig. 1F, top). In addition, the Rap1b−/− pro/pre-B cells showed similar BrdU incorporation as WT cells (Fig. 1F, bottom), thereby excluding a defect in the proliferation as a probable cause for our observations. However, consistent to the role of Rap1b in regulating integrin activation, we observed a significantly decreased adhesion of Rap1b−/− pro/pre-B cells to the mouse stromal cell line M2-10B4, reduced to 60% of WT (Fig. 1G).
Cells from auxiliary and inguinal LNs were stained with anti-B220, anti-IgM, and anti-IgD Abs. The percentage of B220+, IgM+IgD+, and IgM−IgD+ cells was comparable between Rap1b−/− and WT mice (Fig. 2D). Additionally, we examined the T cell development in Rap1b−/− mice. The cellularity of thymus, spleen, and LN from Rap1b−/− mice was comparable to that of WT (thymus, WT: 120.7 ± 8.9 × 106 cells; Rap1b−/−: 119.0 ± 40.8 × 106 cells, spleen, WT: 107.6 ± 24.6 × 106 cells; Rap1b−/−: 99.7 ± 16.2 × 106 cells; and LN: 4.8 ± 1.1 × 106 cells; Rap1b−/−: 4.8 ± 2.4 × 106 cells; n = 6–8 mice). T cell development appears normal in thymus, spleen, and LN based on CD4 and CD8 expression in the absence of Rap1b (data not shown). These results indicate Rap1b plays a role in B but not in T cell development.
FIGURE 2.
Reduced MZ B cells in the spleens of Rap1b−/− mice. A and B, Single-cell suspensions derived from spleens were stained with anti-B220, anti-IgM, and anti-IgD mAbs. Histograms of B220+ cells from the WT and Rap1b−/− mice are shown (A). The staining pattern of IgM and IgD of spleen cells were shown (B). The percentage of subsets is shown (bottom) (n = 7 mice for both experiment). C, Splenocytes were stained with anti-B220, anti-CD21, and anti-CD23 mAbs. B220+ splenocytes were separated into CD21low/−CD23low/− newly forming (NF) B cells, CD23highCD21low/− FO B cells, and CD21highCD23low/− MZ B cells (upper). Splenocytes were also stained with anti-CD21 and anti-IgM (middle) or anti-CD1d and anti-IgM mAbs (lower). D, Single-cell suspensions derived from auxiliary and inguinal LNs were stained with anti-B220, anti-IgM, and anti-IgD mAbs. The percentage of B220+, IgM+IgD+, and IgM−IgD+ cells were shown (right). B–D, n = 6–8 mice.
Rap1b regulates the homeostasis of MZ B cells
Next, we analyzed the B cell maturation in the spleen. The percentage B220+ B cells were comparable between Rap1b−/− and WT mice (Fig. 2A). However, when we stained splenocytes with anti-IgM and anti-IgD Abs, Rap1b−/− mice showed a reduction in IgM+IgDlow/− population (Fig. 2B). B220+ splenic B cells can be further separated into newly forming B cells (CD21low/−CD23low/−), FO B cells (CD23highCD21low/−), and MZ B cells (CD21highCD23low/−). The percentages of FO B cells were comparable between Rap1b−/− and WT mice. However, the MZ B cells were significantly reduced (~55%), whereas the newly forming B cells were relatively increased in Rap1b−/− mice (Fig. 2C, upper panels). MZ B cells can also be defined by their exclusive high-level expression of IgM, CD21, and CD1d (40). Staining for these markers confirmed the reduction in IgMhighCD21high and IgMhighCD1dhigh B cell population (Fig. 2, D, middle and lower panels).
MZ B cells reside in a unique location around the follicles in the spleen. To examine the lymphocyte organization, we performed H&E staining of spleen cryosections. Rap1b−/− mice displayed normal spleen architecture of red and white pulps (Fig. 3A). Staining with anti-CD3ε and anti-B220 mAbs revealed a normal B and T cell organization within the white pulps of the spleens (Fig. 3B). To view the MZ, we used MOMA-1 mAb, which stains the met-allophillic macrophages. The location of metallophillic macrophages defines the boundary between the FO and MZ. The spleen sections from the WT mice revealed a clearly defined band of B220+ MZ area outside of the MOMA-1+ metallophillic macrophages. However, similar regions were greatly reduced and less contiguous in Rap1b−/− mice (Fig. 3C). Staining splenocytes with anti-IgM and anti-IgD mAbs also demonstrated a reduction in IgMhighIgD− MZ B cells in the Rap1b−/− mice (Fig. 3D). The immunostaining results are consistent with the flow cytometry data. The significant alteration in the newly forming B cell to MZ B cell ratio indicates that the Rap1b plays a critical role in the homeostasis of MZ B cells.
FIGURE 3.
Reduced MZ compartment in the spleens of Rap1b−/− mice. A, H&E staining of cryosection of WT and Rap1b−/− spleens. B–D, Immunostaining of spleen cryosections with anti-CD3-PE (red), MOMA-1-FITC (green), and anti-B220-Cy5 (blue) mAbs (B); with anti-IgM-PE (red) and MOMA-1-FITC (green) mAbs (C); and with anti-IgM-PE (red) and anti-IgD-FITC (green) mAbs (D). MZ areas are indicated (open arrowheads). A minimum of 20 white pulps was analyzed and data presented are representatives of two mice in each group. Scale bar represents 100 μM.
MZ B cell defect is cell autonomous
Rap1b is expressed in multiple cell lineages (4). Therefore, the defects we observed in MZ B cells can be cell intrinsic or due to an abnormality in the stromal microenvironment. To distinguish between these two possibilities, we performed bone marrow transplantation experiments. As expected, the transfer of WT bone marrow cells into JAK3−/− mice led to the reconstitution of newly forming, FO, and MZ B cells (Fig. 4A). However, similar reconstitutions using Rap1b−/− bone marrow cells into JAK3−/− mice gave rise to normal FO B cells, but significantly reduced MZ B cells (p = 0.007; n = 4 mice) and slightly increased newly forming B cells, which mimicked the phenotype in Rap1b−/− mice (Fig. 4B). Additionally, transfer of WT bone marrow cells to sub-lethally irradiated Rap1b1b−/− mice showed they were able to reconstitute the MZ population in the Rap1b−/− host (Fig. 4C). Taken together, our results demonstrate that the B cell defects in the spleen of Rap1b−/− mice were cell intrinsic, and the splenic environment in Rap1b−/− mice was able to support the development of WT B cells.
FIGURE 4.

MZ B cell defect in Rap1b−/− mice is cell autonomous. Bone marrow cells derived from Rap1b−/− and WT mice were i.v. transferred to sublethally irradiated JAK3−/− or lethally irradiated Rap1b−/− mice in three groups: WT to JAK3−/− (A), Rap1b−/− to JAK3−/− (B), WT to Rap1b−/− (C) mice. At 8 wk later, reconstituted cells from recipient spleens were stained with anti-B220, anti-CD21, and anti-CD23 mAbs. Gated B220+ cells were separated into CD21low/−CD23low/− newly forming (NF) B cells, CD23highCD21low/− FO B cells, and CD21highCD23low/− MZ B cells. Number indicated is percentage of B cell subsets from one representative mouse of each recipient group (n = 4 mice for all experiments).
Rap1b is dispensable for splenic B cell proliferation
Earlier study indicated a role for Rap1 signaling in lymphocyte proliferation (41, 42). Therefore, we hypothesized that the reduction in MZ B cells in Rap1b−/− mice could be due to defective cell proliferation. We purified B cells from spleens and stimulated them with goat anti-IgM F(ab′)2, goat anti-IgM F(ab′)2 plus IL-4, or LPS. [3H]Thymidine incorporation assay showed comparable proliferation of Rap1b−/− B cells as that of WT (Fig. 5A). To further investigate the proliferation of B cell subsets, FO B cells and MZ B cells were sorted, CFSE-labeled, and stimulated with LPS. Percentages of dividing cells between Rap1b−/− and WT mice were compared and found to be equivalent (Fig. 5B). Therefore, it seems that Rap1b does not regulate splenic B cell proliferation under current stimulation conditions.
FIGURE 5.
Rap1b is dispensable for B cell proliferation, Erk1/2, p38, and AKT phosphorylation. A, Splenic B cells were stimulated with 10 μg/ml goat anti-mouse IgM F(ab′)2, 10 μg/ml goat anti-IgM F(ab′)2 plus 10 ng/ml IL-4, 1 μg/ml LPS, or left untreated for 72 h. [3H]Thymidine incorporation is shown as average of triplicate wells. B, Sorted spleen FO and MZ B cells were labeled with 1 μM CFSE and then stimulated with 10 μg/ml LPS. The cell division was analyzed by flow cytometry after 72 h. The percentage of cells that have divided is shown. C, Activation of Rap1 after BCR cross-linking and SDF-1 stimulation in WT B cells. Purified splenic B cells were stimulated with 50 μg/ml goat anti-mouse IgM F(ab′)2 or 100 ng/ml SDF-1. An aliquot of total lysate was used to detect the total Rap1 expression as a loading control (lower). Remaining lysates were incubated with GST-RalGDS and active Rap1-GTPase was detected with anti-Rap1 polyclonal Ab (upper, short exposure; middle, longer exposure; lower, total Rap1 protein loading control). D, Activation of Rap1 after BCR cross-linking and SDF-1 stimulation was diminished in Rap1b−/− B cells. Purified splenic B cells from WT and Rap1b−/− mice stimulated as in C and 10-min long exposure was shown in the residual Rap1a in Rap1b−/− B cells. E, Activation of Erk1/2 and p38 after BCR cross-linking and SDF-1 stimulation. Purified splenic B cells were stimulated with 50 μg/ml goat anti-IgM F(ab′)2 or 100 ng/ml SDF-1 for 2 min. The active Erk1/2 and p38 were detected by anti-phospho-Erk or phospho-p38 mAbs. The total Erk1/2, p38, and actin were used as protein loading controls. Data in C and D are representative of three independent experiments. F, Purified splenic B cells were stimulated with goat anti-IgM F(ab′)2 polyclonal at indicated time points. The phosphorylated AKT (Ser473) and total AKT protein were detected with anti-phospho-AKT-S473 and anti-AKT Abs, respectively. Data shown are representative of two independent experiments.
Diminished Rap1-GTPase activation in the Rap1b−/− B cells
Earlier studies have shown that BCR engagement and chemokine stimulation lead to increased GTP-bound active Rap1 in B cells (18, 23, 42). However, the specific isoform activated is not known. Therefore, we purified resting B cells from spleens and stimulated them with goat anti-IgM F(ab′)2 and SDF-1. The active Rap1-GTP was analyzed by a pull-down assay. In WT B cells, Rap1-GTP was increased to more than 2-fold compared with the basal level (Fig. 5C, upper). Overexposure led to the diminished difference between control and activated samples (Fig. 5C, middle). As the Rap1a level was extremely low compared with Rap1b in B cells and no good commercial Rap1a specific Ab are available, we used anti-Rap1 Ab that recognizes both the isoforms. The basal level of Rap1-GTP in Rap1b−/− resting B cells was hardly detectable even after a prolonged exposure (Fig. 5D, lane 4), which led to the saturation of Rap1-GTP in WT sample. Upon stimulation, Rap1-GTP was also increased in Rap1b−/− B cells. However, it was much lower than that in WT cells (Fig. 5D, lanes 5 and 6). These results indicate that although both Rap1a and Rap1b can be converted to active GTP-bound forms through BCR or chemokine receptor-mediated activation, the major isoform is Rap1b.
Lack of Rap1b does not affect the phosphorylation of Erk1/2, p38, or AKT
Activation of Erk1/2, p38, and AKT are critical for lymphocytes proliferation and survival (43, 44). Yet the role of Rap1b in the activation of these molecules is not clear. Overexpression of Rap-GAPII in B cells, which inhibited the activation of both Rap1 and Rap2, did not affect Erk1/2 and p38 phosphorylation but led to increased AKT activation (45). To test the role of Rap1b in these events, we examined the BCR or SDF-1-stimulated splenic B cells. Fig. 5E shows that the levels of Erk1/2 and p38 phosphorylation were comparable between Rap1b−/− and WT. It is important to note that the BCR- or SDF-1-mediated activation resulted in only a minimal increase of Rap1-GTP in Rap1b−/− B cells (Fig. 5D). Therefore, Rap1b does not seem to regulate the phosphorylation of Erk1/2 and p38. Furthermore, we stimulated splenic B cells with goat anti-IgM F(ab′)2 Ab and found similar levels of AKT phosphorylation at its Ser473 residue in Rap1b−/− as that of WT. This finding indicates that Rap1b-GTPase does not regulate AKT phosphorylation during BCR signaling in splenic B cells (Fig. 5F).
Rap1b regulates T-dependent but not T-independent humoral immunity
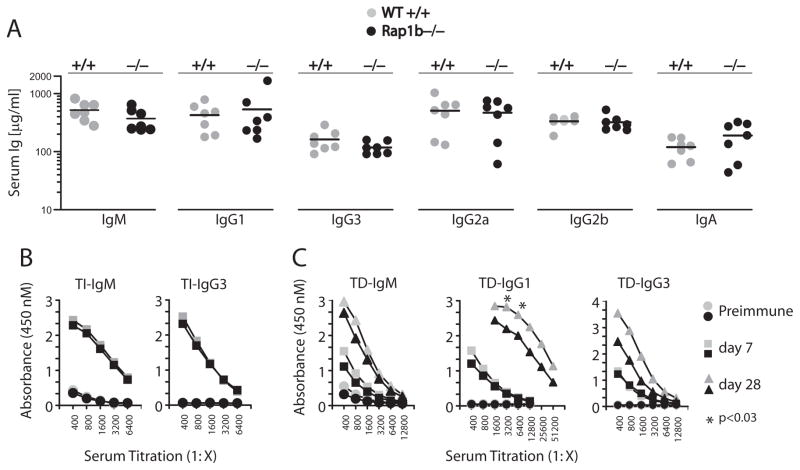
The present study indicates that Rap1b is the dominant isoform in B cells and regulates B cell development. To test whether Rap1b plays a role in B cell-mediated immunity, we first analyzed the basal Ig levels in the sera of Rap1b−/− mice. Fig. 6A shows that the preimmune Ig IgM, IgG1, IgG2a, IgG2b, IgG3, and IgA levels were comparable between WT and Rap1b−/− mice. Because MZ B cells have been shown to play an important role in T-independent Ab generation (46), we challenged the mice with T-independent Ag TNP-Ficoll and collected sera to analyze the presence of Ag-specific Abs. Rap1b−/− mice were able to generate similar amounts of TNP-specific IgM and IgG3, the two major Ig isotypes generated during T-independent responses, as generated by WT mice (Fig. 6B). Optimal Ab response in Rap1b−/− mice indicates that the remaining MZ B cells were sufficient for T-independent Ab generation. Next, we examined the T-dependent immune responses by i.p. injection of TNP-KLH. In the primary response, the generation of TNP-specific IgM, IgG1, and IgG3 was comparable between Rap1b−/− and WT mice (Fig. 6C). However, after Ag boost, the TNP-specific IgG1 was significantly lower in the Rap1b−/− mice compared with that of WT (Fig. 6C). Although not significant, the TNP-specific IgG3 were also lower in Rap1b−/− mice during the secondary responses (Fig. 6C). Therefore, we conclude that Rap1b is required for optimal T-dependent but not T-independent humoral responses.
FIGURE 6.
T-dependent but not T-independent humoral response is impaired in Rap1b−/− mice. A, Basal Ig levels (μg/ml) of 6- to 8-wk-old mice were determined by ELISA (gray circle, WT; ●, Rap1b−/−, n = 7 mice). B, T-independent immune responses were intact in Rap1b−/− mice. Mice were i.p. injected with TNP-Ficoll, and sera were collected at days 0 and 7 after immunization. TNP-specific IgM and IgG3 isotype levels at days 0 and 7 were measured by ELISA. The values were presented as average of absorbance of serial dilution at 450 nm of each group. Preimmune (gray circle, WT; ●, Rap1b−/−) and day 7 (
 , WT; ■, Rap1b−/−) mice are shown for n = 4 mice. C, T-dependent immunization. Mice were i.p. injected with TNP-KLH and were boosted at day 21. TNP-specific Ab responses at day 0, 7, and 28 were determined by ELISA. Day 28 (
, WT; ■, Rap1b−/−) mice are shown for n = 4 mice. C, T-dependent immunization. Mice were i.p. injected with TNP-KLH and were boosted at day 21. TNP-specific Ab responses at day 0, 7, and 28 were determined by ELISA. Day 28 (
 , WT; ▲, Rap1b−/−) mice are shown for n = 4 mice. SD was excluded from the data because of the variability in a few individual sera.
, WT; ▲, Rap1b−/−) mice are shown for n = 4 mice. SD was excluded from the data because of the variability in a few individual sera.
Rap1b regulates B cell chemotaxis, adhesion, and in vivo homing
Rap1 regulates cell adhesion and migration (30). To better understand the functional role of Rap1b in B cell migration, we first examined the chemokines SDF-1 and CXCL13 and lipid S1P-induced chemotaxis in Transwell assays. Rap1b−/− splenic B cells displayed significantly reduced migration toward chemokine gradient (Fig. 7A). S1P is important for lymphocyte migration (47, 48). Similar to SDF-1 and CXCL13, S1P has been shown to activate Rap1 and integrin activation is required for the correct localization of MZ B cells (39, 49). Consistent with published data (50), MZ B cells showed robust response to S1P and showed higher migration toward S1P. In contrast, Rap1b−/− B cells showed dramatically reduced migration compared with WT (Fig. 7B). MZ B cells have higher αL and β2 integrin expression and show higher adhesion to ICAM-1 compared with FO B cells (39). Similarly, S1P-induced adhesion to ICAM-1 was also reduced in Rap1b−/− B cells (Fig. 7C). To further investigate these defects, we performed an in vivo homing assay. Rap1b−/− and WT-derived splenocytes were separately labeled with TAMRA and CFSE-DA, respectively, premixed and i.v. injected into the recipient mice. To provide a Rap1b-sufficient microenvironment, we used the WT mice as the recipients. Three hours later, cells from spleen, auxiliary/inguinal and mesenteric LN were stained with anti-B220 and anti-CD3ε mAbs, and the ratio of Rap1b−/− to WT-derived lymphocytes were quantified (Fig. 7D). The homing of B and T cells from Rap1b−/− and WT donors into the recipient spleens were comparable. However, the ability of Rap1b−/− B cells to migrate into the auxiliary or inguinal LN was significantly reduced (~60%). Similarly, homing of Rap1b−/− B cells into mesenteric LN was also affected (Fig. 7E, left). We also observed reduced ability of Rap1b−/− T cells to migrate into the auxiliary or inguinal LN (Fig. 7E, right). Together, these results demonstrate that Rap1b plays a critical role in chemokine-mediated migration and in the in vivo homing of B cells.
FIGURE 7.
In vitro chemotaxis, adhesion, and in vivo homing of Rap1b−/− B cells are reduced. A, A total of 150 ng/ml SDF-1- or 500 ng/ml CXCL13-induced migration of Rap1b−/− B cells is impaired. The migration rate is presented as a percentage of migrated cells compared with the input cells that were added directly the lower chamber. Data presented are representative of three independent experiments. B, Reduced migration of Rap1b−/− MZ and FO B cell to S1P. RBC-depleted splenocytes were added to Transwell with S1P gradient (0–1000 nM) and allowed to migrate for 3 h. The migrated cells were stained with anti-B220, CD21, and CD23 mAbs. The migration rate is presented as a percentage of migrated cells compared with the input cells that were added directly the lower chambers. Data presented are representative of three independent experiments. C, Reduced S1P-induced adhesion to ICAM-1 of Rap1b−/− B cells. The 96-well plates were coated with BSA or recombinant mouse ICAM-1 10 μg/ml as described in Materials and Methods and cells were incubated with or without 100 nM S1P and incubated at 37°C for 60 min. After washing to remove the nonadherent cells, adherent cells were stained for B220, CD23, and CD21. Adhesion was calculated as a percentage of input cells. D, Lack of Rap1b impairs the in vivo homing of B and T cells. RBC-depleted WT and Rap1b−/− splenocytes were labeled with CFSE and TAMRA, respectively, and injected into WT recipient mice. At 3 h later, cells from spleens, auxiliary or inguinal LN, and mesenteric LN of recipient mice were stained with anti-B220-allophycocyanin or anti-CD3-PE-Cy7. E, A representative recipient mouse for B cell homing is shown (right) for data from n = 7 mice. The homing efficiency is shown as the ratio of homed Rap1b−/− to WT B or T cells after corrected to the ratio of injected Rap1b−/− to WT B or T cells (left) for data obtained from n = 3 mice.
Discussion
In this study, we demonstrate that Rap1b is the dominant isoform in B cells. Using gene knockout mice, we show that Rap1b is essential for pro/pre-B cell development in the bone marrow and for the homeostasis of MZ B cells in the spleen. Furthermore, Rap1b is required for optimal T-dependent humoral immunity, chemokine- and S1P-stimulated migration, S1P-induced adhesion to ICAM-1, and in vivo homing of B cells. Rap1b is also important for B cell progenitor adhesion to stromal cell lines. Rap1b is not required for splenic B cell proliferation and BCR-mediated activation of Erk1/2, p38, or AKT.
One of the key findings in this study is the role of Rap1b in B cell development in the bone marrow. A reduction of pro/pre-B cells and immature B cells indicates that the requirement of Rap1b cannot be compensated by Rap1a or Rap2 isoforms. Several gene knockout mice with altered Rap1 signaling have been recently generated (25, 26, 36). Deletion of SPA-1, a principle Rap1GAP in lineage negative (Lin−) bone marrow cells, resulted in an accumulation of Rap1-GTP and lead to an increase of the hematopoietic progenitors (27). Expression of constitutively active Rap1, RapE63, resulted in the Lin− bone marrow cell expansion, whereas SPA-1 led to the suppression (27). However, in the absence of Rap1b, bone marrow-derived B220+ B cell lineage showed similar proliferation rates as that of WT. Interestingly, lack of Rap1b resulted in reduced adhesion to bone marrow stromal cell lines. The reduced bone marrow pro/pre-B cells in Rap1b−/− may be due to reduced adhesion rather than a decreased expansion of B cell lineage. In addition, although lack of Rap1b did not adversely affect the number of B220+CD43+ B progenitors, there was a delayed maturation as indicated by more c-kit+ and fewer CD25+ cells in Rap1b−/− bone marrow. In peripheral blood, a higher percentage of B cells displayed IgM+IgD− immature phenotype compared with that of WT. Reduced adhesion of pro/pre-B cell to stromal cells could also cause a corresponding decrease of B progenitors in the bone marrow of Rap1b−/− mice. Taken together, these results indicate a possible differential regulation of Lin− and Lin+ bone marrow cells by Rap1b. The reduced bone marrow pro/pre-B cells in Rap1b−/− may be due to reduced adhesion rather than an augmented expansion of B cell lineage.
Spleen has a unique architecture composed of stromal and hematopoietic cells that are vital for B cell maturation and function. A strong reduction of MZ B cells in Rap1b−/− mice may be due to the deficiency in B or splenic stromal cells. Rap1b−/− mice showed normal organization of spleen and the defect of MZ B cells was recapitulated in our bone marrow transplantation experiment. Together, these indicate the MZ B defect is cell intrinsic. A MZ B cell defect is also observed in RapL−/− mice validating the role of Rap1-RapL signaling in MZ B cell development (25). However, the MZ B cells in RapL−/− mice are nearly absent, which is a more severe phenotype compared with Rap1b−/− mice. The multiple upstream RapL activators, Rap1 and Rap2 isoforms, could account for the phenotypic differences in these two mouse models (51). Expression of Rap2 in B cells has been previously established (18). Immature B cells emigrate from the bone marrow and home to the secondary lymphoidal organs such as spleen (40). Once migrated into the spleen, newly formed B cells have the potential to differentiate into either FO or MZ B cells (40). Reduced adhesion of pro/pre-B cell to stromal cells may lead to an augmented egress of immature B cells from bone marrow, which may account for the normal FO B cells in Rap1b−/− mice. Rap1 signaling seems not essential for egressed immature B cells to differentiate into FO B cells in Rap1b−/− spleen. This is partly supported by bone marrow progenitors transfected with constitutively active RapE63 that showed similar differential potential as vector-transfected control cells (26). An increase in the newly forming B cells in Rap1b−/− mice could indicate the specific role of Rap1 in the maturation for MZ but not FO B cells. The role of integrin in the retention of MZ B cells has been demonstrated (39). MZ B cells have higher expression level of αLβ2 and α4β1 integrins than FO B cells. As S1P induced activation of Rap1 (49), the dramatically reduced migration to S1P and S1P-induced adhesion to ICAM-1 of Rap1b−/− splenic MZ B cells may contribute to the defect in MZ B cells. In contrast, in the preleukemic SPA−/− mice, the development of FO and MZ B cells is normal (27). This discrepancy between Rap1b−/− and SPA−/− mice is due to the differential expression of these two proteins in bone marrow and spleen B cells. SPA-1 expression level is considerably higher in Lin− than Lin+ cells, which suggests Rap1 signaling may regulate lineage development differentially at different stages. The reduced MZ B cells in Rap1−/− mice is not due to defective cell proliferation as we did not observe a role of Rap1b in LPS- or BCR-induced proliferation of total spleen B cells or sorted FO and MZ B cell, which is supported by the normal proliferation of SPA-1−/− splenic B cells (27). Thus, the role of Rap1 signaling in cell proliferation seems to be cell type-dependent as it has been shown to positively regulate the proliferation of hematopoietic (26) and peritoneal B1 cells (27). The relatively normal development of T cells in the absence of Rap1b could be due to a compensatory effect from a different member of the Rap family such as Rap2.
Cell proliferation, differentiation and survival can be regulated by Erk1/2, p38, and AKT-mediated signaling pathways (52). Ras is one of the major activator of Erk1/2 and p38 (53). Rap1-GTPase has been shown to negatively regulate Ras-mediated Erk and p38 activation in some studies; however, it is controversial (1, 45, 54, 55). Our present study shows that phosphorylation of Erk1/2 and p38 was not changed after BCR and SDF-1 stimulation in Rap1b−/− B cells, although Rap1-GTP was nearly absent. This finding is consistent with earlier studies in which expression of constitutively active V12Rap1a in thymocytes, activation of Rap1 by cAMP in NIH3T3 cells, or overexpression of RapGAPII in B cells exhibited an unaltered Erk1/2 phosphorylation (19, 45, 56). Regulation of Erk1/2 activation by Rap1 has been shown to be cell context-dependent. In SPA-1−/− mice, Rap1 signaling has been shown to regulate Erk phosphorylation positively in Lin− bone marrow cells and negatively in anergic T cells (26, 57). Earlier studies have also that Rap1 signaling AKT phosphorylation (41, 45). In contrast, the similar BCR-mediated activation of AKT (Ser473) phosphorylation was observed in Rap1b−/− B cells as that in WT. This discrepancy may be due to the dual GAP activity of RapGAPII directed to both Rap1 and Rap2. It is most likely that the Rap2 but not Rap1 that regulates the AKT phosphorylation as Rap2 has its unique effectors (58–60).
Cell migration is controlled by multiple coordinated processes including chemoattraction, adhesion, and transmigration through the blood vessels. Chemokine-directed migration of B cells is essential for B cell development, differentiation, and Ab generation (61). The defect of Rap1b−/− B cells in vitro chemotaxis assay and in vivo short-term homing to LN is consistent with the established role of Rap1b in cell migration. Interestingly, the LN cellularity of Rap1b−/− mice was normal. The defect in the short-term homing assay may reflect only a slower cell migration. During the long developmental period, Rap1b−/− B and T cells may still be able to migrate to LN and display normal cellularity in LN. In RapL−/− mice, the defective migration of lymphocytes leads to atrophic thymus, spleen and LNs (25), which could be due to that RapL relays multiple upstream Rap GTPase signaling (51). The migration of T cells in short-term homing assay was far less severe in Rap1b−/− mice compare with B cells, which may reflect a differential expression pattern of Rap family proteins in B and T cells.
The defect in T-dependent but not T-independent humoral response was impaired in the Rap1b−/− mice. The defects in short-term homing of B and T cells in the absence of Rap1b could contribute to this defect. During the T2-independent immune response, MZ and B1 are two predominant B cell types that are recruited for Ab generation. Although, the MZ B cells are reduced to 40% of WT in our experiments, the residual MZ B cells might be enough for T2-independent Ag-induced Ab production. However, the defects in short-term homing of B and T cells in the absence of Rap1b could have contributed to the suboptimal T-dependent Ab production. In addition to cell migration, several other important steps are required for the successful T-dependent immune response. The presentation of antigenic epitope/MHC class II on APCs (dendritic cells or B cells) to Th cells is crucial, which is dependent on successful stable formation of immunological synapses that contain LFA-1, ICAM-1, CD40, and other molecules (62). Rap1 regulates T cell polarization and thereby the interaction with APCs (22). The dominant negative form of Rap1, Rap1N17, has been shown to block TCR-dependent increase in adhesion to ICAM-1. Furthermore, Rap1a−/− T cells failed to properly polarize LFA-1 upon CD3 stimulation (36). Thus, lack of Rap1b could impair the efficiency of T cell-APC interactions leading to a reduction in the secondary T-dependent Ab generation. Our present observations were supported by the findings in the SPA-1−/− mice (27). T cells from SPA-1−/− mice had selective impairment in memory T cell responses against a T-dependent Ag with normal primary Ab response. Lack of SPA-1 also resulted in an augmented levels anti-dsDNA Abs from B1a B cells leading to lupus-like autoimmune disease (27). Therefore, a regulated Rap1-signaling is critical for optimal immune responses. Of note, during publication of our results, Chen et al. published similar results (63).
However, the precise role of Rap1b in B cell development and memory T-dependent humoral response are still not complete elucidated. Induced deletion of Rap1b in B lineage could address this question more specifically. A conditional ablation of Rap1b will help to dissect the distinct role of Rap1b in B cells, T cells, and dendritic cells, the major cell types that involve in T-dependent immunity. Rap1a and Rap1b have different C-terminal sequences leading to distinct posttranslational modifications, which may contribute to isoform-specific function (31, 32, 34). The incomplete loss of MZ B cells and intact T-independent immunity in Rap1b−/− mice may indicate the compensatory role of Rap1a or Rap2 proteins. Continued research using Rap1a or Rap2-deficient mouse models will greatly help us to understand the function of Rap protein family.
Acknowledgments
We thank Jillian Dargatz for helping with mouse care, Barbara A. Fleming for cryosectioning, and Asanga Samarakoon for technical support.
Footnotes
This work was supported in part by an American Cancer Society Scholar Grant RSG-02-172-LIB (to S.M.), by Roche Organ Transplantation Research Foundation Grant 111662730 (to S.M.), by National Institutes of Health Grants R01 A1064826-01 (to S.M.), U19 AI062627-01 (to S.M.), NO1-HHSN26600500032C (to S.M.), and HL-45100 (to G.C.W.), and by American Heart Association Grant SDG0235127N (to M.C.W.). H.C. is a recipient of Wisconsin Breast Cancer Show House Postdoctoral Fellowship. S.M. is a recipient of The American Society for Blood and Marrow Transplantation Young Investigator Award.
Abbreviations used in this paper: GEF, guanine nucleotide exchange factor; GAP, GTPase-activating protein; MZ, marginal zone; FO, follicular; TNP, trinitrophenol; KLH, keyhole limpet hemocyanin; S1P, sphingosine-1-phosphate; SDF, stromal cell-derived factor; LN, lymph node; WT, wild type.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A Ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 2.Rousseau-Merck MF, Pizon V, Tavitian A, Berger R. Chromosome mapping of the human Ras-related RAP1A, RAP1B, and RAP2 genes to chromosomes 1p12—-p13, 12q14, and 13q34, respectively. Cytogenet Cell Genet. 1990;53:2–4. doi: 10.1159/000132883. [DOI] [PubMed] [Google Scholar]
- 3.Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Stork PJ, Dillon TJ. Multiple roles of Rap1 in hematopoietic cells: complementary versus antagonistic functions. Blood. 2005;106:2952–2961. doi: 10.1182/blood-2005-03-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, et al. Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol Cell Biol. 1995;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 8.Dupuy AJ, Morgan K, Von Lintig FC, Shen H, Acar H, Hasz DE, Jenkins NA, Copeland NG, Boss GR, Largaespada DA. Activation of the Rap1 guanine nucleotide exchange gene, CalDAG-GEF I, in BXH-2 murine myeloid leukemia. J Biol Chem. 2001;276:11804–11811. doi: 10.1074/jbc.M008970200. [DOI] [PubMed] [Google Scholar]
- 9.Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 10.Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Rubinfeld B, Munemitsu S, Clark R, Conroy L, Watt K, Crosier WJ, McCormick F, Polakis P. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–1042. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- 12.Kurachi H, Wada Y, Tsukamoto N, Maeda M, Kubota H, Hattori M, Iwai K, Minato N. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2: segregate expression profiles from a rap1GAP gene product. J Biol Chem. 1997;272:28081–28088. doi: 10.1074/jbc.272.44.28081. [DOI] [PubMed] [Google Scholar]
- 13.Brakebusch C, Fassler R. The integrin-actin connection, an eternal love affair. EMBO J. 2003;22:2324–2333. doi: 10.1093/emboj/cdg245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke B, van Triest M, de Bruijn KMT, van Willigen G, Nieuwenhuis HK, Negrier C, Akkerman JW, Bos JL. Sequential regulation of the small GTPase Rap1 in human platelets. Mol Cell Biol. 2000;20:779–785. doi: 10.1128/mcb.20.3.779-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattori M, Minato N. Rap1 GTPase: functions, regulation, and malignancy. J Biochem. 2003;134:479–484. doi: 10.1093/jb/mvg180. [DOI] [PubMed] [Google Scholar]
- 16.Crittenden JR, Bergmeier W, Zhang Y, Piffath CL, Liang Y, Wagner DD, Housman DE, Graybiel AM. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat Med. 2004;10:982–986. doi: 10.1038/nm1098. [DOI] [PubMed] [Google Scholar]
- 17.Ohba Y, Ikuta K, Ogura A, Matsuda J, Mochizuki N, Nagashima K, Kurokawa K, Mayer BJ, Maki K, Miyazaki J, Matsuda M. Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis. EMBO J. 2001;20:3333–3341. doi: 10.1093/emboj/20.13.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLeod SJ, Li AH, Lee RL, Burgess AE, Gold MR. The Rap GTPases regulate B cell migration toward the chemokine stromal cell-derived factor-1 (CXCL12): potential role for Rap2 in promoting B cell migration. J Immunol. 2002;169:1365–1371. doi: 10.4049/jimmunol.169.3.1365. [DOI] [PubMed] [Google Scholar]
- 19.Sebzda E, Bracke M, Tugal T, Hogg N, Cantrell DA. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling 8. Nat Immunol. 2002;3:251–258. doi: 10.1038/ni765. [DOI] [PubMed] [Google Scholar]
- 20.Tsukamoto N, Hattori M, Yang H, Bos JL, Minato N. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J Biol Chem. 1999;274:18463–18469. doi: 10.1074/jbc.274.26.18463. [DOI] [PubMed] [Google Scholar]
- 21.Bertoni A, Tadokoro S, Eto K, Pampori N, Parise LV, White GC, Shattil SJ. Relationships between Rap1b, affinity modulation of integrin αIIbβ3, and the actin cytoskeleton. J Biol Chem. 2002;277:25715–25721. doi: 10.1074/jbc.M202791200. [DOI] [PubMed] [Google Scholar]
- 22.Katagiri K, Hattori M, Minato N, Kinashi T. Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol Cell Biol. 2002;22:1001–1015. doi: 10.1128/MCB.22.4.1001-1015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLeod SJ, Ingham RJ, Bos JL, Kurosaki T, Gold MR. Activation of the Rap1 GTPase by the B cell antigen receptor. J Biol Chem. 1998;273:29218–29223. doi: 10.1074/jbc.273.44.29218. [DOI] [PubMed] [Google Scholar]
- 24.Kinashi T, Aker M, Sokolovsky-Eisenberg M, Grabovsky V, Tanaka C, Shamri R, Feigelson S, Etzioni A, Alon R. LAD-III, a leukocyte adhesion deficiency syndrome associated with defective Rap1 activation and impaired stabilization of integrin bonds. Blood. 2004;103:1033–1036. doi: 10.1182/blood-2003-07-2499. [DOI] [PubMed] [Google Scholar]
- 25.Katagiri K, Ohnishi N, Kabashima K, Iyoda T, Takeda N, Shinkai Y, Inaba K, Kinashi T. Crucial functions of the Rap1 effector molecule RAPL in lymphocyte and dendritic cell trafficking. Nat Immunol. 2004;5:1045–1051. doi: 10.1038/ni1111. [DOI] [PubMed] [Google Scholar]
- 26.Ishida D, Kometani K, Yang H, Kakugawa K, Masuda K, Iwai K, Suzuki M, Itohara S, Nakahata T, Hiai H, et al. Myeloproliferative stem cell disorders by deregulated Rap1 activation in SPA-1-deficient mice. Cancer Cell. 2003;4:55–65. doi: 10.1016/s1535-6108(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 27.Ishida D, Su L, Tamura A, Katayama Y, Kawai Y, Wang SF, Taniwaki M, Hamazaki Y, Hattori M, Minato N. Rap1 signal controls B cell receptor repertoire and generation of self-reactive B1a cells. Immunity. 2006;24:417–427. doi: 10.1016/j.immuni.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Paganini S, Guidetti GF, Catricala S, Trionfini P, Panelli S, Balduini C, Torti M. Identification and biochemical characterization of Rap2C, a new member of the Rap family of small GTP-binding proteins. Biochimie. 2006;88:285–295. doi: 10.1016/j.biochi.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Boussiotis VA, Freeman GJ, Berezovskaya A, Barber DL, Nadler LM. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science. 1997;278:124–128. doi: 10.1126/science.278.5335.124. [DOI] [PubMed] [Google Scholar]
- 30.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 31.Quarck R, Berrou E, Magnier C, Bobe R, Bredoux R, Tobelem G, Enouf J, Bryckaert M. Differential up-regulation of Rap1a and Rap1b proteins during smooth muscle cell cycle. Eur J Cell Biol. 1996;70:269–277. [PubMed] [Google Scholar]
- 32.McPhee I, Houslay MD, Yarwood SJ. Use of an activation-specific probe to show that Rap1A and Rap1B display different sensitivities to activation by forskolin in rat1 cells. FEBS Lett. 2000;477:213–218. doi: 10.1016/s0014-5793(00)01762-2. [DOI] [PubMed] [Google Scholar]
- 33.Klinz FJ, Seifert R, Schwaner I, Gausepohl H, Frank R, Schultz G. Generation of specific antibodies against the rap1A, rap1B and rap2 small GTP-binding proteins: analysis of Rap and Ras proteins in membranes from mammalian cells. Eur J Biochem. 1992;207:207–213. doi: 10.1111/j.1432-1033.1992.tb17039.x. [DOI] [PubMed] [Google Scholar]
- 34.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC. Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005;115:680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwamborn JC, Puschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- 36.Duchniewicz M, Zemojtel T, Kolanczyk M, Grossmann S, Scheele JS, Zwartkruis FJ. Rap1A-deficient T and B cells show impaired integrin-mediated cell adhesion. Mol Cell Biol. 2006;26:643–653. doi: 10.1128/MCB.26.2.643-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke B, Akkerman JW, Bos JL. Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dingjan GM, Maas A, Nawijn MC, Smit L, Voerman JS, Grosveld F, Hendriks RW. Severe B cell deficiency and disrupted splenic architecture in transgenic mice expressing the E41K mutated form of Bruton’s tyrosine kinase. EMBO J. 1998;17:5309–5320. doi: 10.1093/emboj/17.18.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu TT, Cyster JG. Integrin-mediated long-term B cell retention in the splenic marginal zone. Science. 2002;297:409–412. doi: 10.1126/science.1071632. [DOI] [PubMed] [Google Scholar]
- 40.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 41.Jin A, Kurosu T, Tsuji K, Mizuchi D, Arai A, Fujita H, Hattori M, Minato N, Miura O. BCR/ABL and IL-3 activate Rap1 to stimulate the B-Raf/MEK/Erk and Akt signaling pathways and to regulate proliferation, apoptosis, and adhesion. Oncogene. 2006;25:4332–4340. doi: 10.1038/sj.onc.1209459. [DOI] [PubMed] [Google Scholar]
- 42.Dillon TJ, Carey KD, Wetzel SA, Parker DC, Stork PJ. Regulation of the small GTPase Rap1 and extracellular signal-regulated kinases by the costimulatory molecule CTLA-4. Mol Cell Biol. 2005;25:4117–4128. doi: 10.1128/MCB.25.10.4117-4128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 44.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christian SL, Lee RL, McLeod SJ, Burgess AE, Li AH, Dang-Lawson M, Lin KB, Gold MR. Activation of the Rap GTPases in B lymphocytes modulates B cell antigen receptor-induced activation of Akt but has no effect on MAPK activation. J Biol Chem. 2003;278:41756–41767. doi: 10.1074/jbc.M303180200. [DOI] [PubMed] [Google Scholar]
- 46.Balazs M, Martin F, Zhou T, Kearney J. Blood dendritic cells interact with splenic marginal zone B cells to initiate T-independent immune responses. Immunity. 2002;17:341–352. doi: 10.1016/s1074-7613(02)00389-8. [DOI] [PubMed] [Google Scholar]
- 47.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 48.Cyster JG. Specifying the patterns of immune cell migration. Novartis Found Symp. 2007;281:54–61. doi: 10.1002/9780470062128.ch6. [DOI] [PubMed] [Google Scholar]
- 49.Durand CA, Westendorf J, Tse KW, Gold MR. The Rap GTPases mediate CX. Eur J Immunol. 2006;36:2235–2249. doi: 10.1002/eji.200535004. [DOI] [PubMed] [Google Scholar]
- 50.Cinamon G, Matloubian M, Lesneski MJ, Xu Y, Low C, Lu T, Proia RL, Cyster JG. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat Immunol. 2004;5:713–720. doi: 10.1038/ni1083. [DOI] [PubMed] [Google Scholar]
- 51.Miertzschke M, Stanley P, Bunney TD, Rodrigues-Lima F, Hogg N, Katan M. Characterization of interactions of adapter protein RAPL/Nore 1B with Rap GTPases and their role in T cell migration. J Biol Chem. 2007;282:30629–30642. doi: 10.1074/jbc.M704361200. [DOI] [PubMed] [Google Scholar]
- 52.Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 53.Bouschet T, Perez V, Fernandez C, Bockaert J, Eychene A, Journot L. Stimulation of the ERK pathway by GTP-loaded Rap1 requires the concomitant activation of Ras, protein kinase C, and protein kinase A in neuronal cells. J Biol Chem. 2003;278:4778–4785. doi: 10.1074/jbc.M204652200. [DOI] [PubMed] [Google Scholar]
- 54.Cook SJ, Rubinfeld B, Albert I, McCormick F. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zwartkruis FJ, Wolthuis RM, Nabben NM, Franke B, Bos JL. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. EMBO J. 1998;17:5905–5912. doi: 10.1093/emboj/17.20.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enserink JM, Christensen AE, de Rooij J, van Triest M, Schwede F, Genieser HG, Døskeland SO, Blank JL, Bos JL. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat Cell Biol. 2002;4:901–906. doi: 10.1038/ncb874. [DOI] [PubMed] [Google Scholar]
- 57.Ishida D, Yang H, Masuda K, Uesugi K, Kawamoto H, Hattori M, Minato N. Antigen-driven T cell anergy and defective memory T cell response via deregulated Rap1 activation in SPA-1-deficient mice. Proc Natl Acad Sci USA. 2003;100:10919–10924. doi: 10.1073/pnas.1834525100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem. 2004;279:49488–49496. doi: 10.1074/jbc.M406370200. [DOI] [PubMed] [Google Scholar]
- 59.Machida N, Umikawa M, Takei K, Sakima N, Myagmar BE, Taira K, Uezato H, Ogawa Y, Kariya K. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem. 2004;279:15711–15714. doi: 10.1074/jbc.C300542200. [DOI] [PubMed] [Google Scholar]
- 60.Myagmar BE, Umikawa M, Asato T, Taira K, Oshiro M, Hino A, Takei K, Uezato H, Kariya K. PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochem Biophys Res Commun. 2005;329:1046–1052. doi: 10.1016/j.bbrc.2005.02.069. [DOI] [PubMed] [Google Scholar]
- 61.Melchers F, Rolink AG, Schaniel C. The role of chemokines in regulating cell migration during humoral immune responses. Cell. 1999;99:351–354. doi: 10.1016/s0092-8674(00)81521-4. [DOI] [PubMed] [Google Scholar]
- 62.Reichardt P, Dornbach B, Gunzer M. The molecular makeup and function of regulatory and effector synapses. Immunol Rev. 2007;218:165–177. doi: 10.1111/j.1600-065X.2007.00526.x. [DOI] [PubMed] [Google Scholar]
- 63.Chen Y, Yu M, Podd A, Wen R, Chrzanowska-Wodnicka M, White GC, Wang D. A critical role of Rap1b in B-cell trafficking and marginal zone B-cell development. Blood. 2008;111:4627–4636. doi: 10.1182/blood-2007-12-128140. [DOI] [PMC free article] [PubMed] [Google Scholar]