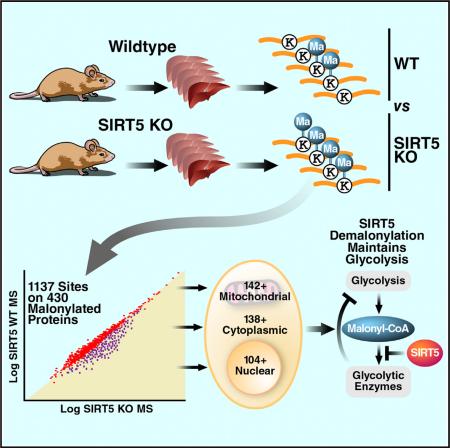
SUMMARY
Protein acylation links energetic substrate flux with cellular adaptive responses. SIRT5 is a NAD+-dependent lysine deacylase and removes both succinyl and malonyl groups. Using affinity enrichment and label free quantitative proteomics, we characterized the SIRT5-regulated lysine malonylome in wild-type (WT) and Sirt5−/− mice. 1,137 malonyllysine sites were identified across 430 proteins, with 183 sites (from 120 proteins) significantly increased in Sirt5−/− animals. Pathway analysis identified glycolysis as the top SIRT5-regulated pathway. Importantly, glycolytic flux was diminished in primary hepatocytes from Sirt5−/− compared to WT mice. Substitution of malonylated lysine residue 184 in glyceraldehyde 3-phosphate dehydrogenase with glutamic acid, a malonyllysine mimic, suppressed its enzymatic activity. Comparison with our previous reports on acylation reveals that malonylation targets a different set of proteins than acetylation and succinylation. These data demonstrate that SIRT5 is a global regulator of lysine malonylation and provide a mechanism for regulation of energetic flux through glycolysis.
Graphical Abstract
INTRODUCTION
Posttranslational modifications (PTMs) play important roles in regulating protein function and are involved in many physiological and pathological processes (Walsh et al., 2005). Lysine acetylation is one of the most studied modifications and regulates many biological processes including transcriptional activation (Berger, 2002), protein degradation (Ito et al., 2002), and enzymatic activity (Shimazu et al., 2010; Bharathi et al., 2013). Recent work has expanded the diversity of acyl modifications modifying the ε-amine of lysine residues beyond acetylation to include succinylation, malonylation, glutarylation, and others (Chen et al., 2007; Du et al., 2011; Peng et al., 2011; Tan et al., 2014). While acetylation neutralizes the positive charge of protonated lysines, succinylation and malonylation are bulkier groups and also carry negatively charged carboxyl moieties that may lead to distinct functional and structural outcomes. Regulation of these recently discovered acylations is carried out in part by members of the sirtuin (SIRT) family of lysine deacylases.
The sirtuins are a highly conserved family of nicotinamide adenine dinucleotide (NAD+-dependent enzymes that regulate a variety of metabolic pathways (Chang and Guarente, 2014). Of the seven mammalian sirtuins, SIRT3 and SIRT4 are located exclusively in mitochondria, whereas SIRT5 is found in both mitochondria and the cytosol (Matsushita et al., 2011). SIRT3 acts as the major deacetylase for mitochondrial proteins (Schwer et al., 2002) and regulates many metabolic enzymes in mitochondria through reversible lysine acetylation (Hallows et al., 2011, Sol et al., 2012, Rardin et al., 2013a). SIRT5 was initially reported to have deacetylase activity (Nakagawa et al., 2009), but recent reports have shown that SIRT5 preferentially removes negatively charged acylcarboxyl moieties (Peng et al., 2011; Du et al., 2011; Tan et al., 2014). SIRT5 removes both glutaryl and succinyl moieties from the urea cycle enzyme carbamoyl phosphatase synthase (CPS1), leading to its enzymatic activation and increased ammonia detoxification (Du et al., 2011; Tan et al., 2014). In addition, SIRT5 desuccinylates lysine residues near the catalytic pocket of the ketogenic enzyme 3-hydroxy-3-methylglutaryl-CoA synthase 2, leading to its activation and increased ketogenesis (Rardin et al., 2013b). In addition to these activities, SIRT5 also catalytically removes malonyl groups from the lysine side chain of protein substrates. This reaction is NAD+ dependent and leads to a demalonylated lysine and the generation of nicotinamide (NAM) and 2′-O-malonyl-ADP-ribose (Figure 1A) (Peng et al., 2011).
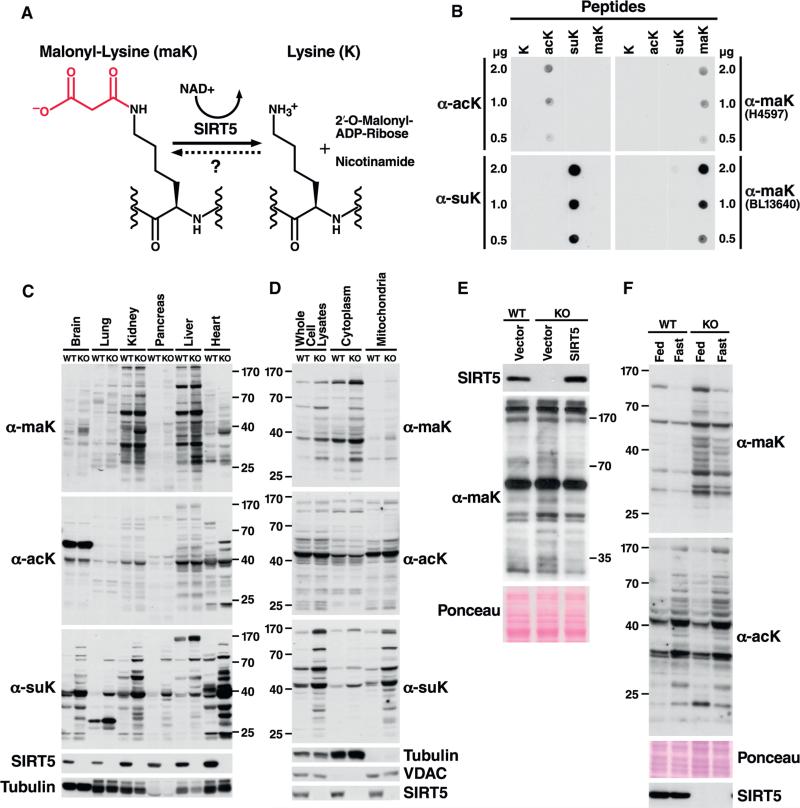
Figure 1. Generation of Malonyl-Lysine Specific Antibodies and Characterization of Malonylation Distribution in Mouse Tissues and Subcellular Liver Compartments.
(A) Malonyl-lysine and the demalonylation reaction catalyzed by SIRT5. The malonyl group on the lysine residue is shown in red.
(B) Malonyl-lysine antibodies specifically identify malonyl-lysine containing peptide, but not acetyl- or succinyl-lysine in a dotplot assay.
(C) Western blot analysis comparing lysine malonylation, succinylation, and acetylation levels in mouse organs derived from WT or Sirt5−/− mice. The absence of SIRT5 in Sirt5−/− mouse tissues was confirmed using anti-SIRT5 antibody.
(D) Western blot analysis for lysine malonylation, succinylation and acetylation in whole cell lysates, cytosolic or mitochondrial fractions derived from WT or Sirt5−/− mouse livers. Purity of subcellular fractionation was confirmed by western blot for the cytosolic protein tubulin and the mitochondrial protein VDAC.
(E) Global hyper-malonylation is reversibly de-malonylated by SIRT5 overexpression. A lentivirus vector was used either as an empty vector (lane 2) or to overexpress SIRT5 (lane3) in SIRT5 KO primary hepatocytes. Lysine malonylation levels were examined in whole cell lysates by western blotting.
(F) Global maK level changes following starvation. Global malonylation levels in whole liver lysates were compared between fed and starved WT or SIRT5 KO mice. Note that malonyl-lysine levels are higher in feeding than fasting conditions both in WT and SIRT5 KO mice whereas acetylation is higher in fasting than feeding and unaffected by SIRT5.
The biological significance of lysine malonylation remains poorly understood, due in part to a lack of knowledge of the proteins and lysine residues modified. Three studies have examined the distribution of lysine malonylation sites: Peng et al. (2011) identified 25 peptides from 17 proteins, Bao et al. (2013) reported the identification of another 361 malonylated proteins using a chemical probe MalAM-yne, and Du et al. (2015) recently reported 573 malonylation sites over 268 proteins in db/db mice commonly used to model type 2 diabetes. The Peng and Bao studies were conducted in HeLa cell lines, while Du reported malonylation sites present in vivo. No information is currently available on the in vivo regulation of malonylation by SIRT5.
From what is known about other acylation reactions, malonyl-Coenzyme A (malonyl-CoA) likely serves as the universal donor for malonylation reactions either via the activity of a specific malonytransferase or via a non-enzymatic mechanism (Peng et al., 2011; Wagner and Payne, 2013).
The levels of various acyl-CoAs fluctuate as a function of cellular energy metabolism, and different metabolic conditions can significantly regulate global protein acylation, which has been studied in detail for acetyl-CoA as substrate for protein acetylation in yeast and mice (Kim et al., 2006; Cai et al., 2011; Hirschey et al., 2011). Malonyl-CoA is an important intermediary metabolite as it serves as both a key building block in fatty acid synthesis and a metabolic sensor and allosteric regulator of fatty acid oxidation (Saggerson, 2008). Malonyl-CoA cellular levels represent the competing activities of two distinct enzymes: acetyl-CoA carboxylase and malonyl-CoA decarboxylase (Saggerson, 2008). Under anabolic conditions, acetyl-CoA is converted by acetyl-coA carboxylase to malonyl-CoA, which then serves as the first building block in fatty acid synthesis. Thus, variations in malonyl-coA levels occur during physiological conditions and may serve as a signal inducing the differential malonylation and altered function or properties of a subset of cellular proteins.
Here, we have used a novel antiserum specific for malonylly-sine to enrich peptides containing malonyllysine. Using a label-free proteomic method called MS1 Filtering (Rardin et al., 2013a), we identified and quantified changes in the whole-cell malonylome in liver lysates from wild-type (WT) and Sirt5−/− mice. Pathway analysis of malonylated proteins regulated by SIRT5 and experiments both in vitro and in vivo revealed an important role for SIRT5 in regulating glycolysis.
RESULTS
Generation of a Specific Malonyllysine Antiserum
First, we immunized rabbits with malonylated BSA and generated polyclonal antibodies specific for malonyllysine. We assayed antibody specificities by dot plot analysis against peptides carrying different acylation modifications. Two rabbit sera containing malonyllysine antibodies, H4597 and BL13640, showed specific reaction against malonyllysine (maK)-containing peptides, but no signal against acetyllysine (acK)- or succinyllysine (suK)-containing peptides (Figure 1B). In contrast, anti-acetyl and anti-succinyl lysine antibodies showed a robust signal for their respective peptides (Figure 1B). These data demonstrate the high specificity of these antibodies.
Increased Protein Malonylation in Mice Lacking SIRT5
Next, we used this antiserum (H4597) to probe key mouse tissues for protein malonylation. Comparison of levels of malonylation in brain, lung, kidney, pancreas, liver and heart from WT and Sirt5−/− (KO) mice by western blot analysis showed a significant increase in protein malonylation in tissues from Sirt5−/− mice (Figure 1C). Among them, kidney and liver showed the most protein malonylation. In agreement with previous reports (Rardin et al., 2013b; Park et al., 2013), protein succinylation was also increased in mice lacking SIRT5, but succinylated proteins were most abundant in the heart (Figure 1C). In contrast, protein acetylation levels were comparable between WT and Sirt5−/− mouse tissues (Figure 1C). As SIRT5 is localized to both mitochondria and the cytosol, we compared the amount of total protein malonylation in mitochondria and cytosolic fractions from WT and Sirt5−/− mouse liver lysates. Interestingly, malonylated proteins appeared to be more abundant in the cytosolic fraction of both WT and KO animals (Figure 1D) than in the mitochondrial fraction. This contrasts with what we had reported for protein succinylation in the absence of SIRT5, which was most abundant in the mitochondrial fraction (Rardin et al., 2013b) (Figure 1D). Taken together, these data demonstrate the in vivo hypermalonylation of proteins across tissues in Sirt5−/− animals and indicate that malonylation appears more abundant in the cytosol than in mitochondria.
Reduction of Malonylation by SIRT5 Expression and during Fasting
To further link the hypermalonylation observed in Sirt5−/− mice with SIRT5 protein function, we used a lentivirus vector to express human SIRT5 in primary hepatocytes from mice lacking SIRT5 (Figure 1E). Reexpression of SIRT5 reduced global protein malonylation to levels comparable to those in WT cells (Figure 1E).
During fasting, malonyl-CoA levels decline as acetyl-CoA is diverted into catabolic pathways. In contrast, malonyl-CoA levels increase during feeding as acetyl-CoA is transformed into malonyl-CoA by acetyl-coA carboxylase for fatty acid synthesis (McGarry et al., 1978; Guynn et al., 1972). Accordingly, we observed that malonylation of global liver protein was lower in fasting than in feeding conditions, both for SIRT5KO and WT mice (Figure 1F). As previously reported, the opposite was observed for acetylation, which was higher under fasting than feeding conditions (Figure 1F). In agreement with the model that SIRT5 does not regulate global acetylation, no differences in liver protein acetylation were observed between wt and SIRT5KO mice (Figure 1F). These results support the model that hypermalonylation in SIRT5KO liver occurs due to failure to remove malonylation.
Defining and Quantifying the Lysine Malonylome in Liver from WT and Sirt5−/y Mice
To determine the extent of malonylation in mouse liver tissue, we developed a robust workflow for the enrichment and identification of peptides from protein digests containing maK modifications (Figure 2A). Liver tissue was isolated and homogenized from 5 WT and 5 Sirt5−/− animals and normalized to total protein concentration. Western blotting of these extracts confirmed the absence of SIRT5 in the Sirt5−/− animals (Figure S1A). Equal amounts of protein from individual animals were digested with trypsin, and 20 μg of protein was removed from each sample to independently assess protein expression levels in both samples. To control for sample process variability 150 fmol of a heavy-isotope-labeled maK peptide, TVDGPSGmaKLWR [13C6 15N4] was spiked into each sample prior to affinity enrichment using two polyclonal anti-maK antibodies. Peptides containing maK were eluted and analyzed in duplicate by liquid chromatography (LC)-MS/MS on a TripleTOF 5600 MS, and data were searched against the mouse proteome.
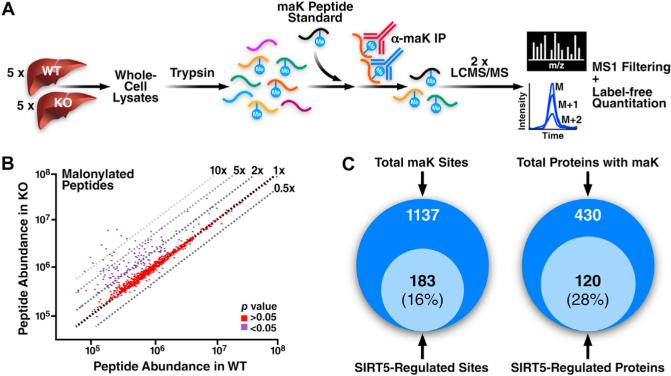
Figure 2. Enrichment and Identification of Whole Liver Lysine Malonylome by Label-Free Quantitation.
(A) Whole cell liver lysates were prepared from 5 WT and 5 Sirt5−/− mice (Sv129, n = 5, 10 months of age, fed). Individual samples were digested with trypsin and spiked with 150 fmol of a malonyllysine peptide standard. Lysine malonylated peptides were immunoprecipitated and samples were analyzed by LC-MS/MS in duplicate. Ion intensity chromatograms were extracted using MS1 Filtering and peptide abundance compared between WT and KO animals.
(B) Scatterplot shows individual malonylated peptide abundances quantified by MS1 Filtering from WT and Sirt5−/− mice. Dashed lines represent fold change between the two conditions (WT versus Sirt5−/− mice). Peptides showing significant difference in abundance (p value < 0.05) are shown in purple while all other peptides are shown in red. (C) Overlap of the malonyl-lysine sites and proteins targeted by SIRT5 in the mouse liver malonylome (KO:WT ≥ 1.5, p value < 0.05). For peptide information and quantification of individual sites, see Tables S1 and S2.
Using this approach we identified 1,078 sites of lysine malonylation from 2,266 unique maK peptides across 421 proteins with a false discovery rate of <1% (Table S1). We manually verified 59 additional malonylation sites due to search engine limitations in efficiently recognizing the neutral loss of CO2 (–44 Da) from fragment ions containing the maK residue (Table S1). This gave us a total of 1,137 sites across 430 proteins (Figures 2B and 2C). These results demonstrated high selectivity in the antibody-based enrichment, as malonylated peptides represented 66% of all peptide identified, as well as a lack of cross-reactivity to lysine-acetylation and succinylation, as none of the 694 non-malonylated peptides identified (34% of total) contained either of these modifications. Of the 1,137 sites identified, 71% of sites were identified in both backgrounds (WT and KO), while 18% and 11% were identified only in the KO or WT, respectively (Figure S1B). At the protein level, 75% were identified in both backgrounds, while 15% and 10% were identified only in the KO or WT, respectively (Figure S1B).
To determine the role of SIRT5 in regulating sites of lysine malonylation, we employed a label-free quantitative technique named MS1 Filtering (Schilling et al., 2012). MS1 Filtering extracts and measures ion intensity chromatograms for peptides obtained during the MS1 scan, allowing direct comparison of relative abundances between WT and KO samples. Using this approach, we observed a coefficient of variation of 29% for the peptide standard, indicating good reproducibility across all acquisitions. In total, 1,484 maK peptides covering 1,037 sites were quantified across all samples. Representative peptides for each maK site were chosen based on the most abundant form observed in the data set (Table S2). Ion abundance for each peptide was normalized to the peptide standard and a ratio was calculated (KO:WT). Of the 1,137 sites identified, we determined that 183 sites on 120 proteins were hypermalonylated in the Sirt5−/− animals based on a ≥1.5-fold increase with a p value < 0.05 (Figure 2B). This represents 16% of all sites identified and 28% (120) of malonylated proteins (Figure 2C). The fold change also varied, with 25 sites having more than a 5-fold increase in malonylation (Table S2).
To control for changes in protein expression, we also used MS1 Filtering to analyze digested liver lysates and quantify unmodified peptides from proteins that were found to be malonylated. Samples were analyzed in duplicates by LC-MS/MS, and 322 of the malonylated proteins were identified (Table S3). Peptide ion abundances were normalized to a peptide from a non-malonylated protein, and a ratio was calculated (KO:WT). The majority of peptides had a ratio of ~1 (Figure S1C), and peptides were averaged for each protein to determine relative levels of protein expression (Table S4). No significant increase was observed across proteins, indicating changes in malonylation are not due to changes in protein expression levels in the Sirt5−/− mice, similar to what we observed in the Sirt3−/− mouse (Rardin et al., 2013a). We then quantified an additional 30 proteins that were not identified as being malonylated, which exhibited no change in expression, suggesting that liver proteome abundance is not significantly altered in the absence of SIRT5 (Table S4).
Distribution, Sequence Motif Features, and Conservation Analysis for Malonylation and SIRT5-Regulated Sites
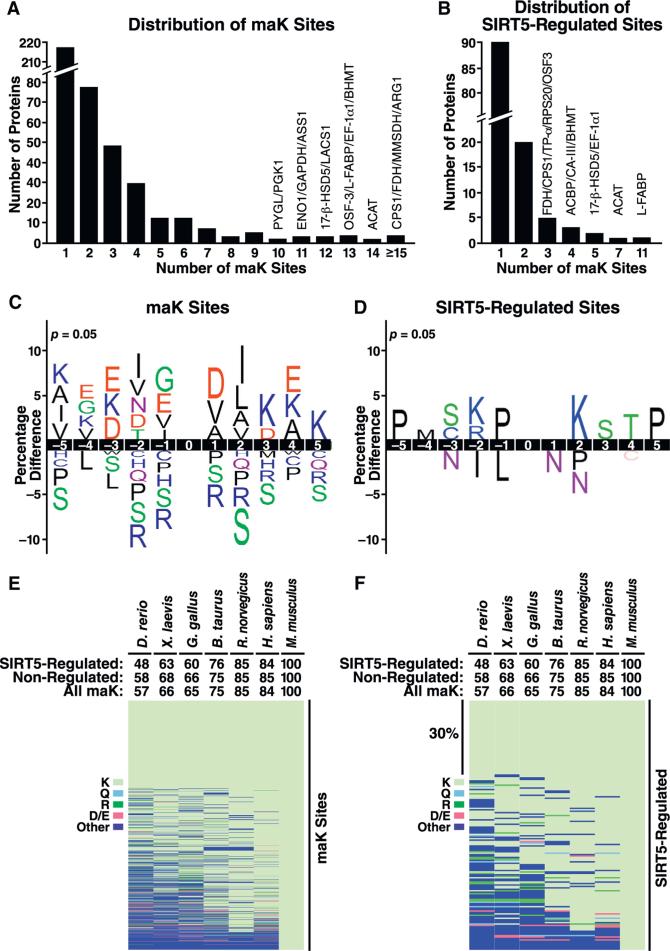
Across the identified proteins, 217 (50.5%) had a single malonylation site and 213 (49.5%) had two or more maK sites (Figure 3A). Carbamoyl-phosphate synthase (CPS1), the first enzyme of the urea cycle, was the most heavily malonylated enzyme with 31 maK sites identified. However, only 3 of 31 sites were significantly increased in Sirt5−/− animals (Figure 3B). In contrast, the liver fatty acid binding protein (L-FABP) was the most dynamically regulated SIRT5-targeted protein, as 11 of 13 sites were significantly increased in Sirt5−/− mice (Figure 3B). Of the 120 SIRT5-regulated proteins that we identified, 73% had a single site significantly changed in the absence of SIRT5 (Figure 3B). The vast majority of sites and proteins demonstrated no signifi-cant change in lysine malonylation in the absence of SIRT5.
Figure 3. Site Distribution, Sequence Logo, and Conservation Analysis.
(A) Distribution of the number of maK sites identified per protein. Most malonylated proteins contain a single site, but some contain multiple sites.
(B) Distribution of the number of SIRT5-regulated sites per protein. Abbreviations include CPS1 (Carbamoyl-phosphate synthase, mitochondrial), FDH (Cytosolic 10-formyltetrahydrofolate dehydrogenase), MMSDH (Methylmalonate-semi-aldehyde dehydrogenase, mitochondrial), ARG1 (Arginase-1), ACAT (Acetyl-CoA acetyltransferase, mitochondrial), OSF-3 (Peroxiredoxin-1), L-FABP (Fatty acid-binding protein, liver), EF-1α1 (Elongation factor 1-alpha 1), BHMT (Betaine-homocysteine S-methyltransferase 1), 17-β-HSD5 (Estradiol 17 beta-dehydrogenase 5), LACS1 (Long-chain-fatty-acid-CoA ligase 1), ENO1 (Alpha-enolase), GAPDH (Glyceraldehyde-3-phosphate dehydrogenase), ASS1 (Argininosuccinate synthase), PYGL (Glycogen phosphorylase, liver form), PGK1 (Phosphoglycerate kinase 1), ACBP (Acyl-CoA-binding protein), CA-III (Carbonic anhydrase 3), BHMT (Betaine-homocysteine S-methyltransferase 1), TP-α (Trifunctional enzyme subunit alpha, mitochondrial), and RPS20 (40S ribosomal protein S20).
(C and D) Consensus sequence logo plots centered on the lysine of all identified maK sites (±5 amino acids) (C) or all identified SIRT5-regulated maK sites (≥1.5-fold and p < 0.05) (D).
(E and F) Heatmap depicting the conservation index of maK (E) or SIRT5-regulated maK sites (F) across seven vertebrate species. The percentage of sites conserved for all sites, target and non-target sites is listed above the heatmap. Lysine (K), glutamine (Q), arginine (R), aspartic acid (D), or glutamic acid (E) and other amino acids are represented by the colors indicated in the heat-map. See Table S5 for individual site conservation details.
Next, we used IceLogo (https://code.google.com/p/icelogo/) to compare the amino acid sequences around all observed malonylated sites to non-malonylated sites from our data set (Figure 3C). We found a modest enrichment for nonpolar amino acids, such as leucine, valine, or isoleucine, in positions close to the modified lysine (Figure 3C). The SIRT5-regulated logo differed from the total malonylation logo: leucine and isoleucine were under-represented at positions close to modified sites, whereas lysine and arginine were enriched at positions −2 and +2 (Figure 3D). However, the low overall changes in amino acid abundance near both all malonylated and SIRT5-regulated sites indicate no major effect of protein sequence on either lysine malonylation or its control by SIRT5.
The sirtuin family of deacylases are conserved from bacteria to human (Hirschey et al., 2011), and lysine malonylation has been identified from bacteria to humans (Du et al., 2011; Peng et al., 2011; Xie et al., 2012). To determine the extent of conservation for lysine malonylation sites across vertebrates, we generated a conservation index using AL2CO (Pei and Grishin, 2001). Of the 1,137 sites identified in mouse, we found that 84% were conserved in humans, while 57% were conserved in zebrafish (Figure 3E). Similarly, 84% of SIRT5-regulated sites were conserved in humans and 48% in zebrafish (Figure 3F). Of the SIRT5-regulated sites, only 30% were conserved across all species, implying that regulated sites are not universally functionally important. Those sites not conserved in humans or rats were most often substituted by an arginine (2%) or glutamic acid (2%). However, SIRT5-target sites were most often changed to glutamic acid and arginine at 4% and 2% of sites in humans and 2% and 2% in rats, respectively.
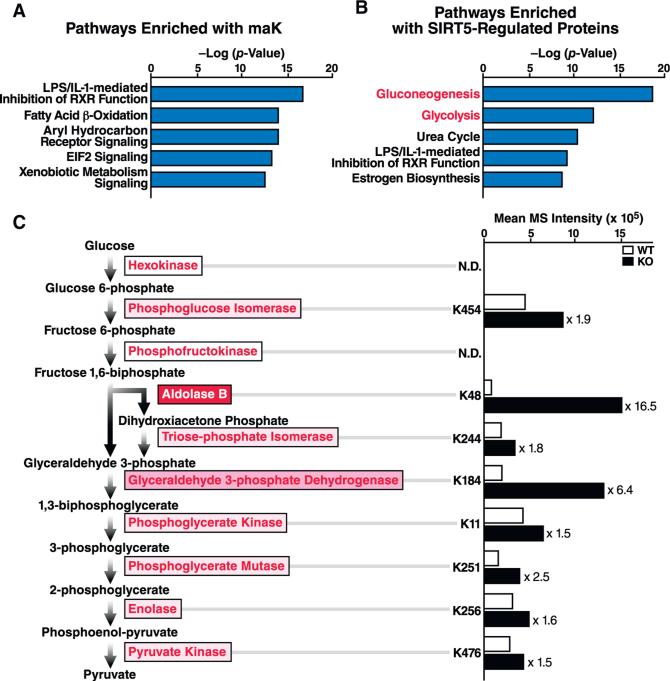
Pathway Analysis Reveals Glycolysis as SIRT5-Regulated Pathway
Canonical pathway enrichment analysis was performed using the Ingenuity Pathway Analysis tool to identify biological pathways enriched with malonylated proteins (Figures 4A and 4B). The pathways most enriched with SIRT5-regulated malonylated proteins were gluconeogenesis and glycolysis. In glycolysis, glucose is broken down into pyruvate in the cytosol through ten enzymatic reactions, seven of which are reversible and shared by gluconeogenesis. We also identified several enzymes in the urea cycle, both mitochondrial (carbamoyl-phosphate synthase and ornithine carbamoyltransferase) and cytosolic (argininosuccinate synthase, argininosuccinate lyase, and arginase). The role of SIRT5 in regulating the urea cycle via reversible deacetylation, desuccinylation, and deglutarylation of carbamoyl-phosphate synthase in mitochondria has been previously documented (Nakagawa et al., 2009; Ogura et al., 2010; Rardin et al., 2013b; Tan et al., 2014). These novel observations suggest that SIRT5 may also regulate the cytosolic component of the urea cycle via reversible protein malonylation.
Figure 4. Glycolytic Enzymes Are Hyper-Malonylated in the Absence of SIRT5.
(A and B) Ingenuity pathway analysis of malonylated proteins (A) and SIRT5-regulated malonylated proteins (B).
(C) Glycolytic enzymes are highlighted next to the reaction they catalyze. The color intensity of boxes (white to red) indicates the the ratio of SIRT5 KO hypermalonylation (Sirt5−/− /WT) with red being high and white corresponding to a value of 1. MaK sites in each enzyme and their regulation by SIRT5 are shown at right as a bar graph of mean MS intensity in the presence and absence of SIRT5.
Increased Malonylation of Glycolytic Enzymes in Mice Lacking SIRT5
Comparing the relative abundance of lysine malonylation in all glycolytic enzymes of Sirt5−/− mice with control animals showed that six out of ten glycolytic enzymes carried one malonylated lysine regulated by SIRT5 (Figure 4C). Among them, Aldolase B, which catalyzes the cleavage of fructose 1,6-bisphosphate into glyceraldehyde 3-phosphate and dihydroxyacetone phosphate, exhibited the highest increase in lysine malonylation in the absence of SIRT5 (WT/KO ratio = 16-fold). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which converts glyceraldehyde 3-phosphate to glycerate 1,3-bisphosphate, was the second most dynamically regulated glycolytic enzyme (WT/KO ratio = 6.4) (Figure 4C). Interestingly, Du et al. (2015) recently reported hypermalonylation of Aldolase B in db/db mouse liver, though its regulation by SIRT5 was not addressed.
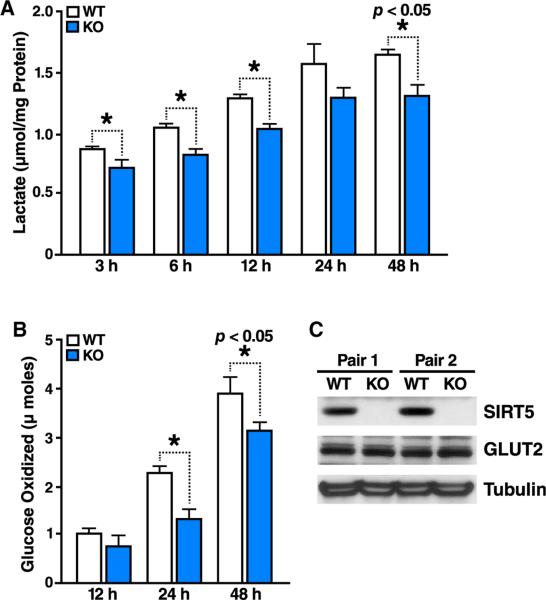
To examine the possible functional role of malonylation of these glycolytic enzymes and their regulation by SIRT5, we directly measured glycolytic flux in primary mouse hepatocytes. The production of lactate via pyruvate, the final prod uct of glycolysis, is a well-established marker of glycolytic flux (Vander Heiden et al., 2009). Primary hepatocytes isolated from WT mice produced lactate when cultured in vitro indicating healthy, glucose-consuming hepatocytes (Figure 5A). However, primary hepatocytes isolated from Sirt5−/− mice showed decreased lactate production relative to controls at all time points, consistent with decreased glycolytic flux in the absence of SIRT5 (Figure 5A). To measure glycolytic flux directly, we cultured primary mouse hepatocytes with 6,6 2D-glucose and measured the release of deuterium-labeled water by mass spectrometric (MS) analysis (Beysen et al., 2007). Confirming results obtained with lactate measurement, glycolytic flux evaluated by the amount of oxidized glucose was significantly lower in SIRT5-KO than in WT (Figure 5B). Expression of the GLUT2 glucose transporter, which is highly expressed in liver (Thorens et al., 1990) and was not detectably malonylated, was unchanged in SIRT5-KO primary hepatocytes (Figure 5C), ruling out differences in glucose intake as the source of this reduction in glycolysis. These results suggest that SIRT5-regulated protein malonylation of glycolytic enzymes controls their activities and glycolytic flux.
Figure 5. Glycolysis Is Suppressed in the Absence of SIRT5.
(A) Lactic acid production is impaired in primary hepatocytes derived from Sirt5−/− versus WT animals. Lactic acid was measured in culture media of primary hepatocytes isolated from WT and Sirt5−/− animals at different time points. Each graph is representative of two independent experiments; n = 3 measurements/sample; mean ± SD.
(B) Glycolytic flux measured by deuterium-labeled glucose is suppressed in KO primary hepatocytes. Oxidized glucose was measured in primary hepatocytes isolated from WT and Sirt5−/− animals at different time points. Graph is representative of two independent experiments; n = 3 measurements/sample, mean ± SD.
(C) Comparison of Glucose transporter 2 expression between WT and SIRT5 KO primary hepatocytes as assayed by western blotting using tubulin as a loading control.
Regulation of GAPDH Activity by Malonylation of Lysine 184
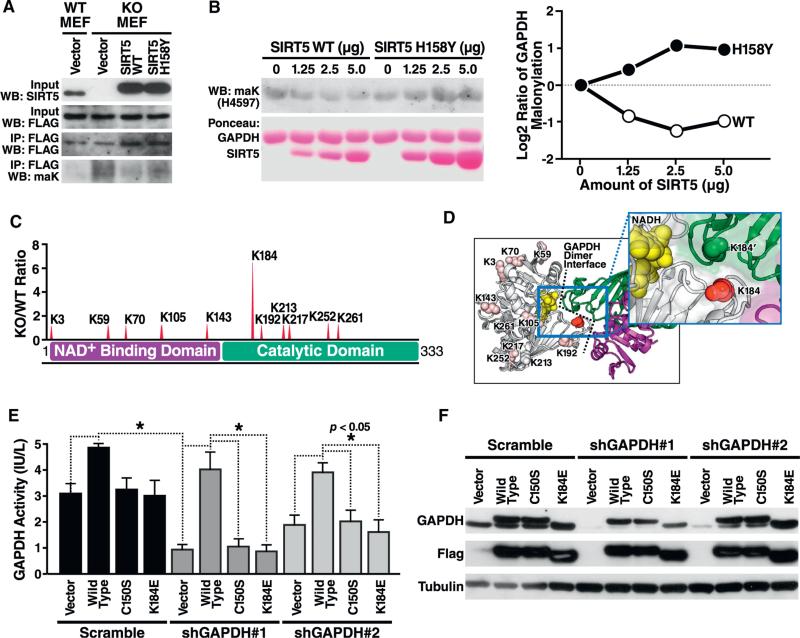
In our proteomic workflow, GAPDH is detectably malonylated at 11 lysine residues. To validate this result and determine the relative abundance of SIRT5-regulated malonylation on GAPDH, we co-expressed Flag epitope-tagged GAPDH and either WT or catalytically inactive H158Y SIRT5 in WT or SIRT5KO mouse embryonic fibroblasts (MEFs) (Figure 6A). Anti-malonyllysine western blotting following immunoprecipitation for GAPDH revealed that a majority of GAPDH malonylation is reversed by WT, but not SIRT5H158Y, expression (Figure 6A). Furthermore, in vitro coincubation of GAPDH with WT or H158Y SIRT5 expressed in HEK293T cells revealed GAPDH demalonylation by WT but not inactive mutant SIRT5 (Figure 6B).
Figure 6. The Major Site of Malonylation in GAPDH by SIRT5 (K184) Is Important for GAPDH Enzymatic Activity.
(A) GAPDH is reversibly de-malonylated by WT but not enzymatically inactive mutant SIRT5 overexpression in cell culture. Flag-tagged GAPDH and SIRT5 were co-overexpressed in WT or SIRT5 KO MEFs, and western blotting against maK was performed following immunoprecipitation by anti-Flag antibody.
(B) GAPDH is reversibly de-malonylated by SIRT5 in vitro. After purifying GAPDH and SIRT5 WT or H158Y enzymatically inactive mutant overexpressed in HEK293T cells, respectively, de-malonylation was performed by adding NAD. MaK level of GAPDH was assayed by western blotting. The quantification of maK levels is shown in the right panel, in which WT but not H158Y SIRT5 effectively demalonylated GAPDH in vitro.
(C) Sites of malonylation in the GAPDH protein with respect to its two functional domains, NAD binding, and catalytic domains. The ratio of malonylation between WT and Sirt5−/− samples are shown above.
(D) Relative malonylation between WT and Sirt5−/− samples is illustrated as a redness color scale, with the most SIRT5-regulated lysine (K184) shown as 100% red.
(E) K184E mutant shows defective GAPDH enzymatic activity. Endogenous human GAPDH was knocked down with two different shRNAs in HEK293T cells after a 4-day selection with puromycin. Expression vector for Flag-tagged mouse GAPDH WT, or mutants C150S (enzymatically inactive) and K184E were overexpressed. GAPDH activity was measured in whole-cell lysates 24 hr after transfection of expression vectors by enzymatic assay.
(F) Knockdown of endogenous GAPDH and expression of WT, C150S, and K184E mutants was confirmed by western blotting using an antiserum against GAPDH or against Flag.
Of 11 malonylated sites on GAPDH, lysine 184 showed the highest differential malonylation in the absence of SIRT5 (6.4-fold) (Figures 4C and 6C). A homology model of murine GAPDH based on the crystal structure of rabbit GAPDH (RCSB ID 1J0X) reveals that K184 is found at the functionally important dimer interface (Figure 6D).
As previously noted, malonylation shifts a lysine's charge by changing the positive charge of its ε-amino group to a negatively charged carboxylic acid. To test whether modification of lysine 184 is sufficient to affect GAPDH enzymatic activity, we generated a mutated mouse GAPDH expression vector in which K184 is replaced by a glutamic acid (K184E), a mutation that, like malonyllysine, substitutes a negatively charged carboxylic acid for the positively charged lysine. We also constructed two different shRNA expression vectors that uniquely targeted human and not mouse GAPDH to knock down endogenous GAPDH in HEK293T cells. These constructs were cotransfected, and GADPH expression and enzymatic activity were measured in cell lysates. The two shRNAs knocked down GAPDH expression and endogenous GAPDH enzymatic activity to different degrees (Figure 6F). Cotransfection of an expression vector for WT GAPDH restored GAPDH expression and activity (Figures 6E and 6F). However, catalytically inactive GAPDH (C150S) (Mustafa et al., 2009) and the K184E mutant did not restore enzymatic activity, despite similar levels of expression (Figures 6E and 6F). Flag-epitope-tagged WT and C150S GAPDH protein migrated more slowly than untagged protein, likely reflecting increased size. However, GAPDH K184E mutant migrated more rapidly, presumably due to its more negative charge (Figure 6F). These data indicate that K184 is important for GAPDH enzymatic activity and that a malonyl-lysine mimic mutation inhibits its enzymatic activity.
Overlap of Lysine Acetylome, Succinylome, and Malonylome in the Mitochondria
We recently reported that nearly 80% of lysine succinylation sites identified in mouse liver mitochondria can also be acetylated (Rardin et al., 2013b). To determine whether malonylation targets the same sites as acetylation and succinylation, we used our two previously reported mouse liver mitochondria data sets (Rardin et al., 2013a, 2013b). However, as these studies examined acetyl and succinyl modifications from mitochondrial enriched fraction, we restricted our comparison to proteins listed as mitochondrial in the MitoMiner data set (Smith et al., 2012). With this filter constraining all three data sets, we identified a total of 2,259 sites of lysine acylation on mitochondrial proteins with 138 sites modified by all three modifications: malonylation, succinylation, and acetylation (Figure S2A). Pathway analysis of proteins bearing all three modifications identified key metabolic pathways such as fatty acid oxidation, glutaryl-CoA degradation, and ketogenesis (Figure S3). An additional 84 lysine residues identified here as malonylation sites could be either acetylated (68) or succinylated (16) representing ~21% of maK sites (Figure S2A). Notably, succinylation sites overlapped with other acylation modifications at 86% of sites. To determine the extent of overlap between SIRT3 and SIRT5 targeted proteins, we compared regulated sites that were increased in individual KO animals by ≥1.5-fold with a p value < 0.05 (Figure S2B). Of 225 regulated proteins identified in this manner, 15 showed increased acylation by all three modifications (Table 1) with an additional four proteins regulated by succinylation and malonylation (Figure S2B). Strikingly, 47% of malonylated mitochondrial proteins were not observed to be regulated by acetylation or succinylation. Finally, we found that 16 sites across 11 proteins were targeted by all three modifications and regulated by SIRT3 and SIRT5 (Table 2). These enzymes represent diverse mitochondrial pathways including the TCA cycle, oxidative phosphorylation, the urea cycle, ketogenesis, and fatty acid metabolism.
Table 1.
Proteins and Lysine Sites Regulated by Three Different Acylation Marks and SIRT3/SIRT5
| Number of Acylation Marks |
|||
|---|---|---|---|
| Protein Name | acK | suK | maK |
| Trifunctional enzyme subunit alpha | 25 | 22 | 3 |
| Carbamoyl phosphate synthase | 6 | 34 | 3 |
| Hydroxymethylglutaryl-CoA synthase | 15 | 13 | 2 |
| Acetyl-CoA acetyltransferase | 6 | 10 | 7 |
| Glutamate dehydrogenase 1 | 7 | 10 | 1 |
| Citrate synthase | 7 | 10 | 1 |
| Acyl-coenzyme A synthetase ACSM1 | 6 | 8 | 1 |
| ADP/ATP translocase 2 | 5 | 7 | 1 |
| Ornithine carbamoyltransferase | 3 | 8 | 1 |
| ATP synthase subunit O | 3 | 6 | 2 |
| Glycine N-acyltransferase-like protein Keg1 | 6 | 3 | 1 |
| 10 kDa heat shock protein | 1 | 7 | 1 |
| 3-hydroxyacyl-CoA dehydrogenase type-2 | 4 | 4 | 1 |
| Malate dehydrogenase | 1 | 5 | 1 |
| Succinate dehydrogenase flavoprotein subunit | 2 | 3 | 1 |
Mitochondrial malonylome compared to SIRT3-regulated acetylome or SIRT5-regulated succinylome numbered by SIRT3/SIRT5-regulated acylation sites per protein.
Table 2.
Proteins and Lysine Sites Regulated by Three Different Acylation Marks and SIRT3/SIRT5
| KO: WT Ratio |
||||
|---|---|---|---|---|
| Protein Name | Sites | acK | suK | maK |
| Acetyl-CoA acetyltransferase | K187 | 1.5 | 5.9 | 2.5 |
| Acetyl-CoA acetyltransferase | K220 | 1.6 | 1.7 | 1.8 |
| Acetyl-CoA acetyltransferase | K260 | 11 | 94 | 1.7 |
| Acyl-coenzyme A synthetase ACSM1 | K534 | 7.9 | 180 | 7.9 |
| ADP/ATP translocase 2 | K147 | 4.5 | 30 | 1.5 |
| ATP synthase subunit O | K97 | 42 | 400 | 1.7 |
| Carbamoyl phosphate synthase | K1291 | 1.5 | 30 | 1.7 |
| Carbamoyl phosphate synthase | K1479 | 1.6 | 4.9 | 1.5 |
| Hydroxymethylglutaryl-CoA synthase | K350 | 1.6 | 57 | 4.3 |
| Malate dehydrogenase | K239 | 28 | 160 | 11 |
| Ornithine carbamoyltransferase | K292 | 4.7 | 4.5 | 4.1 |
| Succinate dehydrogenase flavoprotein subunit | K179 | 5.7 | 3.9 | 2.9 |
| Trifunctional enzyme subunit alpha | K60 | 1.7 | 21 | 1.6 |
| Trifunctional enzyme subunit alpha | K406 | 3.2 | 34 | 2.3 |
| Trifunctional enzyme subunit alpha | K644 | 13 | 89 | 1.6 |
| 3-hydroxyacyl-CoA dehydrogenase type-2 | K212 | 3.8 | 40 | 9.2 |
Malonylated sites in mitochondrial proteins compared to acetylome and succinylome data. Numbers indicate KO:WT ratio in each acylome (Sirt3–/– in acK and Sirt5–/– in suK and maK).
DISCUSSION
Reversible acylation of lysines is emerging as an increasingly diverse set of PTMs of proteins. The structural diversity, widespread abundance, and dynamics of the cellular acyl-proteome suggest its role in regulating networks of cellular function is complex. Thus, understanding this “acylation code” is a critical step toward understanding its implications for health and disease. Toward this goal, we characterized the lysine malony-lome and its regulation by SIRT5 using an affinity purification and label-free quantification approach previously used to define the liver mitochondrial acetyl- and succinyl-proteomes (Rardin et al., 2013a, 2013b). This methodological consistency allowed us to perform meta-analyses of these three prevalent acylation marks. Importantly, we found that malonylation differs significantly compared to acetylation and succinylation in its distribution, in its target proteins, and in the individual lysine residues modified.
The Malonylproteome Is Dynamically Regulated by SIRT5
We have identified 1,137 unique malonyllysine sites on 430 proteins in mouse liver by affinity enrichment and label-free quantification and observed hypermalonylation (≥1.5-fold change) at 183 sites on 120 proteins in the absence of SIRT5 in the liver. These data suggest that 16% of identified maK sites are regulated by SIRT5 and 28% of malonylated proteins are SIRT5 targets. Glycolysis, gluconeogenesis, urea cycle, and fatty acid β-oxidation, crucial components of a cell's metabolic networks, are among the most dynamically regulated pathways. Consistent with these results, we found that glycolytic flux is suppressed in the absence of SIRT5 in primary hepatocytes. We also observed that SIRT5 regulates the activity of GAPDH, a key glycolytic enzyme, via demalonylation of a key residue, K184, located at the enzyme's homodimerization interface. These data provide a comprehensive analysis of the lysine malonylome in vivo and illustrate potential regulatory mechanisms of malonylation and its regulation by SIRT5 on intermediary metabolism.
The Malonylome Is Largely Distinct from the Acetyl- and Succinyl-Proteomes
Changes in lysine malonylation observed by western blot analysis in WT and Sirt5−/− animals indicated that lysine malonylation and its regulation by SIRT5 was more prevalent in the cytosol than mitochondria. This is in contrast to our previous study of lysine succinylation in liver, where comparative staining of multiple cellular compartments with anti-succinyllysine antibody clearly showed predominantly mitochondrial succinylation (Rardin et al., 2013b). However, our MS-based proteomic survey of the mouse liver malonylome also identified 400 maK sites on 119 mitochondrial proteins representing ~35% of sites identified, suggesting that lysine malonylation also significantly affects mitochondrial biology. 56 maK sites on 36 mitochondrial proteins were increased in the absence of SIRT5.
As succinylation and malonylation are both regulated by SIRT5 and both studies were carried out in liver tissue, we compared the overlap of succinylation and malonylation on mitochondrial proteins. Surprisingly, 56% of mitochondrial maK sites overlapped with previously identified suK sites, and 44% of maK sites were distinct from both succinylation and acetylation (Rardin et al., 2013b). This is in sharp contrast to the fact that succinylation sites overlap with other acylations at over 80% of sites (Rardin et al., 2013b). Since we currently do not know how these modifications occur, by the activity of unique enzymes or via a non-enzymatic mechanism, we cannot explain these differing specificities. If maK sites are modified non-enzymatically, these data suggest that local environmental factors at unique lysines residues such as pKa, solvent accessibility, acyl-CoA concentration, and/or tertiary structure may be distinct for malonyl-CoA than for acetyl-CoA and succinyl-CoA. If these modifications are enzymatically driven, it is possible that one acyltransferase can utilize both acetyl-CoA and succinyl-CoA while another acyltransferase with differing specificity may use only malonyl-CoA as a substrate. While our data cannot determine the relative abundances of these three modifications, the number of modified peptides identified in this study and our two previously published studies provides a broad indication. The total number of unique acylation sites is highest for acetylation (2,187), and successively lower for succinylation (1,190) and malonylation (1,137), an ordering in general agreement with the relative concentrations of AcCoA, SuCoA, and MalCoA, respectively. Finally, it is possible that mitochondrial protein acylation reflects both enzymatic and non-enzymatic mechanisms. The identification of specific mitochondrial acyltransferases will be critical to resolve these remaining questions.
Malonyl-CoA as Feedback Metabolite
Malonyl-CoA serves as the building block for de novo fatty acid synthesis, and a critical regulatory role of this metabolite in cellular fuel-switching has been appreciated for five decades (Wakil, 1962). Cytosolic malonyl-CoA is produced from acetyl-CoA by acetyl-CoA carboxylase (ACC) when glycolytic flux to acetyl-CoA exceeds energetic demand. Under these conditions, malonyl-CoA allosterically inhibits carnitine palmitoyl-transferase 1 (CPT1), the rate-limiting mitochondrial fatty acid transporter, and thereby suppresses β-oxidation while directing acetyl-CoA pools toward fat synthesis (Saggerson, 2008). Intra-cellular concentrations of malonyl-CoA are highly dynamic; during fasting or in livers of diabetic rats, levels are reduced by approximately 50% (McGarry et al., 1978; Guynn et al., 1972). Du et al. (2015) recently reported the hypermalonylation of liver proteins in a mouse model of diabetes and the potential for this malonylation to inhibit glycolytic enzyme function. Here, we directly demonstrate both a potential role for malonyl-CoA as a negative feedback regulator of glycolysis and a role for SIRT5 in limiting this negative feedback. According to this model, SIRT5 activity removes malonyl groups from modified glycolytic enzymes and prevents malonylation-mediated inhibition. While primary hepatocytes isolated from mice lacking SIRT5 clearly show decreased glycolytic flux, it is currently not clear how malonylation and SIRT5 contribute to cellular glucose homeostasis in the whole organism of a mouse. However, negative feedback exerted by malonylation on glycolytic flux could redirect glucose away from oxidation in glycolysis toward glycogen synthesis or the pentose-phosphate pathway.
In addition to this role in glycolysis, reversible protein malonylation also regulates other key cellular pathways including the urea cycle and other mitochondrial enzymes. It is tempting to speculate that hyperglycemia causes a substantial built-up of malonyl-CoA levels and subsequently global hypermalonylation in non-lipogenic tissues that increase malonyl-CoA levels in response to glucose, such as cardiac or skeletal muscle (Hamilton and Saggerson, 2000; Saha et al., 1995). The prevalence and potential pathophysiological consequences of chronic hypermalonylation in these and other tissues are currently unclear.
FUTURE DIRECTIONS
The partial overlap among malonylated sites and acetylated/succinylated sites suggests that malonylation can work synergistically with acetylation or succinylation via distinct pathways. As SIRT5 appears to regulate proteins in both the mitochondrial and cytosolic compartments, it is intriguing that the two top cytosolic pathways regulated by malonylation, glycolysis and the urea cycle, both feed directly into critical mitochondrial networks (i.e., TCA cycle and ammonia detoxification [CPS]). Generating Sirt3/Sirt5 double knockout mice may therefore lead to novel, more dramatic phenotypes and provide insights into the role of Sirtuins in regulating stress resistance and health span through modulation of the acylproteome.
The recent demonstration of potentially inhibitory hypermalonylation in db/db mice offers a window into a potential role for SIRT5 in preventing or attenuating disease. Crossing mouse strains used to model diabetes with Sirt5 knockouts may enable us to study the physiological effects of malonylation and demalonylation in detail to a degree not currently available for other SIRT5-regulated acylations such as succinylation and glutarylation.
Finally, the proteins we found to be targeted by SIRT5 extend beyond those directly involved in energy metabolism, including ethanol degradation, steroid synthesis, and urea detoxification. It will be of interest to address how SIRT5 coordinates these diverse pathways and what role the different subcellular locations of SIRT5 play in these processes. For example, the urea cycle contains cytosolic and mitochondrial enzymes that must function in coordination.
Determining the stoichiometries of malonyl and other acyl modifications such as acetylation and succinylation will also be critical to provide insight on the co-regulation of these networks. While two methods have recently been reported for assessing acetylation stoichiometry using stable-isotope-labeled acetic anhydrides (Baeza et al., 2014; Nakayasu et al., 2014)—which in principle could be adapted to succinylation—this approach is not feasible for malonylation, as the analogous anhydride, malonic anhydride, is inherently unstable and unsuitable for differential labeling.
The results presented here will provide a powerful resource to identify the role of malonylation in the regulation of pathways critical for cellular function and survival.
EXPERIMENTAL PROCEDURES
Generation of Novel Antisera Specific for Malonyl-Line
Malonyl-lysine antibodies were generated at Cell Signaling Technology by immunizing New Zealand White rabbits with a KLH-conjugated peptide library containing a central malonyl-lysine residue (XXXXXK*XXXXX, where X is any amino acid except C, W, and Y and K* represents the malonylated lysine). Polyclonal antibody was affinity-purified using the malonyl-lysine peptide library, and specificity for malonyl-lysine was confirmed by ELISA and immunoaffinity enrichment followed by MS.
MS and Chromatographic Parameters
All samples used for MS1 filtering experiments were analyzed by reverse-phase LC-ESI-MS/MS with an Eksigent Ultra Plus nano-LC 2D HPLC system connected to a quadruple time-of-flight TripleTOF 5600 mass spectrometer (AB SCIEX) in direct injection mode. MS/MS spectra for all identified peptides may be viewed using Panorama at the following location using the login gibsonreview@gmail and the password gibson (https://daily.panoramaweb.org/labkey/project/Gibson/SIRT5_Malonylome/begin.view?). See Supplemental Experimental Procedures for technique details.
Bioinformatic Database Searches
MS data sets were analyzed and searched using Mascot server version 2.3.02 (Matrix Sciences) and ProteinPilot (AB SCIEX 4.5beta) with the Paragon algorithm (4.5, 1656). All data files were searched using the SwissProt 2013_01 database with a total of 538,849 sequences but restricted to Mus musculus (16,580 protein sequences). See Supplemental Experimental Procedures for details.
Conservation Index of Lysine Malonylation Sites
Non-redundant malonylated peptides with distinct peaks were mapped to orthologous proteins for conservation analysis (Table S5). UniProt sequences were aligned to the nr database using blastpgp (BLAST suite 2.2.18) (Altschul et al., 1997). Multiple sequence alignments were generated by CLUSTALW (2.0.12) using default settings (Larkin et al., 2007) for mouse proteins with >10 matching sequences of 30%–94% sequence identity. Conservation indices for modified lysines were determined by (i) counting conserved lysines across the seven queried species and (ii) using AL2CO on the alignment (Pei and Grishin, 2001). Sites absent from the multiple alignment in a given species were excluded when calculating rates of mutation to various amino acids.
Pathway Analysis
Canonical pathway analyses were generated through the use of IPA (Ingenuity Systems; www.ingenuity.com).
Sample Preparation for Mouse Liver Protein Lysate
All mouse experiments were performed according to IACUC approved protocols. Livers were collected from five WT and five fed Sirt5−/− male mice 10 months of age in the presence of deacetylase inhibitors (20 mM nicotin-amide and 1 μM trichostatin A), minced, and lysed by dounce homogenization. For additional details see Supplemental Information.
Dot Plot Assay and Western Blot
Dot plot staining was performed to determine newly developed maK antibody specificity against other types of lysine acylation. Levels of global lysine malonylation, succinylation, and acetylation in whole-cell lysates of various organs of WT and Sirt5−/− mice were also tested by western blot analysis. Antibody vendors and item numbers are listed in the Supplemental Information.
Measurement of GAPDH Activity
GAPDH activity was assayed with GAPDH assay kit (Cat No.E-101) following the manufacture's instruction (Biomedical Research Service Center, University at Buffalo, State University of New York). For additional details, see Supplemental Information.
Lentiviral-Overexpression of SIRT5
Human SIRT5 cDNA was cloned into pCDH-IRES-puromycin vector (System Bioscience). After lentivirus production in HEK293T cells using 4-plasmid transfection (Dull et al., 1998), filtered media are condensed by ultracentrifuge. Lentivirus infection of primary hepatocytes was performed following isolation. Following 4-day selection by 1 μg/ml puromycin, cells were collected and prepared for western blotting.
Demalonylation Assay of GADPH by SIRT5 in MEFs
SIRT5 WT or KO MEFs were infected with pseudoretrovirus for SIRT5 or SIRT5 H158Y and grown with hygromycin for 2 weeks, then infected with Flag-tagged GADPH lentivirus 1 day prior to the cell collection. Anti-Flag M2 antibody immunoprecipitation was performed against Flag-GAPDH following cell collection. The level of maK in GAPDH was assessed by western blotting. For additional details see Supplemental Information.
Demalonylation Assay of GADPH by SIRT5 In Vitro
Recombinant GAPDH and SIRT5 overexpressed in HEK293T cells were purified using anti-Flag beads (Rardin et al., 2013b). In vitro de-malonylation was assayed by adapting a SIRT3-mediated deacetylation protocol described previously (Schwer et al., 2006). For additional details see Supplemental Information.
Mouse Hepatocyte Isolation and Deuterium-Glucose Oxidation Assay
See Supplemental Experimental Procedures for detailed information.
Supplementary Material
Highlights.
Lysine malonylation is a protein posttranslational modification removed by SIRT5
Proteomic profiling shows a relationship between lysine malonylation and glycolysis
Malonylation suppresses GAPDH, which is targeted by SIRT5 for removal
Malonylation targets different proteins than acetylation and succinylation
ACKNOWLEDGMENTS
We thank F.W. Alt for generously providing the original Sirt5−/− mouse strain, C. Her for primary hepatocyte preparation, T. Roberts and J. Carroll for figure preparation, and G. Howard for editorial assistance. This work was supported by National Institutes of Health Grants T32AG000266 (M.J.R.), PL1 AG032118 (B.W.G.), R24 DK085610 (E.V.), and P30 DK026743 (E.V.) and institutional support from the J. David Gladstone Institutes (E.V.). This work was also supported in part by the Shared Instrumentation Grant S10 RR024615 (B.W.G.) and the generous access of a TripleTOF 5600 by AB SCIEX at the Buck Institute. Y.N. was supported by the Japanese Society for Promotion of Science and the Uehara Memorial Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
M.J.R., B.W.G., and E.V. designed experiments. Y.N. and M.J.R. performed experiments, analyzed data, and wrote the paper; W.H. and P.G. conceived ideas and oversaw the research program; M.F. and M.H. designed and did experiments and analyzed data; C.C. performed structural analysis; A.K.S. performed bioinformatic analysis; R.N. did experiments.
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures, five tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.05.022.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Zhao Q, Yang T, Fung YME, Li XD. A chemical probe for lysine malonylation. Angew. Chem. Int. Ed. Engl. 2013;52:4883–4886. doi: 10.1002/anie.201300252. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Beysen C, Murphy EJ, McLaughlin T, Riiff T, Lamendola C, Turner HC, Awada M, Turner SM, Reaven G, Hellerstein MK. Whole-body glycolysis measured by the deuterated-glucose disposal test correlates highly with insulin resistance in vivo. Diabetes Care. 2007;30:1143–1149. doi: 10.2337/dc06-1809. [DOI] [PubMed] [Google Scholar]
- Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, et al. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J. Biol. Chem. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H-C, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Cai T, Li T, Xue P, Zhou B, He X, Wei P, Liu P, Yang F, Wei T. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteomics. 2015;14:227–236. doi: 10.1074/mcp.M114.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guynn RW, Veloso D, Veech RL. The concentration of malonyl-coenzyme A and the control of fatty acid synthesis in vivo. J. Biol. Chem. 1972;247:7325–7331. [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C, Saggerson ED. Malonyl-CoA metabolism in cardiac myocytes. Biochem. J. 2000;350:61–67. [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stančáková A, Goetzman E, Lam MM, Schwer B, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai C-H, Kovacs JJ, Higashimoto Y, Appella E, Yao T-P. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, Yanagi S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells. 2011;16:190–202. doi: 10.1111/j.1365-2443.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- McGarry JD, Takabayashi Y, Foster DW. The role of malonyl-coa in the coordination of fatty acid synthesis and oxidation in isolated rat hepatocytes. J. Biol. Chem. 1978;253:8294–8300. [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. H2S signals through protein S-sulfhydration. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000464. http://dx.doi.org/10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu ES, Wu S, Sydor MA, Shukla AK, Weitz KK, Moore RJ, Hixson KK, Kim JS, Petyuk VA, Monroe ME, et al. A method to determine lysine acetylation stoichiometries. Int. J. Proteomics. 2014;2014:730725. doi: 10.1155/2014/730725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Nakamura Y, Tanaka D, Zhuang X, Fujita Y, Obara A, Hamasaki A, Hosokawa M, Inagaki N. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem. Biophys. Res. Commun. 2010;393:73–78. doi: 10.1016/j.bbrc.2010.01.081. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics. 2011;10:M111.012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci. USA. 2013a;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013b;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- Saha AK, Kurowski TG, Ruderman NB. A malonyl-CoA fuel-sensing mechanism in muscle: effects of insulin, glucose, and denervation. Am. J. Physiol. 1995;269:E283–E289. doi: 10.1152/ajpendo.1995.269.2.E283. [DOI] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol. Cell. Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO. Preparation of rat liver cells. I. Effect of Ca 2+ on enzymatic dispersion of isolated, perfused liver. Exp. Cell Res. 1972;74:450–454. doi: 10.1016/0014-4827(72)90400-4. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Blackshaw JA, Robinson AJ. MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 2012;40(Database issue):D1160–D1167. doi: 10.1093/nar/gkr1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sol EM, Wagner SA, Weinert BT, Kumar A, Kim H-S, Deng C-X, Choudhary C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS ONE. 2012;7:e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorens B, Cheng Z-Q, Brown D, Lodish HF. Liver glucose transporter: a basolateral protein in hepatocytes and intestine and kidney cells. Am. J. Physiol. 1990;259:C279–C285. doi: 10.1152/ajpcell.1990.259.2.C279. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Payne RM. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil SJ. Lipid metabolism. Annu. Rev. Biochem. 1962;31:369–406. doi: 10.1146/annurev.bi.31.070162.002101. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr. Protein post-translational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.