Abstract
SHANK3 (also known as PROSAP2) is a postsynaptic scaffolding protein at excitatory synapses in which mutations and deletions have been implicated in patients with idiopathic autism, Phelan–McDermid (aka 22q13 microdeletion) syndrome, and other neuropsychiatric disorders. In this study, we have created a novel mouse model of human autism caused by the insertion of a single guanine nucleotide into exon 21 (Shank3G). The resulting frameshift causes a premature STOP codon and loss of major higher molecular weight Shank3 isoforms at the synapse. Shank3G/G mice exhibit deficits in hippocampus-dependent spatial learning, impaired motor coordination, altered response to novelty, and sensory processing deficits. At the cellular level, Shank3G/G mice also exhibit impaired hippocampal excitatory transmission and plasticity as well as changes in baseline NMDA receptor-mediated synaptic responses. This work identifies clear alterations in synaptic function and behavior in a novel, genetically accurate mouse model of autism mimicking an autism-associated insertion mutation. Furthermore, these findings lay the foundation for future studies aimed to validate and study region-selective and temporally selective genetic reversal studies in the Shank3G/G mouse that was engineered with such future experiments in mind.
Keywords: autism, behavior, Phelan–McDermid syndrome, postsynaptic density, Shank3, synaptic plasticity
Introduction
Shank3 is a postsynaptic scaffolding protein (Boeckers et al., 1999a, b, 2005) with roles in spine shape/maturation (Roussignol et al., 2005; Durand et al., 2012), localization of glutamate receptors (Naisbitt et al., 1999; for review, see Sheng and Kim, 2000; Roussignol et al., 2005; Uchino et al., 2006), and growth cone motility (Naisbitt et al., 1999; Durand et al., 2012). Mutations and copy number variants of SHANK3 have been implicated in patients with idiopathic autism (Durand et al., 2007; Moessner et al., 2007; Gauthier et al., 2009; Chen et al., 2010; Kolevzon et al., 2011; Waga et al., 2011; Soorya et al., 2013; for review, see Uchino and Waga, 2013) and Phelan–McDermid syndrome (Bonaglia et al., 2001, 2011; Wilson et al., 2003; Phelan, 2008; Sarasua et al., 2011; Phelan and McDermid, 2012), making SHANK3 deletion/mutation among the most prevalent genetic causes of autism.
Multiple laboratories have attempted to delete SHANK3 in mice by targeting regions of the gene with none deleting all isoforms (Bozdagi et al., 2010; Peça et al., 2011; Wang et al., 2011; Yang et al., 2012; Kouser et al., 2013; for review, see Jiang and Ehlers, 2013). Failure to delete all isoforms of Shank3 is likely due to the presence of multiple promoters within the gene (Lim et al., 1999; Zhu et al., 2014). Although each of these models has some construct validity, none represents an accurate mimic of a human SHANK3 mutation.
Because of these difficulties, we sought to mimic an autism-associated mutation in the SHANK3 gene in a mouse model by targeting an autism-associated guanine nucleotide insertion mutation (Shank3G) in exon 21 found in two brothers with autism, but not in their unaffected brother (Durand et al., 2007). This mutation causes a frameshift and a premature STOP codon in exon 21, theoretically encoding a truncated Shank3 protein lacking the C-terminal region (Durand et al., 2007, 2012). Overexpression of this theoretical truncated Shank3 has been shown to alter synaptic transmission and dendritic spine morphology in cultured neurons (Durand et al., 2012).
We also sought to create a Shank3 mutant that might be genetically rescued to wild-type at various times during development and in selected brain regions when crossed with inducible or brain-region-specific cre-recombinase lines. Thus, we knocked in the insertion mutant exon 21 upstream of the wild-type exon 21, inserting a transcriptional STOP (neo-STOP) cassette after the mutant exon 21 to prevent exon skipping or splicing out of the mutant exon 21 (Dragatsis and Zeitlin, 2001; Guy et al., 2007). To make this genetically reversible, loxP sites were inserted flanking the insertion mutant exon 21 and neo-STOP cassette, allowing for deletion of the mutant exon/neo-STOP cassette and reversion to wild-type Shank3 upon activation of cre-recombinase. We have identified behavioral, biochemical, and electrophysiologic phenotypes in this novel autism model. These findings set the stage for future studies designed to define the critical periods and brain regions involved in these Shank3 insertion mutant phenotypes.
Materials and Methods
Construction of genetically reversible exon 21 insertion mutant targeting vector.
To create a targeting vector with a single nucleotide insertion mutation in exon 21 of the Shank3 gene followed by a transcriptional stop (Neo-stop) cassette (Dragatsis and Zeitlin, 2001), the e21-Neo-stop cassette was assembled by combining a wild-type Shank3 exon 21 and PGK-Neo gene cassette with the His3-SV40 pA sequences in pBluescript II SK (+/−) plasmid (Fig. 1) (Dragatsis and Zeitlin, 2001; Guy et al., 2007). Two loxP sequences in the same orientation flanked the e21-Neo-stop sequence. Then, the plasmid was subjected to site-directed mutagenesis in vitro using Quick Change XL Site-directed Mutagenesis Kit (Stratagene) with PCR mutation primers of e21PM-sense (PM for point mutant) (CAGCAATGGGCAGGAGCCCAGCAGGCTGGGGGGCTGAAGAGGAC) and e21PM-antisense (GCTCCTCTTGCAGCCCCCCAGAATGCTGGGCTCCTGCCCATTGCTG) to generate the plasmid containing exon 21 with single nucleotide insertion mutation and Neo-stop cassette (G-Neo-stop cassette). The insertion mutation is designed to mimic an insertion mutation identified in two brothers with autism with an extra “G” inserted into exon 21 (Durand et al., 2007) and located at position 3728 of mouse mRNA of Shank3. This results in a frameshift and premature stop codon downstream of the mutation in exon 21. The G-Neo-stop cassette was inserted into a unique BglII site located within intron 20 near exon 21 of a 7.0 kb genomic Shank3 fragment containing a portion of intron 20, exon 21, and a portion of intron 21 in pBluescript II SK (+/−). This insertion made a 1.3 kb 5′ flanking homology fragment and a 5.7 kb 3′ flanking homology fragment. To enrich for targeting clones by negative selection, a diphtheria toxin gene cassette was cloned adjacent to the 5′ end of the 5′ homology in the plasmid. The Neo portion of the Neo-stop cassette provided a positive selection marker for targeting clones (Dragatsis and Zeitlin, 2001; Guy et al., 2007).
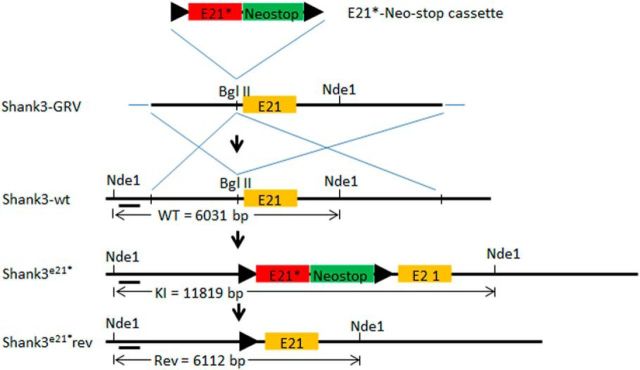
Figure 1.
Schematic diagram of the exon 21 insertion mutation (insert G) mouse model. A gene repair targeting vector (Shank3-GRV) was created via insertion of an insertion mutant exon 21 followed by a NeoStop cassette flanked by loxP sites (“floxed” E21G-NeoStop cassette) upstream of the wild-type exon 21. The resulting Shank3-GRV was targeted in mouse ES cells into the wild-type Shank3 gene (Shank3-wt). This resulted in the creation of mice constitutively expressing a human Shank3 exon 21 insertion mutation associated with autism, the Shank3G mouse model. In future experiments, the Shank3G mice can be genetically reversed to wild-type Shank3 whenever and wherever cre-recombinase is activated or expressed. Nde1 denotes restriction sites with size of diagnostic restriction fragments indicated between the Nde1 sites below.
Gene targeting.
The targeting vector DNA was linearized by Not1 and introduced by electroporation into SM-1 (129Sv/Evs6) embryonic stem (ES) cells grown on mitomycin-C-treated G418-resistant primary mouse embryonic fibroblasts. DNA was purified from the ES cells and analyzed by Southern blotting with a probe that distinguished between the targeting and wild-type Shank3 alleles. ES cells from three correct clones were injected into the blastocoel cavity of E3.5 C57BL6 embryos using standard procedures. The chimeric males with >90% agouti coat color on black background were bred with 6-week-old females of wild-type C57BL6J. The Shank3G mutation mice were crossed with wild-type C57BL6J mice >4 times.
Genotyping.
Routine genotyping was performed using PCR (4′ min denaturation at 98°C, followed by 25 cycles consisting of 30 s denaturation at 98°C, 30 s annealing at 69°C, and 25 s extension at 72°C using iProof kit from Bio-Rad) with the following 2 primers: 21M-loxp1-sequence-sense (CTGTTGGTGTCAGTTCTTGCAGATG, in intron 20) and 21M-sequence-loxp2-antisense (CAAGGATGCTGGCCATTGAATGGCTTC, in exon 21). This PCR generated a 596 bp product for wild-type Shank3 allele, a 638 bp product for Shank3-G-stop allele, and a 680 bp product for recombined Shank3 allele following cre-mediated recombination.
Biochemistry.
Western blotting was performed as previously described (Kouser et al., 2013). Synaptic protein levels from 7 littermate triplets (WT/Shank3WT/G/Shank3G/G, 5–6 months old) were determined by immunoblotting whole hippocampal tissue homogenized in ACSF, 5 mm EDTA, and 1× Halt protease and phosphatase inhibitor mixture (Thermo Scientific) diluted from 100× stock solution; 10 μg of protein was loaded per lane and blotted with antibodies for synaptic proteins and internal loading controls (β-actin). An Image Works film processor was used to develop films, and the chemiluminescence signals were quantified, normalized, and analyzed using ImageJ, Image Studio, and Microsoft Excel.
For synaptosome preparation, all steps were performed at 4°C or on ice, and all buffers contained protease and phosphatase inhibitors (Thermo Scientific), as previously described (Kouser et al., 2013). Mice were killed by live decapitation, and rapidly dissected hippocampi were homogenized in Syn-Per (Thermo Scientific), ∼1 ml/100 mg tissue. Samples were centrifuged at 1200 × g for 10 min. The resulting supernatant was then centrifuged at 15,000 × g for 20 min. Pellets (synaptosomes) were resuspended in buffer B (3 mm sucrose in 6 mm Tris, pH 8.0) with 1% SDS, briefly sonicated, and flash frozen in liquid nitrogen. Samples were stored long-term at −80°C, and protein concentration was quantified by DC Protein Assay (Bio-Rad).
For postsynaptic density (PSD) preparation, hippocampal synaptosome pellets were isolated as described above, resuspended in 1 ml buffer B, and homogenized. Each n represents pooled synaptosome pellets from 4 mice. Homogenates were overlaid on sucrose density gradients, 1.15, 1, and 0.85 M sucrose, and centrifuged at 82,500 × g. The fraction between 1.15 M and 1 M sucrose layer was isolated, resuspended in buffer C (6 mm Tris, pH 8.0, 1% Triton X-100), and incubated for 15 min before centrifugation at 32,800 × g for 20 min. The pellets were resuspended in buffer D (6 mm Tris, pH 8.0, 0.5% Triton X-100) and incubated for 15 min. After centrifugation for 1 h at 201,800 × g, the resulting pellets (PSD-II) were resuspended in a minimal volume of buffer D with 1% SDS and flash frozen in liquid nitrogen.
Behavioral overview.
Behavioral tests were performed on a cohort of age- and sex-matched littermate progeny of heterozygous matings (6 female WT/het/homo, 9 male WT/het/homo, 2 female WT/het, 2 male WT/het, and 4 male WT/homo pairs) (WT, n = 23; Shank3WT/G, n = 19; Shank3G/G, n = 19) during the light cycle of the mice. All mice were born within 10 weeks of each other, and generally only one pair or triplet came from any given individual litter of mice. Behaviors were tested at 2–6 months of age by an experimenter blind to genotype in the following order: elevated plus maze, dark/light, open field, locomotor, grooming, marble burying, rotarod, nesting behavior, Morris water maze, and visible water maze. Mice were weighed immediately before behavioral testing with homozygous mutant mice trending toward weighing less with a strong main effect of sex on weight and a significant main effect of genotype on weight with no significant interaction between weight and genotype. Consistent with a lack of interaction between weight and genotype, on post hoc analysis differences between WT versus homozygous mutant did not reach statistical significance, nor did WT versus heterozygotes (WT, 30.4 ± 1.4 g; Shank3WT/G, 30.8 ± 1.6 g; Shank3G/G, 26.2 ± 1.2 g; two-way ANOVA; main effect of sex: F(1,55) = 15.05, p = 0.0002; main effect of genotype: F(2,55) = 4.107, p = 0.02; sex × genotype interaction: F(2,55) = 0.117, p = 0.89; Tukey HSD: WT vs Het: p = 0.980; WT vs KO: p = 0.060; Het vs KO: p = 0.050).
Three-box social interaction test was conducted in a separate cohort of age- and sex-matched littermate progeny of heterozygous matings (5 female WT/Shank3WT/G/Shank3G/G, 6 male WT/Shank3WT/G/Shank3G/G, 2 female WT/Shank3WT/G littermate pairs, 2 male WT/Shank3WT/G pairs, 3 female WT/Shank3G/G pairs, and 1 male WT/Shank3G/G pair) (WT, n = 19; Shank3WT/G, n = 15; Shank3G/G, n = 15).
Behavioral results are described out of the order in which they were tested to simplify presentation of the data. Statistical analyses of behavioral data were conducted using Statistica software (Version 10, Statsoft) using either two-way ANOVAs or three-way repeated-measures (rm) ANOVA using genotype and sex as the main variables and trial as the repeated measure, where applicable. Post hoc Tukey's Honest Significant Difference (HSD) test for unequal N was applied for significant effects and interactions. For detailed numerical statistical results, see Table 1.
Table 1.
Statistical analysis of behavioral studiesa
| Parameter | Comparison | Results |
|---|---|---|
| Open field (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| Time in center | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.01, p = 0.90; main effect of genotype: F(2,55) = 1.15, p = 0.32; sex × genotype interaction: F(2,55) = 0.30, p = 0.73 |
| Frequency in center | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.07, p = 0.78; main effect of genotype: F(2,55) = 1.11, p = 0.33; sex × genotype interaction: F(2,55) = 0.67, p = 0.51 |
| Time in periphery | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.34, p = 0.55; main effect of genotype: F(2,55) = 0.26, p = 0.76; sex × genotype interaction: F(2,55) = 1.41, p = 0.25 |
| Frequency in periphery | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.80, p = 0.37; main effect of genotype: F(2,55) = 1.43, p = 0.24; sex × genotype interaction: F(2,55) = 2.47, p = 0.09 |
| Time in center/time in periphery | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.06, p = 0.79; main effect of genotype: F(2,55) = 1.20, p = 0.30; sex × genotype interaction: F(2,55) = 0.65, p = 0.52 |
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.01, p = 0.89; main effect of genotype: F(2,55) = 2.56, p = 0.08; sex × genotype interaction: F(2,55) = 2.75, p = 0.07 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.96; WT versus Shank3G/G: p < 0.05; Shank3WT/G versus Shank3G/G: p = 0.06 | ||
| Velocity | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.004, p = 0.94; main effect of genotype: F(2,55) = 2.35, p = 0.10; sex × genotype interaction: F(2,55) = 2.90, p = 0.06 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.96; WT versus Shank3G/G: p < 0.05; Shank3WT/G versus Shank3G/G: p = 0.07 | ||
| Dark/light box (WT, N = 23; Shank3WT/G, N = 19, Shank3G/G, N = 19) | ||
| Total activity | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.15, p = 0.69; main effect of genotype: F(2,55) = 2.96, p = 0.059; sex × genotype interaction: F(2,55) = 1.48, p = 0.23 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.99; WT versus Shank3G/G: p < 0.05; Shank3WT/G versus Shank3G/G: p < 0.05 | ||
| Time in light side | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.06, p = 0.79; main effect of genotype: F(2,55) = 0.87, p = 0.42; sex × genotype interaction: F(2,55) = 1.36, p = 0.26 |
| Crosses | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.20, p = 0.65; main effect of genotype: F(2,55) = 0.66, p = 0.51; sex × genotype interaction: F(2,55) = 2.12, p = 0.12 |
| Latency to enter light side | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.92, p = 0.33; main effect of genotype: F(2,55) = 6.66, p < 0.01; sex × genotype interaction: F(2,55) = 0.69, p = 0.50 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.38; WT versus Shank3G/G: p < 0.05; Shank3WT/G versus Shank3G/G: p < 0.001 | ||
| Time in dark side | Sex, genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.06, p = 0.79; main effect of genotype: F(2,55) = 0.87, p = 0.42; sex × genotype interaction: F(2,55) = 1.36, p = 0.26 |
| Elevated plus maze (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.03, p = 0.85; main effect of genotype: F(2,55) = 4.83, p < 0.05; sex × genotype interaction: F(2,55) = 1.79, p = 0.17 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.99; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p < 0.01 | ||
| Velocity | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.03, p = 0.85; main effect of genotype: F(2,55) = 4.83, p < 0.05; sex × genotype interaction: F(2,55) = 1.79, p = 0.17 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.99; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p < 0.01 | ||
| Time in open arms/time in both arms | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.17, p = 0.68; main effect of genotype: F(2,55) = 0.23, p = 0.79; sex × genotype interaction: F(2,55) = 0.24, p = 0.78 |
| Entries in open/entries in both | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 3.52, p = 0.06; main effect of genotype: F(2,55) = 0.24, p = 0.78; sex × genotype interaction: F(2,55) = 0.62, p = 0.54 |
| Morris water maze: initial training (WT, N = 22; Shank3WT/G, N = 19; Shank3G/G, N = 18) | ||
| Latency to reach platform | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,53) = 0.94, p = 0.33; main effect of genotype: F(2,53) = 3.83, p < 0.05; main effect of day: F(8,424) = 38.03, p < 0.001; sex × genotype interaction: F(2,53) = 1.07, p = 0.34; sex × day interaction: F(8,424) = 0.43, p = 0.90; genotype × day interaction: F(16,424) = 0.76, p = 0.73; sex × genotype × day interaction: F(16,424) = 0.80, p = 0.67 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.30; WT versus Shank3G/G: p < 0.05; Shank3WT/G versus Shank3G/G: p = 0.49 | ||
| Distance traveled to platform | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,53) = 1.22, p = 0.27; main effect of genotype: F(2,53) = 5.61, p < 0.01; main effect of day: F(8,424) = 144.53, p < 0.001; sex × genotype interaction: F(2,53) = 0.71, p = 0.49; sex × day interaction: F(8,424) = 0.49, p = 0.86; genotype × day interaction: F(16,424) = 5.58, p < 0.001; sex × genotype × day interaction: F(16,424) = 0.90, p = 0.56 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.88; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p < 0.05 | ||
| Thigmotaxis | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,53) = 0.20, p = 0.65; main effect of genotype: F(2,53) = 0.99, p = 0.37; main effect of day: F(8,424) = 44.17, p < 0.001; sex × genotype interaction: F(2,53) = 0.52, p = 0.59; sex × day interaction: F(8,424) = 0.21, p = 0.98; genotype × day interaction: F(16,424) = 1.48, p = 0.10; sex × genotype × day interaction: F(16,424) = 0.69, p = 0.79 |
| Average swim speed | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,53) = 0.78, p = 0.37; main effect of genotype: F(2,53) = 36.66, p < 0.001; main effect of day: F(8,424) = 34.05, p < 0.001; sex × genotype interaction: F(2,53) = 0.44, p = 0.64; sex × day interaction: F(8,424) = 0.64, p = 0.74; genotype × day interaction: F(16,424) = 9.79, p < 0.001; sex × genotype × day interaction: F(16,424) = 0.54, p = 0.92 |
| Tukey HSD: WT versus Shank3WT/G: p < 0.001; WT versus Shank3G/G: p < 0.001 Shank3WT/G versus Shank3G/G: p < 0.05 | ||
| Morris water maze: probe trial (WT, N = 22; Shank3WT/G, N = 19; Shank3G/G, N = 18) | ||
| % time in quadrant | Sex, genotype, and quadrant | 3-way rmANOVA; main effect of sex: F(1,53) = 0.00000001, p = 0.99; main effect of genotype: F(2,53) = 0.00000008, p = 0.99; main effect of quadrant: F(3,159) = 20.621, p < 0.001; sex × genotype interaction: F(2,53) = 0.0000002, p = 0.99; sex × quadrant interaction: F(3,159) = 1.91, p = 0.13; genotype × quadrant interaction: F(6,159) = 0.89, p = 0.50; sex × genotype × quadrant interaction: F(6,159) = 1.20, p = 0.30 |
| Target versus other quadrants | Tukey HSD: target NW versus right NE: p < 0.001; target NW versus left SW: p < 0.001; target NW versus opposite SE: p < 0.001 | |
| Target versus other quadrants within WT | Tukey HSD: target NW versus right NE: p = 0.78 target NW versus left SW: p = 0.40; target NW versus opposite SE: p < 0.05 | |
| Target versus other quadrants within Shank3WT/G | Tukey HSD: target NW versus right NE: p < 0.001; target NW versus left SW: p < 0.001; target NW versus opposite SE: p < 0.001 | |
| Target versus other quadrants within Shank3G/G | Tukey HSD: target NW versus right NE: p < 0.01; target NW versus left SW: p < 0.05; target NW versus opposite SE: p < 0.001 | |
| No. of platform crossings | Sex, genotype, and platform | 3-way rmANOVA; main effect of sex: F(1,53) = 2.54, p = 0.11; main effect of genotype: F(2,53) = 1.56, p = 0.21; main effect of quadrant: F(3,159) = 25.05, p < 0.001; sex × genotype interaction: F(2,53) = 0.35, p = 0.70; sex × quadrant interaction: F(3,159) = 2.50, p = 0.06; genotype × quadrant interaction: F(6,159) = 1.36, p = 0.23; sex × genotype × quadrant interaction: F(6,159) = 0.72, p = 0.62 |
| Target versus other platforms | Tukey HSD: target NW versus right NE: p < 0.001; target NW versus left SW: p < 0.001; target NW versus opposite SE: p < 0.001 | |
| Target versus other platforms within WT | Tukey HSD: target NW versus right NE: p = 0.06 target NW versus left SW: p = 0.37; target NW versus opposite SE: p < 0.01 | |
| Target versus other platforms within Shank3WT/G | Tukey HSD: target NW versus right NE: p < 0.001; target NW versus left SW: p < 0.001; target NW versus opposite SE: p < 0.001 | |
| Target versus other platforms within Shank3G/G | Tukey HSD: target NW versus right NE: p < 0.01; target NW versus left SW: p < 0.05; target NW versus opposite SE: p < 0.001 | |
| Average swim speed | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,53) = 0.03, p = 0.85; main effect of genotype: F(2,53) = 1.45, p = 0.24; sex × genotype interaction: F(2,53) = 0.14, p = 0.86 |
| % thigmotaxis | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,53) = 0.21, p = 0.64; main effect of genotype: F(2,53) = 0.07, p = 0.93; sex × genotype interaction: F(2,53) = 1.11, p = 0.33 |
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,53) = 0.02, p = 0.86; main effect of genotype: F(2,53) = 1.49, p = 0.23; sex × genotype interaction: F(2,53) = 0.13, p = 0.87 |
| Visible water maze (WT, N = 22; Shank3WT/G, N = 19; Shank3G/G, N = 18) | ||
| Latency to platform | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,53) = 0.30, p = 0.58; main effect of genotype: F(2,53) = 2.87, p = 0.06; sex × genotype interaction: F(2,53) = 0.71, p = 0.49 |
| % thigmotaxis | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.0008, p = 0.97; main effect of genotype: F(1,32) = 0.98, p = 0.32; sex × genotype: F(1,32) = 0.04, p = 0.82 |
| Distance traveled to platform | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,53) = 0.02, p = 0.88; main effect of genotype: F(2,53) = 9.32, p < 0.001; sex × genotype interaction: F(2,53) = 0.81, p = 0.44 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.70; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p < 0.001 | ||
| Average swim speed | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,53) = 0.12, p = 0.72; main effect of genotype: F(2,53) = 5.12, p < 0.01; sex × genotype interaction: F(2,53) = 0.66, p = 0.51 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.10; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p = 0.52 | ||
| Nesting behavior (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| Increase in nest height | Sex, genotype, and time | 3-way rmANOVA; main effect of sex: F(1,55) = 0.114, p = 0.73; main effect of genotype: F(2,55) = 7.19, p < 0.01; main effect of time: F(2,110) = 21.03, p < 0.001; sex × genotype interaction: F(2,55) = 2.18, p = 0.12; sex × time interaction: F(2,110) = 0.39, p = 0.67; genotype × time interaction: F(4,110) = 1.07, p = 0.37; sex × genotype × time interaction: F(4,110) = 0.85, p = 0.49 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.37; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p = 0.24 | ||
| Increase in nest width | Sex, genotype, and time | 3-way rmANOVA; main effect of sex: F(1,55) = 0.71, p = 0.40; main effect of genotype: F(2,55) = 4.60, p < 0.05; main effect of time: F(2,110) = 23.64, p < 0.001; sex × genotype interaction: F(2,55) = 1.62, p = 0.20; sex × time interaction: F(2,110) = 1.73, p = 0.18; genotype × time interaction: F(4,110) = 0.71, p = 0.58; sex × genotype × time interaction: F(4,110) = 0.18, p = 0.94 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.77; WT versus Shank3G/G: p < 0.05; Shank3WT/G versus Shank3G/G: p = 0.10 | ||
| Three choice interaction test: baseline (WT, N = 19; Shank3WT/G, N = 15; Shank3G/G, N = 15) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,43) = 0.34, p = 0.55; main effect of genotype: F(2,43) = 6.85, p < 0.01; sex × genotype interaction: F(2,43) = 0.24, p = 0.78 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.99; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p < 0.01 | ||
| Velocity | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,43) = 0.0007, p = 0.97; main effect of genotype: F(2,43) = 6.44, p < 0.01; sex × genotype interaction: F(2,43) = 0.26, p = 0.76 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.99; WT versus Shank3G/G: p < 0.01; Shank3WT/G versus Shank3G/G: p < 0.05 | ||
| Time spent sniffing | Sex, genotype, and interaction target (front vs back) | 3-way rmANOVA: main effect of sex: F(1,43) = 0.0001, p = 0.99; main effect of genotype: F(2,43) = 1.37, p = 0.26; main effect of target: F(1,43) = 1.28, p = 0.26; sex × genotype interaction: F(2,43) = 0.004, p = 0.99; sex × target interaction: F(1,43) = 0.14, p = 0.70; genotype × target interaction: F(2,43) = 0.50, p = 0.60; sex × genotype × target interaction: F(2,43) = 0.16, p = 0.84 |
| Three choice interaction test: social preference (WT, N = 19; Shank3WT/G, N = 15; Shank3G/G, N = 15) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,43) = 4.59, p < 0.05; main effect of genotype: F(2,43) = 1.85; p = 0.16; sex × genotype interaction: F(2,43) = 1.25, p = 0.29 |
| Velocity | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,43) = 3.30, p = 0.07; main effect of genotype: F(2,43) = 1.60; p = 0.21; sex × genotype interaction: F(2,43) = 1.46, p = 0.24 |
| Time spent sniffing | Sex, genotype, and interaction target (inanimate vs social) | 3-way rmANOVA: main effect of sex: F(1,43) = 0.001, p = 0.96; main effect of genotype: F(2,43) = 0.144, p = 0.86; main effect of target: F(1,43) = 6.01, p < 0.05; sex × genotype interaction: F(2,43) = 0.46, p = 0.62; sex × target interaction: F(1,43) = 0.06, p = 0.79; genotype × target interaction: F(2,43) = 0.02, p = 0.97; sex × genotype × target interaction: F(2,43) = 2.25, p = 0.11 |
| Effect of target (inanimate vs social) within each genotype | Tukey HSD: WT: p = 0.60; Shank3WT/G: p = 0.77; Shank3G/G: p = 0.80 | |
| Three choice interaction test: social novelty (WT, N = 19; Shank3WT/G, N = 15; Shank3G/G, N = 15) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,43) = 4.59, p < 0.05; main effect of genotype: F(2,43) = 1.85; p = 0.16; sex × genotype interaction: F(2,43) = 1.25, p = 0.29 |
| Velocity | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,43) = 3.30, p = 0.07; main effect of genotype: F(2,43) = 1.60; p = 0.21; sex × genotype interaction: F(2,43) = 1.46, p = 0.24 |
| Time spent sniffing | Genotype and interaction target (familiar vs stranger) | 3-way rmANOVA: main effect of sex: F(1,43) = 0.257, p = 0.61; main effect of genotype: F(2,43) = 0.07, p = 0.93; main effect of target: F(1,43) = 4.52, p < 0.05; sex × genotype interaction: F(2,43) = 0.26, p = 0.76; sex × target interaction: F(1,43) = 1.88, p = 0.17; genotype × target interaction: F(2,43) = 31, p = 0.73; sex × genotype × target interaction: F(2,43) = 0.06, p = 0.93 |
| Effect of target (familiar vs novel) within each genotype | Tukey HSD: WT: p = 0.91; Shank3WT/G: p = 0.48; Shank3G/G: p = 0.92 | |
| Locomotor habituation (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| Total beam breaks | Sex, genotype, and bin | 3-way rmANOVA; main effect of sex: F(1,55) = 0.20, p = 0.65; main effect of genotype: F(2,55) = 0.38, p = 0.68; main effect of trial: F(23,1265) = 105.79, p < 0.001; sex × genotype interaction: F(2,55) = 2.62, p = 0.08; sex × trial interaction: F(23,1265) = 0.94, p = 0.53; genotype × trial: F(46,1265) = 1.72, p < 0.01; sex × genotype × trial interaction: F(46,1265) = 1.21, p = 0.16 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.97; WT versus Shank3G/G: p = 0.49; Shank3WT/G versus Shank3G/G: p = 0.40 | ||
| First bin only | Genotype and sex | 2-way ANOVA; main effect of sex: F(1,55) = 0.38, p = 0.53; main effect of genotype: F(2,55) = 10.85, p < 0.001; sex × genotype interaction: F(2,55) = 0.50, p = 0.60 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.96; WT versus Shank3G/G: p < 0.001; Shank3WT/G versus Shank3G/G: p < 0.001 | ||
| Stereotypy beam breaks | Sex, genotype, and bin | 3-way rmANOVA; main effect of sex: F(1,55) = 5.95, p < 0.05; main effect of genotype: F(2,55) = 1.18, p = 0.31; main effect of trial: F(23,1265) = 50.59, p < 0.001; sex × genotype interaction: F(2,55) = 3.78, p < 0.05; sex × trial interaction: F(23,1265) = 0.70, p = 0.84; genotype × trial: F(46,1265) = 0.84, p = 0.76; sex × genotype × trial interaction: F(46,1265) = 1.30, p = 0.08 |
| Ambulatory beam breaks | Sex, genotype, and bin | 3-way rmANOVA; main effect of sex: F(1,55) = 0.03, p = 0.84; main effect of genotype: F(2,55) = 1.21, p = 0.30; main effect of trial: F(23,1265) = 101.88, p < 0.001; sex × genotype interaction: F(2,55) = 2.03, p = 0.14; sex × trial interaction: F(23,1265) = 1.03, p = 0.41; genotype × trial: F(46,1265) = 2.16, p < 0.001; sex × genotype × trial interaction: F(46,1265) = 0.99, p = 0.47 |
| Rotarod (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| Time to fall off | Sex, genotype, and trial | 3-way rmANOVA; main effect of sex: F(1,55) = 12.36, p < 0.001; main effect of genotype: F(2,55) = 20.37, p < 0.001; main effect of trial: F(7,385) = 23.36, p < 0.001; sex × genotype interaction: F(2,55) = 3.73, p < 0.05; sex × trial interaction: F(7,238) = 0.51, p = 0.82; genotype × trial: F(14,385) = 1.25, p = 0.27; sex × genotype × trial interaction: F(14,385) = 0.60, p = 0.86 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.10; WT versus Shank3G/G: p < 0.001; Shank3WT/G versus Shank3G/G: p < 0.01 | ||
| Marble burying (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| No. of marbles buried | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 1.31, p = 0.25; main effect of genotype: F(2,55) = 9.80, p < 0.001; sex × genotype interaction: F(2,55) = 0.29, p = 0.74 |
| Tukey HSD: WT versus Shank3WT/G: p = 0.10; WT versus Shank3G/G: p < 0.001; Shank3WT/G versus Shank3G/G: p = 0.06 | ||
| Grooming (WT, N = 23; Shank3WT/G, N = 19; Shank3G/G, N = 19) | ||
| Time spent grooming | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.53, p = 0.46; main effect of genotype: F(2,55) = 0.97, p = 0.38; sex × genotype interaction: F(2,55) = 1.27, p = 0.28 |
| No. of bouts | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.004, p = 0.94; main effect of genotype: F(2,55) = 0.73, p = 0.48; sex × genotype interaction: F(2,55) = 0.09, p = 0.91 |
| Time per bout | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,55) = 0.12, p = 0.72; main effect of genotype: F(2,55) = 0.69, p = 0.50; sex × genotype interaction: F(2,55) = 1.25, p = 0.29 |
a WT, Wild-type; rmANOVA, ANOVA (between-subjects factors are generally sex and genotype) with repeated measures (day, time, or trial); F(x, y), F ratio statistic used to determine whether the variances in two independent samples are equal; x, y, degrees of freedom (df), a measure of the number of independent pieces of information on which the precision of a parameter estimate is based. × = number of groups − 1; y = number of animals per group − 1, multiplied by the number of groups.
Elevated plus maze.
Mice were placed in the center of a white Plexiglas elevated plus maze (each arm 33 cm long and 5 cm wide with 15 cm high black Plexiglas walls on closed arms) and allowed to explore for 5 min (Powell et al., 2004). The test was conducted in dim white light (∼7 lux). Mice were monitored using CleverSys TopScan Software, and time spent in, and entries into, the open and closed arms was measured. Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Dark/light.
Dark/light test was performed as described previously (Powell et al., 2004). Briefly, the dark/light apparatus consisted of two chambers (each chamber 25 cm × 26 cm), one brightly lit (∼1700 lux) and the other kept dark with a small door (7 × 7 cm) separating the two. Mice were habituated for 2 min in the dark side, the door was opened, and then mice were allowed to move freely between the two sides for 10 min. Time spent in, and the number of entries into, each side was measured along with locomotor activity using photobeams monitored by Med PC IV data acquisition software. Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Open field.
The open field test was performed as described previously (Powell et al., 2004; Blundell et al., 2009), with the exception of the data acquisition software. Mice were monitored using CleverSys TopScan Software after being placed in a white plastic arena (48 × 48 × 48 cm) for 10 min. Time spent in and number of entries into the center of the arena (15 ×15 cm) as well as locomotor activity were measured. The test was conducted in dim white light (∼7 lux). Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Locomotor.
Locomotor activity was tested by placing the mice in a fresh home cage with minimal bedding and monitoring their activity for 2 h using photobeams linked to a computer with data acquisition software (San Diego Instruments) (Powell et al., 2004). The test was conducted in the dark. Three-way rmANOVA was used to analyze the data with genotype and sex as between-subject factors and time as a within-subject factor.
Grooming.
Mice were placed in a novel home cage without bedding, and time spent grooming the face, head, or body was measured for 10 min. Number of grooming bouts that lasted >1 s was also recorded. Time per bout was calculated by dividing the total time spent grooming by the number of grooming bouts initiated. Each grooming parameter described above was then analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Marble burying.
As previously described (Blundell et al., 2010), 20 marbles were evenly placed around the edges of a novel home cage with 5 cm of bedding, and mice were given 30 min in the cage. After 30 min, the number of marbles buried was recorded. A marble was defined as buried when <25% of the marble was visible. The test room was well lit (∼80 lux). Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Accelerating rotarod.
Coordination and motor learning were tested using a rotarod as previously described (Powell et al., 2004). Mice were placed on a stationary rotarod (IITC Life Sciences) in a well lit room, which was then activated and accelerated from 0 to 45 revolutions per minutes over 5 min. The latency for mice to fall off the rod or take one revolution was measured. Trials were repeated 4 times with intertrial intervals of 30 min over a single day. Data were analyzed using three-way rmANOVA with genotype and sex as between-subject factors and trials as a within-subject factor.
Nesting.
Nesting behavior was performed in a well-lit (∼80 lux) room by first habituating the mouse to a novel home cage with ∼1.5 cm of bedding for 15 min, and then a cotton nestlet (5.5 ×5.5 × 0.5 cm) was put in the cage. Height and width of the nests were measured at 30, 60, and 90 min (Etherton et al., 2009). Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Morris water maze.
The Morris water maze task was conducted as previously described (Powell et al., 2004; Tabuchi et al., 2007). Briefly, a white, circular pool 1.2 m in diameter was filled with water (22 ± 1οC) made opaque with nontoxic, “gothic white” liquid tempera paint, and a circular platform (10 cm in diameter) was submerged ∼1 cm beneath the surface of the water. The testing room was well lit (∼80 lux) and filled with a number of extramaze cues. Training was conducted over 9 consecutive days with 4 trials/d and an intertrial interval of ∼1–1.5 min. Mice were placed pseudo-randomly into each of 4 starting locations for each of 4 daily training trials. In each trial, mice swam until they found the hidden platform or were guided to it by the experimenter if not found within 60 s. Mice remained on the platform for 15 s before being removed to their home cage. Daily data were averaged across the 4 trials. A probe trial was conducted on day 10; the hidden platform was removed, and mice were placed in the pool and allowed to swim for 60 s. For reversal water maze training, training resumed the next day following the probe trial for 5 d with the platform in the opposite quadrant of the maze; on the sixth day, a second probe trial was administered. Data were analyzed using three-way rmANOVA with genotype and sex as between-subject factors and trial days as a within-subject factor for training. For probe trials, quadrant or platform location was used as the within-subject factor.
Three chambered social approach.
Social versus inanimate object preference and preference for social novelty analyses were performed in a three chambered box with small openings connecting the chambers as described previously (Blundell et al., 2009) and based to a large extent on the original descriptions (Moy et al., 2004; Nadler et al., 2004). The test was conducted in dim white light (∼7 lux). The mouse behavior was monitored objectively using CleverSys TopScan Software. This test consisted of three, 10 min trials. During the first trial, the mouse was allowed to explore the entire apparatus with empty cages in each end chamber. In the second trial, the mouse was given a choice between an inanimate cage and a caged, social target. For the third trial, the mouse was given a choice between a caged, novel social target versus a caged, familiar social target. Locations of empty cages and social targets were counterbalanced, and mice were placed back into their home cage for very brief intervals between trials. Data were analyzed using three-way mixed ANOVA with genotype and sex as between-subject factors and target as a within-subject factor.
Electrophysiology.
Male mice (4–6 weeks old) were anesthetized with 8% chloral hydrate (400 mg/kg) and then transcardially perfused with ACSF, and the brains rapidly removed and placed in ice-cold dissection ACSF containing sucrose. Acute coronal slices 300- to 400-μm-thick were made using a VT1000s Vibratome (Leica Biosystems). Slices containing dorsal hippocampus were allowed to recover at 35 ± 0.5°C for 30 min in normal ACSF and slowly cooled to room temperature over a 30–45 min period for holding before recording. A cut was made between CA3-CA1 to prevent recurrent excitation and epileptiform activity during recordings. All recordings were performed at 32 ± 0.5°C with a flow rate of 3–5 ml/min.
Data were collected using Clampex (pClamp software suite, version 10.3; Molecular Devices). CA3-CA1 synapses were stimulated by a 100 μs biphasic pulse placed 400–500 μm laterally from the recording electrode. The distance between the recording electrode and the stimulating electrode was kept constant within these bounds.
Extracellular “field” recordings.
Field EPSPs (fEPSPs) were evoked using a biphasic pulse through a custom-made monopolar nichrome stimulating electrode for highly focal stimulation and a narrow stimulus artifact. Stimulating and recording electrodes (1–3 MΩ) were placed in the stratum radiatum 400–500 μm apart. All extracellular recordings were performed using dual-well submersion slice electrophysiology chambers (Fine Science Tools or Automate Scientific). Data were collected at 10 kHz and filtered at 1 kHz using model 1800 amplifiers (A-M Systems) and Clampex version 10.3 data acquisition software (Molecular Devices). Stimulation was controlled via model 2100 stimulus isolators (A-M Systems).
Input/output (I/O) curves were generated by sequential 50 μA steps from 0 to 350 μA and the amplitude of the resulting fiber volley, as well as the initial fEPSP slope (10%–40%) were measured. Frequency of stimulation was 0.05 Hz, which does not induce plasticity. Each data point is the average of five consecutive traces at the same stimulus intensity.
Paired-pulse ratio (PPR) was measured at 0.05 Hz with interstimulus intervals 30–500 ms in pseudo-random order to prevent induction of plasticity (50, 500, 100, 80, 200, 30 ms). Immediately before each experiment, stimulus intensity was set at 50% of the maximum fEPSP and adjusted to prevent epileptiform activity at the shorter interstimulus intervals. Once set, stimulus intensity did not change during the course of the experiment. The average of 5 trials at each interstimulus interval is reported per slice.
Long-term potentiation (LTP) was induced with either 1 train or 4 trains of 100 Hz for 1 s, separated by 20 s. A 20 min baseline was recorded before stimulation and followed by 60 min of 0.05 Hz stimulation. Stimulus intensity was set to generate 50% of the maximum fEPSP and did not change during the course of the experiment. The average amount (percentage) of LTP was determined at 55–60 min after induction, relative to the 10 min immediately preceding LTP induction.
mGluR-LTD was induced by application of 100 μm (RS)-3,5-dihydroxyphenylglycine (DHPG) for 10 min followed by a 20 min washout. Stimulus intensity was set at 75%–85% of the maximum fEPSP slope according to I/O curves performed immediately before each experiment and did not change during the course of the experiment. The mean magnitude (percentage) of LTD was determined at 55–60 min following the start of DHPG washout and normalized to the last 10 min of the baseline period immediately preceding DHPG application.
Whole-cell patch-clamp recordings.
EPSCs were recorded in voltage-clamp from CA1 pyramidal neuron cell bodies visually identified at 80× magnification using an AxioExaminer D1 Differential interference contrast microscope (Zeiss). Borosilicate glass electrodes (4–6 MΩ) were filled with internal pipette solution containing the following (in mm): 110 Cs methanesulfonate, 15 CsCl, 8 NaCl, 10 TEA-Cl, 2 EGTA, 10 HEPES, 3 N-(2,6-dimethylphenylcarbamoylmethyl)triethylammonium chloride (QX 314), 2 ATP, 0.3 GTP. Observed junction potential was ∼10 mV and was compensated in each experiment. Cells with >25% change in either access resistance (15–25 MΩ) or holding current were not included in the analysis. Data were collected at 10 kHz and filtered at 1–3 kHz using Mutliclamp 700B amplifiers, a Digidata 1440 digitizer, and Clampex version 10.3 data acquisition software (Molecular Devices). Stimulation was controlled via a model 2200 stimulus isolator (A-M Systems) through a monopolar tungsten microelectrode (FHC) with a 0.1 ms biphasic pulse. All experiments were performed in the presence of 100 μm picrotoxin.
For NMDA/AMPA, stimulus intensity (230–250 μA) was set to generate a 200–500 pA EPSC at −70 mV holding potential. Once a stable 5–10 min baseline was achieved, 20 consecutive traces were obtained at −70 mV holding potential and at 40 mV holding potential at 0.1 Hz. The NMDA/AMPA ratio was taken as the ratio of primarily NMDA receptor-mediated EPSCs at 40 s after stimulus onset at 40 mV to the peak of the primarily AMPA receptor-mediated EPSCs 10–15 ms after stimulus onset at −70 mV.
Miniature spontaneous EPSCs (mEPSCs) were measured at −70 mV holding potential in the presence of picrotoxin and 1 μm TTX 5–10 min after break-in. Each cell was recorded for 15–20 min, and a 5 min continuous trace was analyzed for frequency and amplitude (≥3 pA) using manual and automated analysis in Clampfit version 10.3 (Molecular Devices).
Raw traces were analyzed with Clampfit version 10.3 (Molecular Devices). Plotting and statistics were performed in GraphPad version 6 (GraphPad Software). Results were considered significant if p < 0.05.
Solutions.
ACSF contained the following (in mm): 120 NaCl, 3.5 KCl, 1.25 NaH2PO4, 1 MgSO4, 26 NaHCO3, 10 dextrose, and 2 CaCl2. Dissection ACSF consisted of the following (in mm): 75 sucrose, 87 NaCl, 3 KCl, 1.25 NaH2PO4, 7 MgSO4, 26 NaHCO3, 20 dextrose, and 0.5 CaCl2. All solutions were adjusted to pH 7.4 and saturated with 95% O2/5% CO2.
Drugs.
Tetrodotoxin (TTX), picrotoxin, QX 314, and DHPG were obtained from Tocris Bioscience. Cs methanesulfonate and CsCl were obtained from Sigma-Aldrich. All other reagents were obtained from Fisher Scientific.
Generation of reversible Shank3G mouse line.
The progeny from the final backcross of Shank3G mice with the wild-type C57BL6J mice was then crossed with a CreESRT transgenic mouse line from The Jackson Laboratory (strain:B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J). This cross-produced Shank3G mutation mice with and without the CreESRT (Cre+ and Cre−, respectively). The mice with the heterozygous Shank3G mutation with CreESRT (Reversible-Shank3GCre+) were then crossed with heterozygous Shank3G mutation without CreESRT(Irreversible-Shank3GCre−). This final cross yielded the Reversible-Shank3GCre+ mice, which upon tamoxifen administration will allow the Cre recombinase to enter the nucleus and excise the mutated region of the Shank3 gene, resulting in WT Shank3 expression, effectively reversing the mutation.
Tamoxifen dosing.
Adult (8-week-old) Reversible-Shank3GCre+ mutation mice were randomly assigned to the regular diet group or the tamoxifen diet group. Accordingly, mice were fed regular 18% protein chow (vehicle) or tamoxifen diet (500 mg/kg, Harlan Laboratories) for 4 weeks ad libitum. The post-treatment, tamoxifen diet was replaced with regular 18% protein chow for 2 weeks before testing.
Results
The Shank3G mutation results in loss of predominant higher molecular weight Shank3 isoforms at hippocampal synapses
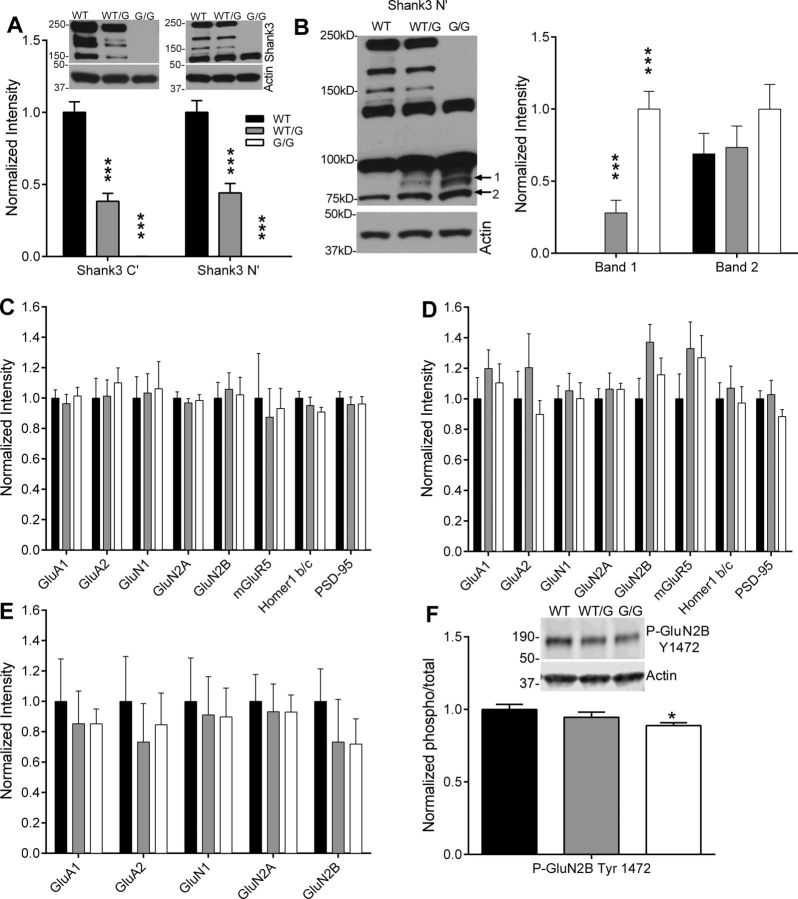
To determine the effect of the Shank3 mutation on Shank3 protein expression in the hippocampus, synaptosomes were isolated from hippocampus and subjected to Western blot analysis (n = 8 mice per genotype). As expected, there was a ∼50% decrease in Shank3 in Shank3WT/G mice and complete loss of Shank3 in Shank3G/G mice (Fig. 2A), as identified with Shank3 C-terminal antibody (one-way ANOVA: F(2,21) = 89.75, p < 0.0001) and Shank3 N-terminal antibody (one-way ANOVA: F(2,21) = 68.31, p < 0.0001). Western blotting with the Shank3 N-terminal antibody revealed the appearance of a single novel band of ∼90 kDa (band 1) in synaptoneurosomes from Shank3WT/G and Shank3G/G mice (Fig. 2B; one-way ANOVA: F(2,21) = 34.92, p < 0.0001), in addition to the smaller ∼75 kDa band (band 2) present in WT mice and unchanged in Shank3WT/G and Shank3G/G mice (Fig. 2B; one-way ANOVA: F(2,21) = 1.18, p > 0.05). All group statistical data shown are normalized to actin loading controls on each respective blot.
Figure 2.
Expression of Shank3 and associated proteins at hippocampal synapses. A, Western blots with both C-terminal and N-terminal Shank3 antibodies reveal that all three major Shank3 isoforms are absent in hippocampal synaptosomes from Shank3G/G mice. B, The Shank3 N-terminal antibody reveals the appearance of one novel Shank3 isoform and a trend toward enhanced expression of another isoform of low molecular weight in Shank3G/G mice compared with WT. Expression of synaptic and scaffolding proteins known to associate with Shank3 is unchanged in hippocampal synaptosomes (C) and in whole hippocampal lysates (D) from Shank3G/G mice. n = 8 mice per genotype. E, Ionotropic glutamate receptor expression remains unchanged in the hippocampal PSD preparations in the Shank3WT/G and Shank3G/G mice. n = 6 sets of hippocampi pooled from 4 mice per genotype. F, A decrease in phosphorylation levels of GluN2B at tyrosine-1472 was observed in the hippocampal synaptosomes of Shank3G/G mice. n = 6 sets of hippocampi pooled from 4 mice per genotype. *p < 0.05. ***p < 0.001.
Synaptic expression of glutamate receptors and Shank3-associated synaptic proteins in the hippocampus was unchanged in Shank3WT/G and Shank3G/G mice (Fig. 2C). By Western blot analysis of hippocampal synaptosomes, we found no change in the expression of AMPA receptor subunits (GluA1 one-way ANOVA: F(2,21) = 0.21, p > 0.05; GluA2 one-way ANOVA: F(2,21) = 0.26, p > 0.05), or in the expression of NMDA receptor subunits (GluN1 one-way ANOVA: F(2,21) = 0.041, p > 0.05; GluN2A one-way ANOVA: F(2,21) = 0.19, p > 0.05; GluN2B one-way ANOVA: F(2,21) = 0.07, p > 0.05). Expression levels of mGluR5, the primary mediator of mGluR-LTD at CA3-CA1 synapses was also unchanged in Shank3WT/G and Shank3G/G mice (one-way ANOVA: F(2,21) = 0.09, p > 0.05). Similarly, expression of the Shank3-associated postsynaptic proteins Homer1 b/c and PSD95 was not significantly changed in Shank3WT/G or Shank3G/G mice (Homer1 b/c, one-way ANOVA: F(2,21) = 1.09, p > 0.05; PSD95, one-way ANOVA: F(2,21) = 0.25, p > 0.05).
We also found no change in glutamate receptor or Shank3-associated Homer1b/c expression in whole hippocampal lysates from Shank3WT/G or Shank3G/G mice (Fig. 2D; GluA1 one-way ANOVA: F(2,21) = 0.59, p > 0.05; GluA2 one-way ANOVA: F(2,21) = 0.84, p > 0.05; GluN1 one-way ANOVA: F(2,21) = 0.09, p > 0.05; GluN2A one-way ANOVA: F(2,21) = 0.23, p > 0.05; GluN2B one-way ANOVA: F(2,21) = 2.29, p > 0.05; mGluR5 one-way ANOVA: F(2,21) = 1.18, p > 0.05; Homer1 b/c one-way ANOVA: F(2,21) = 0.17, p > 0.05; PSD95 one-way ANOVA: F(2,21) = 1.29, p > 0.05; n = 8 mice per genotype). W also looked at glutamate receptor expression in hippocampal PSDs and found no change in Shank3WT/G and Shank3G/G mice (Fig. 2E; GluA1 one-way ANOVA: F(2,15) = 0.16, p > 0.05; GluA2 one-way ANOVA: F(2,15) = 0.27, p > 0.05; GluN1 one-way ANOVA: F(2,15) = 0.05, p > 0.05; GluN2A one-way ANOVA: F(2,15) = 0.06, p > 0.05; GluN2B one-way ANOVA: F(2,15) = 0.49, p > 0.05).
Phosphorylation of key synaptic proteins, particularly of NMDA and AMPA receptors, can alter protein function without a change in expression levels (for review, see Lee, 2006; Chen and Roche, 2007). We analyzed tyrosine and serine kinase phosphorylation sites in hippocampal synaptosomes to identify any post-translational modifications on glutamate receptors. We found decreased phosphorylation of the GluN2B Tyr 1472 site in the Shank3G/G mice (Fig. 2F; one-way ANOVA: F(2,21) = 3.387, p > 0.05; WT vs G/G: p < 0.05; WT vs WT/G: p > 0.05; WT/G vs G/G: p > 0.05), but not in levels of phosphorylation of GluN2B at serine-1480, GluN2A at tyrosine-1246, GluA1 at serine-845, and GluA1 at serine-831 in the hippocampal synaptosomes (data not shown).
Despite a loss of predominant higher molecular weight isoforms of Shank3 at hippocampal synapses, expression and trafficking of glutamate receptors and other scaffolding proteins remain intact. However, altered GluN2B phosphorylation at Tyr 1472 may negatively impact NMDA receptor-mediated synaptic function in Shank3G/G mice.
Homozygous Shank3 mutants have impaired motor learning and coordination
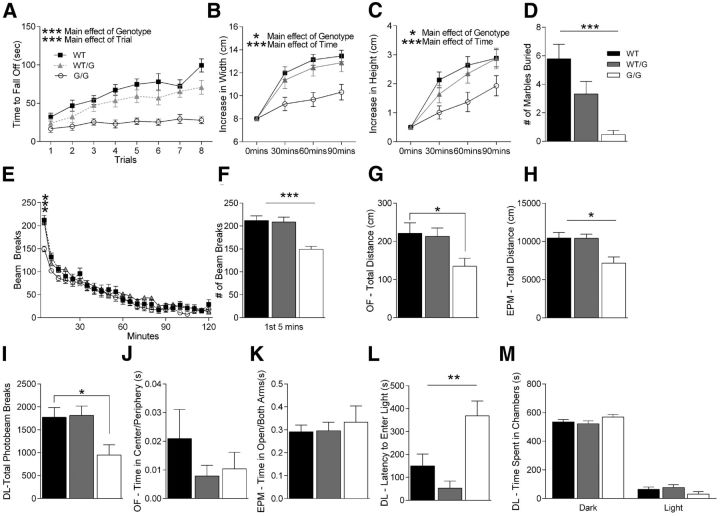
We tested motor learning and coordination in these mice as motor abnormalities are commonly observed in autistic patients (Kopp et al., 2010; Nobile et al., 2011). Shank3G/G mice, but not the Shank3WT/G mice, exhibit impaired motor coordination/learning on the accelerating rotarod. Shank3G/G mice consistently displayed shorter latencies to fall from the rotarod apparatus and showed little to no improvement in motor learning over the duration of the trials, whereas heterozygous mice were unaffected (Fig. 3A; three-way rmANOVA: main effect of genotype: F(2,55) = 20.37, p < 0.001; main effect of trial: F(7,385) = 23.36, p < 0.001; main effect of trial: F(7,385) = 23.36, p < 0.001; genotype × trial interaction: F(14,385) = 1.25, p > 0.05).
Figure 3.
Shank3G/G mice exhibit impairments in behavioral tasks. A, Latency to fall from or to go one full revolution on the rotarod task. Shank3G/G mice exhibit motor coordination impairments in 8 trials of rotarod test conducted over 2 h. Legend in A applies to B, E. B, C, Width and height of nest built as a function of time in a nest-building task. Shank3G/G mice exhibit impairments in nest building behavior over a 90 min period. D, Number of marbles buried during a 30 min marble-burying task. Shank3G/G mice show impaired marble burying behavior. Legend in D applies to F–M. E, Locomotor activity as measured by number of photobeam breaks during successive 5 min intervals over a 2 h period. Shank3G/G mice exhibit normal locomotor habituation over the full 2 h period. F, Number of photobeam breaks during the initial 5 min of the locomotor task shown in E. Shank3G/G mice show decreased activity, initially suggesting abnormal locomotor response to novelty. G, Total distance traveled during the 5 min elevated plus maze task. Shank3G/G mice have decreased locomotor activity in the elevated plus maze task. H, Total distance traveled during the 10 min open field task. Shank3G/G mice have decreased locomotor activity in the open field. I, Number of photobeam breaks during the 10 min dark/light task. Shank3G/G mice have decreased locomotor activity in dark/light. J, Ratio of time spent in the center to time spent in the periphery in an open field task. No differences were observed among genotypes. K, Ratio of time in the open arms versus time in other arms in the elevated plus maze task. Shank3G/G mice spend the same time in open versus closed arms compared with littermate controls. L, Latency to enter the light side of the dark/light apparatus. Shank3G/G mice exhibit an increased latency to enter the light chamber. M, Time spent in dark and light sides of the dark/light apparatus. No difference was observed in the total time spent in the dark versus the light side of the chamber: WT, n = 23; Shank3WT/G, n = 19; Shank3G/G, n = 19 for all panels. *p < 0.05. **p < 0.01. ***p < 0.001.
Homozygous Shank3 mutant mice exhibit an avoidance phenotype toward inanimate objects
We observed a novelty avoidance phenotype in the Shank3G/G mice in several behavioral tasks. When we measured nest building by adding a nestlet to a novel cage following a habituation period, we found that Shank3G/G mice showed very little change in their nestlets, whereas WT and Shank3WT/G littermates readily made nests from the material (Fig. 3B,C; three-way rmANOVA for nest width: main effect of genotype: F(2,55) = 4.60, p < 0.05; main effect of time: F(2,110) = 23.64, p < 0.001; genotype × time interaction: F(4,110) = 0.71, p > 0.05; three-way rmANOVA for nest height: main effect of genotype: F(2,55) = 7.19, p < 0.05; main effect of time: F(2,110) = 21.03, p < 0.001; genotype × time interaction: F(4,110) = 1.07, p > 0.05).
Similar avoidance behavior was observed in the marble burying task which further confirmed our explanation that Shank3G/G mice have a novelty avoidance phenotype (Fig. 3D). Shank3G/G mice showed little to no interest in burying marbles and in many cases left them undisturbed (Fig. 3D; two-way ANOVA: main effect of genotype: F(2,55) = 9.80, p < 0.001; Tukey's HSD: WT vs Shank3WT/G: p > 0.05; WT vs Shank3G/G: p < 0.001), whereas Shank3WT/G mice show a strong trend toward burying less marbles than their WT littermates.
Homozygous Shank3 mutant mice exhibit aberrant locomotor activity in response to novelty
Shank3G/G mice demonstrate decreased locomotor activity in response to novel situations. When examining locomotor activity over 2 h in a novel home cage, Shank3G/G mice demonstrated normal locomotor activity (Fig. 3E; three-way rmANOVA: main effect of genotype: F(2,55) = 0.38, p > 0.05; main effect of trial: F(23,1265) = 105.79, p < 0.001; genotype × trial interaction: F(46,1265) = 1.72, p < 0.01). However, when we analyzed the locomotor activity for the first 5 min in this novel home cage situation, Shank3G/G mice showed a significant decrease in locomotor activity that reverted to the WT level rapidly thereafter (Fig. 3F; two-way ANOVA: main effect of genotype: F(2,55) = 10.85, p < 0.001).
Along the same lines, distance traveled in the novel open field arena over 10 min was decreased in Shank3G/G mice compared with WT littermate controls (Fig. 3G; two-way ANOVA: main effect of genotype: F(2,55) = 2.56, p > 0.05; Tukey's HSD: WT vs Shank3WT/G: p > 0.05; WT vs Shank3G/G: p < 0.05; Shank3WT/G vs Shank3G/G: p > 0.05), and distance traveled in the elevated plus maze over 5 min was also decreased in the Shank3G/G mice (Fig. 3H; two-way ANOVA: main effect of genotype: F(2,55) = 4.83, p < 0.05; Tukey's HSD: WT vs Shank3WT/G: p > 0.05; WT vsShank3G/G: p < 0.01; Shank3WT/G vs Shank3G/G: p < 0.01). Similarly, total number of photobeams interrupted during the 10 min in the dark/light chamber was decreased in Shank3G/G mice (Fig. 3I; two-way ANOVA: main effect of genotype: F(2,55) = 2.96, p > 0.05; Tukey's HSD: WT vs Shank3WT/G: p > 0.05; WT vs Shank3G/G: p < 0.05; Shank3WT/G vs Shank3G/G: p < 0.05). We interpret these data overall as a decrease in locomotor response to novel environments as the mutants clearly habituated normally over time (Fig. 3E).
Shank3 mutant mice do not express an anxiety-like phenotype
Because there are several reports of children with autism suffering from anxiety disorders (Gillott et al., 2001; White et al., 2009), we also tested Shank3G mice in anxiety behaviors and found that these mice do not show an anxiety-like behavior in open field, elevated plus maze, or dark light task. No anxiety-like phenotype was observed in the open field task as both Shank3WT/G and Shank3G/G mice spent the same amount of time in the center as their WT littermates (Fig. 3J; two-way ANOVA: main effect of genotype: F(2,55) = 1.20, p > 0.05). We also did not observe an increase in anxiety-related behavior in the elevated plus maze as all three genotypes spent equal time in the open arms (Fig. 3K; two-way ANOVA: main effect of genotype: F(2,55) = 0.23, p > 0.05).
Interestingly, there was a strong increase in the latency to enter the brightly lit side of the dark/light box for the Shank3G/G mice (Fig. 3L; two-way ANOVA: main effect of genotype: F(2,55) = 6.66, p < 0.01). This is typically interpreted as an increase in anxiety-like behavior; however, there is no difference in the total time spent in either dark or light side of the chamber among the three genotypes (Fig. 3M; time spent in dark side, two-way ANOVA: main effect of genotype: F(2,55) = 0.87, p > 0.05; time spent in light side, two-way ANOVA: main effect of genotype: F(2,55) = 0.87, p > 0.05), and no other anxiety-related task demonstrated a phenotype. Thus, it appears that Shank3G/G mice do not have an anxiety phenotype but may show an avoidance of the novel, brightly lit chamber initially, a finding consistent with their novelty avoidance phenotype observed in other behaviors.
Homozygous Shank3 mutants exhibit minimal spatial learning differences
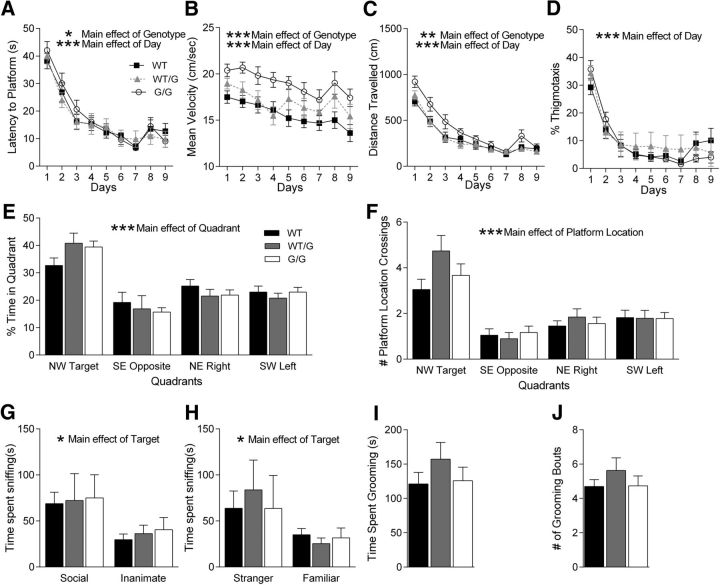
Because SHANK3 mutation/deletion is often associated with intellectual disability (Gong et al., 2012), we tested the Shank3G mice in the Morris water maze task, a test of spatial learning and memory. Shank3G/G mice were significantly impaired in spatial learning compared with WT littermate controls, whereas Shank3WT/G mice showed no deficit using latency to reach the hidden platform as a measure (Fig. 4A; three-way rmANOVA: main effect of genotype: F(2,53) = 3.83, p < 0.05; main effect of day: F(8,424) = 38.03, p < 0.001; genotype × day interaction: F(16,424) = 0.76, p < 0.05; for complete statistical results for all experiments, see Table 1). Interestingly, Shank3WT/G and Shank3G/G mice had significantly higher swim speed during training (Fig. 4B; three-way rmANOVA: main effect of genotype: F(2,53) = 36.66, p < 0.001; main effect of day: F(8,424) = 34.05, p < 0.001; genotype × day interaction: F(16,424) = 9.79, p < 0.001). Using the distance traveled before reaching the hidden platform, a measure that eliminates swim speed as a factor, Shank3G/G exhibited significantly decreased learning, whereas Shank3WT/G mice did not (Fig. 4C; three-way rmANOVA: main effect of genotype: F(2,53) = 5.61, p < 0.01; main effect of day: F(8,424) = 144.53, p < 0.001; genotype × day interaction: F(16,424) = 5.58, p < 0.001). We also looked at the amount of time spent in thigmotaxis (swimming near the maze walls). Both Shank3G/G and Shank3WT/G mice spent the same amount of time in thigmotaxis as their WT littermate controls (Fig. 4D; three-way rmANOVA: main effect of genotype: F(2,53) = 0.99, p > 0.05; main effect of day: F(8,424) = 44.17, p < 0.001; genotype × day interaction: F(16,424) = 1.48, p > 0.05). These findings suggest a deficit in spatial learning in the Shank3G/G mice.
Figure 4.
Shank3G/G mice exhibit mild spatial learning impairment, normal social interaction, and normal grooming behavior. A–D, Training days for the Morris water maze task. For each day of training, data were averaged across four daily trials. A, Latency to reach hidden platform on successive water maze days. Shank3G/G mice take longer to reach the submerged platform. Legend in A applies to B–D. B, Swim speed on successive water maze training days. Shank3G/G mice swim faster in the water maze. C, Distance traveled before reaching the hidden platform on successive water maze training days. Shank3G/G mice travel a longer distance before reaching the submerged platform. D, Percentage time spent in thigmotaxis on successive water maze training days. Shank3G/G mice spend the same amount of time in the thigmotaxis region as their littermate controls. E, Time spent in target quadrant and other quadrants during probe trial in which target platform is removed. Shank3G/G mice spend more time in the target quadrant versus other quadrants. Legend in E applies to F–J. F, Number of target location crossings and corresponding phantom platform location crossings in other quadrants during the probe trial. Shank3G/G mice show a clear preference for the target platform location: WT, n = 22; Shank3WT/G, n = 19; Shank3G/G, n = 18. G, In the three chambered social interaction test, all groups show a normal preference for the social versus the inanimate target and (H) for the stranger mouse versus the familiar mouse: WT, n = 19; Shank3WT/G, n = 15; Shank3G/G, n = 15. I, Shank3G/G mice show no change in total time spent grooming or (J) number of grooming bouts in a 10 min period: WT, n = 23; Shank3WT/G, n = 19; Shank3G/G, n = 19. *p < 0.05. **p < 0.01. ***p < 0.001.
Although Shank3G/G mice were slower to learn the water maze task, their spatial memory performance was unaffected during the initial probe trial. Both Shank3G/G and Shank3WT/G mice showed a significant preference for the target quadrant compared with the three other quadrants (Fig. 4E), indicating that these mice were able to recall a previously learned spatial strategy. WT littermate controls showed a significant preference for the target quadrant compared with the opposite quadrant, demonstrating a spatial strategy to some degree as well, although the difference between the target quadrant and the other two quadrants was not statistically significant (Fig. 4E; three-way rmANOVA: main effect of genotype: F(2,53) = 8.4 × 10−8, p > 0.05; main effect of quadrant: F(3,159) = 20.62, p < 0.001; genotype × quadrant interaction: F(6,159) = 0.89, p > 0.05; Tukey's HSD for WT mice: target NW vs opposite SE: p < 0.05, target NW vs right NE: p > 0.05, target NW vs left SW: p > 0.05; Tukey's HSD for Shank3WT/G mice: target NW vs opposite SE: p < 0.001, target NW vs right NE: p < 0.05, target NW vs left SW: p < 0.001; Tukey's HSD for Shank3G/G mice: target NW vs opposite SE: p < 0.001, target NW vs right NE: p < 0.01, target NW vs left SW: p < 0.001). Similarly, we analyzed the number of times the mice crossed the exact target platform location and corresponding locations in the other 3 quadrants and found similar results (Fig. 4F; three-way rmANOVA: main effect of genotype: F(2,53) = 1.56, p > 0.05; main effect of quadrant: F(3,159) = 25.05, p < 0.001; genotype × quadrant interaction: F(6,159) = 1.36, p > 0.05; Tukey's HSD for WT mice: target NW vs opposite SE: p < 0.01, target NW vs right NE: p > 0.05; target NW vs left SW: p > 0.05; Tukey's HSD for Shank3WT/G mice: target NW vs opposite SE: p < 0.001, target NW vs right NE: p < 0.001; target NW vs left SW: p < 0.001; Tukey's HSD for Shank3G/G mice: target NW vs opposite SE: p < 0.001, target NW vs right NE: p < 0.01, target NW vs left SW: p < 0.05). Following the hidden platform water maze task, the mice were tested in a visible platform version of the water maze to rule out visual issues. The latency to reach a visible platform was unchanged in the mutants compared with WT littermate controls, suggesting no change in visual acuity (data not shown). Together, the training and the probe data suggest that Shank3G/G mice are slightly slower to learn the water maze compared with WT littermate controls but demonstrate significant use of a spatial strategy during subsequent recall.
Shank3 mutant mice do not display social interaction deficits
Social interaction deficits are one of the major characteristic features of autism (Schreibman, 1988; Mahjouri and Lord, 2012). Therefore, we tested social behaviors in Shank3G mice in the three chambered social interaction task. Much like their WT littermate pairs, Shank3WT/G mice and Shank3G/G mice showed no preference for either side in the three chambered social interaction box before introduction of a social target (Table 1). In the test for social versus inanimate preference, all three genotypes spent more time interacting with the social target (Fig. 4G; three-way rmANOVA: main effect of genotype: F(2,43) = 0.144, p > 0.05; main effect of target: F(1,43) = 6.01, p < 0.05; genotype × target interaction: F(2,43) = 0.02, p > 0.05). Interestingly, none of the three genotypes individually demonstrated a significant preference for the social target (Fig. 4G; Tukey's HSD for WT mice: p > 0.05; Tukey's HSD for Shank3WT/G mice: p > 0.05; Tukey's HSD for Shank3G/G mice: p > 0.05), making absolute interpretation of sociability difficult as even the WT mice did not show a preference. Similarly, an overall preference for the novel social target versus familiar social target was observed in the test of preference for social novelty. Again, post hoc tests revealed no significant social novelty preference in the individual groups (Fig. 4H; three-way rmANOVA: main effect of genotype: F(2,43) = 0.07, p > 0.05; main effect of target: F(1,43) = 4.52, p < 0.05; genotype × target interaction: F(2,43) = 31, p > 0.05; Tukey's HSD for WT mice: p > 0.05; Tukey's HSD for Shank3WT/G mice: p > 0.05; Tukey's HSD for Shank3G/G mice: p > 0.05), making interpretation of preference for social novelty difficult.
Shank3 mutant mice show normal grooming
We also characterized grooming behavior in Shank3G mice as a measure of repetitive, stereotyped behavior. Shank3WT/G mice and Shank3G/G mice do not exhibit an increase in total time spent grooming compared with WT mice (Fig. 4I; two-way ANOVA: main effect of genotype: F(2,55) = 0.73, p > 0.05). Similarly, no difference was observed in the number of grooming bouts (Fig. 3J; two-way ANOVA: main effect of genotype: F(2,55) = 0.97, p > 0.05).
Synaptic transmission and plasticity are impaired in mice expressing the Shank3G mutation
We used extracellular (field) electrophysiology to understand how the Shank3G mutation disrupts normal synaptic transmission and plasticity at hippocampal CA3-CA1 synapses. For our initial characterization, we focused on the hippocampus for its well-defined structure/function relationship and its role in learning and memory (Wallenstein et al., 1998).
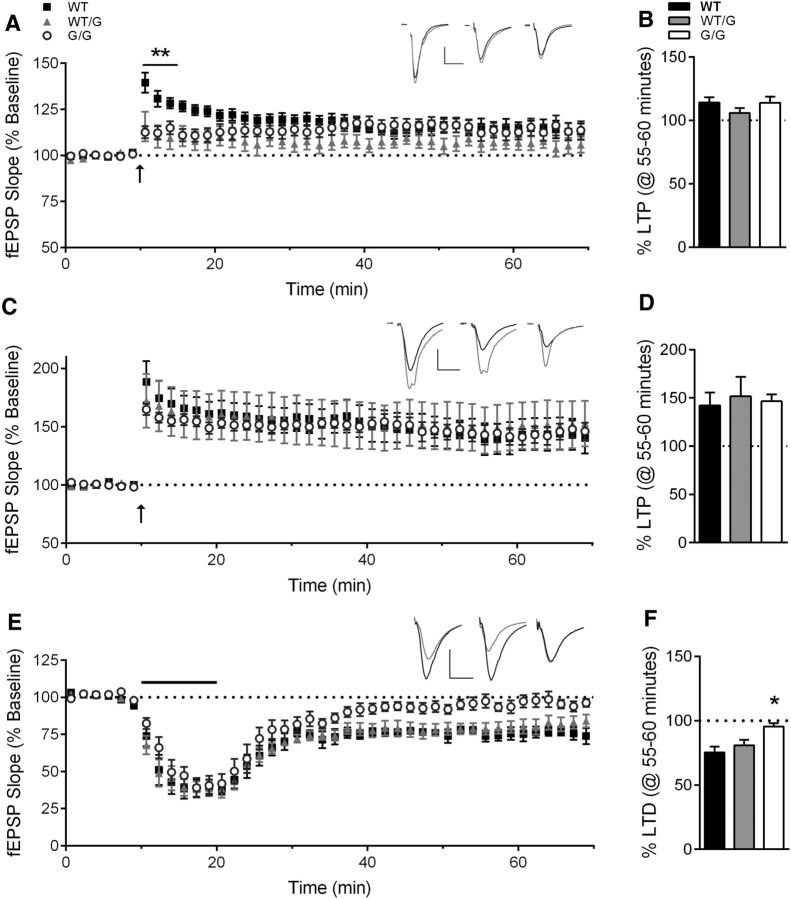
Previously, we found that deletion of exon 21 in the Shank3ΔC/ΔC mouse causes a decrease in LTP in response to a single 1 s train at 100 Hz (Kouser et al., 2013), a finding consistent among other Shank3 mutation mouse models (Bozdagi et al., 2010; Wang et al., 2011; Yang et al., 2012). We anticipated a similar LTP deficit in Shank3G/G mouse, which also lacks the Homer binding region and results in loss of major higher molecular weight Shank3 isoforms. On the contrary, we observed a decrease only in the magnitude of post-tetanic potentiation (PTP) in the first 5 min following the conditioning stimulus (Fig. 5A; one-way ANOVA: F(2,28) = 6.62, p < 0.01; WT, n = 13 slices/8 mice; Shank3WT/G, n = 9 slices/5 mice; Shank3G/G, n = 9 slices/5 mice), but no change in the mean magnitude of LTP at 1 h following the conditioning stimulus (Fig. 5B; one-way ANOVA: F(2,28) = 1.06, p > 0.05). Believing that this alteration in PTP magnitude may indicate a change in threshold for PTP induction, we used a stronger conditioning stimulus of 4 trains at 100 Hz for 1 s each, separated by 60 s (Fig. 5C). In support of this hypothesis, the stronger stimulus elicited LTP that was similar in magnitude across all three genotypes (Fig. 5D; one-way ANOVA: F(2,22) = 0.09, p > 0.05; WT, n = 7 slices/5 mice; Shank3WT/G, n = 10 slices/5 mice; Shank3G/G, n = 8 slices/4 mice).
Figure 5.
Hippocampal synaptic plasticity is impaired in Shank3G/G mice. A, LTP induced by a single 1 s, 100 Hz train (arrow) is normal at 55–60 min after tetanus, summarized in B, although PTP in the first 5 min following conditioning stimulus is decreased in Shank3WT/G and Shank3G/G mice (WT, n = 13 slices/8 mice; Shank3WT/G, n = 9 slices/5 mice; Shank3G/G, n = 9 slices/5 mice). Inset, Average of 10 consecutive raw traces immediately preceding (black) and 60 min (gray) after conditioning stimulus for WT (left), Shank3WT/G (middle), and Shank3G/G (right). Calibration: 0.25 mV, 10 ms. C, LTP induced by four, 1 s, 100 Hz trains (arrow) is not affected by the Shank3G mutation, as summarized at 55–60 min after induction (D) (WT, n = 7 slices/5 mice; Shank3WT/G, n = 10 slices/5 mice; Shank3G/G, n = 8 slices/4 mice). Inset, Average of 10 consecutive raw traces immediately preceding (black) and 60 min (gray) after conditioning stimulus for WT (left), Shank3WT/G (middle), and Shank3G/G (right). Calibration: 0.25 mV, 10 ms. E, mGluR-LTD induced by 10 min application of 100 μm DHPG (bar) is impaired at 55–60 min following DHPG washout (F) in Shank3G/G mice (WT, n = 7 slices/5 mice; Shank3WT/G, n = 13 slices/5 mice; Shank3G/G, n = 8 slices/6 mice). Inset, Average of 10 consecutive raw traces immediately preceding (black) and 60 min (gray) after DHPG application for WT (left), Shank3WT/G (middle), and Shank3G/G (right). Calibration: 0.5 mV, 10 ms. For clarity, scatter plots are represented as the mean ± SEM of 5 consecutive data points. *p < 0.05. **p < 0.01.
Because Shank3 binds to Homer, linking it to mGluRs, we also investigated the effect of the Shank3G mutation on mGluR-LTD, a form of synaptic plasticity known to be disrupted in other mouse models of autism-associated disorders (Bear et al., 2004; Bateup et al., 2011; Bozdagi et al., 2012; Chévere-Torres et al., 2012). Accordingly, Shank3G/G mice exhibit a decrease in the magnitude of mGluR-LTD at 55–60 min after DHPG application (Fig. 5E,F; one-way ANOVA: F(2,25) = 5.35, p < 0.05; WT, n = 7 slices/5 mice; Shank3WT/G, n = 13 slices/7 mice; Shank3G/G, n = 8 slices/6 mice).
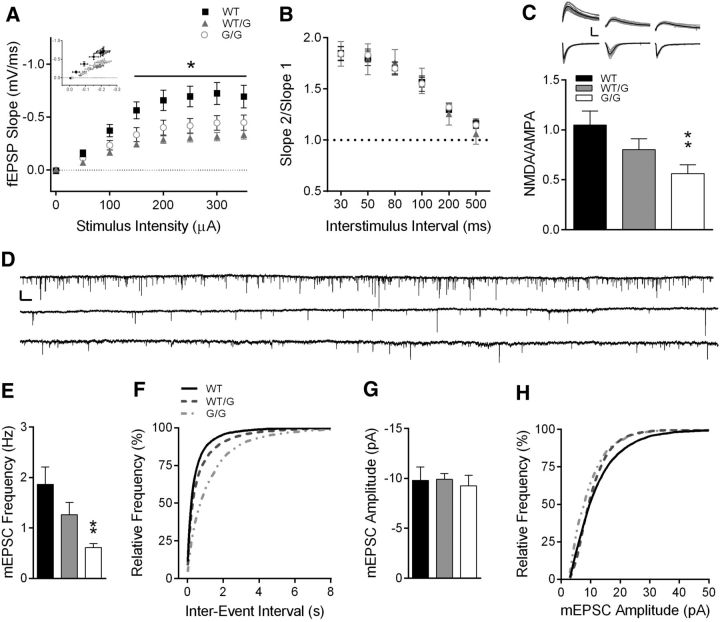
Basal synaptic transmission, in the absence of synaptic plasticity, was also impaired in Shank3WT/G and Shank3G/G mice. I/O curves generated from Shank3WT/G and Shank3G/G mice identified a decrease in fEPSP slope compared with WT (Fig. 6A; two-way RM ANOVA: genotype, F(2,37) = 6.18, p < 0.01; stimulus intensity, F(7,259) = 98.43, p < 0.001; stimulus intensity × genotype, F(14,259) = 6.41, p < 0.001; WT, n = 16 slices/8 mice; Shank3WT/G, n = 12 slices/6 mice; Shank3G/G, n = 12 slices/7 mice). Post hoc analysis further indicates a significant decrease in fEPSP slope at stimulus intensities >100 μA for both Shank3WT/G and Shank3G/G mice (Tukey's multiple comparisons, p < 0.05). Fiber volley amplitude, however, was not affected by the Shank3G mutation (two-way rmANOVA: genotype, F(2,37) = 0.19, p > 0.05; stimulus intensity, F(7,259) = 82.61, p < 0.0001; genotype × stimulus intensity, F(14,259) = 0.61, p > 0.05), suggesting that the decrease in fEPSP slope in Shank3WT/G and Shank3G/G mice is not due to a decrease in the number of axons activated upon stimulation or in axon excitability.
Figure 6.
Hippocampal synaptic transmission is impaired in Shank3WT/G and Shank3G/G mice. A, I/O curves of stimulus intensity versus fEPSP slope indicate a decrease in basal synaptic strength at CA3-CA1 synapses. Inset, fEPSP slope (mV/ms) measured in relation to fiber volley amplitude (mV) (WT, n = 16 slices/8 mice; Shank3WT/G, n = 12 slices/6 mice; Shank3G/G, n = 12 slices/7 mice). B, PPR is not affected by the Shank3G mutation at interstimulus intervals 30–500 ms (WT, n = 12 slices/7 mice; Shank3WT/G, n = 8 slices/5 mice; Shank3G/G, n = 15 slices/8 mice). C, NMDA/AMPA ratio is decreased in Shank3G/G mice (WT, n = 22 cells/10 mice; Shank3WT/G, n = 18 cells/6 mice; Shank3G/G, n = 23 cells/8 mice). Inset, Average of 10 consecutive traces at −70 mV and at 40 mV holding potential from Shank3WT (left), Shank3WT/G (middle), and Shank3G/G (right). Calibration: 200 pA, 25 ms. D, One minute traces of mEPSCs from WT (top), Shank3WT/G (middle), and Shank3G/G (bottom). Calibration: 20 pA, 1 s. Mean mEPSC frequency (E) is decreased in CA1 neurons from Shank3G/G mice and is reflected in the rightward shift in the distribution of interevent frequency (F) in Shank3G/G mice indicating longer interevent intervals in Shank3G/G mice. Mean mEPSC amplitude (G) and distribution of mEPSC amplitudes (H) are not affected by the Shank3G mutation (WT, n = 15 cells/5 mice; Shank3WT/G, n = 21 cells/8 mice; Shank3G/G, n = 14 cells/6 mice). *p < 0.05. **p < 0.01.
Consistent with Shank3's postsynaptic locus, we observed similar PPR in all three genotypes indicating no change in presynaptic function or short-term plasticity (Fig. 6B; two-way rmANOVA: genotype, F(2,32) = 0.02, p > 0.05; interstimulus interval, F(5,160) = 154.4, p < 0.0001; genotype × interstimulus interval, F(10,160) = 0.56, p > 0.05; WT, n = 12 slices/7 mice; Shank3WT/G, n = 8 slices/5 mice; Shank3G/G, n = 15 slices/8 mice). When we measured the relative contribution of NMDA and AMPA receptors to the EPSC, NMDA/AMPA ratio was decreased in Shank3G/G mice compared with WT (Fig. 6C; one-way ANOVA: F(2,60) = 4.88, p < 0.05; Tukey's multiple comparisons: WT vs Shank3WT/G, p > 0.05; WT vs Shank3G/G, p < 0.01; WT, n = 22 cells/10 mice; Shank3WT/G, n = 18 cells/6 mice; Shank3G/G, n = 23 cells/8 mice). Because each experiment was normalized to the AMPA receptor component of the EPSC (200–500 pA), the decrease in NMDA/AMPA ratio was most likely due to a decrease in the amplitude of the NMDA receptor-mediated component of the EPSC. However, there was no difference in decay kinetics of the NMDA receptor-mediated current between WT and Shank3WT/G or Shank3G/G mice to suggest a change in NMDA receptor subunit composition (τ: WT, 77.69 ± 4.76; Shank3WT/G, 86.70 ± 10.81; Shank3G/G, 76.62 ± 5.10; one-way ANOVA: F(2,60) = 0.60, p > 0.05).
Finally, we measured the frequency and amplitude of mEPSCs onto CA1 pyramidal neurons in the presence of 1 μm TTX (Fig. 6D–H). Mean frequency of mEPSCs was decreased in Shank3G/G mice (Fig. 6E; one-way ANOVA: F(2,47) = 5.19, p < 0.01; Tukey's multiple comparisons: p < 0.01; WT, n = 15 cells/5 mice; Shank3WT/G, n = 21 cells/8 mice; Shank3G/G, n = 14 cells/ 6 mice). The cumulative frequency of interevent intervals was shifted rightward, also indicating longer periods between mEPSCs in Shank3G/G mice. Mean amplitude of mEPSCs was unchanged in Shank3WT/G and Shank3G/G mice (Fig. 6G; one-way ANOVA: F(2,47) = 0.12, p > 0.05; WT, n = 15; Shank3WT/G, n = 21; Shank3G/G, n = 14), as reflected in the similar cumulative distribution of mEPSC amplitudes (Fig. 6H). Absence of a change in the largely AMPA receptor-mediated mEPSC amplitude further suggests that the decreased NMDA/AMPA ratio is due to decreased NMDA receptor-mediated responses.
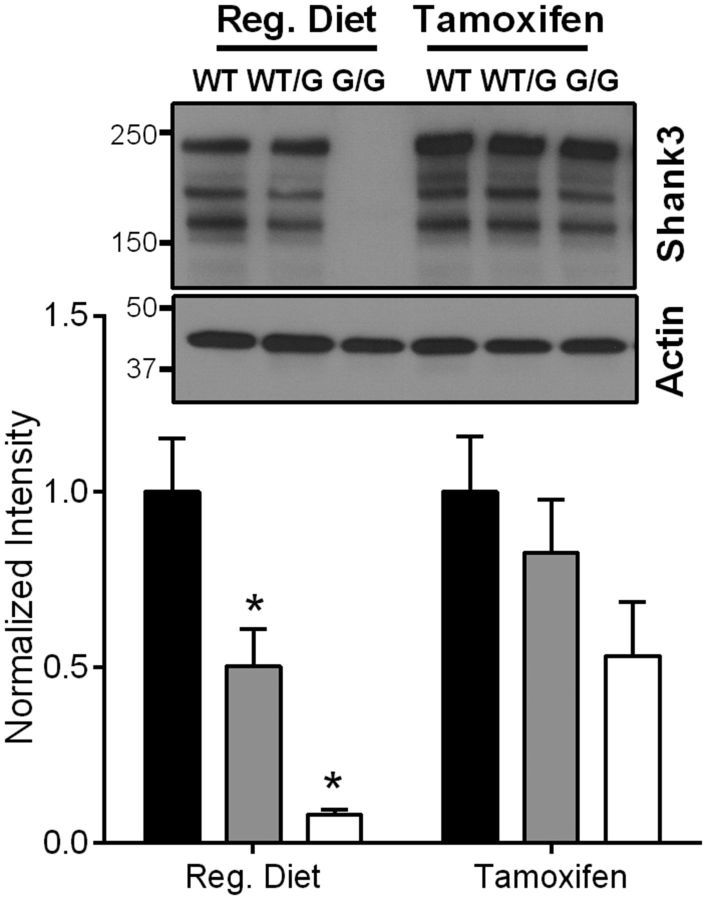
Tamoxifen-induced, genetic reversal results in restoration of Shank3 protein levels
The Shank3G mutation results in loss of predominant higher molecular weight isoforms of Shank3 protein (Fig. 2A) and was designed to be inducibly reversible when crossed with inducible cre recombinase systems, such as the actin promoter driving tamoxifen-inducible cre recombinase transgenic mice. To test the reversibility of this loss in the adulthood, our Shank3G mutant mice were crossed to a CreESRT transgenic mouse line from The Jackson Laboratory (strain:B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J) to produce Reversible-Shank3 WT, heterozygous, and homozygous mice that were CreESRT-positive. Starting at 8 weeks of age, we treated age- and sex-matched mice with tamoxifen diet (500 mg/kg) or regular diet (18% protein chow, vehicle) for 4 weeks and then compared Shank3 levels from whole-brain lysates using Western blot analysis (n = 7 per genotype and treatment group). Cre+ WT mice showed equivalent levels of Shank3 protein in the presence or absence of tamoxifen chow, indicating that tamoxifen alone does not affect Shank3 levels. Untreated Reversible-Shank3+/G/cre+ mice showed 50% reduction in Shank3 protein levels, whereas untreated Reversible-Shank3G/G mice showed nearly complete loss of Shank3 protein (Fig. 7). This indicates no effect of the CreESRT transgene on Shank3 levels in the absence of tamoxifen (no “leaky” genetic reversal). Shank3 protein levels in tamoxifen-treated Reversible-Shank3WT/G mice were indistinguishable from tamoxifen-treated WT levels (Fig. 7). Similarly, Shank3 protein levels in tamoxifen-treated Reversible-Shank3G/G mice were significantly increased up to 55%–60% of the WT level (Fig. 7; two-way ANOVA: main effect of genotype: F(2,36) = 13.70, p < 0.001; main effect of treatment: F(1,36) = 5.72, p < 0.05; genotype × treatment interaction: F(2,36) = 1.54, p > 0.05). Thus, using a tamoxifen-inducible cre recombinase system, we find no leaky genetic reversal as late as age 3.5 months, and Shank3 levels are inducibly reversible in this system upon tamoxifen treatment in adulthood. These findings form the basis for future experiments to test the region and temporal requirement for Shank3 in the phenotypes detailed above.
Figure 7.
Treatment with tamoxifen diet results in biochemical reversal of the Shank3 protein in Reversible-Shank3GCre+ mice. Quantification and representative Western blot of whole-brain lysates showing reversal of Shank3 protein expression with C terminus antibody (JH3025) 2 weeks after administration of tamoxifen diet for 4 weeks (initiated at 8 weeks of age). Data are normalized to the β-actin control and then to the average of WT levels (N = 7). *p < 0.05.
Discussion
The Shank3G/G mouse is the first genetically accurate, reversible Shank3 mouse model of autism (Durand et al., 2007). Rather than a truncated Shank3 protein, the mutation causes loss of multiple higher molecular weight Shank3 protein isoforms (≥150 kDa) using both C- and N-terminal Shank3 antibodies. In addition, there is appearance of a lower molecular weight Shank3 immunoreactive band between 75 and 100 kDa.
The biochemical findings largely mimic alterations in Shank3 isoforms identified in a Shank3 exon 21 deletion model (Kouser et al., 2013). Both the present mutant and the exon 21 deletion result in loss of higher molecular weight Shank3 isoforms. The exon 21 deletion also resulted in loss of a band between 100 and 150 kDa that is not lost in the present model (Kouser et al., 2013). This slight difference in Shank3 isoforms may account for phenotypic differences between the models. We did not identify alterations in Shank3-interacting partners in the hippocampus in either mutant (Kouser et al., 2013), perhaps due to compensatory action of Shank1 and Shank2 in hippocampus.
Behavioral phenotypes in both models are strikingly similar (Table 2), providing replication (Kouser et al., 2013). Both models demonstrated equivalent altered responses to novelty, including avoidance of marbles, and increased initial locomotor response to novelty in locomotor apparatus, open field, and dark/light boxes. Furthermore, both models resulted in long latencies to move from the dark side of dark/light boxes into the novel light side. In addition, both models exhibited significant differences in motor coordination on the accelerating rotarod. Finally, although the Morris water maze abnormality was subtle in the exon 21 insertion mutation, both models demonstrated abnormalities in this task.
Table 2.
Comparison of behavioral and physiological changes between the Shank3ΔC exon 21 deletion mouse model (Kouser et al., 2013) and our present Shank3G exon 21 insertion mutation model
| Behavior | Behavioral parameter | Shank3ΔC (Kouser et al., 2013) (WT/Homo) | Shank3G (WT/Homo) | Shank3G (WT/Het) |
|---|---|---|---|---|
| Body weight | Body weight in grams | ↓ | ↓ | – |
| Open field | Time in center/time in periphery | – | – | – |
| Distance traveled | ↓ | ↓ | – | |
| Velocity | ↓ | ↓ | – | |
| Dark/light | Time spent in dark chamber | ↑ | – | – |
| Time spent in light chamber | ↓ | – | – | |
| Latency to enter light chamber | ↑ | ↑ | – | |
| Distance traveled | ↓ | ↓ | – | |
| Elevated plus maze | Time in open arms/both arms | – | – | – |
| Distance traveled | – | ↓ | – | |
| Velocity | – | ↓ | – | |
| Morris water maze | Training: latency to platform | ↑ | ↑ | – |
| Training: distance traveled | ↑ | ↑ | – | |
| Training: thigmotaxis | ↑ | – | – | |
| Training: velocity | – | ↑ | ↑ | |
| Probe: % time in target quadrant | ↓ | – | – | |
| Probe: frequency of target platform location crossing | – | – | – | |
| Probe: thigmotaxis | ↑ | – | – | |
| Probe: velocity | – | – | – | |
| Nesting | Nest width | ↓ | ↓ | – |
| Nest height | ↓ | ↓ | – | |
| Three choice social interaction test | Front versus back target | – | – | – |
| Social versus inanimate target | – | – | – | |
| Familiar versus novel target | ↓ | – | – | |
| Locomotor test | 2 h | – | – | – |
| First 5 min | ↓ | ↓ | – | |
| Rotarod | Motor learning | – | ↓ | – |
| Motor coordination | ↓ | ↓ | – | |
| Marble burying | No. of buried marbles | ↓ | ↓ | – |
| Grooming | Time spent grooming | – | – | – |
| No. of bouts | – | – | – | |
| Time per bout | – | – | – | |
| Electrophysiology | ||||
| I/O curves | ↓ | ↓ | ↓ | |
| PPR | – | – | – | |
| LTP | ↓ | – | – | |
| mGluR-LTD | – | ↓ | – | |
| mEPSC amplitude | – | – | – | |
| mEPSC frequency | ↓ | ↓ | – | |
| NMDA/AMPA | ↓ | ↓ | – |
Altered nest building could be interpreted as supportive of altered response to novelty or as decreased motor coordination. Anecdotally, Shank3 mutants often completely avoid the nestlets. However, we cannot rule out the possibility that they were simply less effective or motivated nest builders.
Although the model has high construct validity for a human SHANK3 mutation, it does not have face validity for every symptom of Phelan–McDermid syndrome or autism due to SHANK3 mutation/deletion. Behaviorally, the mutant mouse model recapitulates incoordination, intellectual disability, and “insistence on sameness” due to abnormal behavior in response to novelty. It does not show abnormalities in social behavior, a core component of autism and a feature of Phelan–McDermid syndrome. Although it is intellectually satisfying when a genetic mouse model demonstrates strong behavioral face validity with autism or Phelan–McDermid syndrome, we do not feel that absence of social abnormalities in a mouse model provides any information against a gene's involvement in the human disorder. Instead, we use any resulting behavioral abnormality as an outcome measure for future studies linking brain function to mouse behavior.
Shank3 is an integral part of the PSD-95 complex of proteins at excitatory synapses in the hippocampus, yet expression of PSD-95 and Homer was unaffected in Shank3G/G and in Shank3ΔC/ΔC mice (Kouser et al., 2013). Although other Shank3 mouse models lacking exons 4–9 (Bozdagi et al., 2010; Wang et al., 2011) or exon 13 (Peça et al., 2011) have reported decreases in some hippocampal glutamate receptor subunits and other members of the PSD, our results are in agreement with Verpelli et al. (2011) in which no change in PSD-95, Homer, AMPA receptor, or NMDA receptor expression was found at hippocampal synapses following Shank3 knockdown in neuronal culture.
mGluRs bind to Shank3 indirectly through the Homer binding domain, as well as directly to the PDZ domain (Naisbitt et al., 1999), and Homer binding is required for mGluR-LTD (Ronesi and Huber, 2008). Shank3 has also been shown to regulate mGluR5 expression and function in cell culture models (Verpelli et al., 2011). However, we did not observe any alteration in mGluR5 expression in the hippocampus of Shank3G mice. As a consequence, one of the most striking differences in synaptic protein expression between our two Shank3 exon 21 models is the unexpected increase in mGluR5 expression at hippocampal synapses in Shank3ΔC/ΔC mice (Kouser et al., 2013) not present in Shank3G/G mice. Similarly, mGluR-LTD is abolished in Shank3G/G mice but preserved in Shank3ΔC/ΔC mice (Kouser et al., 2013). These findings are difficult to reconcile across the two models, although the slight difference in Shank3 isoform alterations may account for these differences.
Perhaps most puzzling is the lack of a decrease in LTP in Shank3G/G mice, a surprising finding given that decreased hippocampal LTP has been identified in Shank3 mouse models lacking exons 4–9 (Bozdagi et al., 2010; Wang et al., 2011; Yang et al., 2012) and exon 21 (Kouser et al., 2013). Incidentally, we do see a significant decrease in post-tetanic potentiation in the first 5 min following a single 1 s, 100 Hz conditioning stimulus, which may result from decreased NMDA receptor contribution to the EPSC, but is still sufficient for maintaining LTP at 60 min after induction. When the experiments were repeated with a much stronger conditioning stimulus (4, 1 s trains at 100 Hz), this difference in PTP was absent and no change in LTP was identified. This suggests that NMDA receptor function at hippocampal synapses is weaker in Shank3WT/G mice and Shank3G/G mice than in WT mice, consistent with the role of Shank3 in modulation of NMDA receptors at the synapse (Roussignol et al., 2005), but is not detrimental to LTP induced by stronger tetanic stimulation protocols. We leave open the possibility that an intermediate tetanic stimulation protocol under different conditions could lead to a finding of decreased LTP.
The most striking synaptic deficits in both Shank3G/G and Shank3ΔC/ΔC mice (Kouser et al., 2013) are impaired basal synaptic transmission measured via input/output curves, decreased mEPSC frequency, and decreased NMDA/AMPA ratio at CA3-CA1 synapses (Table 2). The alteration in NMDA/AMPA ratio is likely due to a decrease in NMDAR-mediated synaptic responses, which in turn likely contributes to decreased synaptic plasticity in both models. This interpretation is supported by the finding of normal mEPSC amplitude in both models. While at first glance our lack of biochemical alterations in NMDAR subunits in hippocampal synaptosomes and PSDs would seem to contradict this explanation, there are many post-translational mechanisms of decreased NMDA receptor function (for review, see Lee, 2006; Chen and Roche, 2007). Notably, phosphorylation of the GluN2B tyr-1742 residue stabilizes NMDA receptors at the synapse, whereas phosphorylation of the ser-1480 residue results in decreased GluN2B surface expression (Chen and Roche, 2007). Our finding that Shank3G/G mice have reduced GluN2B tyr-1742 phosphorylation, but no change in ser-1480 phosphorylation, is consistent with our decrease in NMDA receptor function. Future studies will be needed to identify the precise mechanism by which NMDAR-mediated synaptic function is decreased in this model.
Interestingly, mEPSC frequency is decreased in Shank3G/G and Shank3ΔC/ΔC mice (Kouser et al., 2013), as was reported in cultured hippocampal neurons expressing the Shank3G mutation (Durand et al., 2012). This could indicate a change in functional synapse number, as levels of Shank3 expression are directly related to mEPSC frequency (Roussignol et al., 2005; Verpelli et al., 2011; Durand et al., 2012) in cultured neurons. Conversely, the decrease in mEPSC frequency could indicate decreased probability of vesicle release, although we observed no changes in PPR to suggest a presynaptic contribution.
Other regions of the brain may also contribute to behavioral deficits identified in mice lacking the C-terminal region of Shank3. Specifically, the striatum is a promising target for future study because locomotor activity in response to novelty and motor coordination are decreased in Shank3G/G mice as well as Shank3ΔC/ΔC (Kouser et al., 2013). Shank3G/G mice also exhibit impaired motor learning, which may further implicate synaptic abnormalities in the cerebellum or prefrontal cortex. Although we cannot completely rule out the ∼3.5 g decrease in weight in the Shank3G/G mice as a contributing factor in the rotarod results, similar decreases in weight seem to correlate with improved rather than decreased rotarod performance (Kovács and Pearce, 2013). The Shank3 exon 21 insertion mutant model highlights a potential pitfall in interpreting and testing functional relevance of human autism mutations. This premature stop codon in exon 21 led to predictions of a truncated Shank3 protein (Durand et al., 2007, 2012). We now see that, at least in a mouse model of this mutation, the effects of this insertion mutation are much more complex and lead to a striking loss of multiple higher molecular weight Shank3 isoforms. While the mechanism of this loss remains unclear, the regulation of the Shank3 gene is much more complex than originally anticipated (Wang et al., 2011; Zhu et al., 2014).
In conclusion, we have generated a genetically accurate and reversible Shank3 exon 21 insertion mutation mouse model of autism. This model results in significant loss of multiple Shank3 protein isoforms and has significant and robust behavioral and electrophysiological abnormalities that are similar to a previous Shank3 exon 21 deletion model. Our findings suggest NMDAR-mediated synaptic transmission as a potential therapeutic target and set the stage for future studies of region-specific and temporal rescue of this mutant.
Footnotes
This work was supported by National Institute of Child Health and Human Development R01HD069560 and R21HD065290, National Institute of Mental Health R01MH093687, the Hartwell Foundation, BRAINS for Autism Foundation, Autism Science Foundation, and Autism Speaks.
The authors declare no competing financial interests.
References
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, Südhof TC, Powell CM. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009;8:114–126. doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Südhof TC, Powell CM. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Winter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED. Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem Biophys Res Commun. 1999a;264:247–252. doi: 10.1006/bbrc.1999.1489. [DOI] [PubMed] [Google Scholar]
- Boeckers TM, Kreutz MR, Winter C, Zuschratter W, Smalla KH, Sanmarti-Vila L, Wex H, Langnaese K, Bockmann J, Garner CC, Gundelfinger ED. Proline-rich synapse-associated protein-1/cortactin binding protein 1 (ProSAP1/CortBP1) is a PDZ-domain protein highly enriched in the postsynaptic density. J Neurosci. 1999b;19:6506–6518. doi: 10.1523/JNEUROSCI.19-15-06506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Liedtke T, Spilker C, Dresbach T, Bockmann J, Kreutz MR, Gundelfinger ED. C-terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J Neurochem. 2005;92:519–524. doi: 10.1111/j.1471-4159.2004.02910.x. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, Bonati MT, Giglio S, Guerrini R, Osimani S, Marelli S, Zucca C, Grasso R, Borgatti R, Mani E, Motta C, et al. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid Syndrome. PLoS Genet. 2011;7:e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7:e42422. doi: 10.1371/journal.pone.0042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BS, Roche KW. Regulation of NMDA receptors by phosphorylation. Neuropharmacology. 2007;53:362–368. doi: 10.1016/j.neuropharm.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Lin SP, Chern SR, Tsai FJ, Wu PC, Lee CC, Chen YT, Chen WL, Wang W. A de novo 7.9 Mb deletion in 22q13.2->qter in a boy with autistic features, epilepsy, developmental delay, atopic dermatitis and abnormal immunological findings. Eur J Med Genet. 2010;53:329–332. doi: 10.1016/j.ejmg.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Chévere-Torres I, Kaphzan H, Bhattacharya A, Kang A, Maki JM, Gambello MJ, Arbiser JL, Santini E, Klann E. Metabotropic glutamate receptor-dependent long-term depression is impaired due to elevated ERK signaling in the DeltaRG mouse model of tuberous sclerosis complex. Neurobiol Dis. 2012;45:1101–1110. doi: 10.1016/j.nbd.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Zeitlin S. A method for the generation of conditional gene repair mutations in mice. Nucleic Acids Res. 2001;29:E10. doi: 10.1093/nar/29.3.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Südhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafrenière RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, Fombonne E, Joober R, Marineau C, Drapeau P, Rouleau GA. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Gillott A, Furniss F, Walter A. Anxiety in high-functioning children with autism. Autism. 2001;5:277–286. doi: 10.1177/1362361301005003005. [DOI] [PubMed] [Google Scholar]
- Gong X, Jiang YW, Zhang X, An Y, Zhang J, Wu Y, Wang J, Sun Y, Liu Y, Gao X, Shen Y, Wu X, Qiu Z, Jin L, Wu BL, Wang H. High proportion of 22q13 deletions and SHANK3 mutations in Chinese patients with intellectual disability. PLoS One. 2012;7:e34739. doi: 10.1371/journal.pone.0034739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolevzon A, Cai G, Soorya L, Takahashi N, Grodberg D, Kajiwara Y, Willner JP, Tryfon A, Buxbaum JD. Analysis of a purported SHANK3 mutation in a boy with autism: clinical impact of rare variant research in neurodevelopmental disabilities. Brain Res. 2011;1380:98–105. doi: 10.1016/j.brainres.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Kopp S, Beckung E, Gillberg C. Developmental coordination disorder and other motor control problems in girls with autism spectrum disorder and/or attention-deficit/hyperactivity disorder. Res Dev Disabil. 2010;31:350–361. doi: 10.1016/j.ridd.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, Liu S, Jaramillo TC, Bangash M, Xiao B, Worley PF, Powell CM. Loss of predominant shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33:18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács AD, Pearce DA. Location- and sex-specific differences in weight and motor coordination in two commonly used mouse strains. Sci Rep. 2013;3:2116. doi: 10.1038/srep02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK. Synaptic plasticity and phosphorylation. Pharmacol Ther. 2006;112:810–832. doi: 10.1016/j.pharmthera.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins: multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Mahjouri S, Lord CE. What the DSM-5 portends for research, diagnosis, and treatment of autism spectrum disorders. Curr Psychiatry Rep. 2012;14:739–747. doi: 10.1007/s11920-012-0327-2. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR, Crawley JN. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004;3:303–314. doi: 10.1111/j.1601-183X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/S0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Nobile M, Perego P, Piccinini L, Mani E, Rossi A, Bellina M, Molteni M. Further evidence of complex motor dysfunction in drug naive children with autism using automatic motion analysis of gait. Autism. 2011;15:263–283. doi: 10.1177/1362361309356929. [DOI] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, McDermid HE. The 22q13.3 deletion syndrome (Phelan–McDermid syndrome) Mol Syndromol. 2012;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MC. Deletion 22q13.3 syndrome. Orphanet J Rare Dis. 2008;3:14. doi: 10.1186/1750-1172-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Südhof TC, Nestler EJ. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/S0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–547. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasua SM, Dwivedi A, Boccuto L, Rollins JD, Chen CF, Rogers RC, Phelan K, DuPont BR, Collins JS. Association between deletion size and important phenotypes expands the genomic region of interest in Phelan–McDermid syndrome (22q13 deletion syndrome) J Med Genet. 2011;48:761–766. doi: 10.1136/jmedgenet-2011-100225. [DOI] [PubMed] [Google Scholar]
- Schreibman L. Diagnostic features of autism. J Child Neurol. 1988;3(Suppl):S57–S64. doi: 10.1177/0883073888003001s11. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Soorya L, Kolevzon A, Zweifach J, Lim T, Dobry Y, Schwartz L, Frank Y, Wang AT, Cai G, Parkhomenko E, Halpern D, Grodberg D, Angarita B, Willner JP, Yang A, Canitano R, Chaplin W, Betancur C, Buxbaum JD. Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Mol Autism. 2013;4:18. doi: 10.1186/2040-2392-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino S, Waga C. SHANK3 as an autism spectrum disorder-associated gene. Brain Dev. 2013;35:106–110. doi: 10.1016/j.braindev.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Uchino S, Wada H, Honda S, Nakamura Y, Ondo Y, Uchiyama T, Tsutsumi M, Suzuki E, Hirasawa T, Kohsaka S. Direct interaction of post-synaptic density-95/Dlg/ZO-1 domain-containing synaptic molecule Shank3 with GluR1 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor. J Neurochem. 2006;97:1203–1214. doi: 10.1111/j.1471-4159.2006.03831.x. [DOI] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Böckers TM, Dityatev A, Sala C. Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J Biol Chem. 2011;286:34839–34850. doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga C, Okamoto N, Ondo Y, Fukumura-Kato R, Goto Y, Kohsaka S, Uchino S. Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr Genet. 2011;21:208–211. doi: 10.1097/YPG.0b013e328341e069. [DOI] [PubMed] [Google Scholar]
- Wallenstein GV, Eichenbaum H, Hasselmo ME. The hippocampus as an associator of discontiguous events. Trends Neurosci. 1998;21:317–323. doi: 10.1016/S0166-2236(97)01220-4. [DOI] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SW, Ollendick T, Scahill L, Oswald D, Albano AM. Preliminary efficacy of a cognitive-behavioral treatment program for anxious youth with autism spectrum disorders. J Autism Dev Disord. 2009;39:1652–1662. doi: 10.1007/s10803-009-0801-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni ML, Wöhr M, Roullet FI, Katz AM, Abrams DN, Kalikhman D, Simon H, Woldeyohannes L, Zhang JY, Harris MJ, Saxena R, Silverman JL, Buxbaum JD, Crawley JN. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3-null mutant mice. J Neurosci. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, Bowman R, Yang H, Goldstein J, Li YJ, Jiang YH. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum Mol Genet. 2014;23:1563–1578. doi: 10.1093/hmg/ddt547. [DOI] [PMC free article] [PubMed] [Google Scholar]