Abstract
The recent discovery of diverse very large viruses, such as the mimivirus, has fostered a profusion of hypotheses positing that these viruses define a new domain of life together with the three cellular ones (Archaea, Bacteria and Eucarya). It has also been speculated that they have played a key role in the origin of eukaryotes as donors of important genes or even as the structures at the origin of the nucleus. Thanks to the increasing availability of genome sequences for these giant viruses, those hypotheses are amenable to testing via comparative genomic and phylogenetic analyses. This task is made very difficult by the high evolutionary rate of viruses, which induces phylogenetic artefacts, such as long branch attraction, when inadequate methods are applied. It can be demonstrated that phylogenetic trees supporting viruses as a fourth domain of life are artefactual. In most cases, the presence of homologues of cellular genes in viruses is best explained by recurrent horizontal gene transfer from cellular hosts to their infecting viruses and not the opposite. Today, there is no solid evidence for the existence of a viral domain of life or for a significant implication of viruses in the origin of the cellular domains.
Keywords: giant viruses, domain of life, long branch attraction, horizontal gene transfer, mimivirus, origin of eukaryotes
1. Introduction
‘The human understanding when it has once adopted an opinion (either as being the received opinion or as being agreeable to itself) draws all things else to support and agree with it. And though there be a greater number and weight of instances to be found on the other side, yet these it either neglects and despises, or else by some distinction sets aside and rejects; in order that by this great and pernicious predetermination the authority of its former conclusions may remain inviolate’.
Francis Bacon (aphorism XLVI from the first book of the Novum Organum [1])
All along the history of biology, scientists have divided the diversity of living beings into a number of discrete major groups that typically received the name of ‘kingdoms’. The first to be recognized were animals and plants (Regnums Animale and Vegetabile in terms of Linnaeus' classification [2]), but others were added subsequently, such as the Protista and Monera by Haeckel [3] and the Fungi by Whittaker in his famous ‘five kingdoms’ classification [4]. These traditional classifications were based on the comparison of phenotypic characters, which can be very problematic for this kind of large-scale analyses as the homology of characters can be difficult to be established for the different taxa and they can also be prone to homoplasy. These limitations are particularly severe in the case of microorganisms, especially for the smallest ones—the bacteria—to the point that as recently as 50 years ago a group of very prominent microbiologists declared that ‘it is a waste of time to attempt a natural system of classification for bacteria … bacteriologists should concentrate instead on the more humble practical task of devising determinative keys to provide the easiest possible identification of species and genera’ [5].
This pessimistic situation changed two decades later, thanks to the development of molecular phylogeny, which finds its grounds in the theoretical work of Zuckerkandl and Pauling, who proposed that the sequences of biological macromolecules (proteins and nucleic acids) contained evolutionary information [6]. The first comprehensive molecular phylogenies including macro- and microorganisms were reconstructed by Woese and Fox [7] in the late 1970s. At that time, the general idea was that life could be divided into two major groups according to basic characteristics of the cellular organization: prokaryotes (a synonym of bacteria) and eukaryotes [8]. However, instead of the expected separation between these two groups, Woese's molecular phylogenetic trees showed the deep divergence of three lineages [7]. On the one hand, the eukaryotes and, on the other hand, two very distant groups of prokaryotic microorganisms, the classical bacteria and a group of species found mostly in extreme habitats. These three groups were first considered to be ‘primary kingdoms’ (Urkaryotes, Eubacteria and Archaebacteria) [7] and later reclassified at the rank of domains (which were dubbed Eucarya, Bacteria and Archaea) [9]. This is probably the most important discovery in the still-short history of molecular phylogenetics. The evolutionary relationships among the three domains remain controversial, in particular in what concerns the origin of eukaryotes [10–12], but their distinctness has been corroborated by the analysis of many phylogenetic markers and complete genome sequences.
2. The elusive fourth domain of life
In the same unexpected way as the archaea were recognized to be one of the primary lineages of life, it is possible to ask a very basic question: do other domains exist? This question can be addressed in different ways. One is by the study of the diversity of living beings in different environments to see whether organisms outside the three domains can be found or not. Today, this type of study greatly benefits from the development of molecular tools that allow a very extensive census of the biodiversity present in environmental samples [13]. Among those techniques, metagenomic analysis using next-generation sequencing technologies is probably the most powerful, as it produces sequences of the whole set of genes of the community under study. Those sequences can then be assigned to specific taxonomic groups based on sequence similarity or phylogenetic analysis, making it possible to assess the taxonomic composition of the community. A recurrent observation in all metagenomes sequenced up to now is the presence of a more or less abundant fraction of sequences very divergent from those of known species or even without any homologue in sequence databases (ORFans) [14]. The origin of these sequences is enigmatic though many of them most likely belong to viruses, which remain largely unknown in most environments [15–18].
It has been much more rarely speculated that some of the divergent sequences retrieved in metagenomic studies may belong to representatives of an unknown number of yet-to-discover domains of life. One example is the identification of divergent sequences of some highly conserved housekeeping protein families (e.g. RecA and RpoB) in the global ocean sampling metagenome data, which occupied a deep-branching position in phylogenetic trees [19]. The possibility that some of these divergent sequences represent divergent paralogues or putative natural chimaeric sequences originated from recombination between distant organisms was considered as very unlikely. The authors favoured the alternative hypotheses that they could belong to new viral lineages or to new domain(s) of life [19]. However, in the same analysis, they failed to observe similar results for the phylogenetic marker par excellence, the small subunit ribosomal RNA (SSU rRNA). This suggests that those divergent protein sequences most likely belong to viral lineages rather than to new cellular domains, for which we should expect to find also the corresponding divergent SSU rRNA sequences.
Besides this infrequent approach, an idea that has gained much more popularity is that certain viruses should be considered as independent lineages in the tree of life, namely new domains of life. This hypothesis rose with the discovery of a new type of viruses characterized by their large size (comparable to that of small prokaryotic cells) and genomes much bigger than those of the classical viruses previously known. The first to be described was the mimivirus [20], and the analysis of its huge genome (1.2 Mbp, the largest characterized in a virus until then) unveiled an unprecedented number of genes involved in transcription and translation homologous to those of cellular organisms [21]. Even more surprising, the phylogenetic analysis of a concatenation of the sequences of seven of those genes appeared to support that the mimivirus might represent a fourth domain of life, sister to the eukaryotes [21]. This virus belongs to the nucleocytoplasmic large DNA viruses family, which was known to contain a large variety of viral lineages, including some of large sizes such as the Phycodnaviridae, but none as big as the mimivirus [22]. Nevertheless, its size record held for just a short time, as other, even bigger, giant viruses were soon discovered [23]. Today, three main lineages of giant viruses are known: Mimiviridae [21,23–25], pithovirus [26] and Pandoraviridae [27]. The latter have the largest genomes, up to 2.77 Mbp [27], but all of them have genomes of more than 500 kbp. In all cases, these genomes are composed of a large amount of ORFans accompanied by a relatively small fraction of genes with homologues in other viruses and/or in cellular organisms.
A few of these genes with cellular homologues have been used to try to place giant viruses in the tree of life. As mentioned above, the first attempt was done using seven mimivirus genes and resulted in a phylogenetic tree where this virus branched as a deeply diverging lineage sister to the eukaryotes [21]. Thus, it appeared that the elusive fourth domain of life had been found and that it was composed of a variety of giant viruses [28–31].
3. Magical viruses?
The discovery of giant viruses and their enigmatic phylogenetic position attracted much scientific attention and, incidentally, served to revive more or less old ideas about the role of viruses in early evolution [32,33]. In parallel with the immense recent improvement of the scientific knowledge on the diversity of viruses infecting members of the three domains of life, speculations on viruses depicted as creative evolutionary agents at the origin of essential traits of cellular organisms have flourished. Among many others, they include viruses as the ‘inventors’ of DNA [34] and viruses at the origin of the eukaryotic nucleus [35,36]. A common point to most of these hypotheses is the absence of clear mechanistic details explaining how the supposed viral inventions were adopted by cellular organisms or gave rise to stable complex cellular structures, such as the nucleus. In fact, in many cases, these hypotheses simply try to provide ad hoc answers to complex evolutionary questions by appealing to completely hypothetical resourceful capacities of viruses (envisaged as multipotent creative agents). An additional problem with these models is the systematic confusion between homology and analogy. This often leads to very unparsimonious proposals.
For example, let us examine the hypothesis that viruses have ‘invented’ DNA. This idea originated from a puzzling observation derived from the first comparative genomic analyses of members of the three domains of life. Bacteria were shown to be endowed with a DNA replication system very different from those of archaea and eukaryotes, as many proteins of the bacterial replication machinery lack homologues in the two other domains [37]. Some authors have speculated that this disparity suggests that the last common ancestor of all living beings had not a DNA but an RNA genome and that DNA evolved twice independently (once in the bacteria and a second time in a lineage leading to archaea and eukaryotes) [38]. It was already known that the RNA polymerase of most mitochondria (organelles derived from ancient endosymbiotic bacteria) was radically different from that of bacteria. It resembled those found in several phages and plasmids, suggesting that mitochondria replaced the original bacterial RNA polymerase by a viral one in all eukaryotes with the exception of the Jakobidae [39]. This was also the case for the mitochondrial DNA primase, probably derived from a T-odd bacteriophage [40]. Some authors were tempted to generalize these observations proposing that the cellular machineries for DNA replication and nucleotide synthesis also evolved first in viruses and were subsequently transferred into the cellular lineages.
This was first claimed for the bacteria [41] and then extended to the eukaryotes [42] and, more recently, to the three domains of life [43]. Thus, the most recent hypothesis of this type posits that the ancestors of the three domains were cells with RNA genomes and that they gained DNA by three independent acquisitions from DNA viruses [43]. However, such a hypothesis has numerous drawbacks. For example, RNA genomes have strong size constraints due to the low fidelity of RNA-dependent RNA polymerases (even those with proofreading activity) which entails a high error rate that cannot be tolerated by big genomes as they would accumulate too many mutations per replication cycle [44]. This is the reason why all RNA viruses have small genomes with a capacity to code for just a few dozen genes [45]. However, using different comparative genomics approaches, it has been estimated that the ancestors of the three domains of life and even their last common ancestor had genomes containing several hundred genes [46]. Such a gene number requires a genome size far beyond the maximum size of RNA genomes. A second important drawback of the ‘viral origin of DNA’ hypothesis is that no known virus has a complete set of genes for all the activities necessary for the synthesis of DNA building blocks and for DNA replication, as all DNA viruses rely partially or totally on their hosts for those activities. It is thus difficult to imagine that three different viruses would have furnished complete DNA nucleotide synthesis and replication machineries to three different RNA cells ancestral to each one of the cellular domains. Moreover, phylogenetic analysis of the genes involved in these activities that were supposedly transferred from the viruses to the cells were shown to have actually followed the opposite way, namely to have been transferred from the cellular hosts to their infecting viruses [47,48]. Finally, it is also hard to envisage that once a cellular lineage had acquired DNA (with all the competitive advantages that DNA provides, including high genome stability and the possibility to have large genome sizes), it did not outcompete all the other, less fit, RNA genome-based lineages. For those genomic and ecological reasons, the hypothesis of three independent DNA acquisitions by the ancestors of the three life domains from viral donors is completely unrealistic.
The same type of criticisms can be addressed to the propositions of a viral origin of the eukaryotic nucleus, which are based on the fact that some large DNA viruses and the nucleus share similarities such as linear chromosomes, mRNA capping and the separation of transcription from translation [35,36]. However, those viruses do not encode the components that build the nuclear membranes or any other nuclear feature. Viruses endowed with a lipid envelope acquire it from the membrane host during viral release [49]. The superficial overall resemblances between the macromolecular complexes formed within the infected cells during the replication of the viruses, known as ‘viral factories’ [50], and bona fide eukaryotic nuclei do not reflect actual homology. This is a similar case to that of the intracellular compartments found in bacterial species of the PVC (Planctomycetes, Verrucomicrobia and Chlamydiales) group, which superficially resemble eukaryotic nuclei, but that have been shown by structural and phylogenetic analysis to be just analogous and not truly homologous structures [51].
These examples show how the hypotheses for the viral origin of revolutionary cellular innovations can, in many cases, be tested (and falsified) by the application of different analyses, in particular molecular phylogeny.
4. Giant viruses, horizontal gene transfer and long branch attraction
Despite the great interest that the discovery of giant viruses attracted in the scientific community and the mass media, it was soon realized that their true phylogenetic status was not a trivial question. The reanalysis of the seven markers that were used to place the mimivirus in the tree of life demonstrated that the ‘fourth domain topology’ initially retrieved was heavily artefactual [52]. There were three reasons for that. The first and most important was that among the markers used (arginyl-, methionyl- and tyrosyl-tRNA synthetases, RNA polymerase II subunits, DNA polymerase sliding clamp protein and 5′–3′ exonuclease), there were some, in particular the three aminoacyl-tRNA synthetases, which have experienced horizontal gene transfer (HGT) events between very distant lineages (even between different domains). This was clearly shown by a gene-by-gene analysis of these markers. For example, the three aminoacyl-tRNA synthetases of one of the species used in the original tree, the well-known gammaproteobacterium Escherichia coli, actually had three different evolutionary origins (bacterial, archaeal and eukaryotic) because of recurrent HGT. Obviously, markers that have been subjected to high HGT levels cannot be analysed together in a multi-marker concatenation as this can only lead to distorted results. Besides the detection of those HGTs, another interesting observation from the gene-by-gene analysis was that mimivirus did no longer emerge as sister group of the eukaryotes but within the eukaryotic domain, in some cases close to amoebal species [52]. Mimivirus is a parasite of amoebas, so these results argued for HGT from the host to the parasite, a common phenomenon observed in many other host–parasite systems, as the mechanism to explain the presence of these genes in the mimivirus' genome. In addition, these observations invalidated the mimivirus as a potential fourth domain of life, because the markers used to support it were inadequate.
In addition to HGT, another factor that undermined the initial phylogenetic analysis was the poor taxonomic sampling. In the original analysis, each domain of life was represented by only three species and, more importantly, representatives of the mimivirus' host group, the amoebas, were missing. Not including host genes can be misleading when studying the evolution of parasites, because host-to-parasite HGT is a frequent phenomenon. The third factor that makes the phylogenetic analysis of viruses a very delicate matter is their high evolutionary rate, with both high mutation and recombination frequencies [53–57]. Viral sequences tend to evolve rapidly and, when they have cellular homologues, they can be very divergent. This divergence becomes visible in phylogenetic trees in the form of long branches. Therefore, because viral sequences evolve fast, they are very prone to be affected by a very well-known phylogenetic reconstruction artefact, the long branch attraction (LBA) described by Felsenstein in the late 1970s [58]. It is also well known that the use of poor taxonomic sampling, simplistic phylogenetic reconstruction methods (such as the non-probabilistic distance- or parsimony-based ones) and/or inadequate substitution models can exacerbate LBA problems [59–64]. Given their high evolutionary rates, viruses are ideal candidates to get trapped in what has been called the ‘Felsenstein zone’, namely the conditions where long branches are misplaced in phylogenetic trees [65]. Thus, if inappropriately analysed, viral sequences most often branch in wrong places, very frequently in basal positions in rooted trees as the outgroups define long branches that artefactually attract those of the viral sequences. In this way, viruses tend to branch far from the slow-evolving sequences, close to the base or at mid-point locations of the trees instead of branching at their true position (e.g. close to their hosts in the case of genes that have experienced host-to-virus HGT).
Several studies have proved empirically that the phylogenetic analyses used to claim the ‘fourth domain of life’ status of giant viruses were artefactual. This was the case for the mimivirus seven-gene phylogeny described above [52], but also for other more recent analyses. For example, a viral fourth domain sister to the eukaryotes has been proposed based on the phylogeny of clamp loader proteins [66] and of the RNA polymerase II (RNAP2), transcription factor II beta, flap endonuclease and proliferating cell nuclear antigen [28]. As in the previous cases, subsequent analyses of those datasets using more robust methods and sequence evolution models as well as more comprehensive taxonomic samplings demonstrated once again that the giant virus sequences were misplaced in the original analyses because of their high evolutionary rate and/or compositional bias and that, actually, these genes had most likely been acquired by the viruses from eukaryotic donors [67,68]. In §5, we revisit one of these examples in more detail.
5. Giant viruses caught in the Felsenstein's trap
In their phylogenetic analysis of the RNAP2 sequences including 80 sequences, giant virus genes appeared at the base of the eukaryotic branch so that Boyer et al. [28] concluded that giant viruses constitute a fourth domain of life, sister to the eukaryotes. This initial analysis was done using relatively simple methods: approximate maximum-likelihood (ML) phylogenetic reconstruction with the single-matrix JTT model (see fig. 2 in [28]). In a detailed reanalysis of this marker, Williams et al. [68] demonstrated that the RNAP2 sequence dataset contained a substantial amount of non-phylogenetic signal, mainly owing to the high evolutionary rate and compositional bias of the viral sequences. They also showed the very poor fit between the sequence data and the JTT model and verified that more complex models exhibited significantly better fit. This was especially the case for the non-homogeneous models tested (UL3, CAT10 and CAT60). Interestingly, phylogenetic trees reconstructed with the same RNAP2 sequence alignment used by Boyer et al. but applying Bayesian inference and any of the above site-heterogeneous mixture models did not support the ‘four domains’ topology. Instead, giant viruses no longer formed a monophyletic group and part of them were located with very long branches within the eukaryotic group suggesting a host-to-virus HGT (see fig. 1 in [68]).
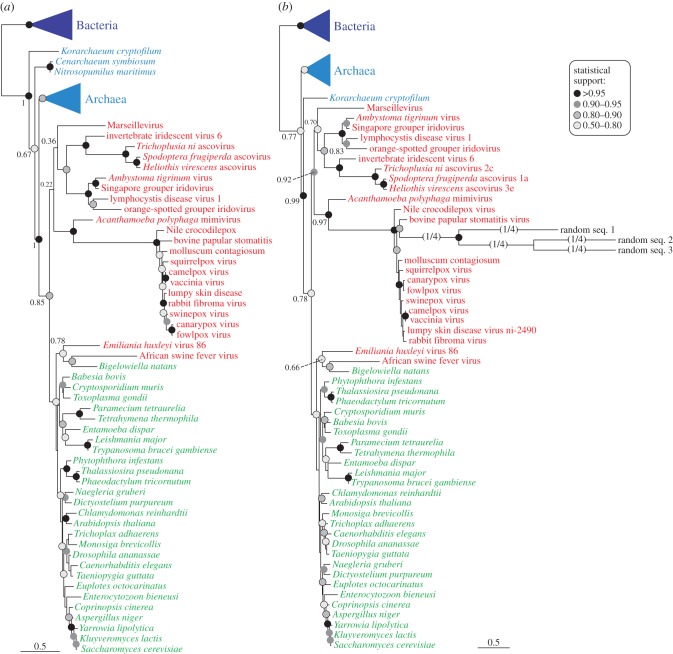
Whereas Williams et al. [68] focused mostly on the impact of using unfit models, we have further investigated other aspects of this sequence dataset as a case study showing the difficulties that the highly divergent viral sequences may generate in phylogenetic analyses. Our first objective was assessing to what extent the RNAP2 viral sequences were divergent and could be affected by LBA. We first tested if they were significantly different from the cellular homologues in terms of amino acid composition. Our test (see the electronic supplementary material) indicated that all the viral sequences had significant compositional bias, which confirmed that they contained a substantial amount of non-phylogenetic signal, as already stated by Williams et al. [68] by means of a homoplasy analysis. This represents a major reason to avoid the use of simple homogeneous models of sequence evolution like the JTT one used by Boyer et al. [28]. We also investigated empirically the possible LBA behaviour of the viral sequences with a classical test based on the use of artificial random sequences. If LBA is at play, random sequences are expected to cluster with the longest branch in a phylogenetic tree [69,70]. To test this, we used the original RNAP2 dataset of Boyer et al. [28] made available by Williams et al. [68]. As in these previous analyses, a phylogenetic tree reconstructed using approximate ML with the JTT model showed the emergence of most viral sequences within a monophyletic group sister to the eukaryotes (figure 1a) though with a statistical support much weaker than the one obtained by Boyer et al. (SH-like local support of 0.22 versus 0.82, respectively). We constructed a set of random sequences with similar amino acid composition to that of those viruses. When included in the phylogenetic analysis (always by approximate ML with the JTT model), either individually or in groups of different size, they invariably branched within the viral clade, which in all cases received a higher statistical support than the initial one without the random sequences (e.g. 0.92 in a tree including three random sequences; figure 1b).
Figure 1.
Phylogenetic trees of RNAP2 sequences calculated by approximated maximum-likelihood with the JTT model based on the sequence dataset of Boyer et al. [28]. (a) Tree reconstructed with 80 taxa and 272 amino acid positions. (b) Tree with the same taxa as in (a) with the inclusion of three random sequences. Statistical support (SH-like local support values) is indicated by filled circles (only values >0.50), except for several important nodes where the actual value is provided. Some branches have been shortened to 1/4 of their actual length (indicated by 1/4). The bar represents 0.5 substitutions per site. Bacteria and archaea are in blue, eukaryotes in green, and viruses in red. For the complete trees see supplementary material, figures S1 and S2.
The experiment of random sequence addition allows extraction of several conclusions. First, the viral sequences were the fastest evolving taxa in the dataset (especially the Poxviridae group, the one with the longest branches), as they strongly attracted the random sequences. Second, the whole group of viral sequences was most likely affected by an LBA artefact, as the inclusion of the random sequences (which increase the noise, but not the phylogenetic signal) always led to higher statistical support for the ‘four domains’ topology (i.e. the monophyly of the viral sequences as an independent group). These results confirm the necessity of using tree reconstruction methods and sequence evolution models apt to deal with this type of highly divergent sequences [67,68]. Thus, we carried out all subsequent analyses applying Bayesian inference with the non-homogeneous sequence evolution model CAT [71].
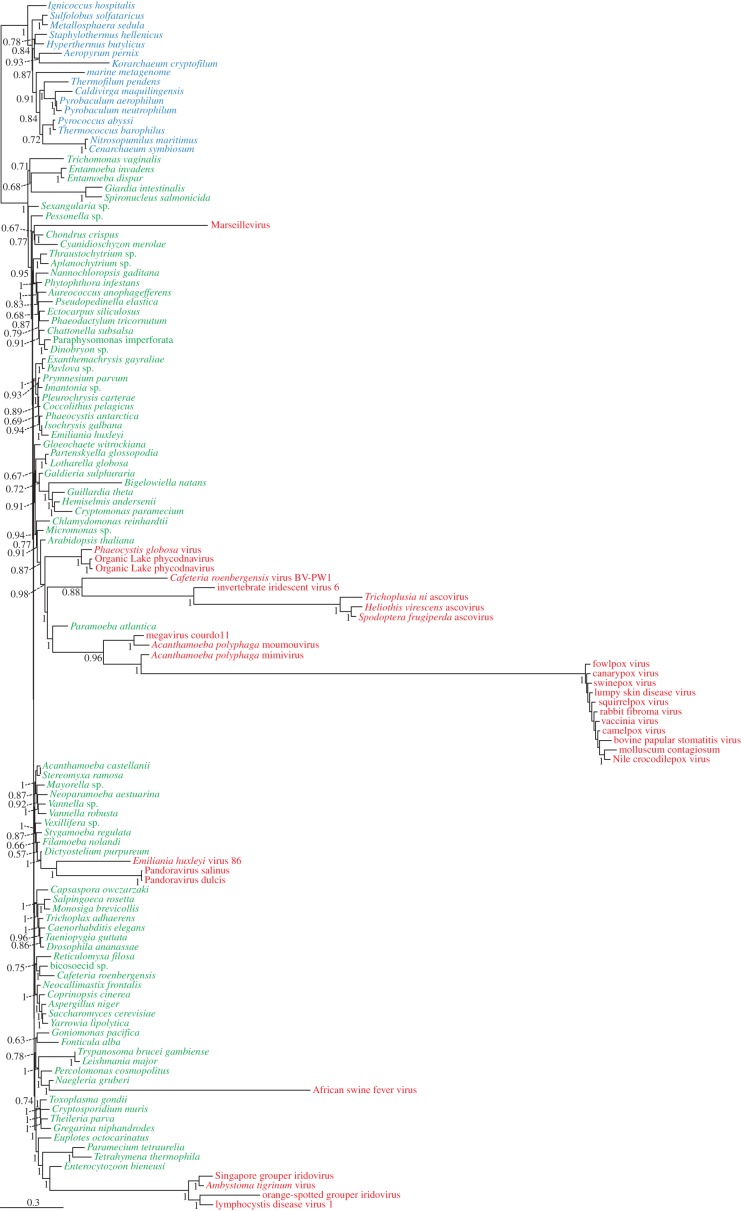
Once we had made evident the fast-evolving nature of the viral sequences and the occurrence of LBA artefacts, we explored taxonomic sampling, an additional factor that, unfortunately, is very often improperly treated in many phylogenetic analyses dealing with viral sequences. It is well known that poor taxonomic sampling may lead to infer spurious phylogenetic relationships [72]. This problem has already been disclosed in the case of phylogenetic analyses that included viruses but not their hosts [21,66], which prevented the accurate detection of host-to-virus HGT events [52,67]. In our example of the RNAP2, the published original analyses appeared to support that the viral sequences were more closely related to the eukaryotic homologues than to the prokaryotic ones. We thus enriched the taxonomic sampling for this marker (up to 127 taxa) with sequences of several large eukaryotic groups that were missing in the original dataset, as well as with a few additional sequences of giant viruses (such as pandoravirus and megavirus). Because the eukaryotic and viral RNAP2 sequences were more similar to the archaeal than to the bacterial ones, we eliminated the latter to avoid the use of very distant outgroup sequences, which is known to intensify LBA problems [61,69,73]. In addition, even when applying very strict site selection criteria (see the electronic supplementary material), the removal of the bacterial sequences allowed increasing substantially the number of conserved alignment positions that could be kept for subsequent phylogenetic inference (from 272 to 427 amino acid positions), thus considerably augmenting the potential amount of phylogenetic signal in our dataset. This improved dataset was submitted to Bayesian phylogenetic analysis with the CAT model. Instead of the ‘four domains’ topology of the initial analyses [28], the resulting tree did not support the monophyly of viral sequences as sister group to the eukaryotes but the emergence of these sequences as very long branches within the eukaryotes (figure 2). This result was already partially found by Williams et al. [68] but in their tree a group of fast-evolving viruses (Marseillevirus, Iridoviridae and Ascoviridae) still emerged at the base of a eukaryotes + viruses group. The use of a closer outgroup (archaea instead of bacteria + archaea), the inclusion of more informative sites and a larger taxonomic sampling allowed us to partially overcome the LBA affecting the very fast-evolving viruses and place all them within the eukaryotes.
Figure 2.
Phylogenetic tree of RNAP2 sequences calculated by Bayesian inference with the CAT model (127 taxa and 427 amino acid positions). Archaea are in blue, eukaryotes in green and viruses in red. Numbers at branches are posterior probabilities. The bar represents 0.3 substitutions per site.
In conclusion, contrary to the original claim that these very divergent viral sequences support the ‘four domains’ hypothesis, the RNAP2, if properly analysed, offers yet another good example that these viruses acquired this gene from their eukaryotic hosts by HGT, as is the case for many other genes shared by viruses and cells [48,52,67,74–79].
6. Conclusions: the vanishing ‘fourth domain’
The passage from a bipartite (prokaryotes versus eukaryotes) to a tripartite (archaea, bacteria and eukaryotes) view of the evolutionary structure of biodiversity is one of the major scientific achievements of the past century [80]. It paved the way for the subsequent discovery that most likely only two primary domains (both prokaryotic, archaea and bacteria) exist [81,82]. The unforeseen discovery of a third domain of life (the archaea) also gave credibility to the possibility that other domains might have gone unnoted too. Different claims for the finding of such additional domains have been published but, as discussed above, the most popular ones arose as a consequence of the discovery of giant viruses. These viruses were considered not only to define a fourth domain of life, but also to have played an important role in the origin of eukaryotes [21,83,84]. However, those conjectures were very often based on rather naive phylogenetic analyses that did not take into account the technical difficulties inherent to working with fast-evolving viral sequences. In fact, whereas some topics concerning viruses (such as their living or non-living nature [33,85–87]) are matter of open speculation and debate, their possible place in the tree of life can be tested by applying rigorous phylogenetic methods. As mentioned above, most claims for the ‘fourth domain of life’ status of viruses were based on simplistic phylogenetic analyses, which ignored the abundant compelling evidence for high evolutionary rate and compositional bias of viral sequences that result in long branches in phylogenetic trees. These characteristics make viral sequences ideal victims for the LBA artefact, leading to flawed phylogenetic trees where viruses branch as an independent group. In addition to the multiple examples of this problem published in recent years [52,67,68], we have presented here an additional case study showing how the application of robust phylogenetic reconstruction methods can alleviate LBA artefacts and retrieve the correct place of the fast-evolving viral sequences.
The results of more accurate phylogenetic analyses have led to systematic rejection of all the re-analysed ‘four domains’ trees. For all of them, host-to-virus HGT events have been demonstrated. Thus, there is no credible phylogenetic evidence nowadays supporting the existence of a fourth domain of life. In this regard, it is noteworthy that some fervent advocates of that hypothesis [21] have radically changed their views to even deny the existence of a tree of life and, consequently, the placement of viruses within it [88]. Likewise, the contribution of viruses to the evolution of fundamental characteristics of cellular organisms remains at best highly speculative (e.g. the origin of cell walls to protect from viral infection [89]) or, when submitted to phylogenetic analysis, simply unsupported (e.g. the viral origin of the DNA replication machinery of bacteria [47]). Several authors have misinterpreted this kind of refutation as equivalent to a radical negation of any role of viruses in cell evolution [66,90]. However, it is important to stress that excluding viruses from the tree of life based on phylogenetic tests does not preclude the major role that viruses have played and still play in the evolution of cellular organisms acting as a strong and dynamic selective pressure on their hosts and fostering a permanent arms race [91]. Viruses are also important actors in cellular evolution by serving as vehicles for gene transfer and as accelerators of gene evolutionary rate (‘mutators’), which may lead to punctual innovations that can be returned back to cells. Some such cases have been convincingly identified, mostly related to organellar nucleic acid metabolism (e.g. mitochondrial RNA polymerase [39] and DNA primase [40]), but, in general, they concern the replacement of activities already present in cells by their viral counterparts rather than the import of truly new functions. Thus, the claimed viral role as the creators of major cellular traits has most likely been overestimated as a consequence of improper (or simply absent) phylogenetic analysis [33,87]. Essential eukaryotic structures such as the nucleus and the nuclear pore or the complex endomembrane system of eukaryotic cells remain fundamentally unexplained by any viral-based model for the origin of eukaryotes.
Despite one decade of phylogeny-based refutation of the fourth domain of life and of several ad hoc hypotheses for the viral origin of essential cellular features, strict phylogenetic criteria remain very often ignored in viral evolutionary studies and those claims are still published [30,92,93]. This evokes what Bacon reproved four centuries ago: ‘The human understanding when it has once adopted an opinion (either as being the received opinion or as being agreeable to itself) draws all things else to support and agree with it’ [1]. In the specific case of the question of the origin of eukaryotes, the incorporation of viruses as a speculative sister domain to eukaryotes or as the origin of the nucleus has not supplied any explanatory power that classical hypotheses based on the interaction of cellular organisms (archaea and bacteria) had not been able to provide already [10,94–97]. Thus, the application of Occam's razor makes it unparsimonious to complicate the hypotheses for the origin of eukaryotes by including viruses in the current absence of mechanistic plausibility and compelling phylogenetic evidence.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the Editors of this special issue of Philosophical Transactions B for the invitation to contribute this article.
Data accessibility
RNAP2 sequence alignments are available in the electronic supplementary material.
Funding
This work was supported by the French CNRS and the European Research Council (under the European Union's Seventh Framework Programme ERC grant agreement no. 322669 ‘ProtistWorld’).
Competing interests
We declare we have no competing interests.
References
- 1.Bacon F. 1620. The new Organon. Aphorisms concerning the interpretation of nature and the kingdom of man. Venice, Italy: Typis Gasparis Gerardi. [Google Scholar]
- 2.Linnaeus C. 1735. Systema naturæ, sive regna tria naturæ systematice proposita per classes, ordines, genera, & species, 12 p. Leiden: Haak. [Google Scholar]
- 3.Haeckel E. 1866. Generelle Morphologie der Organismen: Allgemeine Grundzüge der organischen Formen-Wissenschaft, mechanisch begründet durch die von Charles Darwin reformirte Descendenz-Theorie, 574 p. Berlin: Georg Reimer. [Google Scholar]
- 4.Whittaker RH. 1969. New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science 163, 150–160. ( 10.1126/science.163.3863.150) [DOI] [PubMed] [Google Scholar]
- 5.Stanier RY, Doudoroff M, Adelberg EA. 1957. The microbial world. Englewood Cliffs, NJ: Prentice-Hall Inc. [Google Scholar]
- 6.Zuckerkandl E, Pauling L. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8, 357–366. ( 10.1016/0022-5193(65)90083-4) [DOI] [PubMed] [Google Scholar]
- 7.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA 74, 5088–5090. ( 10.1073/pnas.74.11.5088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanier RY, Van Niel CB. 1962. The concept of a bacterium. Arch. Mikrobiol. 42, 17–35. ( 10.1007/BF00425185) [DOI] [PubMed] [Google Scholar]
- 9.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579. ( 10.1073/pnas.87.12.4576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Embley TM, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440, 623–630. ( 10.1038/nature04546) [DOI] [PubMed] [Google Scholar]
- 11.Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. 2010. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat. Rev. Microbiol. 8, 743–752. ( 10.1038/nrmicro2426) [DOI] [PubMed] [Google Scholar]
- 12.Lombard J, Lopez-Garcia P, Moreira D. 2012. The early evolution of lipid membranes and the three domains of life. Nat. Rev. Microbiol. 10, 507–515. ( 10.1038/nrmicro2815) [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Garcia P, Moreira D. 2008. Tracking microbial biodiversity through molecular and genomic ecology. Res. Microbiol. 159, 67–73. ( 10.1016/j.resmic.2007.11.019) [DOI] [PubMed] [Google Scholar]
- 14.Yooseph S, et al. 2007. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 5, e16 ( 10.1371/journal.pbio.0050016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3, 504–510. ( 10.1038/nrmicro1163) [DOI] [PubMed] [Google Scholar]
- 16.Yin Y, Fischer D. 2006. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol. Biol. 6, 63 ( 10.1186/1471-2148-6-63) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mokili JL, Rohwer F, Dutilh BE. 2012. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2, 63–77. ( 10.1016/j.coviro.2011.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. 2013. Expanding the marine virosphere using metagenomics. PLoS Genet. 9, 12 ( 10.1371/journal.pgen.1003987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, Frazier M, Venter JC, Eisen JA. 2011. Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS ONE 6, 0018011 ( 10.1371/journal.pone.0018011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299, 2033 ( 10.1126/science.1081867) [DOI] [PubMed] [Google Scholar]
- 21.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306, 1344–1350. ( 10.1126/science.1101485) [DOI] [PubMed] [Google Scholar]
- 22.Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 20, 20. [DOI] [PubMed] [Google Scholar]
- 23.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl Acad. Sci. USA 108, 17 486–17 491. ( 10.1073/pnas.1110889108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colson P, Yutin N, Shabalina SA, Robert C, Fournous G, La Scola B, Raoult D, Koonin EV. 2011. Viruses with more than 1,000 genes: mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of mimivirus genes. Genome Biol. Evol. 3, 737–742. ( 10.1093/gbe/evr048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoosuf N, et al. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol. Evol. 4, 1324–1330. ( 10.1093/gbe/evs109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legendre M, et al. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl Acad. Sci. USA 111, 4274–4279. ( 10.1073/pnas.1320670111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippe N, et al. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286. ( 10.1126/science.1239181) [DOI] [PubMed] [Google Scholar]
- 28.Boyer M, Madoui MA, Gimenez G, La Scola B, Raoult D. 2010. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4th domain of life including giant viruses. PLoS ONE 5, e15530 ( 10.1371/journal.pone.0015530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colson P, Gimenez G, Boyer M, Fournous G, Raoult D. 2011. The giant Cafeteria roenbergensis virus that infects a widespread marine phagocytic protist is a new member of the fourth domain of life. PLoS ONE 6, e18935 ( 10.1371/journal.pone.0018935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colson P, de Lamballerie X, Fournous G, Raoult D. 2012. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology 55, 321–332. ( 10.1159/000336562) [DOI] [PubMed] [Google Scholar]
- 31.Legendre M, Arslan D, Abergel C, Claverie JM. 2012. Genomics of megavirus and the elusive fourth domain of life. Commun. Integr. Biol. 5, 102–106. ( 10.4161/cib.18624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-García P. 2012. The place of viruses in biology in light of the metabolism-versus-replication-first debate. Hist. Philos. Lifg Sci. 34, 391–406. [PubMed] [Google Scholar]
- 33.López-García P, Moreira D. 2012. Viruses in biology. Evo Educ. Outreach 5, 389–398. ( 10.1007/s12052-012-0441-y) [DOI] [Google Scholar]
- 34.Forterre P. 2002. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 5, 525–532. ( 10.1016/S1369-5274(02)00360-0) [DOI] [PubMed] [Google Scholar]
- 35.Bell PJ. 2001. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? J. Mol. Evol. 53, 251–256. ( 10.1007/s002390010215) [DOI] [PubMed] [Google Scholar]
- 36.Takemura M. 2001. Poxviruses and the origin of the eukaryotic nucleus. J. Mol. Evol. 52, 419–425. [DOI] [PubMed] [Google Scholar]
- 37.Olsen GJ, Woese CR. 1996. Lessons from an archaeal genome: what are we learning from Methanococcus jannaschii? Trends Genet. 12, 377–379. ( 10.1016/0168-9525(96)30092-9) [DOI] [PubMed] [Google Scholar]
- 38.Mushegian AR, Koonin EV. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA 93, 10 268–10 273. ( 10.1073/pnas.93.19.10268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gray MW, Lang BF. 1998. Transcription in chloroplasts and mitochondria: a tale of two polymerases. Trends Microbiol. 6, 1–3. ( 10.1016/S0966-842X(97)01182-7) [DOI] [PubMed] [Google Scholar]
- 40.Shutt TE, Gray MW. 2006. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 62, 588–599. ( 10.1007/s00239-005-0162-8) [DOI] [PubMed] [Google Scholar]
- 41.Forterre P. 1999. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33, 457–465. ( 10.1046/j.1365-2958.1999.01497.x) [DOI] [PubMed] [Google Scholar]
- 42.Villarreal LP, DeFilippis VR. 2000. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 74, 7079–7084. ( 10.1128/JVI.74.15.7079-7084.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forterre P. 2006. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc. Natl Acad. Sci. USA 103, 3669–3674. ( 10.1073/pnas.0510333103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmes EC. 2003. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 11, 543–546. ( 10.1016/j.tim.2003.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eigen M. 1971. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523. ( 10.1007/BF00623322) [DOI] [PubMed] [Google Scholar]
- 46.Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136. ( 10.1038/nrmicro751) [DOI] [PubMed] [Google Scholar]
- 47.Moreira D. 2000. Multiple independent horizontal transfers of informational genes from bacteria to plasmids and phages: implications for the origin of bacterial replication machinery. Mol. Microbiol. 35, 1–5. ( 10.1046/j.1365-2958.2000.01692.x) [DOI] [PubMed] [Google Scholar]
- 48.Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol. J. 9, 161 ( 10.1186/1743-422X-9-161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahy BWJ, Van Regenmortel MHV. 2009. Desk encyclopedia of general virology, 672 p. Oxford, UK: Elsevier. [Google Scholar]
- 50.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97, 147–172. ( 10.1042/BC20040058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McInerney JO, Martin WF, Koonin EV, Allen JF, Galperin MY, Lane N, Archibald JM, Embley TM. 2011. Planctomycetes and eukaryotes: a case of analogy not homology. Bioessays 33, 810–817. ( 10.1002/bies.201100045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreira D, López-García P. 2005. Comment on ‘The 1.2-megabase genome sequence of Mimivirus’. Science 308, 1114 ( 10.1126/science.1110820) [DOI] [PubMed] [Google Scholar]
- 53.Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148, 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Awadalla P. 2003. The evolutionary genomics of pathogen recombination. Nat. Rev. Genet. 4, 50–60. ( 10.1038/nrg964) [DOI] [PubMed] [Google Scholar]
- 55.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9, 267–276. ( 10.1038/nrg2323) [DOI] [PubMed] [Google Scholar]
- 56.Belshaw R, Sanjuan R, Pybus OG. 2011. Viral mutation and substitution: units and levels. Curr. Opin. Virol. 1, 430–435. ( 10.1016/j.coviro.2011.08.004) [DOI] [PubMed] [Google Scholar]
- 57.Linz B, et al. 2014. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat. Commun. 5, 4165 ( 10.1038/ncomms5165) [DOI] [PubMed] [Google Scholar]
- 58.Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410. ( 10.2307/2412923) [DOI] [Google Scholar]
- 59.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. ( 10.1007/BF01734359) [DOI] [PubMed] [Google Scholar]
- 60.Graybeal A. 1998. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst. Biol. 47, 9–17. ( 10.1080/106351598260996) [DOI] [PubMed] [Google Scholar]
- 61.Moreira D, Philippe H. 2000. Molecular phylogeny: pitfalls and progress. Int. Microbiol. 3, 9–16. [PubMed] [Google Scholar]
- 62.Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F. 2005. Heterotachy and long-branch attraction in phylogenetics. BMC Evol. Biol. 5, 50 ( 10.1186/1471-2148-5-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lartillot N, Brinkmann H, Philippe H. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7(Suppl 1), S4 ( 10.1186/1471-2148-7-S1-S4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodríguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF, Philippe H. 2007. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 56, 389–399. ( 10.1080/10635150701397643) [DOI] [PubMed] [Google Scholar]
- 65.Huelsenbeck JP. 1997. Is the Felsenstein zone a fly trap? Syst. Biol. 46, 69–74. ( 10.1093/sysbio/46.1.69) [DOI] [PubMed] [Google Scholar]
- 66.Claverie JM, Ogata H. 2009. Ten good reasons not to exclude giruses from the evolutionary picture. Nat. Rev. Microbiol. 7, 615 ( 10.1038/nrmicro2108-c3) [DOI] [PubMed] [Google Scholar]
- 67.López-García P, Moreira D. 2009. Yet viruses cannot be included in the tree of life. Nat. Rev. Microbiol. 7, 615–617. ( 10.1038/nrmicro2108-c7) [DOI] [PubMed] [Google Scholar]
- 68.Williams TA, Embley TM, Heinz E. 2011. Informational gene phylogenies do not support a fourth domain of life for nucleocytoplasmic large DNA viruses. PLoS ONE 6, 16 ( 10.1371/annotation/53805ecf-7d10-4d99-9cec-f27f5e0d4166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wheeler W. 1990. Nucleic acid sequence phylogeny and random outgroups. Cladistics 6, 363–367. ( 10.1111/j.1096-0031.1990.tb00550.x) [DOI] [PubMed] [Google Scholar]
- 70.Stiller J, Hall B. 1999. Long-branch attraction and the rDNA model of early eukaryotic evolution. Mol. Biol. Evol. 16, 1270–1279. ( 10.1093/oxfordjournals.molbev.a026217) [DOI] [PubMed] [Google Scholar]
- 71.Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. ( 10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 72.Lecointre G, Philippe H, Le HLV, Le Guyader H. 1993. Species sampling has a major impact on phylogenetic inference. Mol. Phylogenet. Evol. 2, 205–224. ( 10.1006/mpev.1993.1021) [DOI] [PubMed] [Google Scholar]
- 73.Bergsten J. 2005. A review of long-branch attraction. Cladistics 21, 163–193. ( 10.1111/j.1096-0031.2005.00059.x) [DOI] [PubMed] [Google Scholar]
- 74.Tidona CA, Darai G. 2000. Iridovirus homologues of cellular genes: implications for the molecular evolution of large DNA viruses. Virus Genes 21, 77–81. ( 10.1023/A:1008192616923) [DOI] [PubMed] [Google Scholar]
- 75.Shackelton LA, Holmes EC. 2004. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends Microbiol. 12, 458–465. ( 10.1016/j.tim.2004.08.005) [DOI] [PubMed] [Google Scholar]
- 76.Filee J, Siguier P, Chandler M. 2007. I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 23, 10–15. ( 10.1016/j.tig.2006.11.002) [DOI] [PubMed] [Google Scholar]
- 77.Filee J, Pouget N, Chandler M. 2008. Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol. Biol. 8, 320 ( 10.1186/1471-2148-8-320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moreira D, Brochier-Armanet C. 2008. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 8, 12 ( 10.1186/1471-2148-8-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yutin N, Wolf YI, Koonin EV. 2014. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 467, 38–52. ( 10.1016/j.virol.2014.06.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sapp J, Fox GE. 2013. The singular quest for a universal tree of life. Microbiol. Mol. Biol. Rev. 77, 541–550. ( 10.1128/MMBR.00038-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236. ( 10.1038/nature12779) [DOI] [PubMed] [Google Scholar]
- 82.Spang A, et al. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179. ( 10.1038/nature14447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forterre P. 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117, 5–16. ( 10.1016/j.virusres.2006.01.010) [DOI] [PubMed] [Google Scholar]
- 84.Forterre P, Prangishvili D. 2009. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. N.Y. Acad. Sci. 1178, 65–77. ( 10.1111/j.1749-6632.2009.04993.x) [DOI] [PubMed] [Google Scholar]
- 85.Gortner RA. 1938. Viruses—living or non-living? Science 87, 529–530. ( 10.1126/science.87.2267.529) [DOI] [PubMed] [Google Scholar]
- 86.Podolsky S. 1996. The role of the virus in origin-of-life theorizing. J. Hist. Biol. 29, 79–126. [DOI] [PubMed] [Google Scholar]
- 87.Moreira D, López-García P. 2009. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 7, 306–311. ( 10.1038/nrmicro2108) [DOI] [PubMed] [Google Scholar]
- 88.Raoult D. 2009. There is no such thing as a tree of life (and of course viruses are out!). Nat. Rev. Microbiol. 7, 615 ( 10.1038/nrmicro2108-c6) [DOI] [PubMed] [Google Scholar]
- 89.Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 67, 565–585. ( 10.1146/annurev-micro-092412-155633) [DOI] [PubMed] [Google Scholar]
- 90.Forterre P, Krupovic M, Prangishvili D. 2014. Cellular domains and viral lineages. Trends Microbiol. 22, 554–558. ( 10.1016/j.tim.2014.07.004) [DOI] [PubMed] [Google Scholar]
- 91.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836. ( 10.1038/nrmicro2235) [DOI] [PubMed] [Google Scholar]
- 92.Claverie JM, Abergel C. 2013. Open questions about giant viruses. Adv. Virus Res. 85, 25–56. ( 10.1016/B978-0-12-408116-1.00002-1) [DOI] [PubMed] [Google Scholar]
- 93.Forterre P. 2013. Why are there so many diverse replication machineries? J. Mol. Biol. 425, 4714–4726. ( 10.1016/j.jmb.2013.09.032) [DOI] [PubMed] [Google Scholar]
- 94.Martin W, Muller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41. ( 10.1038/32096) [DOI] [PubMed] [Google Scholar]
- 95.Moreira D, López-García P. 1998. Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: The syntrophic hypothesis. J. Mol. Evol. 47, 517–530. ( 10.1007/PL00006408) [DOI] [PubMed] [Google Scholar]
- 96.López-García P, Moreira D. 2006. Selective forces for the origin of the eukaryotic nucleus. Bioessays 28, 525–533. ( 10.1002/bies.20413) [DOI] [PubMed] [Google Scholar]
- 97.Guy L, Ettema TJ. 2011. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587. ( 10.1016/j.tim.2011.09.002) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAP2 sequence alignments are available in the electronic supplementary material.