Abstract
The phylogenetic analysis of 11 CSFV isolates from Karnataka, India obtained during the year 2012–13 was undertaken to obtain the most reliable genetic typing of the CSFV isolates based on E2, NS5B and 5′UTR genomic regions. The study indicated that all the 11 CSFV isolates belonged to subgroup 2.2. The most reliable classification was obtained with sequence data from the NS5B region which separated all the isolates based on the history of outbreak and geographic origin. Analysis of full length E2 amino acid sequences revealed different genetic makeup of Indian 2.2 isolates compared to 2.2 isolates from different countries. The group 2.2 viruses are gradually spreading as confirmed by frequent detection/ isolation of group 2.2 viruses in the recent years and replacing the subgroup 1.1 viruses, which were hitherto predominantly involved in CSF outbreaks in India.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-015-0273-9) contains supplementary material, which is available to authorized users.
Keywords: Classical swine fever virus, Phylogenetic analysis, E2, NS5B and 5′UTR
Introduction
India has one of the largest livestock populations in the world but the pig husbandry is in primitive stage except in north eastern states. The growth and reproduction of pigs is affected by classical swine fever (CSF) that is the major constraints to pig farming in India. Classical swine fever is a disease of domestic pigs and wild boar caused by CSF virus (CSFV) which belongs to genus Pestivirus within the family Flaviviridae. The CSFV genome has a positive polarity RNA of about 12.3 kb in length which contains un-translated regions (UTR) at 5′ and 3′ ends and encodes a single poly protein that is both co and post-translationally processed to yield four structural (C, Erns, E1 and E2) and 8 non structural (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B) viral proteins [11].
Classical swine fever in India was first reported in 1962 and later has been reported from many parts of the country [14, 23, 24]. A compilation of data from OIE indicates that there were 1308 outbreaks of CSF in India during 1996–2008 [16]. It is one of the top five viral diseases of livestock in the country according to data available in NIVEDI [15]. Many outbreaks in the country were undiagnosed because of the sporadic nature of the disease, lack of awareness and scarcity of good diagnostic laboratories. The status of the disease outbreak is largely unknown as the country does not have effective reporting system and a vaccination or eradication policy.
The virus isolates analyzed from different countries, showed that they could be assigned into three main genetic groups (1, 2 and 3) and eleven subgroups including a new subgroup 1.4 from Cuba [19]. Few reports have been documented the occurrence of 1.1 and 2.2 genotype CSF viruses in India [14, 15, 24] using molecular epidemiological techniques. Phylogenetic analysis by Patil et al. [14, 15] indicated the dominance of subgroup 1.1 strains in the country and detailed analysis of subgroup 2.2 viruses indicated the plausible Chinese origin of this subgroup in India using the lone isolate from Karnataka. Their analysis was constrained by the lack of large number of subgroup 2.2 sequences from India and they also suggested the studies involving other regions of the genome for better resolution of the epidemiology of subgroup 2.2. The present study was undertaken for molecular characterization and phylogenetic analysis of CSFV isolates from Karnataka based on E2, NS5B and 5′UTR sequences.
Materials and methods
Animals
Small scale farmers rearing Yorkshire Pigs in different districts of Karnataka state were selected for the study. Blood and tissue samples collected from suspected field cases of CSF in different districts of Karnataka, India were used in this study (Supplementary Table 1). A total of 113 blood and 31 tissue samples like lymph node, spleen, kidney, tonsil, liver and intestine were collected from suspected field cases of CSF (Supplementary Table 1). After collection, tissue samples were kept in viral transport medium and transported on ice. The samples were stored at −70 °C until further use. Details of number of pigs in the farm, number of pigs that were dead, food source, clinical signs, vaccination status, past outbreaks, procurement of pigs and distribution of pigs were noted.
Virus isolation and RT-PCR
The PK-15 (ATCC) cells maintained using Eagle’s minimal essential medium (Sigma) supplemented with 5 % Horse serum were used for virus isolation. The PK-15 cells were infected using the field samples and observed daily for any changes for up to 5 days. After 5 days of infection, total viral RNA was extracted from cell culture supernatant using QIA amp Viral RNA minikit (Qiagen, Germany) according to the protocol supplied by the manufacturer and stored at −80 °C. The concentration and quality of RNA extracted was measured at a dual wavelength of 260 and 280 nm using spectrophotometer. Reverse transcription-PCR (RT-PCR) was performed targeting E2, NS5B and 5′UTR regions using previously published primers (Supplementary Table 2) by One-step RT-PCR kit (Qiagen, Germany). For amplification of NS5B and 5′UTR, reverse transcription was performed at 50 °C for 30 min, followed by initial enzyme activation step at 95 °C for 15 min. Subsequent PCR amplification was carried out with 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, extension at 72 °C for 1 min followed by final extension for 10 min at 72 °C. For amplification of E2 region, reverse transcription was performed with same thermal cycling conditions, whereas subsequent PCR amplification was carried out with 35 cycles of denaturation at 95 °C for 50 s, annealing at 59 °C for 50 s, extension at 72 °C for 1 min and 20 s followed by final extension for 10 min at 72 °C. An expected amplicons of approximately 1274, 449 and 421 bp of E2, NS5B and 5′UTR respectively were obtained for all the isolates.
Sequencing of E2, NS5B and 5′UTR fragments and their phylogenetic analysis
The amplified products were purified from the gel using QIAquick gel extraction kit. After gel purification, all the amplicons of 5′UTR, NS5B and E2 were sent for nucleotide sequencing of both strands at a commercial service (Xcelris India Pvt. Ltd., Ahmedabad). The sequence data generated was received as coloured chromatograph and text files. Phylogenetic tree for E2, NS5B and 5′UTR was constructed as described previously by Greiser-Wilke et al. [7] by using the neighbor-joining method. The reference isolates for which all the three genomic region sequences are available in GenBank or in the nucleotide sequence database of European Union Reference Laboratory (EURL) originated from different parts of world (Supplementary Table 3) were selected and included in the multiple sequence alignment. Briefly, 150 nucleotides of the 5′UTR region, 190 nucleotides of E2 and 409 nucleotides of NS5B were aligned (nucleotide positions corresponding to HCLV India GenBank Acc. No.EU857642) with other reference strain sequences using the Clustal X, Ver. 1.8.3 programme [26] and edited manually using GeneDoc [12]. Same set of 41 reference isolates along with 11 isolates of present study were used for subsequent construction of the Phylogenetic trees based on E2, NS5B and 5′UTR sequences. Nucleotide divergence/similarity was calculated for all the three regions using Megalign module of Lasergene package (DNASTAR Inc., USA). The transition/transversion (Ts/Tv) rate was estimated from the dataset using TreePuzzle, Ver.5.2 [25]. The bootstrap values were calculated with the modules SEQBOOT, DNADIST, NEIGHBOR and CONSENSE of the PHYLIP Ver. 3.59 package [8]. The phylogenetic tree, constructed by using the neighbour-joining method, were computed with the DNADIST, NEIGHBOUR modules with the same parameters. Trees were visualised using the programme, TREEVIEW Ver. 1.6.6 [13]. Each of the data set was repeatedly reanalyzed using the SEQBOOT program to assess the statistical reliability of the dendrograms produced. The phylogeny for each repeat was calculated using DNADIST and then NEIGHBOR. Bootstrap analysis with 1000 replicates was performed. The tree file from NEIGHBOR was then used as the input file in the CONSENSE program. The bootstrap values were then readable in a text format which could be directly correlated to the dendrogram. Bootstrap values more than 500 are numbered along branches for Figs. 1–3 and values in excess of 70% were considered to be significant. For all the phylogenetic trees, congenital tremors isolate (GenBank Acc No. JQ411575) was kept as out group. The nomenclature of the groups and subgroups was done as per Paton et al. [16]. Aligned sequences of all the three regions are available with authors.
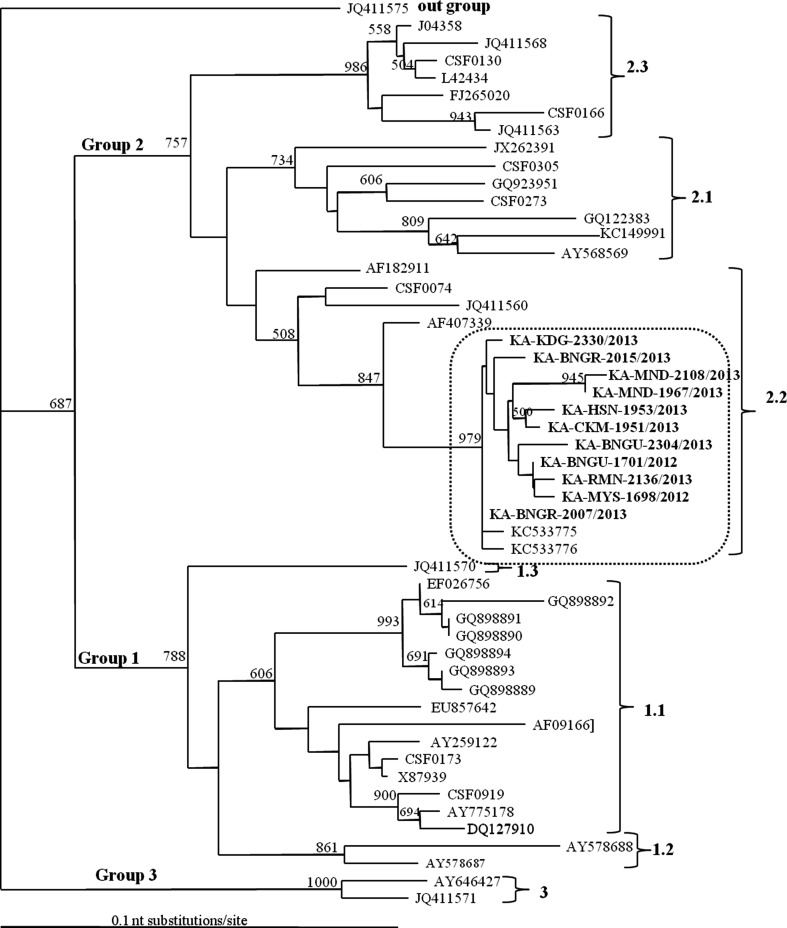
Fig. 1.
Phylogenetic tree based on the analysis of 190 nucleotides within E2 region. The tree is out grouped to the corresponding sequence of the congenital tremor (GenBank Acc. No. JQ411575) isolate and constructed using the neighbour-joining method. Numbers at the node represent the bootstrap values after 500 replicates. Present Karnataka isolates are in bold. Indian isolates cluster on the phylogenetic tree is encircled. The nomenclature of the Groups and subgroups was as per Paton et al. [16]
Fig. 3.
Phylogenetic tree based on the analysis of 150 nucleotides within 5′UTR. The tree is out grouped to the corresponding sequence of the congenital tremor (GenBank Acc. No. JQ411575) isolate and constructed using the neighbour-joining method. Numbers at the node represent the bootstrap values after 500 replicates. Present Karnataka isolates are in bold. Indian isolates cluster on the phylogenetic tree is encircled. The nomenclature of the Groups and subgroups was as per Paton et al. [16]
Analysis of full length E2 amino acid sequences
To get a better insight of variability of the E2 region, its deduced full length amino acid sequences of 11 isolates of present study and 46 isolates retrieved from GenBank and EURL representative of three main groups were further analyzed. The present CSFV isolates were also compared to the reference vaccine strain HCLV India (GenBank Accession no.EU857642).
Results
Phylogenetic analysis based on the E2 (190 nt) sequences
Phylogenetic analysis based on E2, NS5B and 5′UTR sequences of present isolates obtained during 2012–2013 and other reference sequences representing different genogroups and subgroups showed that, all present isolates belong to genogroup 2 and subgroup 2.2 (Figs. 1–3). The transition/transversion (Ts/Tv) rate estimated from the data set using TreePuzzle, Ver. 5.2 was found to be 4.77, 5.06 and 4.19 for phylogenetic trees based on E2, NS5B and 5′UTR respectively. The statistically significant bootstrap values for trees based on E2, NS5B and 5′UTR are 13, 23and 1 respectively (Figs. 1–3).
Percent identities of E2 sequences among present isolates ranged from 95.8–100. The tree based on E2 sequences showed most of the Karnataka isolates were divergent to each other indicating that, they were from different out breaks. Present isolates had 96.6–98.3 % nucleotide similarity with other 2.2 subgroup Indian isolates from Uttar Pradesh state (KC533775 and KC 533776). On the phylogenetic tree, all the Indian isolates were placed in separate cluster from the other members of the 2.2 subgroup from Germany, Austria and Netherlands and had 3.5–10.4 % divergent from them. Interestingly, Indian clade showed a closer phylogenetic relationship to E2-encoding sequences of the Chinese recombinant isolate ‘‘strain 39’’ (possessing an E2 gene from an unknown parental 2.2 genotype isolate [17]) at a genetic distances of 3.3–4.6 % than to those of E2-encoding sequences of isolates from Germany(JQ411560), Austria(CSF00740) and Netherlands(AF1829110). Present isolates were 17.1–19.8 % divergent with vaccine strain (HCLV India) and 17.1–24.0, 19.1–24.8 % divergent from the members of the groups 1 and 3 respectively in E2 sequences (Table 1).
Table 1.
Percent identities and divergence of present Karnataka CSFV isolates with different CSFV genotypes and HCLV India (Vaccine strain) in their E2, NS5B and 5′UTR sequences
| Gene/region | Percent identities | Percent divergence | |||||
|---|---|---|---|---|---|---|---|
| 2.2 subgroup (Present Karnataka isolates) | 2.2 subgroup (global isolates) | 2.1 subgroup isolates | 2.3 subgroup isolates | Group 1 isolates | Group 3 isolates | With HCLV India | |
| E2 (190 bp) | 95.8–100 | 89.5–96.8 | 12.6–17.1 | 13.2–17.8 | 17.1–24.0 | 19.1–24.8 | 17.1–19.8 |
| NS5B | 96.8–100 | 92.4–94.9 | 9.4–13.4 | 10.2–12.8 | 14.0–18.2 | 15.5–17.0 | 16.1–17.3 |
| 5′UTR | 98.0–100 | 94.7–97.3 | 2.7–7.0 | 4.8–8.5 | 3.4–9.2 | 9.2–11.5 | 4.8–7.0 |
Phylogenetic analysis based on the NS5B sequences
In the tree based on NS5B, IND-2007/13 showed 100 % homology with IND-2015/13 indicating the possible single source of virus responsible for both the outbreaks in Devanahalli of Bangalore rural district. This was not surprising, considering the history that both outbreak locations were just 5 km apart from each other and frequent exchange of piglets and materials occur between these two farms. In Hosabudanuru of Mandya district, disease outbreak had occurred twice in the same farm, once in February 2013 and other one in April 2013. The isolates IND-1967/13 and IND-2108/13 which were obtained from this place were 100 % identical in their NS5B sequences (Fig. 2) indicating that both times outbreak has been occurred due to possible single source of virus. The isolate IND-1953/13 which is from Devalkere, Hassan district is 99.5 % identical with IND-1951/13 isolate which is from Halikote, Chikmagalur district in their NS5B sequences (Fig. 2). This similarity of the isolates correlated with history for both outbreaks. The pig farm in Halikote in Chikmagalur district is located in isolated place in forest area where in the source of food is only of vegetarian origin, clearly indicating the only possible source of infection was by recent introduction of pigs purchased from Devalkere, Hassan.
Fig. 2.
Phylogenetic tree based on the analysis of 409 nucleotides within NS5B region. The tree is out grouped to the corresponding sequence of the congenital tremor (GenBank Acc. No. JQ411575) isolate and constructed using the neighbour-joining method. Numbers at the node represent the bootstrap values after 500 replicates. Present Karnataka isolates are in bold. Indian isolates cluster on the phylogenetic tree is encircled. The nomenclature of the Groups and subgroups was as per Paton et al. [16]
The tree based on NS5B sequences clearly separated the present isolates based the history of outbreak and geographic origin as shown in Supplementary Table 1. Percent identities of NS5Bsequences among present isolates ranged from 96.8–100. All 11 isolates were distinct from the IND-294/08, which was isolated from Harohalli, Bangalore in 2008 with divergence of 3.5–4.8 % (Data not shown). But the present isolates were having more identity of 98.5 % with Uttar Pradesh isolate which was isolated in 2006 (KC533775).Present isolates were 16.1–17.3 % divergent with vaccine strain (HCLV India), 14.0–18.2 % divergent with Group1 viruses and 15.5–17.0 % divergent with Group 3 viruses in NS5B sequences (Table 1).
Phylogenetic analysis based on the 5′UTR sequences
The phylogenetic analysis based on the 5′UTR fragment did not clearly revealed the clustering of isolates, which failed to differentiate between some of the individual isolates in present study. The isolates IND-2330/13, IND-2108/13, IND-2007/13, IND-2136/13, IND-2015/13, IND-1967/13 and IND-2304/13 were 100 % identical to each other in their 5′UTR sequences, even though they were from different outbreaks (Fig. 1). This showed that analysis of 5′UTR fragment was not corroborating with history of outbreak and geographic origin of different isolates. The 5′UTR nucleotide sequence of present isolates shared 98–100 % similarity with each other. The percent divergence of 5′UTR sequences of present Karnataka isolates with Group 1 and Group 3 viruses is 3.4–9.2 and 9.2–11.5 respectively (Table 1).
Analysis of full length E2 amino acid (aa) sequences
The deduced full length amino acid sequences of 46 isolates representative of three main groups including11 isolates of this study were compared to know the variability of E2 region. The aa analysis showed that the N-terminal half of E2, which contained four antigenic domains (A–D), was more variable than the C-terminal half (Table 2). The 15 Cysteine residues important in maintaining the conformational structure of E2 [28] were conserved in all 11 isolates along with other reference isolates. No changes were found in 693C and 737C and motif 771LLFD774 in isolates under study, which are essential for structural integrity and conformational recognition of B/C domains [3].The two epitopes recognizable by the CSFV specific and the panpestivirus reactive murine monoclonal antibodies (mAb) at aa 829–839 and 995–998 were highly conserved among present isolates. The epitopes at aa 693–699 and 792–814 were also highly conserved similar to mAb-recognizable epitopes, whereas 844–865 were less conserved than the mAb-recognizable epitopes in the present isolates. A highly conserved linear epitope, 829TAVSPTTLR837 identified in domain A [2] was found to be highly conserved in all 11 isolates and also in the reference isolates.
Table 2.
Summary of variable sites in terms of amino acids in glycoprotein E2 and NS5B between consensus (of 57 sequences), HCLV India (Reference strain) and different genotypes
| Subgroups | N-terminal of E2 | C-terminal of E2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antigenic unit B/C | Overlapping | Unit A/D | |||||||||||||
| 705 | 706 | 729 | 736 | 757 | 779 | 780 | 860 | 867 | 868 | 885 | 939 | 960 | 972 | 1034 | |
| Consensus | N | E | N | I | S | V | T | T | F | H | I | T | R | V | L |
| HCLV India | D | E | N | I | S | S | T | T | F | Y | T | A | R | V | L |
| 1.1 | N/D | E | N | I/S | S | S | T | T | F | Y | T | A/T | R | V | L |
| 1.2 | N | E/K | D | I | S | L | T | T | F | Y | M/T | T | R | V | L |
| 1.3 | N | E | D | I | S | S | T | T | F | Y | A | T | R | V | L |
| 2.1 | N | E | D | I | S | A | I | I/M/ | F | Y/H | M/T/K | T | R | V | L |
| Global 2.2 isolates | N/S | E | D | I | S | V | I | M | F | H/Y | M/V/I | T | R | V | L |
| Indian 2.2 isolates | D/N | G/E | N | T/I | S/F | V | T | T/M | Y | H | I | I | K | I/V | M |
| 2.3 | N | E | D | V | S | A | I | I | F | H/Y | K | T | R | V | L |
| Group 3 | N | E | D | I | S | L/S | S/T/I | I/M | F/Y | N/Y/H | T | T | R | I/V | L |
R Arginine, E Glutamate, I Isoleucine, M Methionine, T Threonine, A Alanine, N Asparginine, G Glycine, L Leucine, F Phenylalanine, Y Tyrosine, D Aspartate, H Histidine, K Lysine, S Serine, V Valine
Among the seven glycosylation sites identified [21], the five sites at aa 805–807, 810–812,874–876, 918–920 and 949–951 were highly conserved in all 11 isolates along with reference isolates. The remaining two potential glycosylation sites at aa 777–779 and 986–988 were conserved in present isolates but varied among the reference isolates. The first potential glycosylation site (777–779) was unique for subgroup 2.2 wherein at aa 779 Valine was present only in the present 11 isolates along with other 2.2 subgroup isolates (Table 2). At aa 705 and aa 706 of B/C domain all the 11 isolates along with other Indian isolates of 2.2 subgroup had Aspartate and Glycine instead of Asparagine and Glutamate respectively. At aa 867 in N terminal half all Indian isolates of 2.2 subgroup had Tyrosine instead of Phenylalanine. At aa 939 and aa 960 in the C-terminal half 11 isolates along with other Indian isolates of 2.2 subgroup have Isoleucine and lysine instead of Threonine and Arginine respectively. The transmembrane domain located in the C-terminal half was having Methionine at aa 1034 instead of Leucine, which is unique in present isolates and also in other Indian isolates of 2.2 subgroup (Table 2).
Discussion
Genetic typing of CSFV isolates and comparison with other Indian isolates and those originating from other countries is important so as to determine the genetic diversity, molecular epidemiology and to establish a control strategy against CSF [16]. In the present study phylogenetic analysis was done for E2, NS5B and 5′UTR to obtain the most reliable genetic typing of the CSFV isolates. It was shown that all the 11 CSFV isolates from Karnataka collected during the year 2012–2013 belonged to genogroup 2 and subgroup 2.2 (Figs. 1–3). To obtain the most reliable genetic typing of the CSFV isolates, phylogenetic analysis was undertaken in all the three regions like E2, NS5B and 5′UTR, which showed that all the 11 CSFV isolates from Karnataka collected during the year 2012–2013 belonged to genogroup 2 and subgroup 2.2 (Figs. 1–3). Percent identity among the 11 isolates ranged from 95.8–100, 96.8–100 and 98–100 in their E2, NS5B and 5′UTR sequences respectively indicating that the isolates were genetically closely related. Those isolates which showed 100 % homology with each other indicated the possible single source of virus responsible for different outbreaks (Figs. 1–3). In the present study, most of the outbreaks were due to recent introductions of new stocks of pigs obtained from other places, as one farm gets infected with CSF, pigs were sold at lesser prices to other pig farmers to reduce the financial losses. The majority of CSF outbreaks in Karnataka occurred in small sized pig farms, where vaccination was not practiced.
In all the phylogenetic trees, Indian isolates were placed in separate cluster from other members of the subgroup 2.2 from Germany, Netherland and Austria (Figs. 1–3).The study also indicated that, viruses isolated from Karnataka were genetically distinct (0–4.3 % in E2, 0–3.3 % in NS5B and 0–2.0 % in 5′UTR). The degree of divergence found between certain isolates in this study could not establish a direct link between the outbreaks. It was hypothesised that the observed sequence divergence was a consequence of independent CSFV introductions or the endemic presence of CSFV in certain districts of Karnataka. Separating grouping of all 11 isolates from the vaccine strain-HCLV India (Figs. 1–3), confirms that present field isolates did not have any vaccine origin. This is further substantiated by the fact that, present isolates were 17.1–19.8 % (E2 190 nt), 16.1–17.3 % (NS5B) and 4.8–7.0 % (5′UTR) nucleotide divergent from the said strain. The E2 sequences of present Karnataka isolates and some of the subgroup 2.2 CSFV isolates recently recovered from North India were only 2.1–5.5 % divergent from Chinese isolate (FJ157197) which was isolated in the year 1999 (Data not shown). Moreover, present isolates had closer phylogenetic relationship (3.3–4.6 %) to E2-encoding sequence of the Chinese recombinant isolate ‘‘strain 39’’. In China, 94 % of the virus strains isolated from field outbreaks during 1986–1999 were belonging to 2.1 and 2.2 subgroups [27].As the present isolates and some of the Indian isolates belonged to subgroup 2.2, it can be inferred that the viruses may have had Chinese origin at some point of time. Recently, Patil et al. [15] have also shown the plausible Chinese origin of the Karnataka isolate IND-294/08 which belongs to subgroup 2.2.
The most reliable classification was obtained with sequence data from the NS5B region which separated present isolates based on the history of outbreak and geographic origin (Supplementary Table 1). The confidence levels indicated by statistically significant bootstrap levels for its branch nodes were highest for the NS5B tree (23 values) and lowest for the 5′UTR tree (only one value). Similar type of reliable classification was obtained with sequence data from the NS5B region by Paton et al. [16].The discrimination possible between isolates depends on the length and variability of the target region of the genome that is used for comparisons [16]. Among all isolates compared, only one pair appears identical in sequences from the E2 region and four pairs from the NS5B region when compared to 31 pairs from the 5′UTR (Figs. 1–3).The N-terminal half of E2 is one of the most variable regions in the CSFV genome [10], hence phylogenetic analysis based on the 190 nt of this region gives better discrimination among different isolates but it had failed to segregate isolates based on the history of outbreak and geographic origin as was in case of NS5B region. The success ofNS5B over E2 has been assumed to be due to the latter’s tendency for greater variability and higher frequency of back mutations, which sometimes may mislead the evolutionary analysis [16]. The lowest nucleotide identity between two CSFV isolates was 78.9 % (Congenital tremor and KA-BNGR3/13), 82.6 % (AY646427 and GQ122383) and 88.7 % (Kanagawa and KC533776) in their E2, NS5B and 5′UTR sequences respectively.
The 5′UTR is highly conserved and its genetic stability makes the region a favourite target for diagnostic Prather than as a phylogenetic target, as it gives poorer discrimination within the CSFV genotype than can be obtained with the other two regions. Grouping of all isolates were same in all three trees except in 5′UTR tree where RUCSFPLUM isolate has grouped along with 1.1 isolates (Fig. 3) whereas in E2 and NS5B tree it has been grouped with 1.2 isolates which shows that grouping of group 1 isolates was not proper. The same observation was also demonstrated by Postel et al. [17] for isolates belonging to individual subgenotypes of 1.1, 1.2, 2.1 and 2.3. Due to advantage and disadvantages of each region, Paton et al. [16] advised to study different genomic regions whenever new isolates are included in the analyses that fall into new or distinct clusters and also when comparing very similar viruses. When bootstrap values are low, it is useful to analyse more than one genomic region, in order to be confident of the validity of the genetic typing [16].
In the present study, deduced full length amino acid (aa) sequences of 11 isolates were analyzed with other 46 reference isolates. All fifteen cysteine residues in E2 region, six in the N-terminal half and nine in the proximal C-terminal half are conserved among pestiviruses [28], indicating that cysteine is important in maintaining the conformational structure of E2. The amino acid (aa) analysis of present isolates showed the 15 Cysteine residues were conserved along with other reference isolates, which confirmed that there was no change in conformational structure of E2. The disulfide bond between 693C and 737C and motif 771LLFD774 are essential for structural integrity and conformational recognition of B/C domains [3]. There were no changes observed in 693C and 737C and motif 771LLFD774 of present isolates indicating structural integrity and conformational recognition of B/C domains. Both conformation dependent [3] and linear epitopes are present on E2 proteins of CSFVs. The two epitopes recognizable by the CSFV specific and the panpestivirus reactive murine monoclonal antibodies (mAb) at aa 829–839 and 995–998 were highly conserved among our isolates, which is in agreement with Chen et al. [4]. The other epitopes at aa 693–699 and 792–814 were also highly conserved, whereas epitope at aa 844–865 was less conserved in few isolates, which indicates that there is no much change in epitope structure in present isolates compared to majority of reference isolates. A highly conserved linear epitope, 829TAVSPTTLR837 identified in domain A play a significant role in CSFV virulence [20] and has been used to develop marker vaccines and for serodiagnostic test [9]. This epitope, 829TAVSPTTLR837 was also highly conserved in the isolates along with other reference isolates.
Neutralizing epitopes in E2 proteins are dependent on the presence of glycosylation. Lack of glycosylation leads to the synthesis of non-immunogenic proteins, that failed to induce protection against CSFV and affects virus virulence and viability [22]. All the seven glycosylation sites were highly conserved in the present isolates indicating that there was no change in virulence pattern and epitopes of E2 proteins. At aa 705, 706, 867, 939 and 960, the present isolates along with other Indian isolates in subgroup 2.2 have unique aminoacid residues viz. Aspartate, Glycine, Tyrosine, Isoleucine and Lysine respectively (Table 2). These changes might have contributed the formation of a separate cluster of Indian isolates (Figs. 1–3), further indicating minor difference in the genetic makeup of Indian isolates.
The involvement of subgroup 2.2 viruses from Karnataka, in one of the CSF outbreaks in Harohalli, near Bangalore during the year 2008 was reported by Patil et al. [14]. Later Chakroborthy et al. [2] recovered three isolates from Bangalore district, further confirmed the presence of 2.2 subgroup viruses in the region. Present and previous studies of genetic typing [15] in Karnataka had indicated that there has been a switch in virus populations from subgroup 1.1 to 2.2 after 2008 and 2.2 viruses has dominated the field infections. Even though subgroup 1.1 viruses were predominant in India, the subgroup 2.2 viruses are gradually spreading as confirmed by frequent detection/ isolation of subgroup 2.2 viruses in the recent years, particularly from the northern India [14, 15] and neighbouring country like Nepal [18]. During 2012-13, the prevalence of subgroup 2.2 isolates was also reported from Andhra Pradesh, Punjab, Orissa, Meghalaya and Arunachal Pradesh states (Unpublished data from NIVEDI, Bangalore). The sequences available from Genbank also suggested the prevalence of subgroup 2.2 isolates in Uttar Pradesh and Uttarakhand states during 2011–2012 (Figs. 1–3) (these sequences were not included for analysis since all the three region sequences were not available for these isolates). Thus, it may be concluded that sub group 2.2 viruses are gradually replacing the subgroup 1.1 viruses, which were hitherto predominantly involved in CSF outbreaks. At present, it is not clear whether subgroup 2.2 viruses have any selective advantages over subgroup 1.1 viruses in infecting the hosts but nevertheless their presence in recent outbreaks is indicating the changing epidemiological scenario. According to Chen et al. [4] the possible reasons could be, the subgroup 2.2 isolates may possess a higher replication rate than the 1.1 isolates in pigs and they may also contain higher affinity to compete the cellular receptors.
Several reports from other countries also pointed out similar situations in the endemic areas, where the historical group i.e. Group 1which persisted for many years have become ‘silent’ and is being replaced by the recent Group 2. There has been a switch from Group 1 to Group 2 after 1970 in Europe [16]. In China, 94 % of the virus strains isolated from field outbreaks during 1986–1999 have changed from Group 1 to 2.1 and 2.2 subgroups [27]. In recent years, subgroup 2.1 has become predominant in China and subgroup 2.2 viruses have become ‘silent’ types that rarely cause epizootics [6, 10]. In Korea, a switch from subgroups 3.2 to 2.1 has been observed after 1999 [1]. In Thailand, subgroup 2.2 became predominant from 1996 onwards. In Taiwan, there has been a switch from subgroups 3.4 to 2.1 after 1996 [5].
Finally, among the three regions analysed, the most reliable classification was obtained with sequence data from the NS5B region which separated present isolates based on the history of outbreak and geographic origin. Analysis of full length E2 amino acid sequences revealed different genetic makeup of Indian 2.2 isolates compare to other 2.2 isolates from different countries. Genetic typing indicated that 2.2 viruses are gradually replacing the subgroup 1.1 viruses in Karnataka, India. Though the present analysis is constrained by the lack of large number of subgroup 2.2 sequences from other states of India, it has revealed the genetic relationships among virus strains from different regions. Future studies should focus on analysis, involving large number of subgroup 2.2 viruses from different states of the India so that reasons for shifting of 1.1 viruses to 2.2 viruses can be found out.
Electronic supplementary material
Acknowledgments
The authors are highly thankful to Dean, Veterinary College, Bengaluru (KVAFSU) and Director, ICAR-NIVEDI, Bengaluru for their guidance, support and for providing necessary facilities for research work.
References
- 1.Cha SH, Choi EJ, Park JH, Yoon SR, Kwon JH, Yoon KJ, Song JY. Phylogenetic characterization of classical swine fever viruses isolated in Korea between 1988 and 2003. Virus Res. 2007;126:256–261. doi: 10.1016/j.virusres.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 2.Chakraborty S, Veeregoda BM, Chandranaik BM, Rathnamma D, Isloor S, Venkatesha MD, Leena G, Veeresh H, Patil SS. Molecular characterization and genogrouping of classical swine fever virus isolated from field outbreaks. Indian J Anim Sci. 2011;81:21–25. [Google Scholar]
- 3.Chang CY, Huang CC, Lin YJ, Deng MC, Chen HC, Tsai CH, Chang WM, Wang FI. Antigenic domains analysis of classical swine fever virus E2 glycoprotein by mutagenesis and conformation-dependent monoclonal antibodies. Virus Res. 2010;149:183–189. doi: 10.1016/j.virusres.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Chen N, Hu H, Zhang Z, Shuai J, Jiang L, Fang W. Genetic diversity of the envelope glycoprotein E2 of classical swine fever virus: recent isolates branched away from historical and vaccine strains. Vet Microbiol. 2008;127:286–289. doi: 10.1016/j.vetmic.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Deng MC, Huang CC, Huang TS, Chang CY, Lin YJ, Chien MS, Jong MH. Phylogenetic analysis of classical swine fever virus isolated from Taiwan. Vet Microbiol. 2005;106:187–193. doi: 10.1016/j.vetmic.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. Phylip: phylogeny inference package (version 3.5c) Cladistics. 1989;5:164–166. [Google Scholar]
- 7.Greiser-Wilke I, Fritzemeier J, Koenen F, Vanderhallen H, Rutili D, de Mia GM, Romero L, Sanchez-Vizcaino JM, Rosell R, San Gabriel A. Molecular epidemiology of a large classical swine fever epidemic in the European Union in 1997–1998. Vet Microbiol. 2000;77:17–27. doi: 10.1016/S0378-1135(00)00253-4. [DOI] [PubMed] [Google Scholar]
- 8.Jiang DL, Gong WJ, Li RC, Liu GH, Hu YF, Ge M, Wang SQ, Yu XL, Tu CC. Phylogenetic analysis using E2 gene of classical swine fever virus reveals a new subgenotype in China. Infect Genet Evol. 2013;17:231–238. doi: 10.1016/j.meegid.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Lin M, Lin F, Mallory M, Clavijo A. Deletions of structural glycoprotein E2 of classical swine fever virus strain Alfort/187 resolve a linear epitope of monoclonal antibody WH303 and the minimal N-terminal domain essential for binding immunoglobulin G antibodies of a pig hyperimmune serum. J Virol. 2000;74:11619–11625. doi: 10.1128/JVI.74.24.11619-11625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowings P, Ibata G, Needham J, Paton D. Classical swine fever virus diversity and evolution. J Gen Virol. 1996;7:1311–1321. doi: 10.1099/0022-1317-77-6-1311. [DOI] [PubMed] [Google Scholar]
- 11.Meyers G, Thiel HJ. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:58–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 12.Nicholas KB, Nicholas Jr HB. 1997. GeneDoc: analysis and visualization of genetic variation (distributed by the authors; http://www.psc.edu/biomed/genedoc/).
- 13.Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 14.Patil SS, Hemadri D, Shankar BP, Raghavendra AG, Veeresh H, Sindhoora B, Chandan S, Sreekala K, Gajendragad MR, Prabhudas K. Genetic typing of recent classical swine fever isolates from India. Vet Microbiol. 2010;141:367–373. doi: 10.1016/j.vetmic.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 15.Patil SS, Hemadri D, Veeresh H, Sreekala K, Gajendragad MR, Prabhudas K. Phylogenetic analysis of NS5B gene of classical swine fever virus isolates indicated plausible Chinese origin of Indian subgroup 2.2 viruses. Virus Genes. 2012;44:104–108. doi: 10.1007/s11262-011-0572-1. [DOI] [PubMed] [Google Scholar]
- 16.Paton DJ, McGoldrick A, Greiser-Wilke I, Parchariyanon S, Song JY, Liou PP, Stadejek T, Lowings JP, Bjorklund H, Belak S. Genetic typing of classical swine fever virus. Vet Microbiol. 2000;73:137–157. doi: 10.1016/S0378-1135(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 17.Postel A, Schmeiser S, Bernau J, Meindl-Boehmer A, Pridotkas G, Dirbakova Z, Mojzis M, Becher P. Improved strategy for phylogenetic analysis of classical swine fever virus based on full length E2 encoding sequences. Vet Res. 2012;43:50. doi: 10.1186/1297-9716-43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postel A, Jha VC, Schmeiser S, Becher P. First molecular identification and characterization of classical swine fever isolates from Nepal. Arch Virol. 2013;158:207. doi: 10.1007/s00705-012-1463-z. [DOI] [PubMed] [Google Scholar]
- 19.Postel A, Schmeiser S, Perera CL, Rodríguez LJ, Frias-Lepoureau MT, Becher P. Classical swine fever virus isolates from Cuba form a new subgenotype 1.4. Vet Microbiol. 2013;161:334–338. doi: 10.1016/j.vetmic.2012.07.045. [DOI] [PubMed] [Google Scholar]
- 20.Risatti GR, Holinka LG, Carrillo C, Kutish GF, Lu Z, Tulman ER, Sainz IF, Borca MV. Identification of a novel virulence determinant within the E2 structural glycoprotein of classical swine fever virus. Virology. 2006;355:94–101. doi: 10.1016/j.virol.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Risatti GR, Holinka LG, FernandezSainz I, Carrillo C, Lu Z, Borca MV. N-linked glycosylation status of classical swine fever virus strain Brescia E2 glycoprotein influences virulence in swine. J Virol. 2007;81:924–933. doi: 10.1128/JVI.01824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sainz IF, Holinka LG, Lu Z, Risatti GR, Borca MV. Removal of N- linked glycosylation site of classical swine fever virus strain Brescia Erns glycoprotein affects virulence in swine. Virology. 2008;370:122–129. doi: 10.1016/j.virol.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 23.Sapre SN, Moghe RG, Bhagwat SV, Chaudhry PG, Purohit BL. A note on observations and investigations into an outbreak of swine fever in Bombay (Maharashtra) Indian Vet J. 1962;39:527–534. [Google Scholar]
- 24.Sarma DK, Mishra N, Vilcek S, Rajukumar K, Behera SP, Nema R, Dubey P, Dubey SC. Phylogenetic analysis of recent classical swine fever virus (CSFV) isolates from Assam, India. Comp Immunol Microbiol Infect Dis. 2011;34:11–15. doi: 10.1016/j.cimid.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu C, Lu Z, Li H, Yu X, Liu X, Li Y, Zhang H, Yin Z. Phylogenetic comparison of classical swine fever virus in China. Virus Res. 2001;81:29– 37. doi: 10.1016/S0168-1702(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 28.Weiland E, Stark R, Haas B, Rümenapf T, Meyers G, Thiel HJ. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfidelinked heterodimer. J Virol. 1990;64:3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.