Abstract
Rabies is caused by negative strand RNA-virus classified in the genus Lyssavirus, family Rhabdoviridae of the order Mononegavirales. The aim of the present study was to identify and analyze nucleotides sequence of nucleoprotein (N) gene of rabies virus (RABV) from two cases of water buffaloes (Bubalus bubalis) bitten by a fox in Egypt, 2013. The diseased buffaloes showed nervous manifestations with fever. Specimens from brains of the buffaloes with suspected rabies were collected. RABV in collected samples was identified using direct fluorescent antibody (dFA) technique, histopathological examination and reverse transcription-polymerase chain reaction (RT-PCR). Also, nucleotides sequence of partially amplified nucleoprotein (N) gene was compared with the other street strains of RABV available on GenBank. The results revealed that RABV antigen was identified in the brains of diseased buffaloes by dFA technique and the characteristic intracytoplasmic inclusions (Negri bodies) and RABV nucleic acid were detected by histopathology and RT-PCR, respectively. The identified virus showed close genetic relationship with street strains identified previously from dogs in different Governorates in Egypt and with strains identified in Israel and Jordan indicating transmission of the virus between Egyptian Governorates with a potential transmission from and/or to our neighboring countries.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-015-0263-y) contains supplementary material, which is available to authorized users.
Keywords: Direct fluorescent antibody (dFA) technique, Egypt, Rabies virus (RABV), Reverse transcription-polymerase chain reaction (RT-PCR), Sequence analysis, Histopathology, Water buffaloes
Introduction
Rabies is a widespread neurological zoonotic disease which affects any warm blooded animals (including humans). Rabies virus (RABV) belongs to the genus Lyssavirus of the family Rhabdoviridae within the order Mononegavirales. The RABV genome is almost 12 kb in size consisting of five monocistronic RNAs, which encodes the nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and RNA-dependent RNA polymerase (large protein). Between the monocistronics there are four intergenic regions (IGRS) with different lengths. Besides, at the end of the genome there are two untranslated regions (UTRS), which play an important role in virus replication. The order of relative conservation of these five genes from high to low could be N > L>M > P>G [16, 24].
There are seven recognized genotypes of lyssavirus defined on the basis of their genetic similarity: rabies virus (genotype 1), Lagos bat virus (genotype 2), Mokola (genotype 3), Duvenhage virus (genotype 4), European bat lyssavirus type 1 (genotype5), European bat lyssavirus type 2 (genotype 6) and Australia bat lyssavirus (genotype 7). Additionally four novel genotypes: Aravan virus, Khujand virus, Irkut virus and West Caucasian [3, 8] were recently recovered from bats in Eurasia. Genotype 1 is responsible for classical rabies in terrestrial mammals throughout the world.
The nucleoprotein (N) gene has been extensively used for genetic typing and evolutionary studies because of its relatively lower variation among reservoir-associated variants and geographic lineages [4, 7, 14, 23].
The original method for typing RABV was a panel of monoclonal antibodies (mAbs) to the viral nucleoprotein (N) [13, 22]. However that method is quite antiquated and was replaced by phylogenetic analysis [11, 14].
Rabies transmission usually begins with the bite of an infected host, although transmission has been documented via mucous membrane contamination, aerosol transmission, and corneal and organ transplants [2, 15, 18].
Reliable laboratory diagnosis of rabies could be done by the direct fluorescent antibody (dFA) technique and the reverse transcription polymerase chain reaction (RT-PCR) as both techniques can detect the virus in 98–100 % of the disease cases. Histopathogical examination could also be used on the diagnosis but with much less sensitivity [12]. The aim of the present study was to diagnose two cases of water buffaloes (Bubalus bubalis) rabies in Egypt as well as comparing the partial nucleotides sequence of the nucleoprotein (N) gene with the other street strains of rabies virus (RABV) available on GenBank.
Materials and methods
Sample collection
Brain specimens were collected from two water buffaloes (Bubalus bubalis) bitten by a fox in Dakahlia Governorate, Egypt in 2013 with suspected rabies clinical signs in form of different nervous manifestations such as paralysis of throat, excessive salivation, extension of neck and grinding of teeth in association with fever. Part of these samples was taken to −20 °C for virus identification by reverse transcription-polymerase chain reaction and nucleotide sequencing. Another part was put in bottles containing 10 % neutral buffered formalin for dFA technique and histopathological testing. Brain sample from normal buffalo were included as a negative control.
Identification of the causative agent
Direct fluorescent antibody (dFA) technique
It was carried out as described previously [6, 19] as follows:
Formalin-fixed, paraffin embedded brain tissues were cut at 5 μm. Slides were heated at 55 °C to melt the paraffin, deparaffinized in xylol, hydrated through graded ethanols and finally rinsed in phosphate buffered saline (PBS) and then the section were left to dry in air for 30 min, fixed with acetone for 10 min and then washed with PBS, pH 7.6. A few drops of 1:100 dilutions of FITC-conjugated anti-RABV antibodies (Chemicon, Temecula, CA, USA) and the slides were kept in a humidified chamber for 1 h at 37 °C. The slides were thoroughly washed with PBS for 15 min three times. They were then mounted with buffered glycerin, covered with a cover slip and examined under a fluorescent microscope (Carl Zeiss, Germany).
Histopathological examination
Histopathological sections were carried out by fixing of brain specimens in 10 % neutral buffered formalin solution. The fixed specimens were trimmed, washed and dehydrated in ascending grades of alcohol, cleaned in xylene, embedded in paraffin then sectioned (5 μm) and stained with hematoxyline and eosin [1]. The stained sections were examined under light microscopy.
Reverse transcription-polymerase chain reaction (RT-PCR)
Oligonucleotide primers used in the RT-PCR reactions were designed by Langoni et al. [9] and synthesized by TIB MOLBIOL Syntheselabor GmbH. The primers were received in lyophilized form and resuspended in Tris/EDTA (TE) buffer to reach a final concentration of 100 pmol/μl and were designed to amplify a specific segment of 295 bp from N protein gene of RABV genome. The primers sequences for RT-PCR amplification were as follows: forward primer, 5´-ATAGAGCAGATTTTCGAGACAGC-3´ and reverse primer, 5´-CCTCAAAGTTCTTGTGGAAGA-3. RNA extraction was done using QIAamp Viral RNA Mini Kit (Cat. No. 52906) according to the manufacturer’s instructions. Normal non-infected brain samples were included as a negative control sample.
RT-PCR amplification was carried out as described before [9] using QIAGEN OneStep RT-PCR Kit (Cat. No. 210212). Briefly, 10 μl 5× QIAGEN OneStep RT-PCR Buffer, 2 μl dNTP Mix (containing 10 mM of each dNTP), 2 μl Forward primer, 2 μ reverse primer, 2 μl ofQIAGEN OneStep RT-PCR Enzyme Mix, 30 μl RNase-free water and 5 μl template RNA. The RT- PCR had reverse transcription step of 50 °C for 30 min then an initial cycle of 94 °C for 10 min, followed by 34 cycles of 94 °C for 1 min, 56 °C for 30 s, 72 °C for 1 min and a final elongation step of 72 °C for 5 min.
Amplified product analysis was carried out as described previously [21]. Briefly 10 μl of the PCR product was mixed with 1 μl 10× gel loading buffer and loaded to the individual wells of a 1 % agarose gel. In addition, 2 μl of a 100 bp DNA molecular weight marker was loaded with 2 μl loading buffer in a single outside well to be used as DNA ladder. The amplified DNA products were detected in comparison with DNA ladder using the U.V. transilluminator. The gel was photographed.
Nucleotide sequencing and analysis of sequencing data
Sequencing of the PCR amplicons were performed by SeqLab Gottingen GmbH (Gottingen, Germany). The obtained sequence data were analysed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and then, the alignment *.aln output file was used for performing the neighbour-joining (N-J) phylogenetic analysis with 1000 repeats of bootstrap tests analysis, and calculation of divergence and identity per cents were performed using MegAlign (DNASTAR, Lasergene®, Madison, WI, USA).
Results
Identification of the causative virus
Direct fluorescent antibody (dFA) technique revealed presence of RABV in the brain specimens from the diseased buffaloes while the control sample (normal brain) showed a negative reaction (Fig. 1). On histopathological examination, the characteristic cytoplasmic inclusions (Negri bodies) was detected in examined section (Fig. 2) beside perivascular infiltration (cuffing) with lymphocytes, few plasma cells, and macrophages. Also, some neurons were also found to be degenerative or necrotic.
Fig. 1.
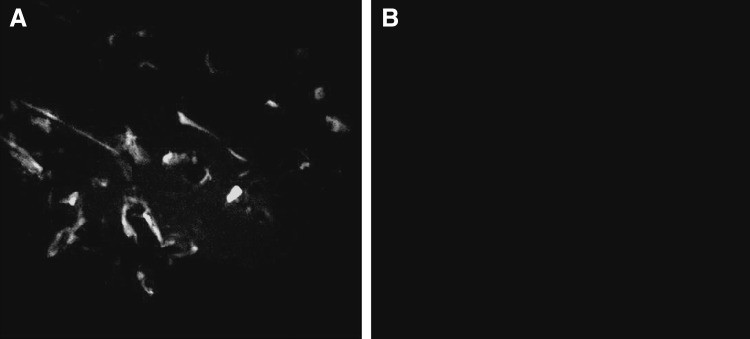
Results of direct flourescent antibody (dFA) technique of specimen from brain of diseased buffalo shows bright apple green viral nucleoprotein (N) antigen targeted by FITC-conjugated anti-RABV antibodies (a) and normal buffalo brain sample shows no viral antigen (b). With ×400 magnification
Fig. 2.
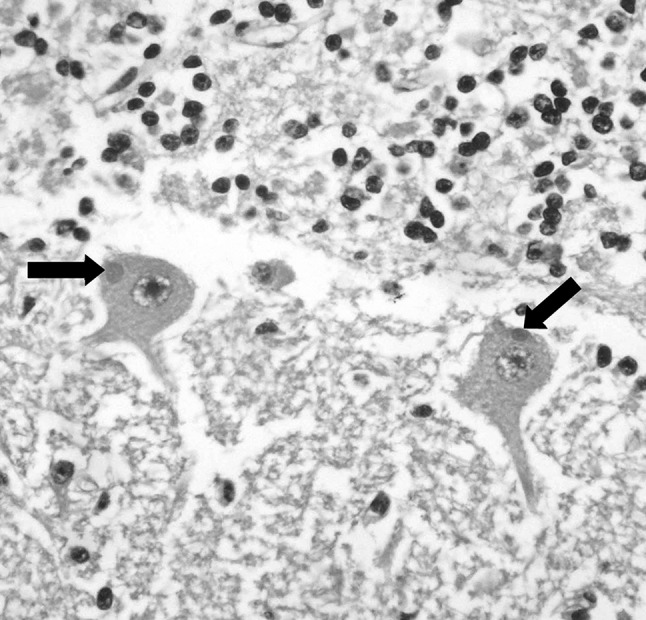
Brain section of diseased buffalo reveals intracytoplasmic eosinophilic inclusions (Negri bodies) in neurons (arrows). HE stain. With ×400 magnification
Analysis of the PCR products obtained from the amplification reaction of extracted RNA by agarose gel electrophoresis revealed the positive amplification of the nucleoprotein (N) gene with correct size (295 bp) (Fig. 3). The negative control sample did not show any amplified product.
Fig. 3.
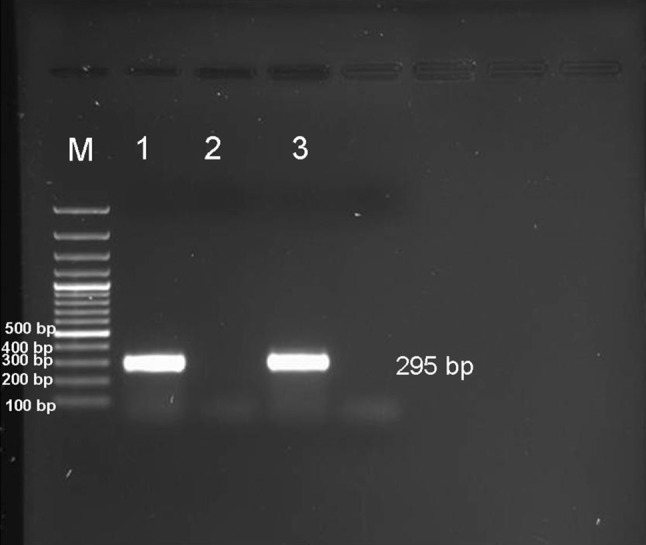
Gel electrophoresis of PCR products of 295 bp of nucleoprotein (N) protein gene of rabies virus genome. M DNA marker, Lane 1 & 3 The amplified products prepared from brain samples of diseased buffaloes, Lane 2 negative control buffalo brain sample
Nucleotide sequencing and analysis of sequencing data
The obtained nucleotide sequence of 295 bp PCR fragments representing the partial sequence of the nucleoprotein (N) gene were analyzed and compared with the published sequences of RABVs available in GenBank (Supplementary Table-1). Amplicons from the two samples were of the same nucleotides sequence and only one sequence was submitted to GenBank (accession no. KP662552).
The multiple alignments revealed that our RABV is found to be different from Egyptian dog RABV identified in cairo (DQ837463 & DQ837461) and from Faium (DQ837462), at about 17 positions. The RABV was showed to have ~93 % identity at the nucleotide level with dog RABVs from cairo and faium (Egypt) with accession no. DQ837463, DQ837461 & DQ837462. Also, Comparing nucleotides sequences of the identified RABV with those from neighboring countries revealed that the virus shared an average identity of 87.3 % with RABVs of Israel (accession no. HM114240, DQ837452, DQ837487, DQ837486 & DQ837483)while shared identity of 86 % with Jordan RABVs (DQ837427 & DQ837422) and 86.7 % with Jordan viruses with accession no. DQ837425& DQ837423.
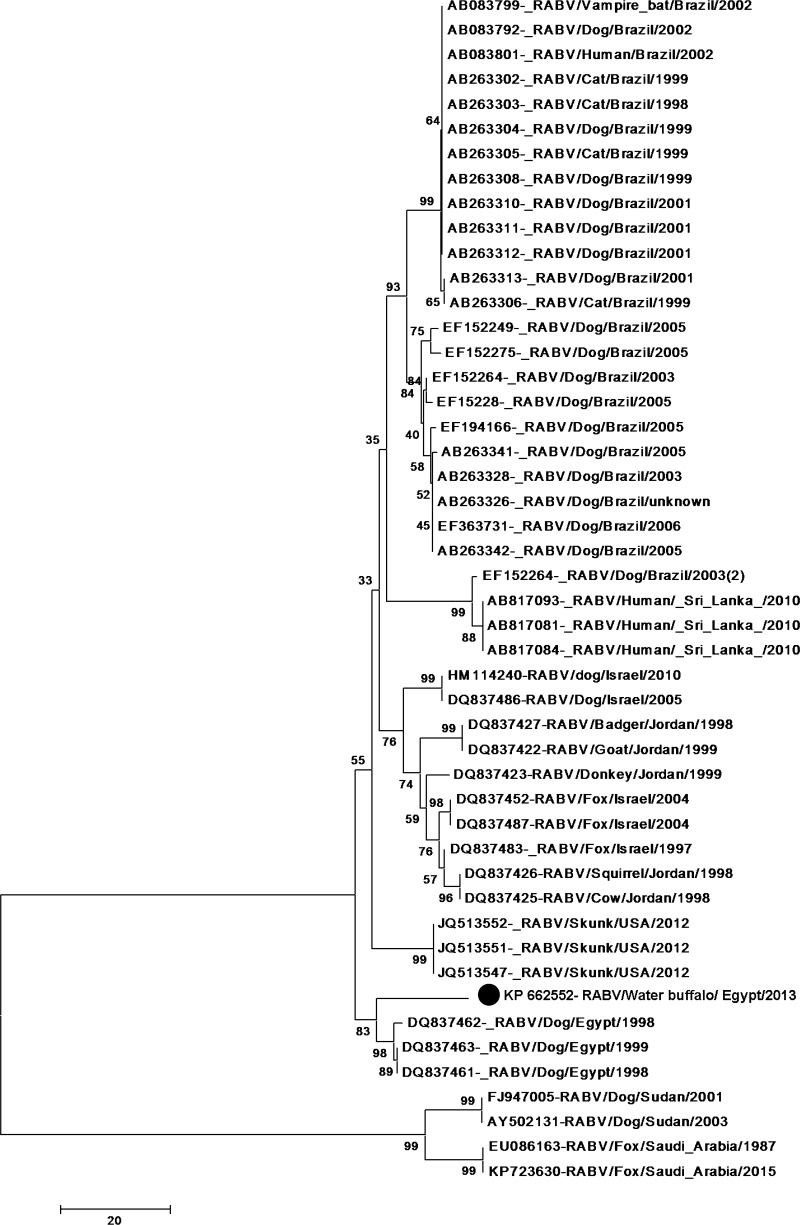
The Phylogenetic tree pattern for the alignment is shown in (Fig. 4).
Fig. 4.
Phylogenetic tree based on nucleoprotein (N) protein gene nucleotide sequences of our rabies virus from water buffalo with others rabies viruses whose N genes were retrieved from the GenBank database sequences. Numbers at the internal nodes represent the bootstrap probabilities (1000 replicates)
Discussion
Rabies is caused by a neurotrophic virus in the genus Lyssavirus. RABV is the causative agent of one of the most widespread, terrifying and deadly zoonotic disease. This virus is highly neurotropic, moving along peripheral nerves from the site of infection to the central nervous system, where it replicates and causes a progressive and invariably fatal neurologic disease [10]. The disease needs a rapid laboratory diagnosis after being suspected for rapid performing of appropriate measures such as slaughter or vaccination.
In this study, dFA and RT-PCR techniques used for detection of RABV in brain samples from two cases of water buffaloes (Bubalus bubalis) bitten by a fox in Egypt serve as a rapid, effective and economic method for laboratory confirmation of the disease [5, 20, 25]. The use of these techniques for direct detection of virus reduces the dependence on tissue culture and the time required to isolate RABV which may delay implementation of appropriate measures for control. Also, histopathological examination was used for diagnosis depending on the characteristic intracytoplasmic inclusion bodies (Negri bodies) but it is time consuming and less sensitive than FAT [12].
Sequence analysis of partial nucleotides sequence of the nucleoprotein (N) gene of our RABV showed close genetic relationship with Egyptian RABVs from dogs in 1998 and 1999 in Cairo and Faium. This indicated that these viruses may have originated from common ancestor and spread between different Governorates. These findings are in agreement with that previously reported [4, 7, 17]. This transgovernmental spread of RABVs occurred in Egypt may be through dog and foxes moving between Governorates without any restrictions. Comparing nucleotides sequences of the identified RABV with those from neighboring countries showed close genetic relationship with those from Jordan & Israel that indicate a potential transmission of RABV from and/or to our neighboring countries.
From this study we could concluded that, dFA and RT-PCR revealed the presence of RABV antigen and nucleic acid, respectively in the brain specimens from the buffalo bitten by fox and could help in rapid diagnosis of the disease. RABV circulate in Egypt among foxes, with occasional transmission to water buffaloes and the RABV from fox in Dakahlia Governorate showed close relationship at nucleotide sequences to RABVs from dogs in different Egyptian Governorates and with strains identified in Israel and Jordan indicating transmission of the virus between Egyptian Governorates with possible transmission from and/or to our neighboring countries through uncontrolled movement of foxes and dogs through the boundaries.
Electronic supplementary material
References
- 1.Bancroft JD, Stevens A, Turner DR. Theory and practice of histological techniques. 4. New York: Churchill Livingstone; 1996. [Google Scholar]
- 2.Blanton JD, Hanlon CA, Rupprecht CE. Rabies surveillance in the United States during 2006. J Am Vet Med Assoc. 2007;231:540–556. doi: 10.2460/javma.231.4.540. [DOI] [PubMed] [Google Scholar]
- 3.Botvinkin AD, Poleschuk EM, Kuzmin IV, Borisova TI, Gazarian SV, Yager P, Rupprecht CE. Novel lyssavirus isolated from bat in Russia. Emerg Infect Dis. 2003;9:1623–1625. doi: 10.3201/eid0912.030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourhy H, Kissi B, Tordo N. Molecular diversity of the Lyssavirus genus. Virology. 1993;194:70–81. doi: 10.1006/viro.1993.1236. [DOI] [PubMed] [Google Scholar]
- 5.Fischman HRS, Chaeffer M. Pathogenesis of experimental rabies as revealed by immunofluorescence. Ann N Y Acad Sci. 1971;177:78–97. doi: 10.1111/j.1749-6632.1971.tb35035.x. [DOI] [PubMed] [Google Scholar]
- 6.Hamir AN, Moser G. Immunoperoxidase test for rabies: utility as a diagnostic test. J Vet Diagn Invest. 1994;6:148–152. doi: 10.1177/104063879400600203. [DOI] [PubMed] [Google Scholar]
- 7.Kissi B, Tordo N, Bourhy H. Genetic polymorphism in the rabies virus nucleoprotein gene. Virology. 1995;209:526–537. doi: 10.1006/viro.1995.1285. [DOI] [PubMed] [Google Scholar]
- 8.Kuzmin IV, Orciari LA, Arai YT, Smith JS, Hanlon CA, Kameoka Y, Rupprecht CE. Bat lyssavirus (Aravan and Khujand) from Central Asia: phylogenetic relationships according to N, P and G gene sequence. Virus Res. 2003;97:65–79. doi: 10.1016/S0168-1702(03)00217-X. [DOI] [PubMed] [Google Scholar]
- 9.Langoni H, Lima K, Menozzi BD, Silva RC. Rabies in the big fruit-eating bat artibeus lituratus from botucatu, southeastern brazil. J Venom Anim Toxins Incl Trop Dis. 2005;2(1):84–87. [Google Scholar]
- 10.Rupprecht CE, Hanlon CA, Hemachudha T. Rabies re-examined. Lancet Infect Dis. 2002;2:337–353. doi: 10.1016/S1473-3099(02)00287-6. [DOI] [PubMed] [Google Scholar]
- 11.Sacramento D, Bourhy H, Tordo N. PCR technique as an alternative method for diagnosis and molecular epidemiology of rabies virus. Mol Cell Probes. 1991;5:229–240. doi: 10.1016/0890-8508(91)90045-L. [DOI] [PubMed] [Google Scholar]
- 12.Shankar BP. Advances in diagnosis of rabies. Vet World. 2009;2(2):74–78. [Google Scholar]
- 13.Smith JS. Rabies virus epitopic variation: use in ecologic studies. Adv Virus Res. 1989;36:215–253. doi: 10.1016/S0065-3527(08)60586-2. [DOI] [PubMed] [Google Scholar]
- 14.Smith JS, Orciari LA, Yager PA, Seidel HD, Warner CK. Epidemiologic and historical relationships among 87 rabies virus isolates as determined by limited sequence analysis. J Infect Dis. 1992;166:296–307. doi: 10.1093/infdis/166.2.296. [DOI] [PubMed] [Google Scholar]
- 15.Swanepoel R, Barnard BJ, Meredith CD, Bruckner GK, Foggin CM, Hubschle OI. Rabies in southern Africa. Onderstepoort J Vet Res. 1993;60:325–346. [PubMed] [Google Scholar]
- 16.Tang Q, Li H. Epidemic situation and related factors analysis of rabies in China. Chin J Epidemiol. 2005;26:223–224. [Google Scholar]
- 17.Tao X, Tang Q, Li H, Mo Z, Zhang H, Wang D, Zhang Q, Song M, Velasco-Villa A, Wu X, Rupprecht CE, Liang G. Molecular epidemiology of rabies in Southern People’s Republic of China. Emerg Infect Dis. 2009;15(8):1192–1198. doi: 10.3201/eid1508.081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans B R Soc Lond. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tepsumethanon V, Lumlertdacha B, Mitmoonpitak C, Fagen R, Wilde H. Fluorescent antibody test for rabies: prospective study of 8,987 brains. Clin Infect Dis. 1997;25:1459–1461. doi: 10.1086/516151. [DOI] [PubMed] [Google Scholar]
- 20.Tsiang H, Ceccaldi PE, Lycke E. Rabies virus infection and transport in human sensory dorsal root ganglia neurons. J Gen Virol. 1991;72:1191–1194. doi: 10.1099/0022-1317-72-5-1191. [DOI] [PubMed] [Google Scholar]
- 21.Viljoen GJ, Nel LH, Crowther JR. Molecular diagnostic PCR handbook. IAEA-FAO, Springer: Dordrecht; 2005. pp. 208–209. [Google Scholar]
- 22.Wiktor TJ, Koprowski H. Monoclonal antibodies against rabies virus produced by somatic cell hybridization: detection of antigenic variants. Proc Natl Acad Sci U S A. 1978;75:3938–3942. doi: 10.1073/pnas.75.8.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiktor TJ, Flamand A, Koprowski K. Use of monoclonal antibodies in diagnosis of rabies virus infection and differentiation of rabies and rabies-related viruses. J Virol Methods. 1980;1:33–46. doi: 10.1016/0166-0934(80)90005-1. [DOI] [Google Scholar]
- 24.Wunner W. Rabies virus. In: Jackson AC, Wunner W, editors. Rabies. London: Academic Press; 2007. pp. 23–68. [Google Scholar]
- 25.Zhang K, Guo J, Xu Z, Xiang M, Wu B, Chen H. Diagnosis and molecular characterization of rabies virus from a buffalo in China: a case report. Virol J. 2011;8:101–105. doi: 10.1186/1743-422X-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.