Abstract
The extent to which gene interaction or epistasis contributes to fitness variation within populations remains poorly understood, despite its importance to a myriad of evolutionary questions. Here, we report a multi-year field study estimating fitness of Mimulus guttatus genetic lines in which pairs of naturally segregating loci exist in an otherwise uniform background. An allele at QTL x5b—a locus originally mapped for its effect on flower size—positively affects survival if combined with one genotype at quantitative trait locus x10a (aa) but has negative effects when combined with the other genotypes (Aa and AA). The viability differences between genotypes parallel phenotypic differences for the time and node at which a plant flowers. Viability is negatively correlated with fecundity across genotypes, indicating antagonistic pleiotropy for fitness components. This trade-off reduces the genetic variance for total fitness relative to the individual fitness components and thus may serve to maintain variation. Additionally, we find that the effects of each locus and their interaction often vary with the environment.
Keywords: epistasis, QTL, fitness, antagonistic pleiotropy, field assay
1. Introduction
Heritable variation in fitness is the basis for evolution by natural selection, and understanding the genetic basis of this variation remains a major frontier in biology [1]. Genetic mapping studies have begun to identify the loci underlying quantitative traits, but the effects of these loci on fitness in nature largely remain unknown. Fitness is a complex function of many traits, few of which are measured in any mapping study. Additionally, the effect of a locus (gene) often depends on the environment experienced by the organism [2]. Field studies, using genetic lines containing known alleles, have shed light on the evolutionary significance of loci identified in controlled settings [3,4]. However, the common observation in mapping studies that genes interact with one another (i.e. exhibit epistasis) [5–7] has received limited attention in natural assays.
Natural assays of gene interactions directly address questions about speciation, adaptation and the evolution of sex [8]. Epistasis is routinely invoked to explain reproductive isolation between species via Dobzhansky–Muller incompatibilities—interactions between heterospecific alleles that cause reduced fitness in hybrids [9,10]. Multiple studies suggest that epistasis contributes to fitness differences between populations within a species [11], including direct evidence from field assays [4,12]. However, evidence for epistasis in fitness for loci that are naturally segregating within populations is limited. Indirect studies, which focus on traits presumably linked to fitness (e.g. flowering time [13]), suggest a role for epistasis, but direct evidence from field assays on the magnitude and pattern of epistasis is currently lacking. These details cannot be inferred from the between-population studies, because the alleles that differentiate populations or species may never have been simultaneously segregating in any population. Even if they had been, their effects would likely have changed substantially after many generations of isolation owing to the accrual of multiple mutations [14]. Natural assays estimating the magnitude and pattern of epistasis between segregating alleles are necessary to inform important issues such as the maintenance of genetic variation, and thus the evolutionary potential of populations.
In this paper, we describe a field assay for epistasis between a pair of naturally segregating loci mapped within a single population of Mimulus guttatus (yellow monkeyflower). In the greenhouse, these loci exhibit epistasis for multiple floral morphology traits (electronic supplementary material, table S1). In the field, these loci interact to determine the developmental timing of the transition to flowering, as well as several non-floral phenotypes. Importantly, we observe sign epistasis for viability (i.e. probability of living to set seed) wherein an allele has a positive effect in one genetic background and a negative effect in alternate backgrounds. We also document a negative correlation among genotypic values for viability and fecundity (antagonistic pleiotropy), which may contribute to the maintenance of these polymorphisms in nature.
2. Material and methods
Two loci (quantitative trait locus (QTL) x10a and QTL x5b), mapped for their effects on flower size, were introgressed into a common isogenic background by intercrossing two nearly isogenic lines (NILs). The isogenic background is IM767, an inbred line derived from approximately 15 generations of self-fertilization of field-collected seed. This line has medium floral trait values and genome sequencing reveals it to be highly homozygous [15]. Controlled crossing of the four possible double-homozygous genotypes produces the nine two-locus genotypes that were assayed. Prior to each field season, we confirmed genotypes performing PCR at diagnostic markers for a subset of individuals [7]. We germinated seed at the University of Oregon greenhouse at a date matching snowmelt in each year (5/1/12 and 5/15/14). Two weeks after germination, we transplanted seedlings into 98-well trays filled with a mixture of potting soil and soil from the transplant site (44.373238 N, 122.130675 W). Trays were placed in three transects of approximately equal length such that each tray was flush with the surrounding soil. In 2012 and 2014, the study consisted of 430 and 1177 individuals, respectively, split evenly across the nine two-locus genotypes.
Seedlings that washed out of the wells or died owing to transplant shock within the first week were removed from subsequent analyses. The remaining individuals were measured at day of first open flower for corolla width, pistil length, length of internode preceding flowering node, pedicle length and node of flower. We averaged measurements across flowers, although approximately 95% of surviving plants produced only one flower. We collected and counted all seed to determine fecundity.
We analysed the data under the following linear model:
where αi is the effect of QTL x10a, βj is the effect of QTL x5b, Yk is the effect of year, Fkl represents the effect of the flat, and all subsequent terms represent interactions between the relevant factors. Fkl was treated as random, while αi, βj and Yk were fixed factors. A significant effect of αβij indicates epistasis between loci. Non-significant interactions involving year were removed from the model. Seed count (fecundity) was natural log transformed prior to analysis owing to the right-skewed distribution, whereas the raw values were used for all other traits (electronic supplementary material, figure S1). Seed count was analysed only among the individuals that set seed (2012: n = 85; and 2014: n = 714). Traits were analysed using mixed effects logistic (viability data; ‘glmer’ function) or linear regression (all other traits; ‘lmer’ function) as implemented in the ‘lme4’ package in R. For viability, significance was determined using ‘anova.merMod’, which contrasts nested models with a likelihood ratio test, and type III Wald χ2-tests as determined by the ‘Anova’ function (‘car’ package) for the remaining traits. We found that Poisson regression for untransformed seed count, which includes individuals that set zero seed, largely mirrors results from viability analysis (electronic supplementary material, table S2); therefore, we opt for analysing viability and fecundity separately. Genotypic values presented in the figures are the least-squares means resulting from the linear model fits (‘lsmeans’ package) for all traits except viability, in which we calculated the survival probability from the logistic regression coefficients. The per cent of explained variance (partial R2) for each term and significance of post-hoc comparisons were determined with the ‘pamer.fnc’ and ‘mcposthoc.fnc’ functions, respectively, (‘LMERConvenienceFunctions’ package). Greenhouse data come from a previous study [16] (details of analysis can be found in the electronic supplementary material table files).
3. Results and discussion
Epistasis had a highly significant effect on viability and on several morphological and developmental traits (table 1). Point estimates for viability suggest sign epistasis (figure 1): the ‘B’ allele at QTL x5b has a negative effect on viability in the ‘aa’ background at QTL x10a, but a positive effect in other backgrounds (‘Aa’ and ‘AA’). The implications of epistatic selection are numerous [17,18], but a simple prediction is that allele frequency change at one locus will change depending on allele frequencies at other interacting loci [19]. In this case, viability selection will favour allele ‘b’ at x5b if the ‘a’ allele at x10a is at high frequency. Conversely, if ‘a’ is at low frequency, the ‘B’ allele at x5b will be favoured. If selection acts purely through viability, the ultimate loss or fixation of either allele at x5b will be determined by the loss or fixation of alleles at x10a.
Table 1.
Summary of linear model fit.
| term | χ2 | d.f. | p-value | partial R2 | term | χ2 | d.f. | p-value | partial R2 |
|---|---|---|---|---|---|---|---|---|---|
| viability | fecundity | ||||||||
| year | 52.56 | 1 | <0.0001 | 24.684 | year | 3.06 | 1 | 0.0804 | 0.331 |
| x10a | 23.97 | 2 | <0.0001 | 4.221 | x10a | 6.20 | 2 | 0.0450 | 0.444 |
| x5b | 13.46 | 2 | 0.0012 | 2.777 | x5b | 0.62 | 2 | 0.7325 | 0.266 |
| x10a × x5b | 24.61 | 4 | <0.0001 | 6.059 | x10a × x5b | 4.69 | 4 | 0.3210 | 0.529 |
| days to flower | pedicle length | ||||||||
| year | 2.85 | 1 | 0.0912 | 0.599 | year | 4.83 | 1 | 0.0279 | 0.055 |
| x10a | 6.64 | 2 | 0.0362 | 0.740 | x10a | 31.53 | 2 | <0.0001 | 0.017 |
| x5b | 0.48 | 2 | 0.7856 | 4.794 | x5b | 6.59 | 2 | 0.0371 | 4.321 |
| x10a × x5b | 12.15 | 4 | 0.0163 | 1.237 | x10a × x5b | 39.28 | 4 | <0.0001 | 1.647 |
| x10a × year | 6.27 | 2 | 0.0436 | 0.093 | x10a × year | 24.88 | 2 | <0.0001 | 0.404 |
| x5b × year | 2.03 | 2 | 0.3626 | 0.650 | x5b × year | 13.69 | 2 | 0.0011 | 1.736 |
| x10a × x5b × year | 11.65 | 4 | 0.0201 | 0.959 | x10a × x5b × year | 26.60 | 4 | <0.0001 | 2.164 |
| corolla width | internode length | ||||||||
| year | 57.43 | 1 | <0.0001 | 5.325 | year | 52.24 | 1 | <0.0001 | 6.851 |
| x10a | 5.17 | 2 | 0.0752 | 0.501 | x10a | 2.64 | 2 | 0.2675 | 0.312 |
| x5b | 1.20 | 2 | 0.5482 | 0.278 | x5b | 3.49 | 2 | 0.1743 | 0.778 |
| x10a × x5b | 2.96 | 4 | 0.5652 | 0.196 | x10a × x5b | 27.60 | 4 | <0.0001 | 1.591 |
| x10a × year | 6.57 | 2 | 0.0374 | 0.436 | x10a × year | 6.57 | 2 | 0.0375 | 0.390 |
| pistil length | node | ||||||||
| year | 37.46 | 1 | <0.0001 | 3.898 | year | 38.71 | 1 | <0.0001 | 9.039 |
| x10a | 4.81 | 2 | 0.0901 | 0.140 | x10a | 1.81 | 2 | 0.4043 | 0.465 |
| x5b | 2.65 | 2 | 0.2656 | 0.352 | x5b | 12.03 | 2 | 0.0024 | 4.335 |
| x10a × x5b | 2.49 | 4 | 0.6464 | 0.170 | x10a × x5b | 55.17 | 4 | <0.0001 | 3.651 |
| x10a × year | 7.15 | 2 | 0.0281 | 0.482 | x10a × year | 12.14 | 2 | 0.0023 | 0.695 |
| x5b × year | 10.38 | 2 | 0.0056 | 0.716 | |||||
Figure 1.
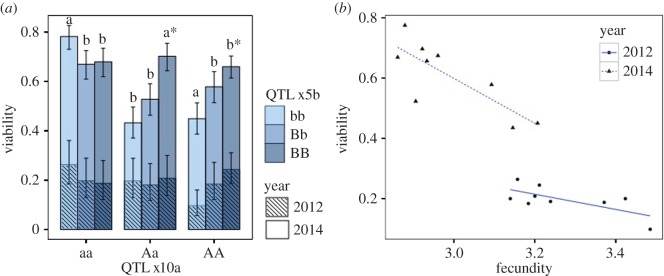
(a) Genetic values for viability of the nine two-locus genotypes for QTL pair x10a–x5b for each year. Different letters indicate significant differences within each ‘A’ background at p < 0.05 for 2014 data. Asterisk indicates p < 0.0001 for the difference between ‘B’ homozygotes within an ‘A’ background. (b) Relationship between viability and fecundity for the nine two-locus genotypic values (2012: r = −0.69; p = 0.039. 2014: r = −0.80; p = 0.008). (Online version in colour.)
The negative correlation between viability and fecundity, the hallmark of antagonistic pleiotropy, should reduce the efficacy of viability selection (figure 1b). Considering total fitness as the product of viability and (female) fecundity, the negative correlation between these components implies that variance in total fitness is reduced relative to the individual components. Since the change in allele frequency is proportional to the fitness variance attributable to a locus, the negative trade-off will slow evolution at these loci, thus promoting the maintenance of polymorphism. In the simplest models, antagonistic pleiotropy between viability and fecundity does not guarantee stable polymorphism unless the total fitness of the heterozygous genotype exceeds both homozygotes (i.e. the locus exhibits overdominance) [20]. Our point estimates for total fitness are not sufficiently precise to determine whether this condition is satisfied for these loci.
The observed viability–fecundity trade-off seems to be driven by the epistatic effects of the loci on development time. The negative correlation between node and viability is accompanied by a positive correlation between node and fecundity, particularly in 2014 (figure 2). Genotypes that flowered at a later node were less likely to survive, but produced more seed if they did survive. This correlation is not significant in 2012, probably because genotypic values are based on very few measurements (approx. 10 individuals per genotype survived to flower). The node at first flower is also positively correlated with the day of flower (r = 0.864, p = 0.0027; based on least-squares means across years). In annual, alpine M. guttatus populations, viability selection primarily acts through the cessation of water availability at the end of the summer (approx. 80% of observed mortality occurred within 10 days in 2014). While individuals that delay flowering to later nodes have the capacity to produce more seed, they run a greater risk of not flowering before senescing.
Figure 2.
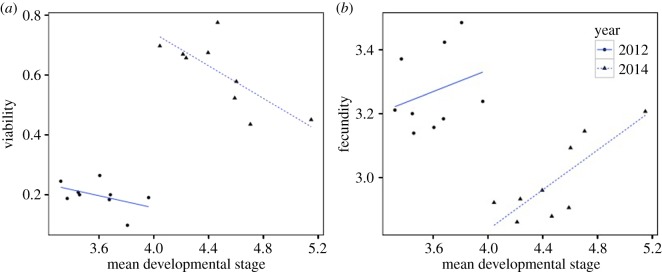
Relationship between fitness components and developmental stage at which plants flower (2012: viability r = −0.47, p = 0.21 and fecundity r = 0.28, p = 0.46; 2014: viability r = −0.76, p = 0.02 and fecundity r = 0.81, p = 0.008). (Online version in colour.)
Other than epistasis and antagonistic pleiotropy, environmental heterogeneity affects polymorphism at these two loci. We observed a strong dependency on environment for both marginal effects (gene-by-environment or G×E interactions) as well as the epistatic/interaction terms (G×G×E) for certain traits (electronic supplementary material, table S3 and figure S2). Of the traits for which significant epistasis was observed in the field, only days to flower exhibits epistasis in the greenhouse. We do not see epistasis for corolla width or pistil length in the field, despite observing strong epistasis in the greenhouse, and the opposite is true for internode and pedicle length (table 1 and electronic supplementary material, table S1). Importantly, only surviving plants manifest the phenotype in the field and prior studies have shown these to be non-random with respect to flower size genotype [21].
Marginal and epistatic effects also varied significantly across years for some traits. The two years of this study differed markedly in terms of the magnitude and pattern of temperature and precipitation. Excluding fitness traits, a significant interaction with year was observed for at least one genetic effect (marginal or epistatic) for each trait (table 1 and electronic supplementary material, figure S2). Genetic interactions with the environment are important because even if selection acts consistently on a trait across time and space, change in allele frequency may not be consistent if marginal and/or epistatic genetic effects depend on environment. Although the genetic effects on viability did not differ significantly across years in this study, spatial and temporal variations in selection have been documented for other polymorphisms in this species and locale [3].
In summary, this experiment provides some of the first evidence that alleles segregating within a natural population generate epistasis in fitness. Alleles interact not only with each other, but also with the environment that the organism experiences. Genetic effects are small relative to environmental effects, but on par with the effects of loci differentiating populations (e.g. comparing the portion of explained variance due to genetic effects relative to [4]). As this study focuses on only two loci, we cannot estimate the quantitative contribution of epistasis to fitness variation. However, this study does demonstrate the importance of natural assays to understand the relevance of genetic mapping studies to evolution in nature.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank L. Hileman and S. Macdonald for comments on the manuscript.
Data accessibility
All data are provided as the electronic supplementary material.
Authors' contributions
P.J.M. and J.K.K. designed the experiment. P.J.M. was responsible for field experiments, analyses, figures and initial drafts. J.K.K. created the NILs and revised drafts of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by NIH grant no. R01 GM073990 (J.K.K.) and the KU Botany Endowment (P.J.M.).
References
- 1.Charlesworth B. 2015. Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc. Natl Acad. Sci. USA 112, 1662–1669. ( 10.1073/pnas.1423275112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheiner SM. 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35–68. ( 10.1146/annurev.es.24.110193.000343) [DOI] [Google Scholar]
- 3.Mojica JP, Lee YW, Willis JH, Kelly JK. 2012. Spatially and temporally varying selection on intrapopulation quantitative trait loci for a life history trade-off in Mimulus guttatus. Mol. Ecol. 21, 3718–3728. ( 10.1111/j.1365-294X.2012.05662.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malmberg RL, Held S, Waits A, Mauricio R. 2005. Epistasis for fitness-related quantitative traits in Arabidopsis thaliana grown in the field and in the greenhouse. Genetics 171, 2013–2027. ( 10.1534/genetics.105.046078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W, et al. 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl Acad. Sci. USA 109, 15 553–15 559. ( 10.1073/pnas.1213423109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore JH. 2003. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 56, 73–82. ( 10.1159/000073735) [DOI] [PubMed] [Google Scholar]
- 7.Kelly JK, Mojica JP. 2011. Interactions among flower-size QTL of Mimulus guttatus are abundant but highly variable in nature. Genetics 189, 1461–1471. ( 10.1534/genetics.111.132423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto SP, Feldman MW. 1997. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 51, 134–147. ( 10.1006/tpbi.1997.1301) [DOI] [PubMed] [Google Scholar]
- 9.Willett C. 2011. The nature of interactions that contribute to postzygotic reproductive isolation in hybrid copepods. Genetica 139, 575–588. ( 10.1007/s10709-010-9525-1) [DOI] [PubMed] [Google Scholar]
- 10.Moyle LC, Nakazato T. 2009. Complex epistasis for Dobzhansky–Muller hybrid incompatibility in Solanum. Genetics 181, 347–351. ( 10.1534/genetics.108.095679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroymann J, Mitchell-Olds T. 2005. Epistasis and balanced polymorphism influencing complex trait variation. Nature 435, 95–98. ( 10.1038/nature03480) [DOI] [PubMed] [Google Scholar]
- 12.Fenster CB, Galloway LF. 2000. Depresión por endogamia y exogamia en poblaciones naturales de Chamaecrista fasciculata (Fabaceae) [Inbreeding and outbreeding depression in natural populations of Chamaecrista fasciculata (Fabaceae)]. Conserv. Biol. 14, 1406–1412. ( 10.1046/j.1523-1739.2000.99234.x) [DOI] [Google Scholar]
- 13.Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl Acad. Sci. USA 101, 15 670–15 675. ( 10.1073/pnas.0406232101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. 2007. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448, 587–590. ( 10.1038/nature05988) [DOI] [PubMed] [Google Scholar]
- 15.Flagel LE, Willis JH, Vision TJ. 2014. The standing pool of genomic structural variation in a natural population of Mimulus guttatus. Genome Biol. Evol. 6, 53–64. ( 10.1093/gbe/evt199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monnahan PJ, Kelly JK. 2015. Epistasis is a major determinant of the additive genetic variance in Mimulus guttatus. PLoS Genet. 11, e1005201 ( 10.1371/journal.pgen.1005201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf JB, Brodie ED, Wade MJ. 2000. Epistasis and the evolutionary process. Oxford, UK: Oxford University Press. [Google Scholar]
- 18.Carter AJR, Hermisson J, Hansen TF. 2005. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor. Popul. Biol. 68, 179–196. ( 10.1016/j.tpb.2005.05.002) [DOI] [PubMed] [Google Scholar]
- 19.Wade M. 2002. A gene's eye view of epistasis, selection and speciation. J. Evol. Biol. 15, 337–346. ( 10.1046/j.1420-9101.2002.00413.x) [DOI] [Google Scholar]
- 20.Hedrick PW. 1999. Antagonistic pleiotropy and genetic polymorphism: a perspective. Heredity 82, 126–133. ( 10.1038/sj.hdy.6884400) [DOI] [Google Scholar]
- 21.Mojica JP, Kelly JK. 2010. Viability selection prior to trait expression is an essential component of natural selection. Proc. R. Soc. B 277, 2945–2950. ( 10.1098/rspb.2010.0568) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided as the electronic supplementary material.


