Abstract
The intercalary segment is a limbless version of the tritocerebral segment and is present in the head of all insects, whereas other extant arthropods have retained limbs on their tritocerebral segment (e.g. the pedipalp limbs in spiders). The evolutionary origin of limb loss on the intercalary segment has puzzled zoologists for over a century. Here we show that an intercalary segment-like phenotype can be created in spiders by interfering with the function of the Hox gene labial. This links the origin of the intercalary segment to a functional change in labial. We show that in the spider Parasteatoda tepidariorum the labial gene has two functions: one function in head tissue maintenance that is conserved between spiders and insects, and a second function in pedipalp limb promotion and specification, which is only present in spiders. These results imply that labial was originally crucial for limb formation on the tritocerebral segment, but that it has lost this particular subfunction in the insect ancestor, resulting in limb loss on the intercalary segment. Such loss of a subfunction is a way to avoid adverse pleiotropic effects normally associated with mutations in developmental genes, and may thus be a common mechanism to accelerate regressive evolution.
Keywords: chelicerata, Hox genes, bodyplan evolution, tritocerebral segment, pedipalp specification, regressive evolution
1. Introduction
The evolution of novel animal body plans requires the evolution of novel traits. Often, however, new bodyplans also emerge by the loss of features and organs. Examples of this so-called regressive evolution include eye loss in cavefish [1], wing loss in ants [2] or the loss of limbs in the snake bodyplan [3]. Another textbook example for regressive evolution is limb loss on the intercalary segment in insects [4]. The limbless intercalary segment is an integral component of the head capsule of all insects and, therefore, must be phylogenetically older than all extant insect groups. The controversy about its origin and the genetic basis for its limblessness ranks among the longest standing unsolved problems in zoology.
Anatomically, the intercalary segment corresponds to the tritocerebral segment in other arthropods [5]. In contrast to insects, several non-insect arthropods (e.g. spiders) have retained the limbs of the tritocerebral segment, thus providing a valuable model for tritocerebral segment development before the onset of regressive evolution in the insect ancestor. We have, therefore, studied the developmental genetic mechanisms of tritocerebral segment formation in the spider Parasteatoda tepidariorum (previously placed in the genus Achaearanea [6]). In spiders, the tritocerebral segment bears the pair of pedipalps [5], a multi-functional appendage type used in feeding, sensory perception and for many specialized purposes like visual communication, stridulation or sperm transfer (reviewed in [7]). The developmental genetic mechanisms needed to form the pedipalpal segment and its appendages are, however, not known.
One gene that is strongly expressed in the tritocerebral segment of all arthropods studied so far is the Hox gene labial (lab) [8]. On the one hand, this strong evolutionary conservation indicates an important role of lab in the arthropod tritocerebral segment. On the other hand, lab expression is conserved in all arthropods, regardless of the presence of limbs on the tritocerebral segment, and therefore, a possible role of lab in the control of limblessness has been dismissed previously [9]. In this work, we have investigated the role of lab in the spider P. tepidariorum and its importance for limb formation in the spider tritocerebral segment, i.e. the pedipalpal segment. We show that apart from a role in head tissue maintenance already known from insects, spider lab has a role in appendage initiation and development that is not present in insects. We propose that the loss of this function in the insect ancestor initiated regressive evolution in the insect tritocerebral segment and thus gave rise to the limbless intercalary segment.
2. Material and methods
(a). Spider colony and gene cloning
Embryos and adults of P. tepidariorum were obtained from our Göttingen strain of P. tepidariorum for all experiments. The animals were kept separately in small plastic vials at 25°C and controlled light cycle (12 L : 12 D cycle). Isolation of gene fragments from P. tepidariorum was performed according to standard molecular cloning techniques. Sequences of primers and accession numbers are included in the electronic supplementary material.
(b). RNA interference
Parental RNA interference (RNAi) was performed as described [10] with minor modifications. To exclude off-target effects non-overlapping fragments were injected separately (electronic supplementary material, figure S4). Full documentation of the injections is also available in the electronic supplementary material.
(c). In situ hybridization, DNA labelling and imaging
In situ hybridization and nuclear staining with Sytox Green were performed as described [11,12] with minor modifications (details are included in the electronic supplementary material). TUNEL detection of fragmented DNA was done as described previously [13]. Images were captured with a Zeiss Axioplan-2 microscope or with a Leica dissection microscope equipped with an Intas digital camera and UV light. Confocal z-stacks of larvae were captured by using a Zeiss LSM 510 microscope.
3. Results
(a). Expression of labial in Parasteatoda tepidariorum
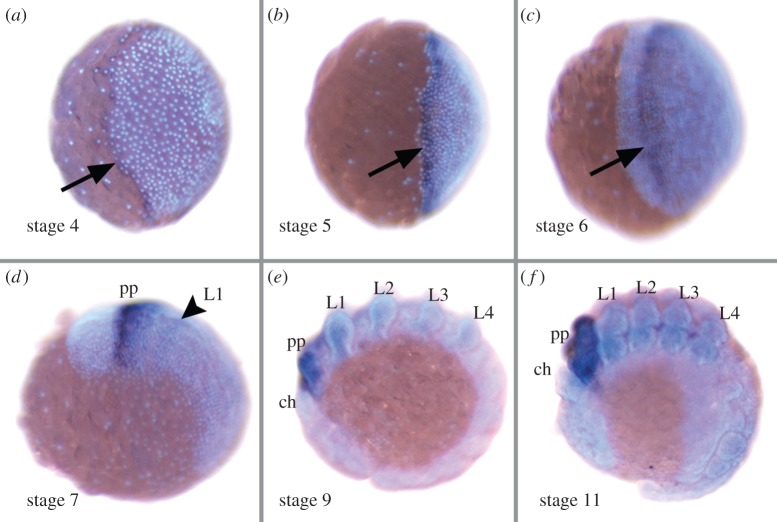
P. tepidariorum has two paralogous copies of the lab gene (see electronic supplementary material for sequence accession numbers). One of these genes, that we denote as lab-1, is expressed in the early embryo (starting at stage 4; staging after [14]) in a circumferential ring along the rim of the germ disc (figure 1a), which becomes broader during stage 5 (figure 1b).
Figure 1.
Developmental expression of lab-1. (a–c) Lateral view of the germ disc at stage 4 (a), stage 5 (b) and stage 6 (c). (d–f) Lateral view of germ band stages, anterior to the left, at stage 7 (d), stage 9 (e) and stage 11 (f). Arrows indicate expression in the presumptive pedipalp segment tissue. The arrowhead indicates beginning weak expression in the L1 segment. Abbreviations: ch, chelicera; L1–L4, walking legs 1–4; pp, pedipalp. (Online version in colour.)
During the transformation of the germ disc into the bilaterally symmetric germ band, which starts at stage 6, the cephalic region as well as the cheliceral and the pedipalpal segments are patterned via a dynamic anterior to posterior wave of gene expression. This dynamic wave is mainly organized by an autoregulatory loop that involves Hedgehog-signalling and the transcription factors orthodenticle and odd-paired [15,16]. The expression domains of several genes that are initially located at the rim of the germ disc during stage 5, are travelling to a more posterior position during stages 6–8. Because lab-1 is co-expressed with several of these ‘travelling genes’ during stage 5, the expression ring of lab-1 is also subject to this wave of gene expression and is relocated from the rim towards the presumptive pedipalpal segment (figure 1c). After the formation of the germ band at stage 7, lab-1 is strongly expressed in the pedipalpal segment and weaker expression is also detected in the first walking leg segment (figure 1d). In later developmental stages (starting with stage 9) there is also very faint expression in the remaining walking leg segments (figure 1e,f). The second lab gene, that we denote as lab-2, is transcribed only late and no expression is detected before stage 6. RNAi with lab-2 did not result in detectable phenotypes.
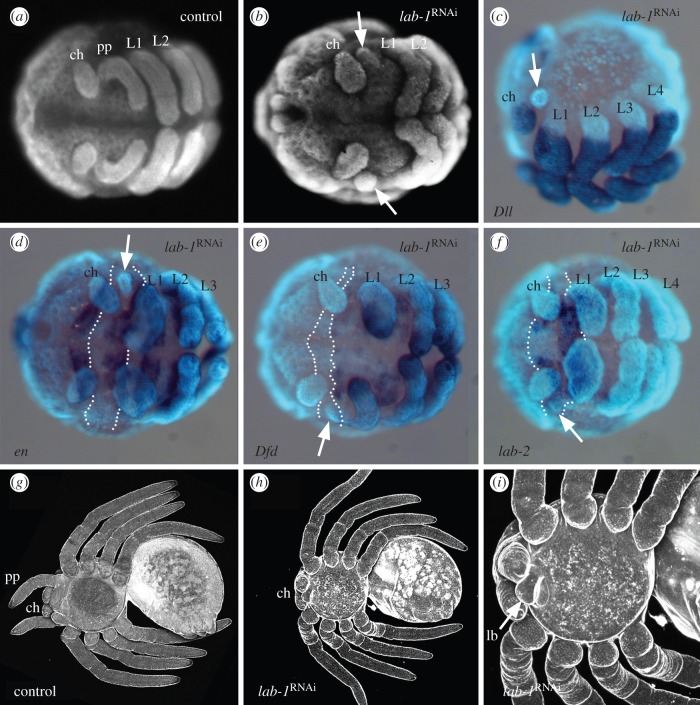
(b). Pt-lab-1 RNAi can create an intercalary segment-like phenotype in the spider
RNAi with lab-1 function led to characteristic phenotypes. The lab-1 RNAi phenotypes comprise defects in the pedipalpal and first walking leg segment, thus corresponding well with the early expression domains of lab-1 in the pedipalpal and first walking leg segments. There are two major categories of phenotypes: in category 1 (denoted in purple in electronic supplementary material, figure S1), only the pedipalpal segment is affected. In category 2 (denoted in green and orange in electronic supplementary material, figure S1), there are additional defects of different severity in the L1 segment. The pedipalp appendages are mostly completely reduced, but small remnants may be present (figure 2b). The expression of the appendage marker gene Distal-less (Dll) is entirely absent from the pedipalpal segment in lab-1 RNAi animals (figure 2c; electronic supplementary material, figure S2), indicating that the specification of appendages in this segment is disrupted. In category 1 and weakly affected category 2 animals the pedipalpal segment itself is reduced in size, but is otherwise morphologically normal (except for the missing appendages). This is also confirmed by genetic marker expression. The segmental borders are specified normally as illustrated by the expression of the segment polarity marker engrailed (en) at the posterior boundary of the limbless pedipalpal segment (figure 2d). The segment also retains its normal identity as indicated by the expression of other Hox genes. In the wild-type, lab-2 and Deformed (Dfd) are expressed in the pedipalpal and in the walking leg segments, respectively, and these patterns are unchanged in lab-1 RNAi animals (figure 2e,f). These results thus demonstrate a role of lab-1 during pedipalp appendage specification.
Figure 2.
Phenotypes after lab-1 RNAi. (a,b) Epifluorescent images stained with Sytox Green. (a) Wild-type embryo. (b) lab-1 RNAi embryo. (c–f) Combined epifluorescent images (light blue) and light microscopy images of lab-1 RNAi embryos stained for expression of Dll (c), en (d), Dfd (e) and lab-2 (f). The arrows in (b–f) point to vestiges of the pedipalp. The dotted lines in (d–f) indicate the approximate area of the pedipalp segment. (g–i) Confocal images of larvae. (g) Wild-type larva. (h) lab-1 RNAi larva. (i) Magnified view of prosoma of a lab-1 RNAi larva, showing the labium (lb). All panels in ventral view and anterior up. See figure 1 for abbreviations. (Online version in colour.)
In category 2 animals, there are additional defects in the L1 segment. In weaker phenotypes, in the L1 segment the legs are only shortened and broadened. In stronger phenotypes, the legs in L1 are entirely missing (electronic supplementary material, figure S3a). Initially, in these embryos the pedipalpal and the L1 segment are physically present (at least partially) as indicated by the gap between the cheliceral segment and the L2 segment (electronic supplementary material, figure S3b). But the absence of segmental gene expression (e.g. stripes of en) in this area points to severe developmental deficiencies, and the rate of cell death is strongly increased in the entire prosomal part of the germ band (electronic supplementary material, figure S3c,d), which is also observed after lab loss in insects [17–19]. This suggests that lab-1 is also generally required for tissue maintenance in the entire anterior portion of the germ band, including the pedipalpal and L1 segments.
Only few lab-1 RNAi embryos survive and reach the larval stage, and no animals survive beyond the first larval instar. These larvae completely lack the pedipalpal appendages (figure 2g,h), but other elements of the pedipalpal segment are present, demonstrated by the presence of the labium (figure 2i), which is a protrusion of the sternum (the ventral armour plate of the spider exoskeleton) and is the modified sternite of the pedipalpal segment [20]. Thus, the morphology of the pedipalpal segment in these lab-1 RNAi larvae strikingly resembles the intercalary segment in insects. We did not observe homeotic transformations in lab-1 RNAi animals, neither in the first larval instar nor in the embryo. The transformation of dorsal head tissue towards thoracic fate has been described for Drosophila melanogaster, but only in the adult, and not in the embryo or larva [17]. Thus, the available data from P. tepidariorum are compatible with the data from D. melanogaster, because in both cases no transformations are present before the adult stage. The late role in homeosis might also be present in P. tepidariorum, but because lab-1 RNAi animals die before reaching the adult stage, we were not able to investigate this possibility.
(c). Pt-lab-1 can transform the L1 segment towards pedipalp identity
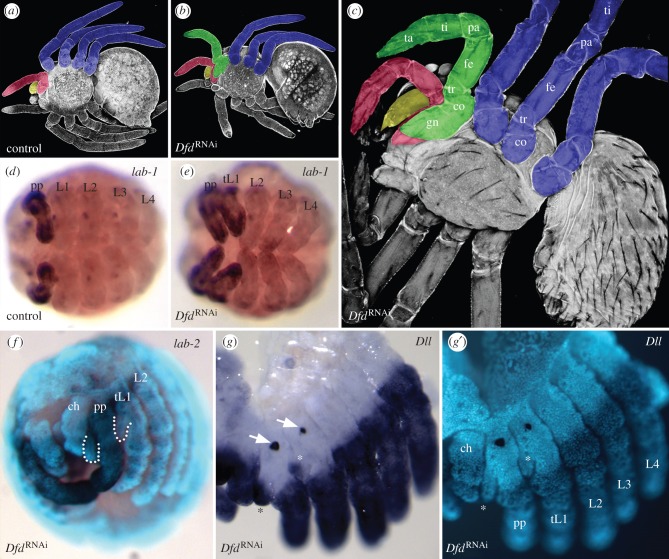
Because lab-1 has a role in pedipalp appendage specification, we also asked whether lab-1 can play this role in other segments, i.e. is ectopic expression of lab-1 able to initiate pedipalp appendage development in other segments? Because targeted misexpression is not yet available in P. tepidariorum, we achieved activation of lab-1 in the L1 segment by interfering with the function of the Hox gene Dfd. Dfd is expressed in all four walking leg segments and thus posteriorly adjacent to the pedipalpal segment [21]. RNAi with Dfd leads to the ectopic expression of lab-1 (figure 3d,e), but not of lab-2 (figure 3f), in the L1 segment. As a consequence, the L1 legs are transformed towards pedipalp identity. Pedipalps can easily be distinguished from legs: they differ from the legs by being shorter and missing one limb segment (the metatarsus). The pedipalps are also characterized by the presence of a ventral outgrowth, the gnathendite, that in the embryo is further identified by its separate expression of Dll. In addition, late embryonic stages possess an egg tooth on the outside of the basal portion of the pedipalps, which is used for penetration of the eggshell. In Dfd RNAi animals the transformation of L1 into pedipalp is either partial or complete (summary in electronic supplementary material, figure S1). The partially transformed L1 leg is shortened and proximally thickened. In complete transformations, the transformed L1 leg is morphologically indistinguishable from the normal pedipalp (figure 3a,b). In the embryo, the gnathendite of the transformed L1 leg expresses Dll and the egg tooth is also present (figure 3g,g′). In nymphs that have completed cuticle differentiation and limb joint formation, it can be demonstrated that the fully transformed L1 leg indeed consists of only six limb segments and is lacking the metatarsus (figure 3c). The fact that only lab-1, but not lab-2 is misexpressed in the L1 segment after Dfd RNAi, indicates that the specification of pedipalp appendage identity observed in the transformed L1 legs is the specific effect of lab-1.
Figure 3.
Phenotypes after Dfd RNAi. (a–c) Confocal images, ventral view. Appendages on the right body half have been colourized to indicate their identity (blue, leg; red, pedipalp; yellow, chelicera; green, first leg transformed into pedipalp identity). Limb segments are first visible in the nymph (indicated in c). (a) Control larva. (b) Dfd RNAi larva. (c) Dfd RNAi first nymphal instar. (d and e) Light microscopical images of stage 11 embryos stained for lab-1, ventral view. (d) Control embryo. (e) Dfd RNAi embryo (tL1, transformed L1). (f) lab-2 expression in Dfd RNAi embryo. Combined epifluorescent and light microscopy image. Dotted lines denote gnathendites in pedipalp and tL1. (g,g′) Expression of Dll in a Dfd RNAi embryo. Light microscopy image (g) and corresponding epifluorescent image (g′). Arrows and asterisks point to the egg tooth and Dll expressing gnathendite, respectively, in the pedipalp and the tL1. co, coxa; fe, femur; gn, gnathendite; pa, patella; ta, tarsus; ti, tibia; tr, trochanter. See figure 1 for other abbreviations. (Online version in colour.)
4. Discussion
The Hox gene labial shows a highly conserved expression pattern in the tritocerebral segment in all arthropods [8] and even in onychophorans [22,23], most probably the arthropod sister group [24,25]. Consistent with this observation, our results identify a highly conserved role of labial in head tissue maintenance that has been known already from functional studies in D. melanogaster and the beetle Tribolium castaneum. In D. melanogaster loss of lab function leads to increased cell death in the head region and eventually to the loss of several head segments including the intercalary, mandibular, maxillary and labial segments, hence the gene name [17]. A similar effect has also been observed in the beetle T. castaneum, where lab RNAi phenocopies show increased cell death in the head including at least the intercalary, mandibular and maxillary segment [18].
However, our results also reveal a second role of lab-1 in the specification and formation of the pedipalp limbs. This role is apparently absent from insect lab homologues, because they lack the corresponding limbs altogether, although lab is strongly expressed in the insect intercalary segment. Intriguingly, the loss of this lab-1 function in spiders removes the genetic cues to form appendages on the pedipalpal segment and thus experimentally recapitulates the origin of limb loss on the tritocerebral segment. The result is a pedipalpal segment phenotype that is strikingly similar to the insect intercalary segment, not only in terms of morphology but also in terms of gene expression. We therefore suggest that the dual role seen in lab-1 in P. tepidariorum represents the ancestral condition of the labial gene in arthropods, and is retained in most chelicerates and probably also in the second antennal segment of crustaceans [26]. The role in head tissue maintenance appears to be indispensable and is highly conserved, but the role in limb formation has been lost in insects. Interestingly, insects are not the only arthropods that lack limbs on the intercalary segment homologue. Myriapods also lack limbs on the tritocerebral segment [27,28]. This limb loss has previously been regarded as homologous in myriapods and insects and served as the key character for uniting the two groups in the taxon Antennata [28], but recent molecular phylogenies do not support the Antennata taxon and place the myriapods at the base of all mandibulate arthropods [29]. Accordingly, limb loss on the tritocerebral segment in insects and in myriapods has evolved by convergence. Another intriguing case is the Pycnogonida (sea spiders), an enigmatic arthropod group usually considered as relatives of spiders and other chelicerates [29,30]. Some species of Pycnogonida lack the pedipalp limbs, while other species have them fully formed [31]. Thus, in pycnognids limb loss on the tritocerebral segment apparently evolved several times within the group. The role of lab in myriapods and pycnogonids, however, is not yet known and it will therefore be interesting to study, whether their limblessness is also linked to evolutionary changes in the role of lab.
The role of selection in the regressive evolution of organs was already discussed by Charles Darwin in ‘On the origin of species’ [32]. Using the loss of eyes in cavefish as an example, he realized that the loss of eyes is neither particularly advantageous, nor would the preservation of eyes be a major disadvantage. It appeared to Darwin that cavefish have lost their eyes simply because it does not matter whether they have eyes or not. Owing to this line of reasoning, it is generally assumed that regressive evolution is caused by neutral evolution and genetic drift, rather than natural selection. However, recent studies provide evidence for strong positive selection for the reduction of traits [33,34], especially if the reduced trait is genetically coupled to another highly adaptive trait (antagonistic pleiotropy) [35,36]. Although limb loss on the tritocerebral segment has apparently evolved several times, the selectional benefit of a limbless tritocerebral segment is unclear at the moment. At any rate, losing the function of a gene is especially difficult for developmental genes, because they usually have more than one function and their loss would lead to multiple defects with negative rather than positive or neutral effect. Our results with lab-1 suggest that the loss of only a particular subfunction of a developmental gene is one way to avoid adverse pleiotropic effects, and may thus be a common mechanism to facilitate regressive evolution in animals.
Supplementary Material
Acknowledgements
We thank Marco Winkler for technical assistance, Beate Preitz for help with confocal microscopy and Matt Benton for critical reading of the manuscript. We also thank Wim Damen, Sara Khadjeh, Alistair McGregor and Ernst Wimmer for discussions.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
M.P. performed the molecular cloning work and the RNAi analyses and commented on the manuscript. E.E.S. performed parts of the cloning and in situ hybridization analysis. N.T. performed parts of the lab-2 RNAi analysis. M.P. and N.M.P. designed the study and analysed the data. N.M.P. wrote the paper. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work has been funded by the Deutsche Forschungsgemeinschaft (DFG; grant nos. PR 1109/1-1, PR 1109/4-1, and PR 1109/6-1 to N.M.P.). Additional financial backing has been received from the Göttingen Graduate School for Neurosciences, Biophysics and Molecular Biosciences (GGNB), the Göttingen Center for Molecular Biosciences (GZMB) and the University of Göttingen (GAU). N.T. is supported by a Christiane-Nüsslein-Volhard-Foundation fellowship and a ‘Women in Science’ award by L'Oréal Deutschland and the Deutsche UNESCO-Kommission.
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. 2007. Regressive evolution in the Mexican Cave tetra, Astyanax mexicanus. Curr. Biol. 17, 452–454. ( 10.1016/j.cub.2007.01.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abouheif E, Wray GA. 2002. Evolution of the gene network underlying wing polyphenism in ants. Science 297, 249–252. ( 10.1126/science.1071468) [DOI] [PubMed] [Google Scholar]
- 3.Cohn MJ, Tickle C. 1999. Developmental basis of limblessness and axial patterning in snakes. Nature 399, 474–479. ( 10.1038/20944) [DOI] [PubMed] [Google Scholar]
- 4.Snodgrass RE. 1935. Principles of insect morphology. New York, NY: McGraw-Hill Book Company. [Google Scholar]
- 5.Damen WGM, Hausdorf M, Seyfarth EA, Tautz D. 1998. The expression pattern of Hox genes in the spider Cupiennius salei suggests a conserved mode of head segmentation in arthropods. Proc. Natl Acad. Sci. USA 95, 10 665–10 670. ( 10.1073/pnas.95.18.10665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida H. 2008. A revision of the genus Achaearanea (Araneae: Theridiidae). Acta Arachnol 57, 37–40. ( 10.2476/asjaa.57.37) [DOI] [Google Scholar]
- 7.Pechmann M, Khadjeh S, Sprenger F, Prpic NM. 2010. Patterning mechanisms and morphological diversity of spider appendages and their importance for spider evolution. Arthropod. Struct. Dev. 39, 453–467. ( 10.1016/j.asd.2010.07.007) [DOI] [PubMed] [Google Scholar]
- 8.Hughes CL, Kaufman TC. 2002. Hox genes and the evolution of the arthropod body plan. Evol. Dev. 4, 459–499. ( 10.1046/j.1525-142X.2002.02034.x) [DOI] [PubMed] [Google Scholar]
- 9.Abzhanov A, Kaufman TC. 2004. Hox genes and tagmatization of the higher Crustacea (Malacostraca). Crustacean Issues 15, 43–74. [Google Scholar]
- 10.Akiyama-Oda Y, Oda H. 2006. Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development 133, 2347–2357. ( 10.1242/dev.02400) [DOI] [PubMed] [Google Scholar]
- 11.Pechmann M, Prpic NM. 2009. Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: Mygalomorphae). Dev. Genes Evol. 219, 189–198. ( 10.1007/s00427-009-0279-7) [DOI] [PubMed] [Google Scholar]
- 12.Prpic NM, Schoppmeier M, Damen WGM. 2008. Whole-mount in situ hybridization of spider embryos. CSH Protoc. 3, 933–936. ( 10.1101/pdb.prot5068) [DOI] [PubMed] [Google Scholar]
- 13.Prpic NM, Damen WGM. 2005. Cell death during germ band inversion, dorsal closure and nervous system development in the spider Cupiennius salei. Dev. Dyn. 234, 222–228. ( 10.1002/dvdy.20529) [DOI] [PubMed] [Google Scholar]
- 14.Mittmann B, Wolff C. 2012. Embryonic development and staging of the cobweb spider Parasteatoda tepidariorum C. L. Koch, 1841 (syn.: Achaearanea tepidariorum; Aranaeomorphae; Theridiidae). Dev. Genes Evol. 220, 89–105. ( 10.1007/s00427-012-0401-0) [DOI] [PubMed] [Google Scholar]
- 15.Pechmann M, McGregor AP, Schwager EE, Feitosa NM, Damen WGM. 2009. Dynamic gene expression is required for anterior regionalization in a spider. Proc. Natl Acad. Sci. USA 106, 1468–1472. ( 10.1073/pnas.0811150106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanayama M, Akiyama-Oda Y, Nishimura O, Tarui H, Agata K, Oda H. 2011. Travelling and splitting of a wave of hedgehog expression involved in spider-head segmentation. Nat. Commun. 2, 500 ( 10.1038/ncomms1510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrill VKL, Diederich RJ, Turner FR, Kaufman TC. 1989. A genetic and developmental analysis of mutations in labial, a gene necessary for proper head formation in Drosophila melanogaster. Dev. Biol. 135, 376–391. ( 10.1016/0012-1606(89)90187-5) [DOI] [PubMed] [Google Scholar]
- 18.Schaeper ND, Pechmann M, Damen WGM, Prpic NM, Wimmer EA. 2010. Evolutionary plasticity of collier function in head development of diverse arthropods. Dev. Biol. 344, 363–376. ( 10.1016/j.ydbio.2010.05.001) [DOI] [PubMed] [Google Scholar]
- 19.Posnien N, Bucher G. 2010. Formation of the insect head involves lateral contribution of the intercalary segment, which depends on Tc-labial function. Dev. Biol. 338, 107–116. ( 10.1016/j.ydbio.2009.11.010) [DOI] [PubMed] [Google Scholar]
- 20.Snodgrass RE. 1948. The feeding organs of Arachnida, including mites and ticks. Smithson. Misc. Coll. 110, 1–93. [Google Scholar]
- 21.Khadjeh S, Turetzek N, Pechmann M, Schwager EE, Wimmer EA, Damen WGM, Prpic NM. 2012. Divergent role of the Hox gene Antennapedia in spiders is responsible for the convergent evolution of abdominal limb repression. Proc. Natl Acad. Sci. USA 109, 4921–4926. ( 10.1073/pnas.1116421109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksson BJ, Tait NN, Budd GE, Janssen R, Akam M. 2010. Head patterning and Hox gene expression in an onychophoran and its implications for the arthropod head problem. Dev. Genes Evol. 220, 117–122. ( 10.1007/s00427-010-0329-1) [DOI] [PubMed] [Google Scholar]
- 23.Janssen R, Eriksson BJ, Tait NN, Budd GE. 2014. Onychophoran Hox genes and the evolution of arthropod Hox gene expression. Front. Zool. 11, 22 ( 10.1186/1742-9994-11-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giribet G, Edgecombe GD. 2012. Reevaluating the arthropod tree of life. Annu. Rev. Entomol. 57, 167–186. ( 10.1146/annurev-ento-120710-100659) [DOI] [PubMed] [Google Scholar]
- 25.Campbell LI, et al. 2011. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc. Natl Acad. Sci. USA 108, 15 920–15 924. ( 10.1073/pnas.1105499108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abzhanov A, Kaufman TC. 1999. Homeotic genes and the arthropod head: expression patterns of the labial, proboscipedia, and Deformed genes in crustaceans and insects. Proc. Natl Acad. Sci. USA 96, 10 224–10 229. ( 10.1073/pnas.96.18.10224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson DT. 1973. Embryology and phylogeny in annelids and arthropods. Oxford, UK: Pergamon Press. [Google Scholar]
- 28.Dohle W. 1997. Myriapod–insect relationships as opposed to an insect–crustacean sister group relationship. In Arthropod relationships (eds Fortey RA, Thomas RH), pp. 305–315. London, UK: Chapman & Hall. [Google Scholar]
- 29.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463, 1079–1083. ( 10.1038/nature08742) [DOI] [PubMed] [Google Scholar]
- 30.Giribet G, Edgecombe GD, Wheeler WC. 2001. Arthropod phylogeny based on eight molecular loci and morphology. Nature 413, 157–161. ( 10.1038/35093097) [DOI] [PubMed] [Google Scholar]
- 31.Bamber RN. 2010. Sea-spiders (Pycnogonida) of the north-east Atlantic. Synopses of the British fauna new series no. 5 , 2nd edn Shrewsbury, UK: Field Studies Council. [Google Scholar]
- 32.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray. [PMC free article] [PubMed] [Google Scholar]
- 33.Klaus S, Mendoza JC, Liew JH, Plath M, Meier R, Yeo DC. 2013. Rapid evolution of troglomorphic characters suggests selection rather than neutral mutation as a driver of eye reduction in cave crabs. Biol. Lett. 9, 20121098 ( 10.1098/rsbl.2012.1098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Wang Z, Xu S, Zhou K, Yang G. 2013. Characterization of hairless (Hr) and FGF5 genes provides insights into the molecular basis of hair loss in cetaceans. BMC Evol. Biol. 13, 34 ( 10.1186/1471-2148-13-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshizawa M, Yamamoto Y, ÓQuin KE, Jeffery WR. 2012. Evolution of an adaptive behaviour and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 10, 108 ( 10.1186/1741-7007-10-108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunter H, Meyer A. 2013. Trade-offs in cavefish sensory capacity. BMC Biol. 11, 5 ( 10.1186/1741-7007-11-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.