Abstract
Orchids are the most diverse family of angiosperms, with over 25 000 species, more than mammals, birds and reptiles combined. Tests of hypotheses to account for such diversity have been stymied by the lack of a fully resolved broad-scale phylogeny. Here, we provide such a phylogeny, based on 75 chloroplast genes for 39 species representing all orchid subfamilies and 16 of 17 tribes, time-calibrated against 17 angiosperm fossils. A supermatrix analysis places an additional 144 species based on three plastid genes. Orchids appear to have arisen roughly 112 million years ago (Mya); the subfamilies Orchidoideae and Epidendroideae diverged from each other at the end of the Cretaceous; and the eight tribes and three previously unplaced subtribes of the upper epidendroids diverged rapidly from each other between 37.9 and 30.8 Mya. Orchids appear to have undergone one significant acceleration of net species diversification in the orchidoids, and two accelerations and one deceleration in the upper epidendroids. Consistent with theory, such accelerations were correlated with the evolution of pollinia, the epiphytic habit, CAM photosynthesis, tropical distribution (especially in extensive cordilleras), and pollination via Lepidoptera or euglossine bees. Deceit pollination appears to have elevated the number of orchid species by one-half but not via acceleration of the rate of net diversification. The highest rate of net species diversification within the orchids (0.382 sp sp−1 My−1) is 6.8 times that at the Asparagales crown.
Keywords: Andes, BAMM, BiSSE, New Guinea Highlands, Pleurothallidinae, speciation
1. Introduction
Orchids form the largest family of flowering plants, with over 880 genera and 25 000 species; they comprise roughly 8% of all vascular plant species and grow in a wide range of habitats worldwide [1,2]. Essentially all temperate orchids are terrestrial, but most orchids inhabit tropical forests and over 80% of those are epiphytes. All orchids rely on fungi for germination and carbon capture in the protocorm stage, and in many taxa this mycorrhizal association remains obligate for life [3]. Orchids in more than 30 small mycoheterotrophic lineages have lost all photosynthetic capacity and rely entirely on fungi for energy [4]. Finally, orchids display extraordinary floral diversity, with striking adaptations to different pollinators among close relatives, partitioning of individual pollinators by precise placement of pollen packets (pollinia) on different parts of their bodies, and extreme convergence and divergence among crossable taxa. The evolution of pollinia, specialization on individual pollinators or mycorrhizal fungi, pollination via deceit, euglossine bees or Lepidoptera, epiphytism per se or associated traits such as CAM photosynthesis, and predominant distribution in the tropics (and especially in extensive cordilleras) have all been proposed as drivers of the extraordinary species richness of orchids [5–18].
A well-resolved, strongly supported, time-calibrated phylogeny is fundamental to any attempt to test such hypotheses and assess the impact of individual traits and the history of geographical and ecological spread on net rates of species diversification. Over the past two decades, molecular phylogenetics has greatly advanced our understanding of orchid relationships. Investigators have used sequences of one to five plastid loci, segments of nuclear ribosomal DNA, or single mitochondrial or low-copy nuclear genes to identify five orchid subfamilies and their relationships to each other, identifying Apostasioideae as sister to all other orchids, Vanilloideae or Cypripedioideae next divergent, and the two largest subfamilies, Orchidoideae and Epidendroideae, sister to each other [19–25]. These advances have overturned many classic views of orchid relationships based on morphology alone [7,26] and led to a new phylogenetic classification of the orchids [1,2], including 17 tribes and 44 subtribes of orchidoids and epidendroids. Molecular phylogenetic studies to date have, however, failed to agree in the placement of Cypripedioideae and Vanilloideae, or to resolve and strongly support relationships among many of the 13 tribes of subfamily Epidendroideae [25–28], which comprises around 80% of all orchid species.
To clarify relationships across orchids and test theories about the impact of various traits on their net rate of species diversification, we adopted a phylogenomic approach, using massively parallel sequencing to amass data on a large fraction of the coding regions present in the plastid genome, including a far greater number of characters per taxon than any prior broad-scale study of orchid phylogeny. We extended this phylogeny using a supermatrix approach to include representatives of 40 of 43 orchid subtribes, and calibrated this tree against the ages of several angiosperm fossils to produce a new timeline for orchid evolution, identify points in the family's history at which the rate of net species diversification accelerated significantly and determine whether such accelerations are correlated significantly with characters that have been proposed as likely drivers of orchid speciation.
2. Material and methods
(a). Phylogenetics
We aligned data for 75 plastid genes from 39 species representing all orchid subfamilies and 16 of 17 tribes, as well as 73 species stratified across all monocot orders and 23 placeholders for major groups of eudicots and basal angiosperms employed as outgroups (electronic supplementary material, table S1). A maximum-likelihood (ML) analysis of the plastome data was conducted using RAxML v. 8.0.9 using the automatic bootstrap option [29]; we compared results obtained from unpartitioned data with those using an optimal partitioning of the 75 loci identified using AIC in PartitionFinder v. 1.1.1 [30]. We conducted a similar analysis of a second dataset that added three plastid genes (atpB, psaB and rbcL) for 162 orchid species, including placeholders for all five orchid subfamilies, 18 of 19 tribes and 40 of 43 subtribes, representing all but 0.4% of described orchid species (electronic supplementary material, table S2).
(b). Time calibration
We calibrated the supermatrix tree against time using branch lengths based on atpB, psaB and rbcL in a Bayesian framework using BEAST v. 1.8.0 [31] (see electronic supplementary material). Seventeen fossils were used as calibration priors, with offsets corresponding to their minimum estimated ages (electronic supplementary material, table S3). All fossil priors were assigned a lognormal distribution (s.d. = 2), accounting for uncertainty in both absolute fossil age estimation and phylogenetic placement. Priors were also placed on the crowns of rosids, magnoliids and Caryophyllales + asterids. Due to a lack of fossils easily attributed to these clades, normal priors were placed on them with mean offsets and 95% confidence intervals mirroring the posterior ages from the exponential clock analysis of Bell et al. [32], with the expectation that the true ages of these nodes would be captured by these wide, conservative priors. Uniform priors were placed on the root node and the stem of Illicium following [33]. Two chains of 100 million generations were run. Tree files from these independent chains were combined after removing 25% as burn-in to construct the maximum clade credibility chronogram.
(c). Rates of net species diversification
To test for significant shifts in net diversification regimes across lineages, we used the Bayesian approach implemented in BAMM v. 2.0 using Bayes factors [34] (see the electronic supplementary material). Analyses were conducted on a chronogram limited to Orchidaceae and other lineages of Asparagales, for Poisson rate priors of 0, 0.2, 0.4, 0.6, 0.8, 1, 5 and 10. For each prior value, two independent MCMC chains of 50 million generations were run; after removing 15% as burn-in, we analysed the output to identify the single best shift configuration with the highest posterior density and the maximum shift credibility configuration. The relative probabilities of less likely shift configurations were also calculated for each value of the Poisson prior.
(d). Correlates of net diversification rates
Based on existing hypotheses, we assessed correlations between six sets of character states and apparent rates of speciation and extinction in orchid lineages using BiSSE [35]. Rationales for these hypotheses are given in the Discussion. Sources of phenotypic data are provided in the electronic supplementary material. BiSSE cannot calculate likelihoods on unresolved tips representing more than 190 taxa, so all tips were down-weighted by a factor of 25, with small clades rounded up to 1 [36].
Gains and losses of each character-state were mapped onto the chronogram using MP and ML. For each character, an unconstrained model for diversification was compared to models where speciation (λ), extinction (μ) and character-state transition rates (q) were individually constrained (λ0 = λ1, μ0 = μ1, q01 = q10). Likelihoods of the constrained models were compared to the unconstrained model, with significance of likelihood scores assessed with ANOVA tests. Net rates of diversification for different character-states were calculated as λ − μ. For individual traits, we measured the advantage in net diversification per million years within lineages conferred by a character-state as 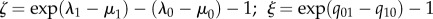 measures the advantage in net diversification per million years per lineage across lineages. ξ is a heuristic measure, given that the realized shifts in character-states (i.e. in births of lineages marked by those states) depends on the standing number of lineages with each alternative state at any one time. Previous studies have used ζ as the rate of net species diversification; essentially, it measures the average rate of diversification within lineages marked by a particular character-state, such as red flowers. We are coining the term ‘across-lineage diversification’ for ξ; it essentially measures the rate at which new lineages marked by, say, red flowers arise via mutation of character-states.
measures the advantage in net diversification per million years per lineage across lineages. ξ is a heuristic measure, given that the realized shifts in character-states (i.e. in births of lineages marked by those states) depends on the standing number of lineages with each alternative state at any one time. Previous studies have used ζ as the rate of net species diversification; essentially, it measures the average rate of diversification within lineages marked by a particular character-state, such as red flowers. We are coining the term ‘across-lineage diversification’ for ξ; it essentially measures the rate at which new lineages marked by, say, red flowers arise via mutation of character-states.
We conducted a phylogenetically unstructured comparison of differences among tropical terrestrial clades, tropical epiphytic clades and temperate terrestrial clades in net diversification rate D = (ln S)/T, where S = number of species in a clade and T = stem age of that clade [36].
3. Results
ML analysis of 77 880 aligned bases (including 31 691 informative characters, of which 9315 were informative within orchids) produced a fully resolved tree in which 26 of 38 nodes within Orchidaceae have bootstrap support values more than or equal to 98% (figure 1). The same topology, with negligible differences in branch lengths and support values, was recovered with an analysis based on the 50 partitions identified by PartitionFinder; for simplicity, we focus here on the tree obtained from the unpartitioned data. This is the first fully resolved, strongly supported backbone phylogeny for Orchidaceae, including representatives of all tribes except the small mycoheterotrophic Gastrodieae (six genera, 70 species). This phylogeny places Apostasioideae sister to all other subfamilies, then Vanilloideae, then Cypripedioideae sister to Orchidoideae and Epidendroideae, and resolving relationships among the tribes of the latter. The exceptionally short branches along the spine of Epidendroideae may help explain why previous analyses based on only one or a few genes have been unable to resolve and strongly support many of the intertribal relationships within that subfamily (figure 1). The plastome tree strongly supports the positions of two previously unplaced subtribes, with Collabiinae sister to Podochileae and Dendrobiinae sister to Malaxideae (figure 1). As expected, the mycoheterotrophic taxa examined (Corallorhiza, Neottia, Rhizanthella) all exhibited accelerated rates of nucleotide substitution. Rhizanthella has one of the most highly reduced plastomes of any angiosperm sequenced to date, with only 59 910 bp and 33 genes [37], and shows a huge acceleration of nucleotide substitution (figure 1).
Figure 1.
Plastome phylogeny for Orchidaceae, based on an ML analysis of sequence variation in 75 genes from the plastid genome of 39 orchid species and 100 angiosperm outgroups (latter not shown). Bootstrap support values are shown above each branch. Orchid genus, subtribe, tribe and subfamily are indicated for each placeholder. Inset: Phylogram shows branch lengths based on inferred genetic substitutions down each branch. Asterisks indicate mycoheterotrophic taxa (Corallorhiza, Rhizanthella and Neottia, top to bottom).
Our supermatrix analysis indicates that the highly specialized Rhizanthella (which flowers underground and has fleshy fruits) diverged from its closest relatives 31 Mya (figure 2; electronic supplementary material, figure S2). This analysis largely preserved relationships among subfamilies, tribes and subtribes seen in the plastome study, but greatly increased the numbers of species and subtribes represented. The supermatrix tree identifies the previously unplaced subtribe Agrostophyllinae as sister to the unplaced genus Coelia, with both sister to Epidendreae, and Calypsoeae sister to all three lineages (figure 1; electronic supplementary material, figure S1).
Figure 2.

Backbone chronogram for Orchidaceae, based on a supermatrix analysis including a total of 201 orchid species and 100 outgroups. Relationships are summarized by subtribes (-inae) and tribes (-eae); subfamilies are indicated on the right. Branch lengths are proportional to time in millions of years. Bootstrap support values are shown above each branch.
Orchids appear to have diverged from the common ancestor of all other members of Asparagales ca 112 Mya, and extant orchid lineages began diverging from each other 90 Mya. Orchids diverged from the common ancestor of all other Asparagales ca 112 Mya, and extant orchid lineages began diverging from each other 90 Mya (see electronic supplementary material, figure S1, and figure 2 for ages ±95% confidence intervals). The stem age of Vanilloideae is 84 My; of Cypripedioideae, 77 My; and of Orchidoideae and Epidendroideae, 64 My, at the dawn of the Tertiary. The eight tribes and three previously unplaced subtribes of so-called ‘upper’ epidendroids—the clade spanned by Arethuseae and Cymbidieae—diverged rapidly from each other between 37.9 and 30.8 Mya (figure 2). A series of short branches also separate four of the five tribes of the ‘lower’ epidendroids (Nervilieae, Tropidieae, Sobralieae and Triphoreae). Our age estimates push back the stem and crown ages of Orchidaceae by 7 and 10 My, respectively, relative to the estimates by Gustafsson et al. [24], but are supported by a much wider set of fossil calibration points.
(a). Evolution of characters hypothesized to affect net diversification
Pollinia characterize Orchidoideae and Epidendroideae and evolved no later than 64 Mya (electronic supplementary material, figure S2a). Epiphytism appears to have evolved once at the base of the upper epidendroids, no later than 35 Mya, and to have been lost at least three times, in Bletiinae, Calypsoeae and Arethusinae (electronic supplementary material, figure S2b). A very few cases of epiphytism are also known from scattered species of Cypripedioideae (e.g. Paphiopedilum, Phragmipedium) and Orchidoideae (e.g. Dispersis, Eurystyles, Pseudoeurystyles) [2]. CAM photosynthesis appears to have arisen at least four times, in the upper epidendroids (with several subsequent losses and reappearance in Epidendreae minus Ponerinae), in Sobralieae, and in scattered species of Vanilleae (electronic supplementary material, figure S2c). Orchids initially had tropical distributions, lost them in ancestral Orchidoideae, secondarily regained them in Cranichideae, and within the latter again lost them in Pterostylidinae and Cranichidinae (electronic supplementary material, figure S2d).
Deceit pollination evolved at least once, in the stem group of all orchids except Apostasioideae, and then was lost in Codonorchideae, Goodyerinae–Spiranthinae–Cranichidinae, Neottieae, Sobralieae–Triphorieae, Tropideae, Thelasinae, Podochilinae, Agrostophyllinae, Bletiinae, Ponerinae, Aerangidinae–Angraecinae, Cymbidiinae, Catasetinae and Zygophyllinae–Stanhopeinae–Coeliopsidinae–Maxillariinae (electronic supplementary material, figure S2e). Deceit appears to involve Hymenoptera in cypripedioids, and many orchidoids in Orchideae and especially Diurideae, except for fungus gnats in subtribe Acianthinae. Dipterans are involved in pollination of many species of the pleurothallid alliance and others, whereas oil-gathering Centris bees appear to be duped by many Oncidiinae. Pollination by male euglossine bees appears to have arisen at least twice, in Catasetinae and Zygophyllinae–Stanhopeinae–Coeliopsidinae, in subtribes that evolved 16–22 Mya, with a later loss in Maxillariinae (electronic supplementary material, figure S2f). Pollination by Lepidoptera appears to have arisen at least five times, in Disinae and Orchidinae, Collabinae, Eriinae, Epidendreae and allies, and Angraecinae–Aerangidinae, with losses in Bletiinae and Pleurothallidinae–Ponerinae (electronic supplementary material, figure S2g).
Invasion of the Andes at more than 1300 m elevation appears to have arisen in at least 10 lineages, including Vanilleae, Orchidinae, Goodyerinae–Spiranthinae–Cranichidinae, Malaxideae, Calypsoeae, Epidendreae, Angraecinae, and Cymbidieae minus Cymbidiinae (electronic supplementary material, figure S2h). Invasion of the New Guinea Highlands at more than 1000 m elevation occurred in at least 15 lineages (electronic supplementary material, figure S2i). Although MP and ML reconstructions suggest many fewer invasions, deep sharing of a distribution in the New Guinea Highlands is impossible given their recent uplift ca 12 Mya [38].
(b). Rates of net diversification within orchids and their close relatives
Apparent rates of speciation and extinction at the crown of Asparagales are λ = 0.088 sp sp−1 My−1 and μ = 0.031 sp sp−1 My−1, respectively. A wide range of Poisson priors—from 0.6 to 10.0—each identified significant shifts of net diversification within Asparagales as sampled, all within the orchids, with one acceleration in the orchidoids, a second acceleration in the upper epidendroids, a nested deceleration in Agrostophyllinae plus Calypsoeae, and a further nested acceleration in Laeliinae–Pleurothallidinae–Ponerinae (figure 3). Initial rates of speciation and extinction in the last, most rapidly diversifying clade—endemic to the Neotropics—are λ = 0.510 My−1 and μ = 0.128 My−1. Rates of net species diversification per million years (λ—μ) in that clade were thus 6.8 times that at the Asparagalean crown; that across the orchidoids, 1.3 times that rate; and that at the base of the upper epidendroids, 2.9 times that rate. Low rates of net diversification persisted in the lower epidendroids, coupled with a drop by two-thirds in the speciation rate. Low values of the Poisson prior (0.0–0.4) led to two inferred shifts in net diversification, in the orchidoids and the upper epidendroids. This same set of shifts was identified as much less likely (52–82%) than the four-shift set emerging from Poisson priors of 0.6, 0.8, 1.0, 5 and 10.
Figure 3.
Diversigram for Orchidaceae, showing three significant shifts of net diversification rate, inferred from BAMM analysis. Initial inferred rates of speciation (λ) and extinction (μ) are shown for each of the numbered clades. Warmer branch colours represent faster rates of net diversification for each lineage (see inset scale). Triangle heights are proportional to the number of present-day taxa in each lineage; triangle lengths reflect crown ages. Insets: rates of speciation (green), extinction (black) and net species diversification (red, with blue probability distribution) projected forward from each of the four critical nodes.
(c). Correlates of net diversification rate
Evolution of pollinia significantly accelerated both speciation and extinction rates, as predicted, and resulted in a 4.9% higher rate of net diversification per million years within lineages (table 1). Only one transition to pollinia occurred, so rates of net diversification across lineages per million years caused by initiation of new lineages with pollinia present or absent was quite low (0.5%). Epiphytism significantly accelerated both speciation and extinction rates relative to the terrestrial habit, yielding a 8.8% edge in net diversification per million years within lineages; q01 and q10 did not differ significantly, so ξ = 0.0%. Tropical distributions significantly accelerated speciation and extinction rates relative to extratropical distributions, resulting in a 2.1% higher rate in net diversification per million years within lineages and 1.9% across lineages. In orchids, this last difference, however, appears to be driven primarily by habit, not latitude per se. A phylogenetically unstructured comparison among subtribes and similar taxonomic units showed that the average net diversification rate did not differ significantly between extratropical and tropical terrestrial lineages (0.115 ± 0.057 My−1 versus 0.118 ± 0.061 My−1, p > 0.92), and that the average rate for tropical epiphytic lineages (0.266 ± 0.077 My−1) was significantly greater than that for both temperate and tropical terrestrial lineages (p < 0.001) (table 2). CAM photosynthesis accelerated speciation and extinction rates relative to C3 photosynthesis, yielding a 20.3% advantage in net diversification per million years within lineages and an 11.8% disadvantage across lineages (ξ = −11.8%, table 1). This calculation overstates CAM's disadvantage across lineages, because CAM characterizes a minority of lineages (electronic supplementary material, table S4), and the actual flux between CAM and non-CAM lineages depends on both ξ and the relative abundance of CAM and non-CAM taxa. Tropical distributions significantly increased speciation and extinction rates, and boosted diversification rates within lineages by 2.1% and across lineages by 1.9%.
Table 1.
Apparent rates of speciation (λ), extinction (μ) and character-state transition (q) associated with particular character-states based on BiSSE analysis. Significantly larger rates are indicated by asterisks. 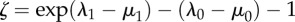 measures the advantage in net diversification (r = λ − μ) per million years within lineages conferred by a character-state;
measures the advantage in net diversification (r = λ − μ) per million years within lineages conferred by a character-state; 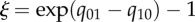 measures the advantage in net diversification per million years per lineage across lineages; it is a heuristic measure, given that the realized shifts in character-states (i.e. in births of lineages marked by those states) depends on the standing number of lineages with each alternative state at any one time.
measures the advantage in net diversification per million years per lineage across lineages; it is a heuristic measure, given that the realized shifts in character-states (i.e. in births of lineages marked by those states) depends on the standing number of lineages with each alternative state at any one time.
| character-state | λ1 | λ0 | μ1 | μ0 | ζ (%) | q01 | q10 | ξ (%) |
|---|---|---|---|---|---|---|---|---|
| pollinia | 0.355** | 0.025 | 0.281*** | 0.000 | 5.1 | 0.0046** | 5.6 × 10−10 | 0.5 |
| epiphytism | 0.574*** | 0.071 | 0.474* | 0.056 | 8.8 | 0.0007 | 0.0010 | 0.0 |
| tropical | 0.465*** | 0.067 | 0.381** | 0.005 | 2.1 | 0.0179 | 0.0006*** | 1.9 |
| CAM | 1.486** | 0.362 | 1.356* | 0.381 | 20.3 | 0.0073 | 0.1323*** | −11.8 |
| deceit | 0.881* | 0.420 | 0.851* | 0.367 | −2.2 | 0.0183*** | 1.3 × 10−9 | 1.9 |
| euglossine | 0.409 | 0.319 | 0.361 | 0.493 | 5.2 | 1.8 × 10−4 | 0.0158** | −1.6 |
| lepidoptera | 0.694 | 0.426 | 0.577 | 0.378 | 7.1 | 0.0016 | 6.0 × 10−7 | 0.2 |
| tropical cordilleras | 0.858*** | 0.152 | 0.663* | 0.179 | 24.9 | 0.0148 | 0.197*** | −21.3 |
*p < 0.05, **p < 0.01, ***p < 0.001.
Table 2.
Diversification rate D (mean ± s.d.) for orchid clades composed of more than 90% temperate terrestrial species, tropical terrestrial species and tropical epiphytes. Values with different superscripts are significantly different (p < 0.01) based on a t-test with unequal variances; values with the same superscript do not differ significantly.
| clade type | D |
|---|---|
| temperate terrestrial clades (n = 6) | 0.115 ± 0.057a |
| tropical terrestrial clades (n = 6) | 0.118 ± 0.061a |
| tropical epiphytic clades (n = 5) | 0.266 ± 0.077b |
Deceit pollination—via mimicry of food sources, nesting sites or potential mates—significantly increased speciation and extinction rates but resulted in a small decrease in net diversification within lineages (ζ = −2.2%), balanced by a comparable increase across lineages (ξ = 1.9%). Provision of chemical allurants for male euglossine bees had no significant impact on λ or μ but increased net diversification rates within lineages by 5.2% with a smaller negative effect (−2.0%) across lineages (table 1). Pollination by Lepidoptera increased speciation and extinction rates within lineages, albeit non-significantly, resulting in a large positive effect (7.1%) on net diversification within lineages and a negligible effect on diversification across lineages (table 1). Life in extensive tropical cordilleras—as exemplified by the Andes and New Guinea Highlands—significantly increased speciation and extinction rates, yielding a large 24.9% advantage in net diversification rate within lineages, but also an apparently large 21.3% disadvantage across lineages (table 1). However, the latter effect is overstated, because most lineages did not invade these cordilleras (electronic supplementary material, table S4) and because diversification within and across montane lineages is underestimated by overestimates of the age of montane lineages in New Guinea (see above). Epiphytism and pollination via deceit, euglossine bees or Lepidoptera accelerated net diversification by large amounts, but their overall effect is less than that of CAM photosynthesis and (especially) life in extensive tropical cordilleras. A 5% advantage in net diversification per million years translates, via compound interest, into a 165% advantage in species numbers in 20 My; a 15% advantage translates into a 1536% advantage over the same period.
4. Discussion
The evolution of pollinia should accelerate speciation by permitting precise placement of pollen and allowing specialization on individual pollinators (e.g. moths versus bees) or parts thereof [6], and by increasing the importance of genetic drift, perhaps resulting in an alternation of drift with strong selection on sexual characteristics [16]. In addition, pollinia allow very small numbers of variants to produce large numbers of offspring, promoting speciation from small numerical bases (perhaps due to highly inefficient pollination), which might otherwise lead to demographic collapse and extinction.
Epiphytism should accelerate speciation in several ways. First, epiphytism is a key innovation that allows the invasion of a new adaptive zone—the branches and boles of trees—largely unoccupied by other vascular plants. Second, epiphytism should help generate and maintain high levels of plant diversity because the habitable surface of bark, branches and twigs is much greater than the ground occupied by a forest; because variation in insolation and humidity within tree crowns allows local, fine-scale niche partitioning; and because altitudinal and topographic variation in fog deposition and evaporation rates creates a range of conditions that can be partitioned at broader spatial scales and may serve to isolate populations and foster local speciation [12–15,36]. Third, epiphytism is associated with high rainfall and humidity, and thus often with tropical montane conditions; the latter can provide large-scale barriers to gene flow (e.g. deep valleys, high ridges) that can isolate populations at larger spatial scales and further accelerate speciation [13,36]. Finally, the tiny seeds associated with epiphytism can provide occasional long-distance dispersal, permitting genetic differentiation to proceed in parallel at many sites along the length of extensive montane areas, such as the Andes [36]. Indeed, epiphytism in Bromeliaceae—the angiosperm family with the second largest number of epiphytic species after Orchidaceae—is associated with an acceleration of speciation rates by 2.5- to 5.3-fold relative to terrestrial lineages [36]. Almost all bromeliads have a Neotropical distribution, so they show no confounding of latitude with epiphytism. Tropical distributions should increase rates of speciation relative to those outside the tropics as a result of greater habitat area, more stable climates, lack of glaciation, and greater opportunity for coevolution of plants and their mutualists and specialized herbivores [17]. Within orchids, temperate versus tropical latitudes per se appear to have had no significant effect on diversification, but the epiphytic habit appears to have accelerated net diversification rates by 8.8% per million years (table 1). Gravendeel et al. [12] used a phylogenetically unstructured analysis to show that epiphytic orchid genera contain more species, on average, than terrestrial genera. Our results generalize their findings and are the first rigorous demonstration, for orchids, that epiphytism accelerates differentiation when phylogeny at various levels is taken account.
CAM photosynthesis had no significant effect on net diversification in bromeliads—in which it occurs both in terrestrial lineages on dry sites and in epiphytic lineages [36]—but it did accelerate speciation, extinction and net diversification in orchids, perhaps because it is so closely tied to epiphytism in orchids [15] and may permit invasion of the most exposed perches, as well as drier forests at lower elevations.
Deceit pollination characterizes one-third of all orchids [9,39]. Mimicry of potential mates is much less common than mimicry of food or nesting sites, but is thought likely to accelerate speciation because subtle changes in floral morphology or volatile compounds can attract different pollinators and lead to reproductive isolation [9], and often results in high efficiency of pollen transfer to conspecifics [40]. We propose that pollination via deceit inevitably involves a density-dependent advantage of new mimetic morphs that are rare relative to models; such negative density-dependence is a potentially strong but previously overlooked force that could favour high rates of diversification. Pollination by male euglossine bees should accelerate speciation by allowing small chemical changes in the allurants provided by orchids to attract bees that are reproductively isolated from each other and by permitting partitioning of individual bee species by placing pollinia on different parts of their bodies [6,18]. Pollination by Lepidoptera often involves nectar storage in corolla tubes or spurs whose lengths can be easily modified and, in so doing, lead to the rapid recruitment or evolution of different pollinators and, ultimately, to accelerated rates of speciation. Floral spurs are not, however, always associated with nectar production [41], so that the argument favouring the evolution of progressively longer spurs and pollinator mouthparts [42] does not necessarily hold, and is difficult to test given the possibility that some orchids (e.g. Disa) use empty spurs as a deceitful means of attracting pollinators.
This study shows that orchids are remarkably species-rich partly as a result of three likely accelerations of net diversification rates, apparently driven in part by the acquisition of pollinia, epiphytism, tropical distributions, CAM photosynthesis, pollination via deceit, male euglossine bees or Lepidoptera, and life on extensive tropical cordilleras. It must be recognized, however, that shifts in net diversification are scale-dependent. Our sampling is well suited to detect shifts in diversification at tribal or subtribal levels within orchids, but inadequate to identify such shifts within genera. Nevertheless, our findings provide the first quantitative support for several earlier hypotheses regarding the genesis of orchid diversity and identify specific points in orchid evolution where these factors played a role. They show that multiple factors—several of them interconnected—have contributed to orchid diversification.
The first significant acceleration of net diversification is at the base of Orchidoideae, one node removed from that at which pollinia evolved (figure 3). The second acceleration coincides with the origin of epiphytism in the upper epidendroids and, less precisely, with the origin of several montane clades in the Andes. A deceleration of diversification occurs in Epidendreae–Agrostophyllinae–Calypsoeae, as expected because this group does not consist of montane epiphytic lineages: Calypsoeae are temperate terrestrials, most Agrostrophyllinae are epiphytes from lowland tropical forests, and—uniquely within tribe Epidendreae—early-divergent Bletiinae have regained the terrestrial habit. The final acceleration coincides with the later divergent elements of tribe Epidendreae and a large concentration of Andean taxa pollinated via deceit or Lepidoptera. Pleurothallids, the most diverse element of this clade, are largely pollinated by dipterans. Low rates of net diversification persisted in the lower epidendroids, which are marked (as are orchidoids) by a lack of epiphytism but, unlike orchidoids, largely lack pollination via deceit, euglossine bees or Lepidoptera. We suspect that this last distinction accounts for low diversification in the lower epidendroids and the displacement of the first acceleration of diversification from the orchidoid–epidendroid crown to the orchidoid crown.
The synergistic effects of epiphytism and life in extensive, topographically complex, tropical cordilleras on geographical speciation at small spatial scales are likely to have had quite large effects on orchid diversity, given the very large number of species in certain clades centred on the Andes (e.g. Cymbidieae minus Cymbidiinae, Epidendreae) and the New Guinea Highlands (e.g. Dendrobiinae), as well as the high local diversity, narrow endemism and rapid spatial turnover of orchid species in these areas [43,44]. The effects of epiphytism per se, CAM photosynthesis and life in tropical cordilleras cannot be teased apart using current techniques, and will always be hard to separate given the causal links among these traits. Surprisingly, epiphytism appears to have had a stronger effect than tropical distribution in accelerating species diversification in orchids. There is no doubt that sexual selection and floral diversification have had a large impact on orchid diversification (table 1). The fact that approximately one-third of all orchids engage in deceit pollination—a mechanism almost unknown in other plants—is especially striking, and suggests that the evolution of such pollination may have increased orchid diversity by 50% over what it otherwise would have been, given non-deceit pollination in the remaining two-thirds of all orchids. Yet deceit, considered alone, appears to have augmented orchid diversity not by accelerating net diversification, but simply by adding more species at roughly the same rate.
The fact that orchids show no significant acceleration of diversification relative to other Asparagales among the three earliest divergent subfamilies (figure 3) implies that the defining characteristics of the orchids as a whole—the column, mycoheterotrophic germination, minute seeds lacking endosperm and a specialized labellum—did not, by themselves, accelerate diversification. But these traits almost surely acted in concert with the evolutionary triggers identified above to promote high levels of speciation. The ability of pollinia to allow small numbers of variants (based on limited pollination or recent mutation) to produce large numbers of offspring, promoting speciation from small numerical bases, depends on the production of large numbers of (therefore) small seeds, and such seeds would be favoured by mycoheterotrophic germination [45]. The dispersal of pollinia and extreme specialization on individual pollinators or pollinator body parts clearly were facilitated by the orchid column and labellum [6]. Evolution of epiphytism was surely favoured by the possession of tiny, dust-like seeds that could settle on twigs and branches regardless of orientation.
The extent and resolution of our phylogenetic reconstruction is inadequate currently to detect the likely diversifying influence of repeated, small-scale adaptive radiations in pollinators or mycorrhizal fungi. Within Platanthera, Disa, Corycium and Drakaea, closely related species often diverge in pollinators or the placement of pollinia on the same pollinator, suggesting that adaptive radiation in pollinators or pollinia position may be an important driver of diversification among closely related orchids [6–8,46]. By contrast, the evidence for narrow specialization of closely related orchids on different mycorrhizal fungi is limited to mycoheterotrophic taxa; photosynthetic species often overlap strongly in their fungal partners [46]. Pollinators but not mycorrhizal fungi are likely to provide a mating barrier between orchid species.
Finally, invasion of extensive tropical cordilleras per se should accelerate speciation, extinction and net diversification, given the abundance of natural barriers to gene flow, dynamic shifts in the location of favourable habitats, and the possibility for geographical speciation proceeding in parallel along the lengths of mountain chains [13,14,17,36]. Invasion of such cordilleras as the Andes and the New Guinea Highlands had by far the largest positive effect on net species diversification within lineages (see above).
We suggest that three additional factors likely to contribute to orchid diversification should be explored, as follows.
(a). Dispersal, time and familial range
The small seeds of orchids and the long time since their initial diversification almost surely fostered greater diversity by permitting them to spread to every continent and most latitudes. Across angiosperm families, species richness and age are unrelated, but families with broader geographical and latitudinal ranges are more diverse [47]. Analyses of orchid historical biogeography (Givnish et al., in preparation) suggest that intercontinental dispersal in orchids is relatively uncommon, but alone appears to have greatly increased orchid species number over what it would have been had orchids remained on a single continent.
(b). Karyotypic evolution
Chromosomal evolution is often overlooked as a diversifying influence in orchids. Yet orchids show a remarkable amount of polyploid and aneuploid variation in chromosome number across subfamilies and within many of the largest genera (e.g. Bulbophyllum, Dendrobium, Epidendrum, Malaxis, Pleurothallis s.l.) [48]. The combination of small seeds, early mycoheterotrophy and pollinia in orchids allows demographic recovery from repeated bottlenecks in effective population size; such bottlenecks are likely to fix mutations via drift, and fixation of karyotypic mutants would quickly generate post-mating isolation barriers.
(c). Limited gene flow or local pollinators in montane epiphytes
A number of epiphytic groups from tropical cordilleras (e.g. Bulbophyllum, Lepanthes) show substantial genetic differentiation among populations even though most temperate orchids surveyed do not (see data of Phillips et al. [49]). The evolution of dozens of closely related Teaguiea species over only a few kilometres in Andean Ecuador [50] and rapid geographical turnover of species identity with distance in Andean epiphytic orchids more generally [43,44] are both consistent with gene flow over only short distances. Whether this is a result of rapid rainout of seeds over a short distance in wet tropical cordilleras, or limited physiological tolerance or dispersal ability in small, soft-bodied, desiccation-intolerant dipterans and other weak-flying pollinators specific to individual species or clades, is currently unknown. The roles of limited dispersal and species ranges of such pollinators, and of intermittent population bottlenecks and drift on chromosome number and speciation in epiphytic orchids, deserve further study.
Supplementary Material
Acknowledgements
We thank Joel McNeal, Erica Wilson and Laura Williams for access to additional plant material.
Data accessibility
All data are presented or cited in the electronic supplementary material.
Authors' contributions
T.J.G. and K.M.C. conceived the study; M.A., K.M.N., W.M.W., N.H.W., J.L.-M. and K.M.C. obtained the molecular data; S.P.L. and L.E. contributed biogeographic data; D.S., A.Z., W.J.D.I., R.K., S.J.H. and T.J.G. participated in data analysis; M.A.C. and M.T.K.A. provided critical plant material; T.J.G. drafted the manuscript; all authors provided comments and final approval.
Competing interests
We have no competing interests.
Funding
This research was supported by grant no. DEB-0830836 to T.J.G. from the NSF Assembling the Tree of Life (AToL) Program; collection efforts in Chile were supported by grant ICM P02-005 to M.T.K.A.
References
- 1.Chase MW, Cameron KM, Barrett RL, Freudenstein JV. 2003. DNA data and Orchidaceae systematics: a new phylogenetic classification. In Orchid conservation (eds Dixon KW, Kell SP, Barrett RL, Cribb PJ), pp. 69–89. Kota Kinabalu, Malaysia: Natural History Publications. [Google Scholar]
- 2.Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds). 2001–2014 Genera Orchidacearum, vol. 1–6 New York, NY: Oxford University Press. [Google Scholar]
- 3.McCormick MK, Taylor DL, Juhaszova K, Burnett RK Jr, Whigham DF, O'Neill JP. 2012. Limitations on orchid recruitment: not a simple picture. Mol. Ecol. 21, 1511–1523. ( 10.1111/j.1365-294X.2012.05468.x) [DOI] [PubMed] [Google Scholar]
- 4.Merckx V, Freudenstein JV. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytol. 185, 605–609. ( 10.1111/j.1469-8137.2009.03155.x) [DOI] [PubMed] [Google Scholar]
- 5.Darwin C. 1885. On the various contrivances by which orchids are fertilized by insects. London, UK: John Murray. [Google Scholar]
- 6.Dressler RL. 1993. Phylogeny and classification of the orchid family. New York, NY: Cambridge University Press. [Google Scholar]
- 7.Hapeman JR, Inouye K. 1997. Plant-pollinator interactions in Platanthera (Orchidaceae). In Molecular evolution and adaptive radiation (eds Givnish TJ, Sytsma KJ), pp. 433–454. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 8.Johnson SD, Linder JP, Steiner KE. 1998. Phylogeny and radiation of pollination systems in Disa (Orchidaceae). Am. J. Bot. 85, 402–411. ( 10.2307/2446333) [DOI] [PubMed] [Google Scholar]
- 9.Cozzolino S, Widmer A. 2005. Orchid diversity: an evolutionary consequence of deception? Trends Ecol. Evol. 20, 487–494. ( 10.1016/j.tree.2005.06.004) [DOI] [PubMed] [Google Scholar]
- 10.Chase MW, Williams NH, de Faria AD, Neubig KM, Amaral MDE, Whitten WM. 2009. Floral convergence in Oncidiinae (Cymbidieae; Orchidaceae): an expanded concept of Gomesa and a new genus Nohawilliamsia. Ann. Bot. 104, 387–402. ( 10.1093/aob/mcp067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiestl FP, Schlüter PM. 2009. Floral isolation, specialized pollination, and pollinator behavior in orchids. Annu. Rev. Entomol. 54, 425–446. ( 10.1146/annurev.ento.54.110807.090603) [DOI] [PubMed] [Google Scholar]
- 12.Gravendeel R, Smithson A, Slik FJW, Schuiteman A. 2004. Epiphytism and pollinator specialization? Phil. Trans. R. Soc. Lond. B 359, 1523–1535. ( 10.1098/rstb.2004.1529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentry AH, Dodson CH. 1987. Diversity and biogeography of Neotropical vascular epiphytes. Ann. MO Bot. Gard. 74, 205–233. ( 10.2307/2399395) [DOI] [Google Scholar]
- 14.Benzing DH. 1990. Vascular epiphytes: general biology and associated biota. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 15.Silvera K, Santiago LS, Cushman JC, Winter K. 2009. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in Orchidaceae. Plant Physiol. 149, 1838–1847. ( 10.1104/pp.108.132555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremblay RL, Ackerman JD, Zimmerman JK, Calvo RN. 2005. Variation in sexual reproduction in orchids and its evolutionary consequences: a spasmodic journey to diversification. Biol. J. Linn. Soc. 84, 1–54. ( 10.1111/j.1095-8312.2004.00400.x) [DOI] [Google Scholar]
- 17.Givnish TJ. 2010. Ecology of plant speciation. Taxon 59, 1326–1366. [Google Scholar]
- 18.Ramírez SR, Eltz T, Fujiwara MK, Gerlach G, Goldman-Huertas B, Tsutsui ND, Pierce NE. 2011. Asynchronous diversification in a specialized plant–pollinator mutualism. Science 333, 1742–1746. ( 10.1126/science.1209175) [DOI] [PubMed] [Google Scholar]
- 19.Waterman RJ, Bidartondo MI, Stofberg J, Combs JK, Savolainen V, Barraclough TG, Pauw A. 2011. The effects of above- and belowground mutualisms on speciation and coexistence. Am. Nat. 177, E54–E68. ( 10.1086/657955) [DOI] [PubMed] [Google Scholar]
- 20.Chase MW, Cameron MW, Hills H, Jarrell D. 1994. DNA sequences and phylogenetics of the Orchidaceae and other lilioid monocots. In Proc. fourteenth world orchid conference (ed. Pridgeon A.), pp. 67–73. Glasgow, UK: Her Majesty's Stationery Office. [Google Scholar]
- 21.Cameron KM, Chase MW, Whitten WM, Kores PJ, Jarrell DC, Albert VA, Yukawa T, Hills HG, Goldman DH. 1999. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot. 86, 208–224. ( 10.2307/2656938) [DOI] [PubMed] [Google Scholar]
- 22.Cameron KM. 2007. Molecular phylogenetics of Orchidaceae: first decade of DNA sequencing. Mem. NY Bot. Gard. 95, 163–200. [Google Scholar]
- 23.Ramírez SR, Gravendeel B, Singer RB, Marshall CR, Pierce NE. 2007. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature 448, 1042–1045. ( 10.1038/nature06039) [DOI] [PubMed] [Google Scholar]
- 24.Gustafsson ALS, Verola CF, Antonelli A. 2010. Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evol. Biol. 10, 177 ( 10.1186/1471-2148-10-177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Górniak M, Paun O, Chase MW. 2010. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56, 784–795. ( 10.1016/j.ympev.2010.03.003) [DOI] [PubMed] [Google Scholar]
- 26.van den Berg C, Goldman DH, Freudenstein JV, Pridgeon AM, Cameron KM, Chase MW. 2005. An overview of the phylogenetic relationships within Epidendroideae inferred from multiple DNA regions and recircumscription of Epidendreae and Arethuseae (Orchidaceae). Am. J. Bot. 92, 613–624. ( 10.3732/ajb.92.4.613) [DOI] [PubMed] [Google Scholar]
- 27.Neubig KM, Whitten WM, Carlsward BS, Blanco MA, Endara L, Williams NH, Moore M. 2009. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst. Evol. 277, 75–84. ( 10.1007/s00606-008-0105-0) [DOI] [Google Scholar]
- 28.Xiang XG, Li DZ, Jin WT, Zhou HL, Li JW, Jin XH. 2012. Phylogenetic placement of the enigmatic orchid genera Thaia and Tangtsinia: evidence from molecular and morphological characters. Taxon 61, 45–54. [Google Scholar]
- 29.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenetic data sets. BMC Evol. Biol. 14, 82 ( 10.1186/1471-2148-14-82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms revisited. Am. J. Bot. 97, 1296–1303. ( 10.3732/ajb.0900346) [DOI] [PubMed] [Google Scholar]
- 33.Sytsma KJ, Spalink D, Berger B. 2014. Calibrated chronograms, fossils, outgroup relationships, and root priors: re-examining the historical biogeography of Geraniales. Biol. J. Linn. Soc. 176, 1–14. ( 10.1111/boj.12193) [DOI] [Google Scholar]
- 34.Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543 ( 10.1371/journal.pone.0089543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. ( 10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]
- 36.Givnish TJ, et al. 2014. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Mol. Phylog. Evol. 71, 55–78. ( 10.1016/j.ympev.2013.10.010) [DOI] [PubMed] [Google Scholar]
- 37.Delannoy E, Fujii S, Colas des Francs-Small C, Brundett M, Small I. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28, 2077–2086. ( 10.1093/molbev/msr028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldwin SL, Fitzgerald PG, Webb LE. 2012. Tectonics of the New Guinea region. Annu. Rev. Earth Planet Sci. 40, 495–520. ( 10.1146/annurev-earth-040809-152540) [DOI] [Google Scholar]
- 39.Jersáková J, Johnson SD. 2006. Mechanisms and evolution of deceptive pollination in orchids. Biol. Rev. 81, 219–235. ( 10.1017/S1464793105006986) [DOI] [PubMed] [Google Scholar]
- 40.Scopece G, Cozzolino S, Johnson SD, Schiestl FP. 2009. Pollination efficiency and the evolution of specialized deceptive pollination systems. Am. Nat. 175, 98–105. ( 10.1086/648555) [DOI] [PubMed] [Google Scholar]
- 41.Johnson SD, Hobbhahn N, Bytebier B. 2013. Ancestral deceit and labile evolution of nectar production in the African orchid genus Disa. Biol. Lett. 9, 20130500 ( 10.1098/rsbl.2013.0500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittall JB, Hodges SA. 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447, 706–709. ( 10.1038/nature05857) [DOI] [PubMed] [Google Scholar]
- 43.Kreft H, Köster N, Küper W, Nieder J, Barthlott W. 2004. Diversity and biogeography of vascular epiphytes in western Amazonia, Ecuador. J. Biogeogr. 31, 1463–1476. ( 10.1111/j.1365-2699.2004.01083.x) [DOI] [Google Scholar]
- 44.Küper W, Kreft H, Nieder H, Köster N, Barthlott W. 2004. Large-scale diversity patterns of vascular epiphytes in Neotropical montane rain forests. J. Biogeogr. 31, 1477–1487. ( 10.1111/j.1365-2699.2004.01093.x) [DOI] [Google Scholar]
- 45.Eriksson O, Kainulainen K. 2011. The evolutionary ecology of dust seeds. Perspect. Plant Ecol. Evol. Syst. 13, 73–87. ( 10.1016/j.ppees.2011.02.002) [DOI] [Google Scholar]
- 46.Phillips RD, Peakall R, Hutchinson MF, Linde CC, Xu T, Dixon KW, Hopper SD. 2014. Specialized ecological interactions and plant species rarity: the role of pollinators and mycorrhizal fungi across multiple spatial scales. Biol. Conserv. 169, 285–295. ( 10.1016/j.biocon.2013.11.027) [DOI] [Google Scholar]
- 47.Ricklefs RE, Renner SS. 1994. Species richness within families of flowering plants. Evolution 48, 1619–1636. ( 10.2307/2410252) [DOI] [PubMed] [Google Scholar]
- 48.Brandham P. 1999. Cytogenetics. In Genera Orchidacearum, vol. 1 (eds Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN), pp. 67–80. New York, NY: Oxford University Press. [Google Scholar]
- 49.Phillips RD, Dixon KW, Peakall R. 2012. Low population genetic differentiation in Orchidaceae: implication for the diversification of the family. Mol. Ecol. 21, 5208–5220. ( 10.1111/mec.12036) [DOI] [PubMed] [Google Scholar]
- 50.Jost L. 2004. Explosive local radiation of the genus Teagueia (Orchidaceae) in the Upper Pastaza watershed of Ecuador. Lyonia 7, 42–47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented or cited in the electronic supplementary material.




