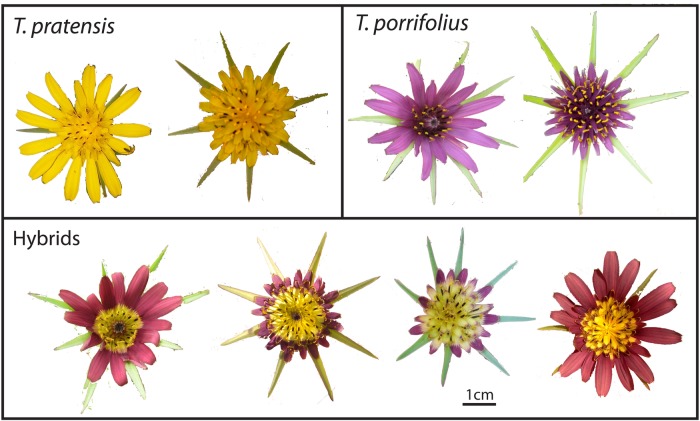
In 1759, Linnaeus convinced his followers that plants could hybridise by crossing flowers in the daisy family and producing intermediate offspring. These hybrids, between Tragopogon pratensis and T. porrifolius, exist naturally today in London, to all appearances the same. We find that most of the London hybrids are in their first generation, though we provide chromosomal evidence that one is a little older. These hybrids do not seem to have given rise to a new species, even though both parents have produced new hybrid species in the last century when crossed with T. dubius.
Keywords: Homoploid, hybridization, invasive, speciation, Tragopogon
Abstract
Hybridization between plant species can generate novel morphological diversity and lead to speciation at homoploid or polyploid levels. Hybrids between biennial herbs Tragopogon pratensis and T. porrifolius have been studied in experimental and natural populations for over 250 years. Here we examine their current status in natural populations in southeast England. All hybrids found were diploid; they tended to grow taller and with more buds than their parental species; many showed partial fertility; a few showed evidence of backcrossing. However, we found no evidence to suggest that the hybrids are establishing as a new species, nor can we find literature documenting speciation of these hybrids elsewhere. This lack of speciation despite at least 250 years of hybridization contrasts with the fact that both parental species have formed new allopolyploid species through hybridization with another diploid, T. dubius. Understanding why hybrids often do not speciate, despite repeated opportunities, would enhance our understanding of both the evolutionary process and risk assessments of invasive species.
Introduction
‘I obtained Tragopogon hybridum two years ago about autumn, in a small enclosure of the garden, where I had planted Tragopogon pratense and Tragopogon porrifolius, but the winter supervening destroyed the seeds. Early the following year, when Tragopogon pratense flowered, I rubbed off the pollen early in the morning, and at about eight in the morning I sprinkled the pistils with pollen from Tragopogon porrifolius and marked the calices with a thread bound around them. From these, towards autumn, I collected the mature seeds, and sowed them in a separate place, where they germinated, and in this year 1759, gave purple flowers with yellow bases, the seeds of which I now send.’ (pp. 126–127.)
Carolus Linnaeus (1760) Disquistitio de Sexu Plantarum, translation from Roberts (1929) Plant Hybridisation before Mendel, Princeton University Press, p. 22.
Since 1760, when Linnaeus published his Disquistitio de Sexu Plantarum, taxonomists have known that hybridization is an evolutionary source of novel morphological variation in plants (Linnaeus 1760; Roberts 1929; as Zirkle 1934, notes, the first artificial plant hybrids are credited to Thomas Fairchild of Hoxton in 1717, but his findings were not widely accepted). Even though hybrid novelties did not fit neatly with Darwin's later emphasis on gradualism and divergence in evolution (Darwin 1859), the evolutionary importance of hybridization has continued to be demonstrated and advocated by successive generations of biologists since Darwin (e.g. Lotsy 1916; Anderson and Stebbins 1954; Arnold 1992; Rieseberg et al. 2003) and is widely accepted today (Soltis and Soltis 2009; Abbott et al. 2013). Hybridization is now a well-attested mechanism for speciation, particularly when accompanied by genome doubling to produce allopolyploids (Rieseberg 1997; Mallet 2007; Soltis and Soltis 2009).
Linnaeus’ (1760) work was largely based upon crossing experiments between Tragopogon pratensis and T. porrifolius. These early experiments were repeated by Focke in Bremen, Germany (Focke 1890, 1897, 1907), by Winge in 1921 in Denmark (Winge 1938) and by Lotsy in the Netherlands (Lotsy 1927). Focke's and Winge's crosses yielded hybrids of similar morphology to Linnaeus', with purple outer ligules and yellow inner florets in the inflorescence, but Lotsy's showed a range of phenotypes not having a yellow centre to the inflorescence (Clausen 1966). Focke noted that some achenes produced by the hybrids germinated (Focke 1890) and that T. pratensis was normally the maternal parent of the hybrids (Focke 1897). The cross has also been repeated twice using North American accessions (Ownbey and McCollum 1953; Fahselt et al. 1976; Tate et al. 2009), derived from European parental accessions introduced to North America by settlers, yielding a range of inflorescence phenotypes in the F1, some having yellow central flowers, and some not. Several of the above studies harvested viable seeds from the F1 hybrids, and showed segregation of traits in F2 generations (Linnaeus 1760; Lotsy 1927; Winge 1938; Clausen 1966).
Hybrids between T. porrifolius and T. pratensis have been observed in the wild in Scandinavia for over 150 years. The PhD thesis of Carl Gosselman (1864) is reported to contain notes of this hybrid near Karlskrona in Sweden (Rouy 1890). Wilhelm Focke (1881, p. 222) reported that Johan Lange found spontaneous hybrids between T. porrifolius and T. pratensis on the Danish islands of Laaland and Funen: ‘the outer flowers brown-violet, the inner yellow’. In 1885, Knut Thedenius (1885) reported the hybrid in Stockholm. In 1890, Rouy named the hybrid T.×mirabilis and noted that it had been found by Gosselman (see above), and later by Foucard, Termonia and Maire in three locations in northern France (Rouy 1890). A population of Tragopogon diploids found in the Czech Republic were initially identified as hybrids between T. porrifolius and T. pratensis (Krahulec et al. 2005), but molecular investigations throw this identification into question (Malinska et al. 2011; Mavrodiev et al. 2013). Intriguingly, earlier than any of these reports, the 14th Volume of the Flora Danica, published in 1780, contains a colour plate (DCCXCVII) labelled as T. porrifolius in which the inflorescence has a yellow centre surrounded by purple outer ligules (Müller 1780); this may have been a hybrid found within a T. porrifolius population. Natural hybrids between T. porrifolius and T. pratensis have also been observed in North America since 1890 (Halsted 1890; Sherff 1911; Farwell 1930; Ownbey 1950; Clausen 1966; Novak et al. 1991).
Recent phylogenetic investigations of the genus Tragopogon confirm that T. pratensis and T. porrifolius are separate species, found in different phylogenetic clades. These investigations also suggest that purple-flowered European diploids (2n = 12) identified as T. porrifolius are polyphyletic with the most widespread lineage being the ‘salsify’ lineage (Mavrodiev et al. 2007). The yellow-flowered T. pratensis may also be non-monophyletic (e.g. Mavrodiev et al. 2012), but its most widespread lineage together with its sister species T. minor (or T. pratensis subsp. minor) consistently appears in the sub-clade Tragopogon. We cannot therefore be certain which lineages were involved in experimental crosses in the past, or which ones naturally hybridized, though the most widespread lineages would seem to be the most likely candidates. It seems probable that at least those hybrids with inflorescences appearing purple with a yellow centre have been repeatedly formed from the same parental lineages.
Tragopogon has become established as a model system for the study of hybridization, due to the discovery of various allopolyploid species (Ownbey 1950; Diaz De La Guardia and Blanca 2004; Mavrodiev et al. 2008a, b, 2015), documentation of further homoploid hybrids (reviewed in Buggs et al. 2008), a thorough phylogenetic framework (Mavrodiev et al. 2004, 2005), the recent resynthesis of hybrids and allotetraploids from their diploid progenitors (Tate et al. 2009) and transcriptome sequencing (Buggs et al. 2010). One intriguing feature of hybrid evolution in Tragopogon is a natural crossing ‘triangle’ among T. dubius, T. porrifolius and T. pratensis (all 2n = 12) within which natural allopolyploids (2n = 24) have formed repeatedly in the last 80 years from hybridizations involving T. dubius, but only homoploid hybrids have been recorded in nature between T. porrifolius and T. pratensis. In recent studies, much progress has been made on understanding the origin and rapid genome evolution of the two allopolyploids (T. mirus and T. miscellus) within this triangle (from T. dubius×T. porrifolius and T. dubius×T. pratensis, respectively) (reviewed in Soltis et al. 2012), but we know comparatively little about the homoploid hybrids between T. porrifolius and T. pratensis.
Both T. pratensis and T. porrifolius have been recorded in Britain since the 16th century to the present, T. pratensis being a native, and T. porrifolius considered to be a horticultural introduction (Gerard 1597; Stace and Crawley 2015). The oldest vouchered record of T. porrifolius in Britain is dated 1721, Bobart Hortus Siccus Sectio VIIA, p. 56 (OXF), collected from Sherard's garden at Eltham, London; T. porrifolius is also noted growing wild there (Dillenius 1732). Archaeobotanical records show Tragopogon sp. seed from Mid-Roman middens in York, dated between 150 and 200 AD (Hall and Kenward 1990). Hybrids have also been reported in Britain (Britton and Todd 1910; Ellis 1929; Clausen 1966; Burrow and Burrow 1978; Stace 2010). Druce curated T. porrifolius×T. pratensis specimens, including hybrids occurring naturally (F. Stratton, 1877; Dixon and Druce, 1907; Todd and Britton, 1910; H.E. Green, 1922; H. Wallis Kew, 1942, (OXF)) and hybrids produced by experimental crosses (C.E Britton, 1916, (OXF)).
This study aims to lay the foundations for the genetic and genomic study of T. porrifolius×T. pratensis hybrids, by identifying and characterizing natural populations. Here, we sampled six sites in southeast England reported to contain populations of T. porrifolius×T. pratensis hybrids, also sampling T. porrifolius and T. pratensis, if present at the sites. By analysing genome sizes, DNA sequences, morphology and seed fertility we explored the nature of the hybrids. Having confirmed their parentage, we asked: (i) Is there evidence for allopolyploid or homoploid hybrid speciation? (ii) Is there potential for gene flow between the two parental species?
Methods
Sampling
Sites in southeast England reported to contain putative T. porrifolius×T. pratensis hybrids were located by examination of botanical records and conversations with local botanists and county recorders for the Botanical Society of the British Isles. Six potential sites (Fig. 1) were visited between May and September 2011, with initial identification of Tragopogon species made using inflorescence morphology according to Stace (2010). At each site we aimed to collect equal numbers of plants of T. porrifolius, T. pratensis and putative hybrids, but this was rarely possible, and the collections made roughly reflected the overall frequency of each taxon at each site. Only plants with inflorescences were collected.
Figure 1.
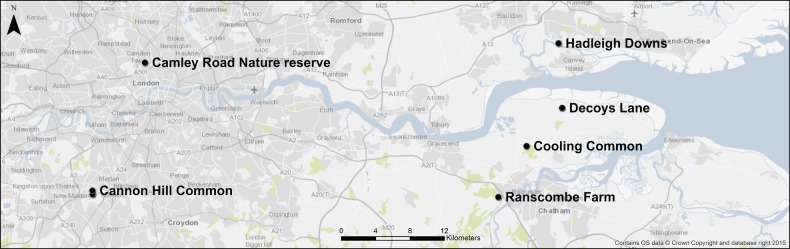
Location of the six populations sampled in southeast England, UK.
At least one parental species was found at each of these sites, and at two of the sites no hybrids were found. All putative hybrids had inflorescences with purple outer flowers and yellow inner flowers (Fig. 2), except for very rare cases with other intermediate morphology where purple and yellow coloration was irregularly mixed in each floret (e.g. Fig. 4). Collections of plants were made as follows: at Cannon Hill Common, 36 T. porrifolius, 15 T. pratensis and 16 hybrids; at Cooling Common, 13 T. porrifolius, 19 T. pratensis and 92 hybrids; at Hadleigh Downs, nine T. porrifolius, five T. pratensis and six hybrids; at Camley Road Nature Reserve, nine T. porrifolius; at Ranscombe Farm seven T. pratensis; at Decoys Lane, 75 T. porrifolius, two T. pratensis and two hybrids. A further 34 T. porrifolius, 42 T. pratensis and 12 hybrids were collected from among these six populations, whose location of origin was lost. Within the two parental species, both long and short ray floret morphs were present and sampled (Fig. 2).
Figure 2.
Typical inflorescences of T. pratensis, T. porrifolius and hybrids found in this study.
Flow cytometry
Fresh leaf samples were collected from plants identified by inflorescence morphology and stored in cool, moist conditions for up to 3 days. Flow cytometry was conducted on these fresh samples at the Jodrell Laboratory, Royal Botanic Gardens, Kew, to measure genome size (2C-value). Tragopogon leaf sections of ∼0.5 cm2 in size were co-chopped with leaves of the internal standard Petroselinum crispum ‘Curled Moss’ parsley [2C = 4.50 pg (Obermayer et al. 2002)], using a clean razor blade, in 1.5 mL of ice-cold ‘general plant isolation buffer’ (GPB, Loureiro et al. 2007) supplemented with 3 % polyvinylpyrrolidone (PVP-40). The homogenate was filtered through a 30-μm nylon mesh filter. The resulting nuclei suspension was stained with 0.5 μL of propidium iodide solution and supplemented with 0.8 μL of RNase to a final concentration of 50 μg mL−1. Samples were stored on ice for 15 min. The relative fluorescence of 1000 nuclei per sample was measured using a Partec GmbH PAII flow cytometer (Münster, Germany) fitted with a 100 W mercury arc lamp. The resulting histograms were analysed with the FlowMax software (v. 2.0, Partec GmbH) and the nuclear DNA contents estimated with the following formula: 2C-target = (target fluorescence peak/standard fluorescence peak) × 2C-standard.
Statistical analyses on morphometric data
For each plant collected, we measured the following morphological traits: length of root, width of root at the top and at midpoint, length from the top of the root to the first branching node of the shoot and length from the top of the root to the tip of the main shoot. Counts of buds, inflorescences, secondary stems and tertiary stems were also recorded. Statistical analyses were performed on samples collected from Cannon Hill Common and Cooling Common as these both had sufficient numbers of T. pratensis, T. porrifolius and hybrids for site to be treated as a random effect. Each trait was investigated for normality with parametric and non-parametric tests of homogeneity of variances with Bartlett tests and Fligner–Killeen tests. The number of tertiary stems and the number of buds per plant were square-root transformed to meet assumptions of normality. Linear mixed effect (LME) models were fitted by residual maximum likelihood (REML) with the function lmer in the lme4 package (Bates et al. 2014). Each trait was fitted individually as a response variable, with plant group treated as a fixed effect and site treated as a random effect. An LME model with all nine traits simultaneously as response variables was also fitted to test for an overall significant difference among the three groups (i.e. T. pratensis, T. porrifolius and hybrids). Statistical analyses were implemented in R (R Development Core Team 2008).
After morphological data were collected, whole plants from each sampling site were pressed and dried using standard herbarium-sized blots and folders.
Estimation of seed set
Where seed heads were available, we counted the number of plump and hollow (i.e. non-viable) achenes within each head. Plump achenes collected from some hybrids were tested for viability by placing them on damp filter paper in petri dishes, and germinants were planted into soil and grown to seedling stage.
DNA sequence analyses
We extracted DNA from a subset of plants from our collections: 15 T. porrifolius plants, 14 T. pratensis plants and 6 hybrids. We also extracted DNA from a plant collected by Foucaud in France in 1889 and identified as T. porrifolius but may represent the syntype of T.×mirabilis by Rouy (1890) held by the Paris Herbarium (PO3290423); a T. porrifolius plant collected (R. Buggs) in Tuscany, Italy in 2011; a T. pratensis plant collected (K. Emelianova) in the French Alps in 2012; and both T. porrifolius (2677-5, collected in Pullman, WA, USA, by C. Cody 27 June 2005) and T. pratensis (2609-24, collected in Spangle, WA, USA in 15 July 1999 by D. & P. Soltis).
Extraction of DNA was done using a modified CTAB method (Doyle and Doyle 1987). Using PCR we amplified the internal transcribed spacer (ITS1 and ITS2) and external transcribed spacer (ETS) sequences (located between the 18S and 26S ribosomal RNA genes), alcohol dehydrogenase (ADH) and three plastid regions (Taberlet et al. 1991), and sequenced them using Sanger sequencing. The ETS, ITS and ADH sequences were placed into multi-species alignments of Tragopogon (Mavrodiev et al. 2005, 2007, 2015), and all sequences were compared on a site-by-site basis. The strategy of amplification of ITS, ETS, ADH and plastid loci followed that described in Mavrodiev et al. (2008b, 2012), including the listed primers. The ADH locus was amplified using the primer pair: ADH_F and ADH_R from Mavrodiev et al. (2012). Maximum likelihood (ML) analyses of the ITS and ETS datasets were conducted separately using PhyML v. 3.1 (Guindon et al. 2010) following the strategy described in Mavrodiev et al. (2014) using sequence data from Mavrodiev et al. (2008b, 2012).
All sequenced samples of T. porrifolius and T. pratensis were included in the phylogenetic analyses to check their identification. For hybrid plants, we compared all sequences on a site-by-site basis and presented all results in the format of Tables (see for example Mavrodiev et al. 2015 for a similar approach), because the presence of multiple polymorphic single-nucleotide polymorphisms (SNP) in the raw nuclear chromatograms may bias phylogenetic tree topologies (reviewed in Soltis et al. 2008).
Karyotype of a putative backcross
The chromosomal composition of a putative hybrid individual from Cannon Hill Common with an unusual morphology and low genome size (see Results) was investigated. The plant was pulled from the soil and its roots were wrapped in wet sterile tissue paper and placed in a polythene bag. The plant was kept at room temperature in a well-lit lab and water added to the roots when the tissue paper began to dry. The terminal 2 cm of growing roots were harvested and pretreated in an aqueous solution of 2 mM 8-hydroxyquinoline for 16 h at 4 °C. Pretreated roots were then fixed in ice-cold 90 % acetic acid for 10 min and transferred to 70 % ethanol for −20 °C storage (Kato et al. 2011). Mitotic chromosome preparations and in situ hybridization were conducted with modifications to Kato et al. (2011) as described in Chester et al. (2012). Chromosome preparations were first subjected to fluorescence in situ hybridization (FISH) and then to genomic in situ hybridization (GISH). For FISH, probes comprised the following repetitive sequences: Cy5-labelled TPRMBO (Pires et al. 2004), Cy3-labelled TGP7 (Pires et al. 2004), Cy3- and fluorescein-labelled 18S rDNA and fluorescein-labelled TTR3 (Chester et al. 2013). For GISH, probes comprised Cy5-labelled total genomic DNA of T. pratensis (Colton, WA, USA; ID: 3939) and Cy3-labelled total genomic DNA of T. porrifolius (Pullman, WA, USA; ID: 3932). Image acquisition and processing, and karyotype construction were carried out as described previously (Chester et al. 2012). The parental origin of chromosomes was based on GISH signals in the centromeric and pericentromeric regions.
Results
Genome sizes
Flow cytometry was used to estimate genome sizes (2C) of 41 T. pratensis, 110 T. porrifolius and 94 putative hybrid plants. The genome sizes of T. pratensis plants ranged from 4.97 to 5.22 pg (mean = 5.14 pg; SD = 0.042), those of T. porrifolius ranged from 6.16 to 6.43 pg (mean = 6.28 pg; SD = 0.047) and those of the hybrids ranged from 5.38 to 5.83 pg (mean = 5.72 pg; SD = 0.064). The means of the three groups differed significantly (F2,242 = 7326, P < 0.00001). If the hybrids were F1 hybrids between these two species, we would expect them to have genome sizes between 5.57 and 5.83 pg, based on the maximum and minimum additive genome sizes of the parents. The lower than expected minimum range of the hybrids' distribution, and their higher standard deviation, was caused by two plants with smaller genome sizes than the others, being 5.38 and 5.54 pg. When these were excluded, the 2C-values of hybrids ranged from 5.61 to 5.83 pg (mean = 5.72 pg; SD = 0.050), as expected. One of the hybrids with a low genome size (5.54 pg), plant 1000 from Cannon Hill Common, was investigated further using cytogenetic methods (see below). All plants tested had genome sizes within the known diploid range of Tragopogon, so none of the plants sampled was polyploid.
Morphometric analysis
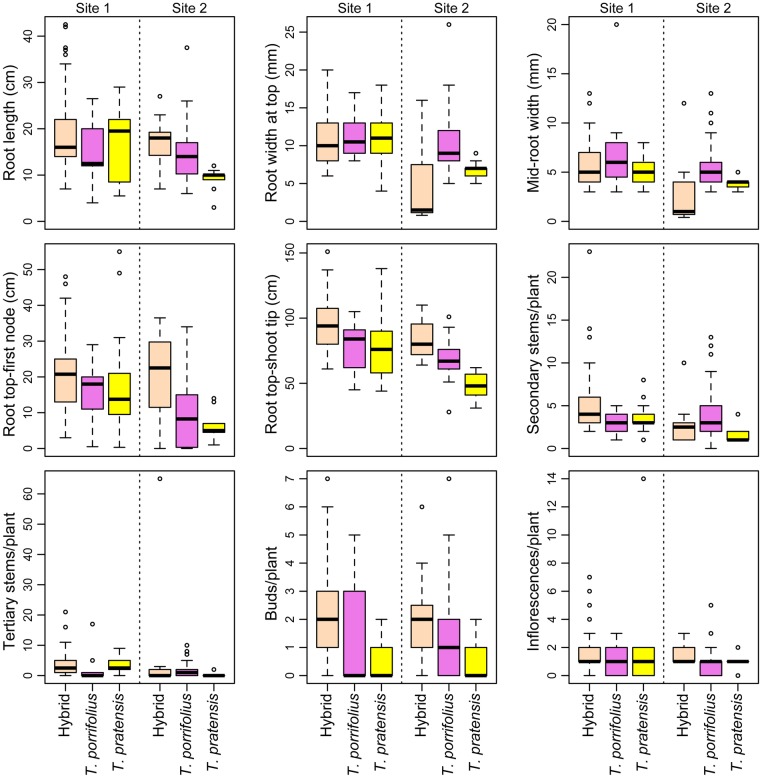
Linear mixed effect models comparing T. porrifolius and T. pratensis with the hybrid taxon, with site as a random effect, were carried out on 9 T. pratensis plants, 42 T. porrifolius plants and 16 hybrids from Cannon Hill Common and 18 T. pratensis plants, 13 T. porrifolius plants and 86 hybrids from Cooling Common. After Bonferroni correction, these showed the hybrids to differ from T. porrifolius in having smaller root width at top and middle, and in being greater in length from the top of the root to the first stem node (Table 1 and Fig. 3). The hybrids were greater than both parental species in length from the top of the root to the tip of the shoot, and in the number of buds per plant (Table 1 and Fig. 3). An LME examining the response of all nine traits shown in Fig. 3, to species, with site as a random effect, showed a significant difference among the three plant groups, with the hybrid significantly different from both parents (T. porrifolius: P = 0.0172, T. pratensis: P = 0.0463) (Table 1).
Table 1.
Linear mixed effect models showing T. porrifolius and T. pratensis compared with the hybrid taxon. Number of tertiary stems and number of buds per plant were square-root transformed. d.f., degrees of freedom.
| Response variable | Species | Value | Standard error | d.f. | t-value | P-value |
|---|---|---|---|---|---|---|
| Root length | (Intercept) | 17.691 | 1.375 | 171 | 12.862 | <0.0001 |
| T. porrifolius | −2.362 | 1.448 | 171 | −1.632 | 0.1046 | |
| T. pratensis | −3.886 | 1.600 | 171 | −2.429 | 0.0162 | |
| Root width at top | (Intercept) | 8.432 | 1.906 | 177 | 4.424 | <0.0001 |
| T. porrifolius | 3.004 | 0.765 | 177 | 3.927 | 0.0001 | |
| T. pratensis | 0.479 | 0.827 | 177 | 0.580 | 0.5627 | |
| Mid-root width | (Intercept) | 4.448 | 0.924 | 171 | 4.813 | <0.0001 |
| T. porrifolius | 2.185 | 0.514 | 171 | 4.249 | <0.0001 | |
| T. pratensis | −0.146 | 0.550 | 171 | −0.265 | 0.7911 | |
| Root base-first node | (Intercept) | 18.754 | 2.299 | 180 | 8.157 | <0.0001 |
| T. porrifolius | −6.897 | 1.933 | 180 | −3.568 | 0.0005 | |
| T. pratensis | −5.535 | 2.156 | 180 | −2.567 | 0.0111 | |
| Root base-shoot tip | (Intercept) | 88.747 | 7.835 | 180 | 11.327 | <0.0001 |
| T. porrifolius | −14.314 | 3.553 | 180 | −4.028 | <0.0001 | |
| T. pratensis | −23.504 | 3.889 | 180 | −6.043 | <0.0001 | |
| Secondary stems | (Intercept) | 4.467 | 0.571 | 180 | 7.821 | <0.0001 |
| T. porrifolius | −0.362 | 0.561 | 180 | −0.645 | 0.5195 | |
| T. pratensis | −1.454 | 0.633 | 180 | −2.295 | 0.0229 | |
| sqrt(Tertiary stems) | (Intercept) | 1.365 | 0.257 | 180 | 5.315 | <0.0001 |
| T. porrifolius | −0.332 | 0.225 | 180 | −1.477 | 0.1416 | |
| T. pratensis | −0.261 | 0.251 | 180 | −1.036 | 0.3015 | |
| sqrt(Buds/plant) | (Intercept) | 1.332 | 0.071 | 180 | 18.692 | <0.0001 |
| T. porrifolius | −0.625 | 0.120 | 180 | −5.194 | <0.0001 | |
| T. pratensis | −0.870 | 0.156 | 180 | −5.583 | <0.0001 | |
| Inflorescences/plant | (Intercept) | 1.696 | 0.155 | 180 | 10.910 | <0.0001 |
| T. porrifolius | −0.660 | 0.263 | 180 | −2.512 | 0.0129 | |
| T. pratensis | −0.141 | 0.340 | 180 | −0.414 | 0.6797 | |
| All nine morphometric | (Intercept) | 68889155 | 15339444 | 172 | 4.491 | <0.0001 |
| variables | T. porrifolius | −62878560 | 26130961 | 172 | −2.406 | 0.0172 |
| T. pratensis | −65969696 | 32872942 | 172 | −2.007 | 0.0463 |
Figure 3.
Box-and-whisker plots of morphological traits of T. porrifolius, T. pratensis and their hybrid collected from Cooling Common (Site 1) and Cannon Hill Common (Site 2). Outliers are shown as circles.
Achene production
We counted the number of achenes in one complete head from each of 69 T. porrifolius plants, 17 T. pratensis plants and 65 hybrids; the mean numbers of achenes per head were 73.5 (SD = 25.7), 47.1 (SD = 19.4) and 62.2 (SD = 17.1), respectively. For a further 25 T. porrifolius, 45 T. pratensis and 21 hybrid plants, we could only count achenes from heads that were incomplete because of loss of achenes due to dispersal or disturbance. In all heads, we calculated the proportions of achenes that were plump versus those that were hollow (i.e. aborted). The mean percentage of aborted achenes was found to be: 24.1 % (n = 94, SD = 37.4) in T. porrifolius plants, 7.9 % (n = 62, SD = 16.3) in T. pratensis plants and 90.4 % (n = 86, SD = 11.3) in hybrids; these results were significantly different (Kruskal–Wallis test: K = 3, H = 111.1, P < 0.0001). The notably high standard deviation for the percentage of aborted seeds in T. porrifolius was due to a subset of 15 T. porrifolius plants in which all seeds were aborted. It should be noted that poor reproductive success in the hybrid due to seed abortion was partly mitigated by higher production of inflorescence buds (Table 1).
Some of the achenes produced by plants whose hybrid status was suggested by their genome size and morphology were successfully germinated and grown to seedling stage. Hybrid plant 312 from Cannon Hill Common produced 28 achenes, of which 7 were plump, and 3 germinated, producing seedlings. Hybrid plant 403 from Cooling Common (which was also shown to be hybrid by DNA sequence analysis; see below) had 80 achenes, of which 6 were plump, and only 1 produced a seedling. Hybrid plant 404 from Cooling Common had 94 achenes, of which 13 were plump and 7 produced seedlings.
DNA sequence analyses
The five hybrids for which we obtained ADH sequences all showed the presence of double peaks at sites that differentiate the parents, corresponding to the bases present in both T. porrifolius and T. pratensis plants (Table 2): this is consistent with an F1 hybrid status of these plants. For their plastid loci, the hybrids only had haplotypes found in T. pratensis (Table 2), which indicates that T. pratensis is the maternal parent of all five of the hybrids analysed. Plant 3447, which had T. porrifolius morphology, showed two unusual base variants in its ADH sequence that were not found in any other plants analysed.
Table 2.
Molecular analyses showing plant collection details, genotypes at plastid (maternally inherited) and ADH (nuclear) variable sites and whether the samples were included in the ITS and ETS analyses.
| Morphology | Location | DNA number | Collection number | Nucleotide calls at variable plastid sites |
Nucleotide calls at variable ADH sites |
ETS | ITS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 205–211 | 1184–1193 | 1221 | 93 | 261 | 406 | 441 | 631 | 639 | 667 | 685 | ||||||
| Porrifolius | Camley Road | 3481 | CRNR13 | – | – | T | ||||||||||
| Porrifolius | Camley Road | 3486 | CRNR14 | – | – | T | ||||||||||
| Porrifolius | Camley Road | 3480 | CRNR3 | – | – | T | ||||||||||
| Porrifolius | Cannon Hill Common | 3449 | 311 | A | T | T | A | C | T | C | A | Y | ||||
| Porrifolius | Cannon Hill Common | 3447 | 329 | A | C | C | A | C | T | C | A | Y | Y | |||
| Porrifolius | Cannon Hill Common | 3477 | 307 | – | – | T | ||||||||||
| Porrifolius | Cannon Hill Common | 3478 | 310 | – | – | T | ||||||||||
| Porrifolius | Cannon Hill Common | 3440 | 322 | – | – | T | Y | Y | ||||||||
| Porrifolius | Cannon Hill Common | 3441 | 809 | – | – | T | Y | |||||||||
| Porrifolius | Cannon Hill Common | 3442 | 817 | Y | ||||||||||||
| Porrifolius | Cannon Hill Common | 3474/77 | 321 | – | – | T | ||||||||||
| Porrifolius | Cooling Common | 3450 | 555 | A | T | T | A | C | T | C | A | Y | Y | |||
| Porrifolius | Cooling Common | 3448 | 621 | A | T | T | A | C | T | C | A | Y | ||||
| Porrifolius | Cooling Common | 3475 | 556 | – | – | T | ||||||||||
| Porrifolius | Decoys Lane | 3444 | 643 | A | T | T | A | C | T | C | A | Y | ||||
| Porrifolius | Decoys Lane | 3443 | 651 | Y | ||||||||||||
| Porrifolius | Hadleigh Downs | 3445 | 427 | A | T | T | A | C | T | C | A | Y | ||||
| Porrifolius | Tuscany, Italy | 3483 | Tuscany | – | – | T | Y | |||||||||
| Porrifolius | France, Foucaud, 1889 | 3790/09 | PO3290423 | Y | ||||||||||||
| Pratensis | Cannon Hill Common | 3454 | 339 | G | T | T | G | A | G | T | A | Y | ||||
| Pratensis | Cannon Hill Common | 3485 | 317 | ATTTTTG | TTATACAAAT | T | ||||||||||
| Pratensis | Cannon Hill Common | 3451 | 337 | ATTTTTG | TTATACAAAT | G | Y | |||||||||
| Pratensis | Cannon Hill Common | 3470 | 302 | ATTTTTG | TTATACAAAT | T | ||||||||||
| Pratensis | Cannon Hill Common | 3473 | 304 | ATTTTTG | TTATACAAAT | T | ||||||||||
| Pratensis | Cannon Hill Common | 3469 | 305 | ATTTTTG | TTATACAAAT | G | ||||||||||
| Pratensis | Cooling Common | 3453 | 551 | G | T | T | G | A | G | T | A | Y | Y | |||
| Pratensis | Cooling Common | 3452 | 552 | G | T | T | G | A | G | T | A | Y | ||||
| Pratensis | Cooling Common | 3455 | 553 | G | T | T | G | A | G | T | A | Y | ||||
| Pratensis | Cooling Common | 3462 | 554 | ATTTTTG | TTATACAAAT | G | Y | |||||||||
| Pratensis | Cooling Common | 3467/82 | 550 | ATTTTTG | TTATACAAAT | G | Y | |||||||||
| Pratensis | Hadleigh Downs | 3468 | 443 | ATTTTTG | TTATACAAAT | T | ||||||||||
| Pratensis | Ranscombe Farm | 3460 | 600 | ATTTTTG | TTATACAAAT | G | ||||||||||
| Pratensis | Ranscombe Farm | 3463 | 601 | ATTTTTG | TTATACAAAT | G | ||||||||||
| Pratensis | French Alps | 3464 | KE001 | ATTTTTG | – | T | Y | |||||||||
| Hybrid | Cooling Common | 3446 | 63 | ATTTTTG | TTATACAAAT | G | A/G | T | T | A/G | M | T/G | C/T | A | Y | Y |
| Hybrid | Cooling Common | 3458 | 65 | ATTTTTG | TTATACAAAT | G | A/G | T | T | A/G | M | T/G | C/T | A | Y | Y |
| Hybrid | Cooling Common | 3456 | 403 | ATTTTTG | TTATACAAAT | G | A/G | T | T | A/G | M | T/G | C/T | A | Y | |
| Hybrid | Cooling Common | 3457 | 409 | ATTTTTG | TTATACAAAT | G | A/G | T | T | A/G | M | T/G | C/T | A | Y | |
| Hybrid | Cooling Common | 3459 | 408 | ATTTTTG | TTATACAAAT | G | A/G | T | T | A/G | M | T/G | C/T | A | Y | |
| Hybrid | Cannon Hill Common | 3484 | RB186 | Y | ||||||||||||
Phylogenetic analyses of ITS and ETS sequence data showed that in almost full agreement with previous results (e.g. Mavrodiev et al. 2005, 2008b) non-hybrids sequenced from southeast England appeared to be T. pratensis or T. porrifolius subspecies porrifolius sensu Flora Europaea (Richardson 1976): i.e. they likely come from the most widespread lineages of each species (see Introduction). Some samples of T. porrifolius possessed from one to few polymorphic SNPs in their ITS (Table 3) and/or ETS (Table 4) sequences, perhaps due to the incomplete homogenization of the repeats between two rDNA loci; this may be evidence for past hybridization. The 1889 collection from France (PO3290423), named as T. porrifolius by Foucaud, and possibly as T.×mirabilis by Rouy (1890) showed numerous double peaks at ETS sites (Table 4) that may suggest a hybrid origin, but as it contained six SNPs in the ETS region that were not found in any of the other plants we sampled, it is unlikely to be a hybrid between T. pratensis and T. porrifolius unless considerable nucleotide divergence has occurred in space and/or time within the species. In the ETS phylogenetic reconstruction, it was found in the Brevirostres clade [see Supporting Information—Fig. S2].
Table 3.
Summary of ITS base calls at variable sites.
| 26 | 34 | 58 | 88 | 90 | 101 | 107 | 411 | 425 | 439 | 497 | 519 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. porrifolius Pullman (USA), 3440, 3442, 3443, 3444, 3445, 3448, 3449 (UK) | A/T | T | A | A | C | C | T | G | T/G | T | C | C |
| T. porrifolius 3248 (UK) | A/T | T | A | A | C | C | T | G | T | T | C | C |
| T. porrifolius 3450 (UK) | A | T | A | A | C | C | T | G | T | T | C | C |
| T. porrifolius 3447, 3441 (UK) | A/T | T | A/G | A/G | C/A | C | C/T | A/G | T | C/T | C/T | C/T |
| Hybrids 3446, 3456, 3457, 3459 (UK) | A/T | T | A/G | A/G | C/A | C | C/T | A/G | T/G | C/T | C/T | C/T |
| Hybrids 3458 (UK) | A/T | T/G | A/G | A/G | C/A | C | C/T | A/G | T/G | C/T | C/T | C/T |
| T. pratensis Spangle (USA) | A | T | G | G | A | T | C | A | T | C | T | T |
| T. pratensis 3455, 3451, 3452, 3453, 3454, 3245 | A | T | G | G | A | C | C | A | T | C | T | T |
Table 4.
Summary of ETS base calls at variable sites.
| 18 | 19 | 44 | 55 | 71 | 147 | 150 | 195 | 200 | 202 | 203 | 212 | 219 | 223 | 278 | 309 | 361 | 362 | 411 | 417 | 428 | 509 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. porrifolius 3483 (Italy) | C | T | C | G | G | T | T | C | A | A | G | G | G | G | A | G | T | G | T | C | A | T |
| T. porrifolius Pullman (USA) and 3440, 3442, 3443, 3444, 3445, 3448, 3449, 3450 (UK) | C | T | C | G | G | T | T | C | A | A | A/G | T/G | G | G | A | G | T | G | T | C | A | T |
| T. porrifolius 3447 and 3441 (UK) | C/T | G/C | C/T | G | A/G | C/T | C/T | C | A/T | A/T | G | T/G | T/G | G | C/A | A/G | T | A/G | T | C/T | A | T |
| Foucaud collection 3709 PO3290423 (France, 1889) | C | T | C | G | G | C/T | T | C/A | A | A/T | G | T/G | G | A/G | A | G | C | G | C | C | T | A/T |
| Hybrid 3484 (Cannon Hill Common, UK) | C | T | C/T | T/G | A/G | C/T | C/T | C | A/T | A/T | G | T/G | T/G | G | C/A | A/G | T | A/G | T | C | A | T |
| Hybrid 3446, 3456, 3457, 3458, 3459 (Cooling Common, UK) | C | T | C/T | T/G | A/G | C/T | C/T | C | A/T | A/T | A/G | T/G | T/G | G | C/A | A/G | T | A/G | T | C | A | T |
| T. pratensis Spangle (USA), 3464 (France), 3460, 3461 (UK) | C | T | T | G | A | C | C | C | T | T | G | G | T | G | C | A | T | A | T | C | A | T |
| T. pratensis 3462, 3463, 3465, 3466, 3467, 3451, 3452, 3453, 3454, 3455, 3467, 3485 (UK) | C | T | T | T | A | C | C | C | T | T | G | G | T | G | C | A | T | A | T | C | A | T |
Cytogenetic analysis of a putative backcross
The putative hybrid plant (1000) from Cannon Hill Common had inflorescence morphology (Fig. 4) and 2C-value (5.54 pg) intermediate between that of a typical hybrid and T. pratensis. It was investigated using in situ hybridization to resolve its genomic composition (Fig. 5). This revealed the chromosome number to be 2n = 12, but the complement did not match that expected of an F1 hybrid, confirming the likely backcross status of the plant. Only one of the six chromosomes (belonging to group D sensu Chester et al. 2013) showed the expected 1 : 1 (T. pratensis:T. porrifolius) parental ratio. The other chromosomes were found in either a 2 : 0 (Group A, B, C and E) or 0 : 2 ratio (Group F), resulting in a bias in chromosome composition towards T. pratensis chromosomes. Several non-reciprocal intergenomic translocations were also observed (see arrows, Fig. 5), with one breakpoint on chromosome A originating from T. pratensis (APr) and two breakpoints on the single D chromosome originating from T. porrifolius (DPo) chromosome. Although GISH differentiation was poor due to the high amount of cross-hybridization between genomes, chromosome identification was supported by the FISH signals [i.e. by the presence of diagnostic TPRMBO, TTR3 and 18S rDNA signals that differ in their distribution between progenitor chromosomes (Chester et al. 2013)]. For both the A and D chromosome translocations, FISH signals were also consistent with the translocations involving homeologous exchanges.
Figure 4.

Inflorescence of plant 1000 from Cannon Hill Common, showing a morphology intermediate between that of hybrids and T. pratensis; the genome size of this plant showed a similar intermediacy.
Figure 5.
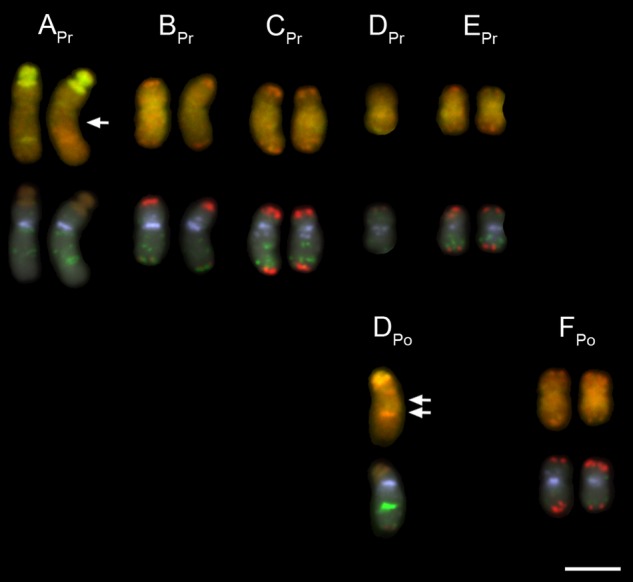
Karyotype of plant 1000. Each chromosome is shown twice, with signals resulting from either GISH (above) or FISH (below). Genomic in situ hybridization produced considerable cross-hybridization between genomes; chromatin of T. pratensis chromosomes appeared green/brown and chromatin of T. porrifolius chromosomes appeared red/orange. Fluorescence in situ hybridization allowed the chromosomes to be assigned to each homeologous group (A–F). Together, FISH and GISH revealed a skewed chromosome composition, with a bias towards T. pratensis. Chromosomes APr (A from T. pratensis) and DPo (D from T. porrifolius) showed intergenomic translocations (breakpoint positions are indicated by arrows). Fluorescence in situ hybridization probes were pseudo-coloured as follows: TGP7 (red), 18S rDNA (brown), TPRMBO (light blue), TTR3 (green). Chromosomes were counterstained with DAPI (grey). Scale bar: 5 μm.
Discussion
We found natural hybrids between T. pratensis and T. porrifolius in four populations in southeast England. At least one parental species was also found in each of these populations, and two sites that had previously been reported as containing hybrids were found to contain only plants with the morphology of the parental species. The hybrid plants had a morphology and fertility that fitted with the descriptions published by Linnaeus for this cross in 1760, and by numerous field and experimental botanists since (see Introduction). The F1 hybrid status of these plants was strongly supported by the flow cytometry data and confirmed by Sanger sequencing of genomic DNA regions. All F1 hybrids from which we sequenced plastid genes showed T. pratensis to be their maternal parent. None of the plants were polyploid. Together, these results suggest that the hybrids we found between T. pratensis and T. porrifolius are mainly ephemeral first-generation hybrids that have not speciated either at the homoploid or allopolyploid levels.
We found some evidence that backcrossing of the F1 hybrids may be occurring. Some hybrid plants produced low numbers of viable seeds, which germinated to produce seedlings. We investigated one plant with a genome size intermediate between those of F1 hybrids and T. pratensis, and found it to have a genome composition that could only have arisen via further rounds of meiosis since an F1. Unexpected DNA base variation in some plants that we initially identified as T. porrifolius may also be a consequence of hybridization followed by backcrossing. Thus, the hypothesis that gene flow may occur between T. pratensis and T. porrifolius via their hybrids merits further investigation.
Due to the low sample sizes and restricted sampling area of this study, our conclusions that speciation has not occurred but that some backcrossing is possible are obviously preliminary and restricted to those populations we sampled. However, we can find no records of natural homoploid hybrid species or allopolyploids between T. pratensis and T. porrifolius, despite frequent reports of hybridization both in their native European range (Gosselman 1864; Focke 1881; Thedenius 1885; Rouy 1890; Clausen 1966; Burrow and Burrow 1978; Stace 2010) and their introduced range in North America (Halsted 1890; Sherff 1911; Farwell 1930; Ownbey 1950; Clausen 1966). While broader surveys of larger numbers of individuals in both Europe and North America will be needed to fully confirm these conclusions, the fact that no new homoploid hybrid or allopolyploid species have been reported for this cross, despite the extensive botanical literature for Europe and North America suggests that our conclusions for southeast England may be true globally. Our findings are also remarkably similar to those of a succession of botanists who have identified T. pratensis, T. porrifolius and their hybrids in the field, or experimented on them, over the last 250 years. In 1966, Jens Clausen was struck by the stability of characters in T. pratensis, T. porrifolius and their hybrid over 200 years, concluding that there is ‘a high degree of permanence of the basic genetic structure of species’ (Clausen 1966, p. 157).
Assuming that these conclusions prove to be correct, hybrids between T. pratensis and T. porrifolius may be a useful study system to address the question of why three other possible outcomes have not evolved: (i) allopolyploid speciation; (ii) homoploid hybrid speciation; and (iii) divergence between T. pratensis and T. porrifolius to prevent hybridization. We outline these research questions below.
Why have allopolyploids not formed?
The lack of allopolyploids between T. pratensis and T. porrifolius is perhaps notable given that natural hybridization of both T. pratensis and T. porrifolius with T. dubius has yielded allopolyploid species on several independent occasions in the last 100 years (Ownbey 1950; Soltis et al. 2004; Symonds et al. 2010), and allopolyploids between T. pratensis and T. porrifolius have been produced by artificial hybridization and colchicine treatment in the glasshouse from American diploid plants (Tate et al. 2009).
One factor may be that T. pratensis and T. porrifolius are more closely related to one another than either of these species is to T. dubius [see Supporting Information—Figs S1 and S2]. As reviewed in Buggs et al. (2011a), it has long been suggested that hybridization between divergent parental species may promote polyploidization. The relationship between parental divergence, hybridization and polyploidy has been discussed in the last decade (Chapman and Burke 2007; Buggs et al. 2008, 2009, 2011a; Paun et al. 2009, 2011), mainly relying on statistical comparisons of parental divergence of homoploid hybrids and allopolyploids in several plant genera. The findings of this paper do not directly add to this discussion, as the discussion's various statistical analyses have already included the different outcomes of crossing in the Tragopogon triangle investigated here. However, the Tragopogon triangle might provide a useful study system to investigate mechanical hypotheses for how divergence might affect the outcomes of hybridization: for example, the possibility that patterns of divergence at particular loci or in particular chromosomal arrangements in T. pratensis, T. porrifolius and T. dubius may be affecting the outcomes of hybridization.
Another possibility that might be investigated is that T. dubius carries alleles that cause it to have a greater proclivity for allopolyploidization than T. pratensis and T. porrifolius. The existence of genetic variants promoting polyploidization was suggested by Grant (1981) and is shown by the success of selective breeding for rates of 2n gamete formation in Medicago and Trifolium (Grant 1981; Ramsey and Schemske 1998).
The influence of historical or biogeographic factors on hybridization in this system also warrants further investigation as they are likely to have a major role. Tragopogon pratensis and T. porrifolius are rarer in the Palouse area of Washington and Idaho in the USA than T. dubius, so there may have been fewer opportunities for hybridization between T. porrifolius and T. pratensis. The fact that Tragopogon allopolyploids have formed in Washington and Idaho but not in Europe may be because occasional environmental shocks such as extreme frosts during flowering (Hagerup 1932; Ramsey and Schemske 1998) have occurred more in Washington and Idaho and induced chromosome doubling. It could also be the case that ecological niches suitable for T. pratensis×T. porrifolius allopolyploids have not been available.
Why has homoploid hybrid speciation not occurred?
This question is perhaps easier to answer because although new homoploid hybrid species may evolve, the conditions required for their establishment are more stringent than the conditions for allopolyploid species establishment (Buerkle et al. 2000), as homoploid hybrids do not benefit from the immediate escape from parental gene flow that polyploids usually enjoy (Stebbins 1950). Models suggest that homoploid hybrid species can only evolve if they have sufficient ecological and spatial isolation from their parental species (Buerkle et al. 2000), due to ecological selection (Gross and Rieseberg 2005), as seems to be the case for homoploid hybrid species of Helianthus (Rieseberg et al. 2003) and Senecio (Abbott et al. 2010). The hybrids found in this study were all growing in similar habitats to the parental species and in close proximity to them; they differed in some morphological traits, being on average transgressive in height and number of buds produced, and were significantly different in an LME model that took all of our morphological measurements into account. Homoploid hybrids are most likely to have evolutionary independence from their parents if they have chromosomal rearrangements that cause them to be reproductively isolated from their parental species (Buerkle et al. 2000; Yakimowski and Rieseberg 2014). The backcross hybrid that we karyotyped shows chromosomal variations that may provide incompatibilities with the parents, but isolation from parents by itself is not sufficient to cause speciation (Buerkle et al. 2000; Yakimowski and Rieseberg 2014).
Why have T. pratensis and T. porrifolius not diverged further?
If two species can hybridize to form low-fitness hybrids, there should be selective pressures causing reinforcement of pre-zygotic isolation mechanisms between the hybridizing species (Dobzhansky 1940). Although this hypothesis has been criticized (e.g. Howard 1993; Marshall et al. 2002), other evidence supports it (e.g. Hopkins and Rausher 2011; Andrew and Rieseberg 2013). In the present study, we find no evidence for ongoing reinforcement as T. pratensis and T. porrifolius appear to have been hybridizing and producing low-fertility hybrids over at least the last 250 years. This may be because 250 years is too short a timespan for further divergence to have evolved, being only 125 generations of these biennial species (c.f. Buerkle and Rieseberg 2008). It may be that the rate of hybrid production is too low to be of significant reproductive cost to either species, or that sites of hybrid production have been ephemeral so that selection has not acted consistently on particular populations for extended periods. Alternatively, it is well known that gene flow can prevent the divergence of species (Slatkin 1987), and it could be that hybridization and concomitant gene flow between T. porrifolius and T. pratensis, though very low, are sufficiently high to hinder increased divergence between the two species. If this were the case, it would appear that levels of gene flow are low enough not to cause merging of the species. It may also be that natural selection is maintaining the two species in the face of gene flow (Nosil 2008; Abbott et al. 2013). Thus, it could be worth investigating whether the two species appear stable due to a dynamic process of gene flow that, in balance with natural selection on the two parental morphs, is holding the system in a dynamic equilibrium.
Conclusions
Tragopogon has been extensively developed as a model system to study the genomics and transcriptomics of allopolyploid speciation, where rapid change has been shown to occur both in the formation of the allopolyploids and in their subsequent generations (Tate et al. 2006; Buggs et al. 2011b, 2012; Chester et al. 2012; Soltis et al. 2012; Lipman et al. 2013). In contrast, although hybrids between T. pratensis and T. porrifolius have been studied scientifically for a longer period than any other plant hybrid, over this 250-year period of experimentation and observation there appears to have been little outward change in the dynamics of this interaction and its morphological consequences. In this paper we speculate as to why this is so, but thorough understanding of the interaction, and particularly of the dynamics of gene flow, which may be critical to the apparent stability of the parental species, will only come through genome-wide analyses of variation in natural populations. The present study lays the foundations for such future research. Understanding why hybrids do not speciate, despite repeated opportunities, would enhance our understanding of both the evolutionary process and risk assessments of invasive species. The apparent stasis of the diploid species and their hybrids in the present study underlines the importance of polyploidy in the promotion of rapid evolution in this genus.
Accession Numbers
Herbarium samples of representative material are deposited with the British Museum Herbarium with accession numbers BM001139296–BM001139307 [see Supporting Information—Table S1].
The DNA sequences have been deposited in GenBank with accession numbers as follows: KT167073–KT167093 (ITS sequences), KT167094–KT167124 (ETS sequences), KT167125–KT167149 (plastid sequences) and KT167150–KT167161 (ADH sequences).
Sources of Funding
This work was principally supported by SYNTAX grant 2010/11 number 13, administered by the Linnean Society of London and funded by NERC and BBSRC, and by NERC Fellowship NE/G01504X/1. Molecular and cytogenetic work was supported by NSF grants DEB-0922003 and DEB-1146065.
Contributions by the Authors
R.J.A.B., E.V.M., A.R.L., I.J.L., M.C., D.E.S. and P.S.S. formed the research questions. A.M., K.E., M.C., A.A.H., J.P., K.S.A., M.S.G., E.V.M. and R.J.A.B. implemented the project in the field and laboratory. All authors contributed to the development, analysis of data and manuscript drafting.
Conflict of Interest Statement
None declared.
Supporting Information
The following additional information is available in the online version of this article —
Figure S1. Maximum likelihood ITS tree of the genus Tragopogon showing hybrid samples forming a clade with T. porrifolius. Samples from the present study have a four-digit ID number in their label.
Figure S2. Maximum likelihood ETS tree of the genus Tragopogon showing hybrid samples forming a clade with T. pratensis, and unexpected placement of the accession labelled T. porrifolius from the Paris herbarium. Samples from the present study have a four-digit ID number in their label.
Table S1. List of herbarium specimens deposited at the British Museum Herbarium with accession numbers.
Acknowledgements
We thank Mark Spencer and Ian J. Kitching of the Natural History Museum, London, Geoffrey Kitchener, Sue Poyser, Kenneth Adams, Ann Sankey, of the Botanical Society of the British Isles, and Jonathan Turner and Dusty Gedge for information on the location of sites containing Tragopogon hybrids. We would like to thank Dr Malika Ainouche (Université de Rennes, France) for the help with collection and images of Tragopogon from Museum national d'Histoire naturelle (France). We thank Elaine Charwat for research help at the Linnean Society Library and Serena Marner at the Fielding-Druce Herbarium, Oxford.
Literature Cited
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Vainola R, Wolf JBW, Zinner D. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26:229–246. 10.1111/j.1420-9101.2012.02599.x [DOI] [PubMed] [Google Scholar]
- Abbott RJ, Hegarty MJ, Hiscock SJ, Brennan AC. 2010. Homoploid hybrid speciation in action. Taxon 59:1375–1386. [Google Scholar]
- Anderson E, Stebbins GL Jr. 1954. Hybridization as an evolutionary stimulus. Evolution 8:378–388. 10.2307/2405784 [DOI] [Google Scholar]
- Andrew RL, Rieseberg LH. 2013. Divergence is focused on few genomic regions early in speciation: incipient speciation of sunflower ecotypes. Evolution 67:2468–2482. 10.1111/evo.12106 [DOI] [PubMed] [Google Scholar]
- Arnold ML. 1992. Natural hybridization as an evolutionary process. Annual Review of Ecology and Systematics 23:237–261. 10.1146/annurev.es.23.110192.001321 [DOI] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7 http://CRAN.R-project.org/package=lme4. [Google Scholar]
- Britton CE, Todd WA. 1910. Tragopogon porrifolium × pratense. Journal of Botany, British and Foreign XLVIII:204. [Google Scholar]
- Buerkle CA, Rieseberg LH. 2008. The rate of genome stabilization in homoploid hybrid species. Evolution 62:266–275. 10.1111/j.1558-5646.2007.00267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. 2000. The likelihood of homoploid hybrid speciation. Heredity 84:441–451. 10.1046/j.1365-2540.2000.00680.x [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Soltis PS, Mavrodiev EV, Symonds VV, Soltis DE. 2008. Does phylogenetic distance between parental genomes govern the success of polyploids? Castanea 73:74–93. 10.2179/0008-7475(2008)73[74:DPDBPG]2.0.CO;2 [DOI] [Google Scholar]
- Buggs RJA, Soltis PS, Soltis DE. 2009. Does hybridization between divergent progenitors drive whole-genome duplication? Molecular Ecology 18:3334–3339. 10.1111/j.1365-294X.2009.04285.x [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Chamala S, Wu W, Gao L, May GD, Schnable PS, Soltis DE, Soltis PS, Barbazuk WB. 2010. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Molecular Ecology 19:132–146. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Soltis PS, Soltis DE. 2011a. Biosystematic relationships and the formation of polyploids. Taxon 60:324–332. [Google Scholar]
- Buggs RJA, Zhang L, Miles N, Tate JA, Gao L, Wei W, Schnable PS, Barbazuk WB, Soltis PS, Soltis DE. 2011b. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Current Biology 21:551–556. 10.1016/j.cub.2011.02.016 [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Chamala S, Wu W, Tate JA, Schnable PS, Soltis DE, Soltis PS, Barbazuk WB. 2012. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Current Biology 22:248–252. 10.1016/j.cub.2011.12.027 [DOI] [PubMed] [Google Scholar]
- Burrow B, Burrow J. 1978. Tragopogon × mirabilis Rouy in west Kent, VC 16. Watsonia 12:157. [Google Scholar]
- Chapman MA, Burke JM. 2007. Genetic divergence and hybrid speciation. Evolution 61:1773–1780. 10.1111/j.1558-5646.2007.00134.x [DOI] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, da Silva AVC, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE. 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proceedings of the National Academy of Sciences of the USA 109:1176–1181. 10.1073/pnas.1112041109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Lipman MJ, Gallagher JP, Soltis PS, Soltis DE. 2013. An assessment of karyotype restructuring in the neoallotetraploid Tragopogon miscellus (Asteraceae). Chromosome Research 21:75–85. 10.1007/s10577-013-9339-y [DOI] [PubMed] [Google Scholar]
- Clausen J. 1966. Stability of genetic characters in Tragopogon species through 200 years. Transactions of the Botanical Society of Edinburgh 40:148–158. 10.1080/03746606608685138 [DOI] [Google Scholar]
- Darwin C. 1859. The origin of species. London: John Murray. [Google Scholar]
- Diaz De La Guardia C, Blanca G. 2004. A new Spanish species of Tragopogon (Asteraceae: Lactuceae). Botanical Journal of the Linnean Society 146:505–511. [Google Scholar]
- Dillenius JJ. 1732. Hortus elthamensis. London. [Google Scholar]
- Dobzhansky T. 1940. Speciation as a stage in evolutionary divergence. The American Naturalist 74:312–321. 10.1086/280899 [DOI] [Google Scholar]
- Doyle J, Doyle JL. 1987. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochemical Bulletin 19:11–15. [Google Scholar]
- Ellis AE. 1929. Tragopogon porrifolius × pratensis, var. minor. The Botanical Society and Exchange Club of The British Isles 9:125. [Google Scholar]
- Fahselt D, Ownbey M, Borton M. 1976. Seed fertility in Tragopogon hybrids (Compositae). American Journal of Botany 63:1109–1118. 10.2307/2441656 [DOI] [Google Scholar]
- Farwell OA. 1930. Botanical gleanings in Michigan, VI. American Midland Naturalist 12:113–134. 10.2307/2419908 [DOI] [Google Scholar]
- Focke WO. 1881. Die pflanzen-mischlinge; ein beitrag zur biologie der gewächse. Berlin: Gebrüder Borntraeger. [Google Scholar]
- Focke WO. 1890. Versuche und Beobachtungen über Kreuzung und Fruchtensatz bei Blutenpflanzen. Abhandlungen herausgegeben vom Naturwissenschaftlichen Verein zu Bremen 11:413–421. [Google Scholar]
- Focke WO. 1897. Neue Beobachtungen über Artenkreuzung und Selbsterilitat. Abhandlungen herausgegeben vom Naturwissenschaftlichen Verein zu Bremen 14:297–304. [Google Scholar]
- Focke WO. 1907. Betrachtungen und Erfahrungen uber Variation und Artenbildung. Abhandlungen herausgegeben vom Naturwissenschaftlichen Verein zu Bremen 19:68–87. [Google Scholar]
- Gerard J. 1597. The Herball, or, Generalle historie of plantes. London: John Norton. [Google Scholar]
- Gosselman CA. 1864. Zoologiska och botaniska iakttagelser inom Blekinge. Unpublished PhD thesis, Lund. [Google Scholar]
- Grant V. 1981. Plant speciation. New York: Columbia University Press. [Google Scholar]
- Gross BL, Rieseberg LH. 2005. The ecological genetics of homoploid hybrid speciation. Journal of Heredity 96:241–252. 10.1093/jhered/esi026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hagerup O. 1932. Über polyploidie in Beziehung zu klima, ökologie und phylogenie. Hereditas 16:19–40. 10.1111/j.1601-5223.1932.tb02560.x [DOI] [Google Scholar]
- Hall AR, Kenward HK. 1990. Environmental evidence fromthe colonia: general accident and Rougier Street. London: Council for British Archaeology. [Google Scholar]
- Halsted BD. 1890. A possible natural hybrid. Bulletin of the Torrey Botanical Club 17:176–177. [Google Scholar]
- Hopkins R, Rausher MD. 2011. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature 469:411–414. 10.1038/nature09641 [DOI] [PubMed] [Google Scholar]
- Howard DJ. 1993. Reinforcement: origin, dynamics, and fate of an evolutionary hypothesis. In: Harrison R, ed. Hybrid zones and the evolutionary process. Oxford: Oxford University Press, 46–69. [Google Scholar]
- Kato A, Lamb JC, Albert PS, Danilova T, Han F, Gao Z, Findley S, Birchler JA. 2011. Chromosome painting for plant biotechnology. Plant chromosome engineering. New York: Humana Press, 67–96. [DOI] [PubMed] [Google Scholar]
- Krahulec F, Kaplan Z, Novak J. 2005. Tragopogon porrifolius × T. pratensis: the present state of an old hybrid population in Central Bohemia, the Czech Republic. Preslia 77:297–306. [Google Scholar]
- Linnaeus C. 1760. Disquistitio de Sexu Plantarum. St Petersburg: Academia Imperiali Scientiarum Petropolitana. [Google Scholar]
- Lipman MJ, Chester M, Soltis PS, Soltis DE. 2013. Natural hybrids between Tragopogon mirus and T. miscellus (Asteraceae): a new perspective on karyotypic changes following hybridization at the polyploid level. American Journal of Botany 100:2016–2022. 10.3732/ajb.1300036 [DOI] [PubMed] [Google Scholar]
- Lotsy JP. 1916. Evolution by means of hybridization. The Hague: Martinus Nijhoff. [Google Scholar]
- Lotsy JP. 1927. What do we know of the descent of man? Genetica 9:289–328. 10.1007/BF01508294 [DOI] [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany 100:875–888. 10.1093/aob/mcm152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska H, Tate JA, Mavrodiev E, Matyasek R, Lim KY, Leitch AR, Soltis DE, Soltis PS, Kovarik A. 2011. Ribosomal RNA genes evolution in Tragopogon: a story of New and Old World allotetraploids and the synthetic lines. Taxon 60:348–354. [Google Scholar]
- Mallet J. 2007. Hybrid speciation. Nature 446:279–283. [DOI] [PubMed] [Google Scholar]
- Marshall JL, Arnold ML, Howard DJ. 2002. Reinforcement: the road not taken. Trends in Ecology and Evolution 17:558–563. 10.1016/S0169-5347(02)02636-8 [DOI] [Google Scholar]
- Mavrodiev EV, Tancig M, Sherwood AM, Gitzendanner MA, Rocca J, Soltis PS, Soltis DE. 2005. Phylogeny of Tragopogon L. (Asteraceae) based on internal and external transcribed spacer sequence data. International Journal of Plant Sciences 166:117–133. 10.1086/425206 [DOI] [Google Scholar]
- Mavrodiev EV, Soltis PS, Gitzendanner MA, Baldini RM, Soltis DE. 2007. Polyphyly of Tragopogon porrifolius L. (Asteraceae), a European native with intercontinental disjuncts. International Journal of Plant Sciences 168:889–904. 10.1086/518258 [DOI] [Google Scholar]
- Mavrodiev EV, Albach DC, Speranza P. 2008a. A new polyploid species of the genus Tragopogon (Asteraceae, Cichorieae) from Russia. Novon 18:229–232. [Google Scholar]
- Mavrodiev EV, Soltis PS, Soltis DE. 2008b. Putative parentage of six Old World polyploids in Tragopogon L.(Asteraceae: Scorzonerinae) based on ITS, ETS, and plastid sequence data. Taxon 57:1215–1215. [Google Scholar]
- Mavrodiev EV, Gitzendanner M, Calaminus AK, Baldini RM, Soltis PS, Soltis DE. 2012. Molecular phylogeny of Tragopogon L. (Asteraceae) based on seven nuclear loci (Adh, GapC, LFY, AP3, PI, ITS, and ETS). Webbia 67:111–137. 10.1080/00837792.2012.10670912 [DOI] [Google Scholar]
- Mavrodiev EV, Krahulec F, Soltis DE, Soltis PS. 2013. A cryptic taxon rather than a hybrid species of Tragopogon (Asteraceae) from the Czech Republic. Kew Bulletin 68:133–141. 10.1007/s12225-012-9431-z [DOI] [Google Scholar]
- Mavrodiev EV, Martínez-Azorín M, Dranishnikov P, Crespo MB. 2014. At least 23 genera instead of one: the case of Iris L. s.l. (Iridaceae). PLoS ONE 9:e106459 10.1371/journal.pone.0106459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodiev EV, Chester M, Suárez-Santiago VN, Visger CJ, Rodriguez R, Susanna A, Baldini RM, Soltis PS, Soltis DE. 2015. Multiple origins and chromosomal novelty in the allotetraploid Tragopogon castellanus (Asteraceae). New Phytologist 206:1172–1183. 10.1111/nph.13227 [DOI] [PubMed] [Google Scholar]
- Müller OF. 1780. Flora DanicaVol. 14. Copenhagen: Danish Crown. [Google Scholar]
- Nosil P. 2008. Speciation with gene flow could be common. Molecular Ecology 17:2103–2106. 10.1111/j.1365-294X.2008.03715.x [DOI] [PubMed] [Google Scholar]
- Novak SJ, Soltis DE, Soltis PS. 1991. Ownbey's Tragopogons: 40 years later. American Journal of Botany 78:1586–1600. 10.2307/2444984 [DOI] [Google Scholar]
- Obermayer R, Leitch IJ, Hanson L, Bennett MD. 2002. Nuclear DNA C-values in 30 species double the familial representation in pteridophytes. Annals of Botany 90:209–217. 10.1093/aob/mcf167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownbey M. 1950. Natural hybridization and amphiploidy in the genus Tragopogon. American Journal of Botany 37:487–499. 10.2307/2438023 [DOI] [Google Scholar]
- Ownbey M, McCollum GD. 1953. Cytoplasmic inheritance and reciprocal amphiploidy in Tragopogon. American Journal of Botany 40:788–796. 10.2307/2438276 [DOI] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. 2009. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytologist 182:507–518. 10.1111/j.1469-8137.2009.02767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. 2011. Parental divergence and hybrid speciation in angiosperms revisited. Taxon 60:1241–1244. [PMC free article] [PubMed] [Google Scholar]
- Pires JC, Lim KY, Kovarik A, Matyasek R, Boyd A, Leitch AR, Leitch IJ, Bennett MD, Soltis PS, Soltis DE. 2004. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. American Journal of Botany 91:1022–1035. 10.3732/ajb.91.7.1022 [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29:467–501. 10.1146/annurev.ecolsys.29.1.467 [DOI] [Google Scholar]
- R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; ISBN 3-900051-07-0 http://www.R-project.org. [Google Scholar]
- Richardson I. 1976. Tragopogon. Flora Europaea 4:322–325. [Google Scholar]
- Rieseberg LH. 1997. Hybrid origins of plant species. Annual Review of Ecology and Systematics 28:359–389. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. 10.1126/science.1086949 [DOI] [PubMed] [Google Scholar]
- Roberts HF. 1929. Plant hybridisation before Mendel. Princeton: Princeton University Press. [Google Scholar]
- Rouy MG. 1890. Remarques sur la synonymie de quelques plantes occidentales. Bulletin de la Société Botanique de France 37:XIV–XX. 10.1080/00378941.1890.10831526 [DOI] [Google Scholar]
- Sherff EE. 1911. Tragopogon pratensis × porrifolius. Torreya 11:14–15. [Google Scholar]
- Slatkin M. 1987. Gene flow and the geographic structure of natural populations. Science 236:787–792. 10.1126/science.3576198 [DOI] [PubMed] [Google Scholar]
- Soltis D, Buggs RA, Barbazuk WB, Chamala S, Chester M, Gallagher J, Schnable P, Soltis P. 2012. The early stages of polyploidy: rapid and repeated evolution in Tragopogon. In: Soltis PS, Soltis DE, eds. Polyploidy and genome evolution. Berlin: Springer, 271–292. [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. 2004. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biological Journal of the Linnean Society 82:485–501. 10.1111/j.1095-8312.2004.00335.x [DOI] [Google Scholar]
- Soltis PS, Soltis DE. 2009. The role of hybridization in plant speciation. Annual Review of Plant Biology 60:561–588. 10.1146/annurev.arplant.043008.092039 [DOI] [PubMed] [Google Scholar]
- Stace CA. 2010. New flora of the British Isles. Cambridge: Cambridge University Press. [Google Scholar]
- Stace CA, Crawley MJ. 2015. Alien plants. Collins New Naturalist Library, Book 128. London: HarperCollins Publishers. [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Symonds VV, Soltis PS, Soltis DE. 2010. The dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution 64:1984–2003. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17:1105–1109. 10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Tate JA, Ni ZF, Scheen AC, Koh J, Gilbert CA, Lefkowitz D, Chen ZJ, Soltis PS, Soltis DE. 2006. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173:1599–1611. 10.1534/genetics.106.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JA, Symonds VV, Doust AN, Buggs RJA, Mavrodiev E, Majure LC, Soltis PS, Soltis DE. 2009. Synthetic polyploids of Tragopogon miscellus and T. mirus (Asteraceae): 60 years after Ownbey's discovery. American Journal of Botany 96:979–988. 10.3732/ajb.0800299 [DOI] [PubMed] [Google Scholar]
- Thedenius KF. 1885. Tragopogon porrifolio-minor Thed. en ny hybrid, funnen i Stockholm. Botaniska Notiser 1885:156–158. [Google Scholar]
- Winge Ø. 1938. Inheritance of species characters in Tragopogon. Compte Rendu des Travaux Laboratoire de Carlsberg, Serie Physique 22:155–193. [Google Scholar]
- Yakimowski SB, Rieseberg LH. 2014. The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology. American Journal of Botany 101:1247–1258. 10.3732/ajb.1400201 [DOI] [PubMed] [Google Scholar]
- Zirkle C. 1934. More records of plant hybridization before Koelreuter. Journal of Heredity 25:3–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.