Abstract
Plasmodium berghei was identified as a parasite of thicket rats (Grammomys dolichurus) and Anopheles dureni mosquitoes in African highland forests. Successful adaptation to a range of rodent and mosquito species established P. berghei as a malaria model parasite. The introduction of stable transfection technology, permitted classical reverse genetics strategies and thus systematic functional profiling of the gene repertoire. In the past 10 years following the publication of the P. berghei genome sequence, many new tools for experimental genetics approaches have been developed and existing ones have been improved. The infection of mice is the principal limitation towards a genome-wide repository of mutant parasite lines. In the past few years, there have been some promising and most welcome developments that allow rapid selection and isolation of recombinant parasites while simultaneously minimising animal usage. Here, we provide an overview of all the currently available tools and methods.
Keywords: Malaria, Plasmodium berghei, Experimental genetics, In vivo model, Transfection, Recombination, Murine malaria model
Introduction
Since the first description of the malaria parasite by Laveran,1 researchers have been trying to gain insights into the biology of Plasmodium parasites. While initial studies solely focussed on observation of wild-type parasites, the ability to genetically manipulate Plasmodium spp., revolutionised the field of malaria research. Successful transfection was first demonstrated in the avian pathogen Plasmodium gallinaceum.2 Since then, a diverse repertoire of Plasmodium parasites proved to be accessible to genetic manipulation, including human,3,4 primate,5,6 and rodent7–9 malaria parasites. The availability of many complete or near-complete genome sequences10–15 has been another huge advance towards a more profound understanding of Plasmodium biology. Genome sequence data have been a key to the scope and success of experimental genetics approaches in malaria research.
Despite the ability to introduce foreign DNA molecules into a variety of malaria parasite species, there are profound differences in the level of accessibility, ease and efficiency of genetic manipulation. For example, despite recent advances that enable the use of zinc-finger nucleases to modify the P. vivax genome more effectively,16 all genetic manipulation of this human malaria parasite is severely hampered by the inability to continuously culture these parasites in vitro, thus necessitating in vivo infections in non-human primates.17Plasmodium falciparum is the deadliest and most devastating human malaria parasite and has been adapted to long-term in vitro growth. As such, it has become the most extensively studied Plasmodium species. For long, inefficiency of transfection technology slowed down progress as the generation of stable genetic mutants could easily last many months (see Ref. 18 for a comprehensive overview). The recent successful adaptation of the CRISPR/Cas9 system,19 however, has the potential to revolutionise again the field by providing an unprecedented ease and speed of generating recombinant P. falciparum lines. On the other hand, experimentation with P. falciparum is predominantly performed in in vitro blood-stage cultures. This is because of obvious issues with maintenance of the complete in vivo life cycle as well as the inability to complete the life cycle in vitro. Suitable in vivo models that highlight the relevance of the findings during an infection should complement the in vitro model.
Rodent malaria parasites, in particular P. berghei and P. yoelii, provide such model systems. They combine fast and efficient experimental genetics techniques with access to the complete in vivo life cycle (Fig. 1). In addition, the evolutionary distances of the rodent malaria parasite clade to either P. falciparum or P. vivax are in the same order of magnitude as the evolutionary distance between P. falciparum and P. vivax.13 All these factors render P. berghei and P. yoelii practical and relevant model species to study common principles of Plasmodium biology. They allow the examination of parasite–host interactions in vivo, including clinically relevant phenomena like parasite sequestration,20 experimental cerebral malaria,21,22 host immune responses23 and parasite immune evasion.24 This is exemplified by the use of intravital imaging techniques, which have proven useful for the investigation of sequestered blood-stage parasites in the brain,25 sporozoite migration26 and liver-stage development.27 Another great advantage, especially when working with infected anopheline mosquitoes, is the inability of P. berghei and P. yoelii to cause malaria in humans. Indeed, much of our knowledge on the Plasmodium mosquito stages stems from findings in P. berghei. For the purpose of this review, we will focus on P. berghei, a versatile and highly amendable malaria model parasite species and the first malaria parasite for which stable genetic manipulation was established.
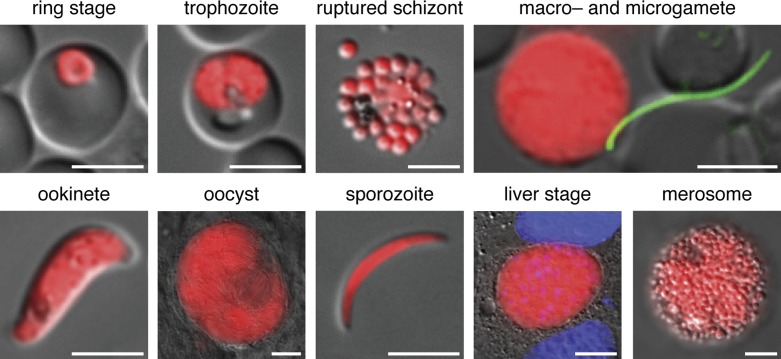
Figure 1.
Live imaging of the complete Plasmodium berghei life cycle using the Berred reference strain.64 Berred expresses high levels of the red fluorescent protein mCherry in all stages under the control of the P. berghei heat shock protein 70 promoter. Shown are asexual blood stages (a ring-stage parasite, a trophozoite, and mature blood-stage merozoites from a ruptured schizont); in vitro activated sexual stage parasite (a male Bergreen microgamete – which expresses high levels of GFP – attaching to a female Berred macrogamete); a cultured ookinete; mosquito stages from in vivo infections (an oocyst at day 14 after the mosquito blood meal and a salivary gland-associated sporozoite); and in vitro cultured liver stages (a liver-stage trophozoite at 48 hours after infection and mature liver-stage merozoites in a merosome released from the hepatocyte). Bars for oocyst, liver stage and merosome, 10 μm; all others, 5 μm.
Transfection
Successful genetic manipulation relies on efficient transfer of modifying DNA constructs into the nucleus and on sufficient parasite survival during the transfection procedure. Asexual blood-stage parasites are the most straightforward to accumulate in large quantities and are haploid negating the need for crossing heterozygotes to achieve homozygote mutants. However, to modify the parasite genome, the targetting DNA construct would have to pass four membranes: (i) the erythrocyte plasma membrane, (ii) the parasitophorous vacuolar membrane, (iii) the parasite plasma membrane and (iv) the nuclear envelope. This is further complicated by the blood-stage parasite's dependence on the integrity of the host erythrocyte for survival. During P. berghei transfections, both matters are overcome by electroporating mature merozoites, the invasive, briefly extracellular forms that establish infection of new erythrocytes. This may partly explain the differences in transfection efficiencies between P. berghei and P. falciparum. For the latter, either developing, intracellular ring-stage parasites are used or uninfected erythrocytes are preloaded with targetting DNA prior to parasite invasion.28 The parasite purification for transfection is based on the phenomenon, that P. berghei parasites develop normally into merozoites in in vitro culture but cannot egress from the erythrocyte without additional mechanical shear stress.29,30 Consequently, ex vivo cultivation of mixed blood stages for 16–18 hours is sufficient for the accumulation of large numbers of enclosed viable merozoites, which can be purified by a subsequent one-step density gradient centrifugation.31 Since the first successful transfections of P. berghei, nearly 20 years ago,7 electroporation protocols have steadily improved. The latest method uses the Nucleofector® technology, which yields transfection efficiencies in the range of 10− 3 to 10− 2.32
Integration
There are different strategies available to genetically manipulate P. berghei: (i) episomal transfections, (ii) single crossover/ends-in, and (iii) double crossover/ends-out homologous recombination (Fig. 2). The choice of which strategy to employ is determined by the required genetic stability of the recombinant parasites and whether the loss of genetic information is unwanted or rather desirable. Even the anticipated difficulties in cloning the parasite's extremely AT-rich DNA, in particular larger fragments of non-coding regions like promoter and terminator sequences, influence the choice of strategy.
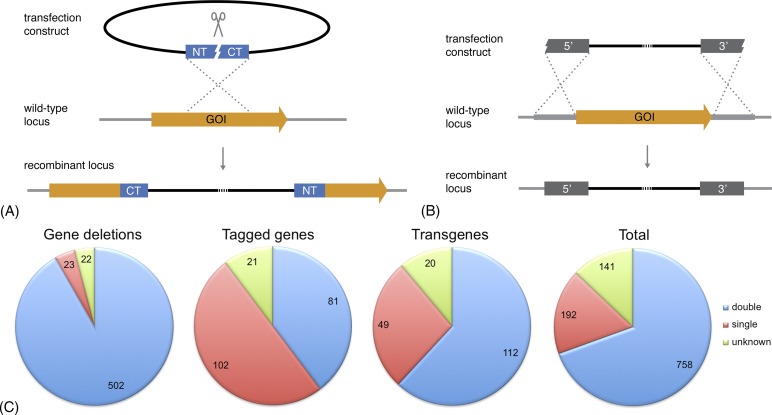
Figure 2.
Generation of recombinant parasites using homologous recombination. (A) Schematic representation of a gene-deletion approach by single crossover/ends-in homologous recombination. The transfection plasmid is linearised roughly in the middle of the single targetting sequence. Successful integration into the wild-type locus leads to the disruption of the gene of interest (GOI) and partial sequence duplication. Two gene fragments remain that are truncated at the carboxy-terminus (CT) or the amino-terminus (NT). This strategy can also be used to introduce tag fusions or to duplicate complete coding sequences. (B) Schematic representation of a gene-deletion approach by double crossover/ends-out homologous recombination. The linearised transfection construction harbours two targetting sequences, one upstream (5′) and one downstream (3′) of the target gene. Successful integration into the wild-type locus leads to deletion of the GOI. This strategy can also be used to introduce tag fusions. (C) Distribution of the approaches used (when known) to generate recombinant parasites harbouring gene deletions, endogenously tagged genes, or transgenes. Data have been extracted from the Rodent Malaria genetically modified Parasites database, RMgmDB, as on 28 November 2014 (A. van Wigcheren & C.J. Janse, personal communications).38 The vast majority of gene-deletion mutants (>90%) were generated using double crossover recombination (blue), whereas at least half of the recombinant parasites expressing endogenously tagged genes were created through single crossover recombination (red). Numbers indicate the total number of mutant parasite lines in each category.
Circular transfection plasmids will not be integrated in the parasite genome but instead will be maintained episomally as long as drug pressure is applied. Such transfections do not lead to the loss of any genetic information; however, the episomes will be lost rapidly in the absence of a selecting drug. Furthermore, the introduction of episomally coded sequences for protein expression and/or localisation harbours the risk of artefacts, e.g. variant plasmid copy numbers between individual parasites. An early study has demonstrated maintenance of as many as 15 plasmid copies per parasite during drug pressure.33 Recently, this concept was utilised to control expression levels of a green fluorescent protein (GFP) and actin 2 through drug-regulated episomal copy numbers.34 The use of a Plasmodium artificial chromosome, harbouring functional centromere and telomere sequences, allows the introduction of a multitude of transgenes simultaneously in a more controlled manner, i.e. with efficient replication and transfer of single copies to daughter cells.35 Using these artificial chromosomes, a high-coverage genomic library has previously been cloned and transfected in P. berghei, with fragment sizes ranging from 10 to 50 kb.36
Stable integration can be achieved using linearised DNA constructs with left and right homology arms that target the construct to a specific locus in the parasite genome. Potential issues with varying copy numbers and the need for continuous drug pressure may be avoided with this technique. The most efficient approach is through single crossover/ends-in homologous recombination (Fig. 2A), for which 250–300 bp of homologous sequence can be sufficient for integration.37 Typically, however, homology arms of 0.5–1 kb are used to increase the efficiency of recombination. For the generation of the transfection construct, a single fragment of target DNA is cloned into a suitable vector and a unique restriction site in the fragment is used for plasmid linearisation. To study gene function, this strategy can be used to disrupt the coding sequence of a gene of interest (GOI). The downside of such an approach is that both amino- and carboxy-terminal truncated versions of the gene remain present in the genome. Hence, this approach also harbours the danger of recombination-mediated reversion in the absence of a positive selection drug. This is especially true in cases where insertion of the transfection vector leads to a reduced fitness of the parasites. Although less appropriate to generate gene-deletion mutants, the single crossover approach has been applied extensively to generate at least 50% of the endogenously tagged parasite lines (Fig. 2C).38 Advantages of this strategy include the requirement for only a single molecular cloning step and a ∼10-fold increase in integration efficiency resulting in the transgenic parasites to emerge at least 1 day earlier (personal observations and CJ Janse, personal communications). Another possible advantage is that one can apply insertional mutagenesis to duplicate a gene at its endogenous locus, e.g. when tagging of an endogenous gene is detrimental for its function but the presence of an additional tagged copy is tolerated.39 Naturally, one should always be cautious interpreting such results as the requirement for an untagged copy of the protein may well suggest that functionality and localisation of the tagged protein are affected.
Despite marginally lower integration efficiency and the need for at least two cloning steps to generate the transfection construct, double crossover/ends-out homologous recombination is nonetheless the method of choice (Fig. 2B and C). The efficiency of integration is greatly dependent on the length of the homologous sequence.40 Interestingly, homology arms that differ ∼4% from the targetted nucleotide sequence are still sufficient to drive integration.41 Double crossover/ends-out homologous recombination is the only way to permanently and stably modify the P. berghei genome since it completely removes the entire coding sequence of a GOI. Hence, parasites cannot revert to wild-type genotypes. This is particularly important in cases where parasites are to be cycled through mosquito stages where no drug pressure can be applied, when generating reference parasite lines, or when studying loss-of-function mutants. It is therefore not surprising that >90% of the reported gene-deletion mutants have been generated using a double crossover strategy (Fig. 2C). The generation of stable genetic mutants is furthermore a prerequisite when aiming to generate parasite lines with multiple genetic modifications, e.g. through recycling of the drug-selectable cassette (see below).
Another important consideration when manipulating the parasite genome is the site of integration. The most direct way to study the function of a gene is by targetted disruption or deletion of the endogenous locus. Alternatively, genetic material may be introduced elsewhere as a transgene. For the latter, genes have been employed that were empirically identified to have dispensable roles during normal life cycle progression; e.g. the gene encoding the gamete surface antigen P230P42 and the P. yoeliiS1 locus.43 Furthermore, the loci of the ribosomal RNA C- and D-units have been used extensively in P. berghei. However, a small defect in oocyst development was observed after gene disruption, which should be considered when planning experiments with transgenic mosquito-stage parasites.41 Although disruptions of several loci had no detectable effects on life cycle progression, it is conceivable that new phenotypic methods may reveal as yet undetected deficits. Furthermore, synergistic effects of additional genetic modification cannot be excluded. To avoid these issues, we have started to employ a silent intergenic locus on P. berghei chromosome 6 (SIL6) that is devoid of genes and transcriptionally silent for stable integration through double crossover.44
Selection
After electroporation, the parasites are injected intravenously into naïve recipient mice. Successfully transfected parasites are usually selected by applying drug pressure. Positive selection of transgenic P. berghei parasites is based on the antifolates pyrimethamine or WR99210.45,46 Both serve as inhibitory substrate analogues of the parasite's bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS) enzyme.47 Pyrimethamine can be administered orally with drinking water, whereas WR99210 needs to be injected repeatedly intraperitoneally or subcutaneously. The most commonly used selection cassettes encode drug-insensitive variants of DHFR-TS from Toxoplasma gondii or P. berghei, which confer resistance to pyrimethamine.7,48–50 Human DHFR confers resistance to pyrimethamine and WR99210,51 thus allowing its use as a second selectable marker. An additional reason why hDHFR has become more commonly used is its relative small size. This facilitates the generation of more complex44,52,53 or PCR-based54 transfection vectors. To date, these two drugs are the only efficient compounds for positive selection in P. berghei that can be used sequentially without the need to recycle the selection cassette (see below). Development of novel selection markers is hampered by two closely related problems: (i) positive selection of transfected parasites cannot be performed in vitro because of the inefficient reinvasion in culture and (ii) drugs must therefore be suited for in vivo application and should be non-toxic to the rodent host.
The P. falciparum chloroquine resistance transporter gene (CRT) has been tested as a potential new resistance marker. It has been demonstrated that mutations in PfCRT promote resistance towards the antimalarial chloroquine in vitro55 and in vivo,56 resulting in elevated IC50 values of up to 17-fold.57 Unfortunately, cross-species complementation in P. berghei did not increase resistance towards the drug during infection, omitting the use of mutant PfCRT as an additional positive selection marker.58 There are no reports on the application, successful or unsuccessful, of any other selection markers functional in P. falciparum.59,60
Isolation
Positive drug-selection is usually not completely effective, resulting in mixtures of transgenic, spontaneously mutated, and wild-type parasites, thus necessitating the purification of transgenic parasites. Traditionally, transfectants have been isolated by limiting dilution,32 through the injection of single parasites from the parental population into several naïve mice (usually 10). The success of this method relies strongly on the ratio of wild-type to mutant parasites and is therefore very inefficient when working with slow growing mutants. To circumvent this problem, parasites may be passaged through several animals under pyrimethamine pressure, resulting in the favoured growth and enrichment of the transfectants prior to cloning. However, this method is labour intensive and requires a large number of experimental animals.
The development of flow cytometry-based isolation methods significantly reduced workload and animal usage (Fig. 3A). The method depends on the introduction of a fluorescent protein expression cassette and subsequent isolation of the fluorescent transgenic parasites by flow cytometry. Initial methods employed the eEF1-alpha promoter to drive GFP expression.61,62 However, owing to regular wild-type contaminations of sorted populations, repeated cycles of flow-cytometric isolation or a subsequent cloning step by limiting dilution were recommended. A novel approach uses the significantly stronger HSP70 promoter, resulting in cytosolic fluorescence levels that are an order of magnitude higher, thus resulting in an almost absolute separation of wild-type and fluorescent mutant parasites.44,63 This method has been shown to be efficient even in the presence of a 100-fold excess of wild-type parasites, provided that the parasitaemia of the donor mice does not exceed 1%. At higher parasitaemias, sorting efficiencies decline because of the presence of double-infected erythrocytes harbouring a transfected and a wild-type parasite. Complications resulting from the maintenance of episomal transfection vector copies were overcome through more efficient linearisation protocols.63
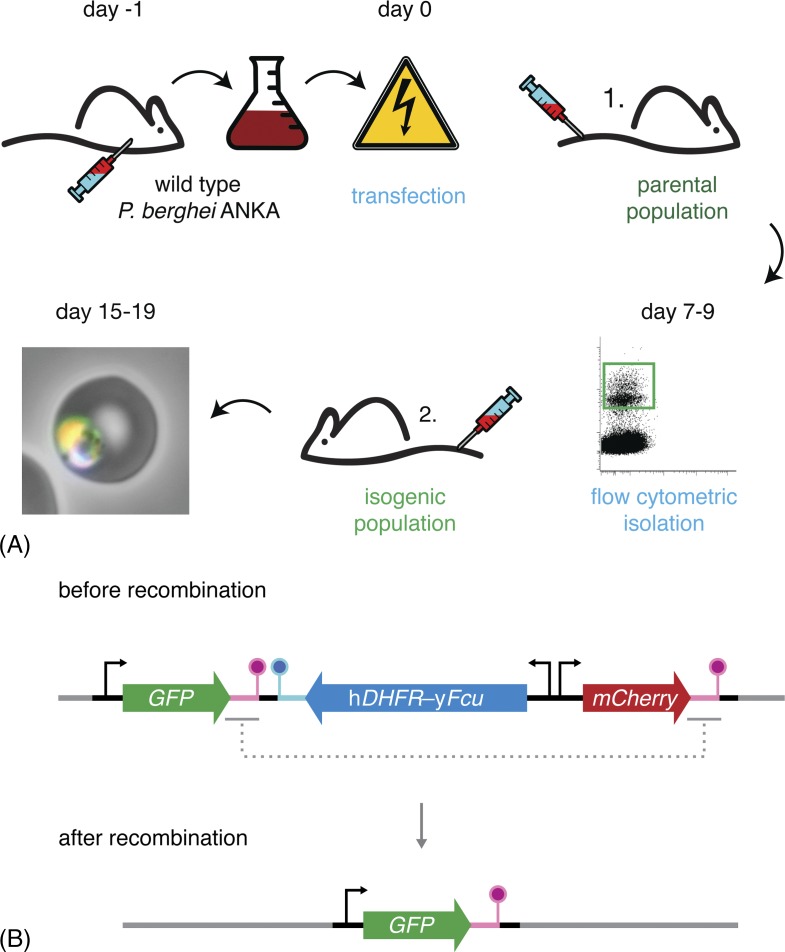
Figure 3.
Flow cytometry-based methods reduce workload and animal usage. (A) Schematic representation of the P. berghei transfection and mutant isolation protocol. Blood of a mouse infected with wild-type parasites is harvested by cardiac puncture and cultured overnight. Following maturation, the parasites fail to egress and arrest at the schizont stage. These schizonts are purified and transfected with the targetting constructs. Transfected merozoites are injected intravenously into a naïve mouse. Administration of pyrimethamine in the drinking water favours the growth of successfully modified parasites, which now also express a fluorescent protein. When the parasitaemia is 0.1–1.0% (typically 7–9 days after transfection), 50 mutant parasites are isolated by flow cytometry and transferred to a naïve mouse. 8–10 days after injection, the isogenic parasite line can be harvested, stored or transferred and tested. (B) Schematic representation of the ‘gene-out, marker-out’ strategy exemplifying recycling of the hDHFR-yFcu drug-selectable cassette.68 Successfully transfected and isolated parasites harbour a fluorescent cassette (GFP; green), a drug-selectable cassette (hDHFR-yFcu; blue), and a second fluorescent cassette (mCherry; red). After intravenous injection into a naïve mouse, 5-fluorocytosine (5-FC) is administered in the drinking water. This favours the growth of parasites that have successfully lost yFcu by homologous recombination of the duplicated sequences flanking the drug-selectable cassette and the mCherry marker (magenta). Finally, recycled parasites can be isolated using flow cytometry through the selection of GFP-positive, mCherry-negative parasites.
In addition to GFP, other fluorophores may be used to isolate parasites, including yellow (YFP), red (mCherry)64 and cyan (CFP) fluorescent proteins (unpublished data). The high levels of fluorescence also facilitate imaging of live and fixed parasites in all life cycle stages, even of the extremely thin and highly motile male microgametes.44 Most importantly though, the flow-cytometric isolation of mutant parasite lines leads to an 80–90% reduction in the use of experimental animals.
Sequential Genetic Modification
The availability of just two selectable markers prevents repeated rounds of genetic manipulation. The generation of double mutants is feasible, although the requirement for repeated intraperitoneal or subcutaneous administration of WR99210 is undesirable. Nonetheless, the sequential introductions of (i) Pb or TgDHFR-TS with pyrimethamine-selection and (ii) hDHFR with WR99210 can verify loss-of-function phenotypes through recovery of wild-type behaviour by complementation. Thus, the circumsporozoite gene was reintroduced after deletion.51 Such a strategy is desirable not only for the generation of revertant strains but also for providing definitive proof for gene essentiality. This could be achieved by introducing an additional copy of the GOI either episomally or as a transgene by means of positive selection with pyrimethamine and insensitive Pb or TgDHFR-TS. If the subsequent deletion of the endogenous locus using WR99210 and hDHFR is successful in this strain but not in wild-type parasites, this provides proof for the accessibility of the genomic locus and essential functions of the gene. However, when attempting to delete an essential gene following introduction of a compensatory second copy of the gene one has to consider the requirement to use heterologous regulatory sequences. Failure to replace at least one of the up- or downstream flanking regions may lead to the preferred deletion of the transgene instead of the original target gene. It is also important to keep in mind that the hDHFR cassette must always be used as the second selectable marker when attempting the generation of double mutant parasites, since it also confers resistance to pyrimethamine. An additional issue limiting the use of WR99210 is its selective capacity. Introduction of hDHFR in the P. berghei genome only resulted in a fivefold increase in WR99210 resistance,51 demonstrating the limitations of this positive selection marker when targetted integration is attempted. In contrast, WR99210 resistance was increased 1000-fold when the hDHFR cassette was maintained episomally, because of an elevated copy number per parasite. Perhaps, this is one of the reasons why so far only two studies have reported the use of sequential gene deletions using this strategy.65,66
Alternatively, flow-cytometric isolation has been used as selection method, i.e. without the use of a drug-selectable cassette.32,62 Despite being relatively inefficient compared to traditional methods using drug selection, this method was employed to generate the widely used reference strain GFPCON. Although largely untested, one might speculate that the improved isolation tools and methods44,63 might facilitate a more reliable use of flow cytometry-based isolation in the absence of drug pressure. Such an approach could even be expanded to multiple manipulation rounds by using multiple fluorescent markers with different colours.64 Still, it remains questionable whether slow growing mutants can be isolated without additional drug pressure.
Recycling of the drug cassette would allow a virtually unlimited number of subsequent transfections. To achieve this, a positive/negative selection cassette was created containing a fusion of hDHFR and the yeast cytosine deaminase/uracil phosphoribosyltransferase gene (yFcu). yFcu metabolises 5-fluorocytosine (5-FC) into a toxic metabolite.53 Thus, this selection cassette first allows the positive selection of successfully transfected parasites by pyrimethamine or WR99120 selection and may next be completely removed again following selection with 5-FC. This loss occurs by means of homologous recombination, using duplicated sequences flanking the selectable marker (Fig. 3B). Although, 5-FC had previously to be administered through intraperitoneal injection, a protocol has been established enabling oral administration through the drinking water, thus facilitating the procedure for both experimenter and mouse.67 A disadvantage is the need for a subsequent cloning step by limiting dilution to isolate parasites that have lost their resistance cassette to use them as a recipient strain in a subsequent transfection. This issue has been elegantly solved by introducing a second, red fluorescent marker in the drug-selectable cassette in an approach that was termed ‘gene-out, marker-out’.68 Following successful integration of the transfection construct, parasites are both GFP- and mCherry-positive and can be isolated by flow cytometry. Subsequent negative selection leads to a loss of the drug-selectable cassette along with the mCherry expression cassette. Hence, successfully recycled parasites can now be isolated by sorting green-only fluorescent parasite while excluding double fluorescent parasites (Fig. 3B).
The ‘gene-insertion, marker-out’ (GIMO) strategy allows the fast generation of drug-selectable marker-free parasites expressing a transgene.69 This method relies on the loss of a stably integrated hDHFR/yFcu drug-selectable cassette from a reference strain or gene-deletion mutant through its replacement by a new marker-free transfection construct. Parasites that have successfully replaced the drug-selectable cassette are selected by administration of 5-FC. The method allows the fast generation of marker-free P. berghei or P. yoelii mutants expressing transgenes. Unfortunately, the application possibilities of GIMO are limited to recipient strains with the adopted drug-selectable cassette integrated without flanking repeat sequences.
Despite the advances and successes in generating double mutant parasite lines, we observed that off-target integrations are relatively common, when using the same vector system repeatedly, favouring integration into the sites of the initial recombination (unpublished data). We would therefore recommend the use of different vector systems for the subsequent transfections. When using an episomal construct in the second transfection, such problems should not be observed.
Double mutants may also be obtained by a classical in vivo cross-fertilisation of two transgenic parasite lines. Mice infected with two different genetically engineered parasite lines are fed to female anopheline mosquitoes where homozygous and heterozygous fertilisations occur. In heterozygous offspring, chromosomal reorganisation or recombination may yield double mutant parasites, which can be isolated after transmission to a naïve recipient mouse. This method was employed to enable conditional mutagenesis using site-specific recombination by combining two transgenic lines, one harbouring the Flp recombinase, and the other containing the FRT recombination sites (see below).52 At least three double gene-deletion mutant parasite lines have been generated using in vivo genetic crossing of two single gene-deletion clones.70,71 The efficiency of this method is highly dependent on the loci of integration. New combinations of two loci on separate chromosomes should occur at high frequencies because of chromosomal redistribution. However, loci on the same chromosome are recombined less frequently. Their uncoupling relies solely on crossing-over and interchromosomal recombination and depends largely on the distance between the two loci. Recently, we demonstrated the generation of a double mutant parasite line, expressing the endogenously tagged translocon components heat shock protein 101 (HSP101) and PTEX88, both of which are located at chromosome 9 and separated by ∼360 kb (unpublished data). Loci that are associated more closely may prove difficult to recombine by meiotic crossing. Flow cytometry may help to isolate recombined parasites when crossing mutant lines that have different fluorescent markers integrated in their respective genetically engineered loci.
In vivo crossing also offers the possibility of employing a larger number of subsequent recombination rounds, thus facilitating the generation of parasites with a multitude of mutations. However, with increasing numbers of mutations, the number of possible combinations increases even more rapidly. Hence, repeated rounds of in vivo crossing require diligent isolation and testing procedures of the offspring after transmission. The application of in vivo crossing is further limited by potential synergistic effects of the mutations that might lead to an impairment of the parasites' ability to complete the life cycle. This restriction renders meiotic crossing useless for the generation of a much desired, safe, late liver-stage arrested vaccine candidate strain.72–74 Ideally, a safe genetically attenuated parasite line would lack a number of genes, e.g. LISP1,75PALM,76,77 and ZIPCO,78 that would allow a nearly complete maturation of liver-stage parasites including the expression of blood-stage antigens. Parasites lacking any one of these genes can cause breakthrough infections; however, one can hope to eliminate these by combining multiple gene deletions. Hence, when behaving as desired, no blood-stage parasites can be obtained upon sporozoite infection. To generate such double mutant parasites, recycling of the drug-selectable cassette is required. Following negative selection, parasites deficient in the liver-stage gene B969 were rendered drug-sensitive. Next, deletion of a second liver-stage gene, SLARP,79 was achieved. This proof of concept provided a safe, but early arrested genetically attenuated whole parasite vaccine strain.80
Conditional Approaches
The possibilities of analysing genes essential for P. berghei blood-stage development have been limited. The establishment of a robust inducible knockdown system has been hindered by the requirements of an in vivo system, e.g. the administered inducers should not be toxic, be taken up efficiently, and have a prolonged systemic half-life. RNAi-mediated knockdown strategies, one of the most powerful inducible systems, cannot be exploited, because of the lack of functioning RNAi machinery in Plasmodium parasites.81 Systems that reduce protein stability by fusion of a destabilisation domain have been successfully applied to in vitro cultures of P. falciparum,82 but thus far no functioning system for P. berghei has been established.
Despite these limitations, there are a number of strategies to further characterise genes with a crucial role during blood-stage growth. Two approaches rely on differential endogenous promoter activities without the need for exogenous inducing or repressing compounds. These approaches are well suited to study gene function in an in vivo model, notably during transmission stages. First, conditional gene ablation can be achieved by exploiting the Flp/FRT system of site-specific recombination (Fig. 4A).52 This technology has been applied to study the function of the essential merozoite surface protein 1 during liver-stage development.83 The recombinase gene was integrated into the genome under the control of a promoter that is silent during blood-stage development and is active when DNA excision is required. By using the thermolabile FlpL recombinase, undesired activity during blood-stage development was further minimised. In a second transfection, FRT sites are inserted at both sides of the target sequence to be deleted from the genome. When the promoter is active, the recombinase is expressed and excises the FRT flanked locus. Therefore, the choice of promoter sequence is critical for the success of this approach.84
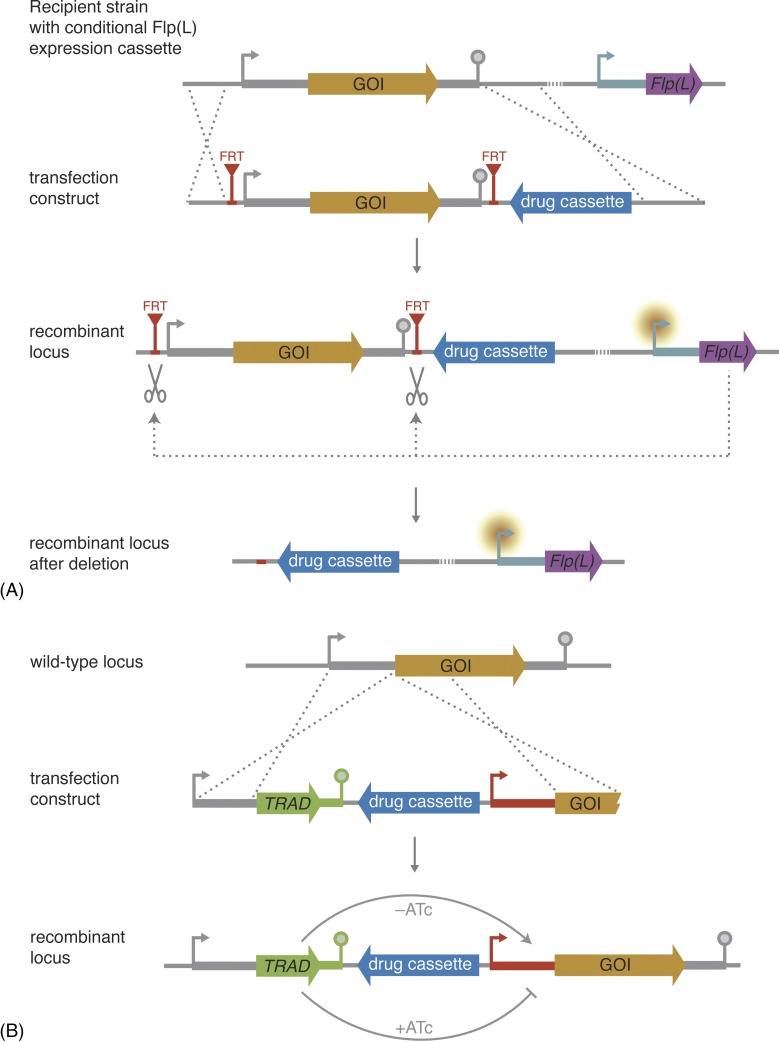
Figure 4.
Conditional gene deletion and knockdown strategies. (A) Schematic representation of conditional gene deletion using the Flp/FRT system. This approach requires the previous integration of a cassette driving the expression of Flp recombinase or its thermolabile variant FlpL under the control of a differentially active promoter (turquoise). By targetted integration, FRT sites are inserted upstream and downstream of the gene of interest (GOI). During asexual blood-stage propagation, the promoter driving Flp(L) transcription is silent and the recombinase is not expressed. When the promoter becomes active during other life cycle stages, Flp(L) is expressed and excises the GOI at the FRT sites. The resultant mutants have now lost the gene and its function can be analysed in subsequent life cycle stages. (B) Schematic representation of conditional knockdown using a tetracycline-repressible system. An inducible promoter (red) is inserted upstream of the GOI. In addition, the coding sequence of a tetracycline repressor (TetRep) protein containing a parasite-specific activating domain (TRAD) is inserted adjacent to the endogenous promoter of the GOI. Expression of TRAD in the absence of anhydrotetracycline (ATc) leads to a transactivation of the inducible promoter by means of TRAD binding, and the GOI is transcribed. However, when ATc is present it induces conformational changes in TRAD, thereby preventing transcription.
Second, gene function in specific stages can also be studied by exchanging the promoter sequence using homologous recombination. The strategy harnesses promoters that display expression during blood-stage development, but which are inactive during other phases of the life cycle. This has been demonstrated for the unconventional class XIV myosin A,85 which is essential during asexual blood-stage development. The endogenous promoter was replaced with the apical membrane antigen 1 (AMA1) promoter, which is active in blood stages but not in ookinetes. This results in functional levels of myosin A permitting normal blood-stage development. During the ookinete stage, however, the promoter swap resulted in a complete loss of myosin A and an impaired ookinete motility.85 Following a similar strategy, central roles in the formation of fertile male gametes and during early mosquito-stage development were demonstrated for the putative histone chaperone FACT-L, which is essential during blood-stage development.86
To study genes refractory to gene deletion during a blood-stage infection, a tetracycline repressor (TetRep)-based system has been established (Fig. 4B).87 It is based on an inducible promoter containing a tet operator. The endogenous promoter of the target gene drives the expression of a fusion of a TetRep protein and a parasite-specific activating domain (TRAD4). This fusion protein can transactivate the tet operator upstream of the GOI. Anhydrotetracycline (ATc) binds to the transactivator thus preventing transcription of the target gene. Administration of ATc in the drinking water led to a ∼90% downregulation of transcription, as shown for the knockdown of the essential actin-binding protein profilin.87 The significance of this technical achievement notwithstanding, downregulation during mosquito-stage development was not observed. Potentially, this was because of problems with ATc administration or because of the inefficiency of the described genetic components during this parasite life cycle stage. During liver-stage development, however, the system proved to be fully functional again. Very recently, this system has been used to confirm the perceived essentiality of the putative PTEX translocon component HSP101 in the translocation and export of cargo proteins.88
Despite all recent advances, the described technologies share a common problem: actual proof of gene essentiality during blood-stage development is difficult to obtain. The possibility to first introduce a second copy of a gene and then remove the endogenous gene is hampered by (i) the unavailability of multiple selection markers, (ii) the need to use heterologous regulatory elements, and (iii) potential dominant negative effects of the genetic duplication. Apart from that, conditional systems all have their own limitations, e.g. incomplete levels of knock down and restrictions on the stages that can be studied.
Visualization
To date, the most efficient way to test essentiality is the independent transfection with a disruptive and a non-disruptive construct, providing proof for the accessibility of the locus and thus indirectly of gene essentiality.
Such a non-disruptive construct can be used for protein localisation studies. One homology arm is derived from the 3′ sequence, equivalent to that used for the disruptive construct. The second arm is derived from the carboxy-terminal coding sequence of the target gene, cloned directly adjacent to and in frame with a tag sequence. Thus, in addition to providing a positive control for transfection efficiency and accessibility of the target locus, protein expression levels and subcellular localisation can be studied. Recently, we have introduced the Berghei Adaptable Transfection (pBAT) plasmid system that allows the generation of deletion and tagging constructs from the same intermediate construct.44 These plasmids have been optimised for size and ease of cloning and combine (i) a recyclable, drug-selectable cassette and (ii) a bright fluorescence cassette flanked by multiple cloning sites and transgene targetting sequences. A variety of plasmids are available that include amino- or carboxy-terminal tagging sequences. One of these transfection plasmids was used to provide support for the apicoplast localisations and essential roles of four of the five sulphur utilisation factors (SUFs).89 Using a comparable strategy but an alternative transfection vector system,90,91 the distinct spatio-temporal distribution of two alveolins was visualised.92 Refractoriness to gene-deletion in combination with accessibility of the loci provided convincing evidence for essential functions during asexual blood-stage development, while conversely the efficient establishment of a blood infection following the endogenous tagging of the proteins leans support to the correct localisation and functionality of these GFP-tagged proteins. This may be further confirmed by assuring that parasite growth rates and protein integrity also remain unaltered.64
Needless to say that microscopy is central to the studying of malaria parasites. Arguably some of the most exciting recent developments are in the intravital imaging of murine malaria parasites during an infection, e.g. using bioluminescence93 or fluorescence. The latter has been used to visualise mosquito-to-mouse transition,94–96 liver-stage development,26,97 the transition from liver to blood-stage infection,27 and parasites in the brain.25,98 Such studies can provide profound insights into pathology and virulence of the parasites, the host immune defences against the pathogen and host–parasite interactions. The continuous developments in experimental genetics aid these studies by providing a range of genetically engineered parasite lines expressing luciferases or fluorescent proteins.
Furthermore, there are reference parasite lines that have fluorescent markers targetting to specific organelles and other subcellular localisations such as the parasitophorous vacuole (unpublished data).99,100 These lines facilitate cell biological studies of this ancient eukaryotic single cell pathogen. For much more extensive reviews of the many exciting developments in various aspects of visualising malaria parasites, we refer the readers to a special issue of Parasitology International.101–106
Next Generation
Ten years after the publication of the nearly complete genome sequence, and 20 years after the development of techniques for stable genetic engineering, the time has come to make the next big leap in our understanding of Plasmodium biology. One way of doing so is by utilising random mutagenesis through site-specific transposable elements. The first method in Plasmodium was developed using a mini-Tn5 derivative to mutagenise an Escherichia coli library of P. berghei DNA.107 More recently, a system relying on the insertion of the lepidopteran transposable element piggyBac developed in P. falciparum108 has been adapted to P. berghei and employed to generate 127 insertions.109 The nature of the target sequence, i.e. AATT, leads to the transposon integrating predominantly in non-coding sequences, making this method particularly useful for regulatory mutations.
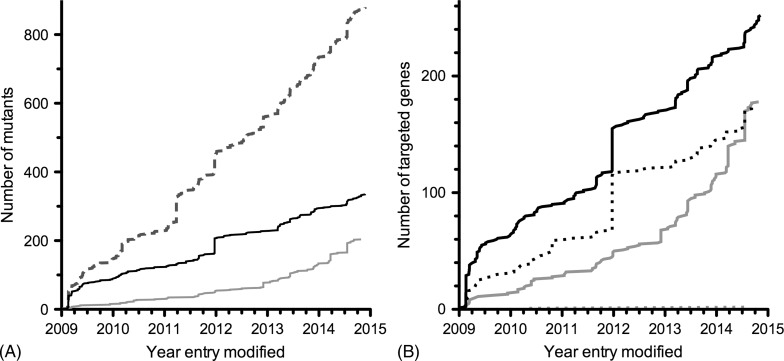
There have been a number of studies employing standard P. berghei transfection technology to study multiple genes. The first and still the third largest study to date targetted 20 genes encoding putative secreted proteins of the ookinete.110 In 2010, the landmark paper by Tewari et al.111 described the largest functional characterisation of 66 putative protein kinases encoded in the P. berghei genome. The generation and characterisation of 23 loss-of-function mutants enabled the identification of some essential regulators of mosquito transmission. In a complementary approach, published 4 years later, the essentiality of 16 of 30 targetted phosphatases during asexual blood-stage growth was shown, with distinct roles of the other phosphatases during life cycle progression and differentiation.112 No more than nine additional studies attempting the deletion of ≥ five genes have provided insights in the functions of a variety of gene/protein families, pathways and complexes. These include studies of genes encoding exported proteins,113,114 components of the Plasmodium translocon of exported proteins,64,115 6-Cys proteins,42 rhomboid proteases,116 protein S-acyl transferases,117 SUFs of the apicoplast89 and P. yoelii early transcribed membrane proteins.118 Currently, 427 genes have been targetted of which 175 (39%) proved refractory to gene deletion and the remaining 252 (59%) have been deleted successfully (Fig. 5).38 Recent years have also seen a steady increase of parasite lines expressing endogenously tagged genes bringing the total at 178 while two genes were refractory to tagging (Fig. 5). Despite all the progress made, it would take until the end of the century to target all P. berghei genes if continued at the current pace.
Figure 5.
Progress of experimental genetics efforts in rodent malaria parasites. Data have been extracted from the Rodent Malaria genetically modified Parasites database, RMgmDB, as on 28 November 2014 (A. van Wigcheren & C.J. Janse, personal communications).38 Note that the date indicated for each entry is the date when the entry was last modified in the database. Therefore, no entries precede the year 2009 in which the database was initiated. (A) Total number of successfully generated mutants (dashed line) including loss-of-function (black) and endogenously tagged (grey) parasite lines. (B) Number of targetted unique genes. Indicated are successful (solid) and unsuccessful (dotted) attempts to delete (black) or endogenously tag (grey) the genes of interest.
Before we can bring targetted, non-random experimental genetics to a truly genome-wide scale, a number of hurdles need to be overcome. The first is the construction of the thousands of transfection constructs needed to target the ∼5000 genes.15 Some advances have been made to facilitate the transfection vector construction. PCR-based construction of replacement vectors can be scaled relatively easily, but is limited to small constructs.54 Another PCR-based solution was used to generate P. yoelii transfection vectors, however, still requires a single enzymatic cloning step.119 A huge advance has been the establishment of the PlasmoGEM system, a pipeline for the genome-scale production of linear replacement vectors with large homology arms.40,120 Utilisation of these vectors in combination with signature-tagged mutagenesis enables the reproducible generation of a pool of up to 90 gene-deletion mutants in a single mouse.121 Each individual mutant can be identified by a unique barcode sequence flanked by a standard sequencing primer annealing site. Thus dispensable gene functions during asexual blood-stage replication and growth rates of the gene-deletion mutants can be assessed; however, the method is not compatible with the isolation of the generated mutants. Therefore a combination of the PlasmoGEM system with flow cytometry-assisted sorting procedures that have tackled the bottleneck of isolating successfully modified parasites would be particularly useful.63,68
Transfection efficiency improved substantially since the establishment of the AMAXA transfection technology.32 Genetic manipulation efficiency has increased to the point where there is little room for further improvement. Nonetheless, the recent report that the CRISPR/Cas9 system is also functional in P. yoelii122 is a useful step forward. CRISPR/Cas9 will be particularly useful to introduce a multitude of small point mutations, which are much harder to generate with the more conventional methods. Perhaps this technology can also be utilised when considering a genome-wide gene-deletion effort.
However, in spite of all these improvements, the phenotypic characterisation of the generated mutants arguably remains the main limiting factor of Plasmodium genome-wide functional genetics. Streamlining analysis and standardising the various checkpoints of life cycle progression would be a first step to expedite phenotypic characterisation. The introduction of a number of bioluminescence-based methods enables faster and less subjective quantification of the P. berghei burden when compared with traditional microscopy-based approaches.123–126 These methods, however, do not permit the comparison of multiple mutants simultaneously. A flow cytometry-based in vivo competition assay has been developed that enables the analysis of blood-stage development of three differently coloured fluorescent parasite lines within a single mouse.64 Introduction of additional fluorescent markers should further increase the number of mutants that might be analysed simultaneously. Unfortunately, no methods enabling the quantification of multiple mutant parasite lines simultaneously in other life cycle stages are yet to be available.
Concluding Remarks
The malaria research community has made some major advances in experimental genetics approaches in the in vivo murine malaria model systems. These improvements had a profound impact on our understanding of malaria parasite biology and infections. Continuous progress, e.g. through the development of methods to generate and isolate multiple recombinant parasite lines simultaneously, will hopefully bring a genome-wide repository of mutant and transgenic parasite lines within our grasp. The efficiency of generating multi-mutant parasite strains including the analyses of gene essentiality through complementation approaches needs improvement. Perhaps the biggest challenge will be the design of efficient and informative methods to analyse these recombinant parasite lines throughout the complex Plasmodium life cycle.
Disclaimer Statements
Contributors JMM and TWAK contributed equally to this work.
Funding None.
Conflicts of interest The authors have no conflicts of interest.
Ethics approval Not applicable.
Acknowledgements
The authors would like to thank Auke van Wigcheren and Chris J. Janse for providing details from the RMgmDB database on the generation of the different mutants and Markus Ganter for critically reading the manuscript. JMM and TWAK are supported by the Netherlands Organization for Scientific Research (NWO-VIDI 864·13.009).
References
- 1.Laveran A. Un nouveau parasite trouvé dans le sang de malades atteints de fièvre palustre. Origine parasitaire des accidents de l'impaludisme Bull Mem Soc Med Hop Paris. 1881;17:158–64. [Google Scholar]
- 2.Goonewardene R, Daily J, Kaslow D, Sullivan TJ, Duffy P, Carter R et al. Transfection of the malaria parasite and expression of firefly luciferase. Proc Natl Acad Sci USA. 1993;90:5234–6. doi: 10.1073/pnas.90.11.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE.Transfection of Plasmodium falciparum within human red blood cellsProc Natl Acad Sci USA. 199592973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfahler JM, Galinski MR, Barnwell JW, Lanzer M. Transient transfection of. Plasmodium vivax blood stage parasites Mol Biochem Parasitol. 2006;149:99–101. doi: 10.1016/j.molbiopara.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 5.van der Wel AM, Tomas AM, Kocken CH, Malhotra P, Janse C, Waters AP et al. Transfection of the primate malaria parasite Plasmodium knowlesi using entirely heterologous constructs. J Exp Med. 1997;185:1499–503. doi: 10.1084/jem.185.8.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kocken CH, Van Der Wel A, Thomas AW. Plasmodium cynomolgi: transfection of blood-stage parasites using heterologous DNA constructs. Exp Parasitol. 1999;93:58–60. doi: 10.1006/expr.1999.4430. [DOI] [PubMed] [Google Scholar]
- 7.van Dijk MR, Waters AP, Janse C. Stable transfection of malaria parasite blood stages. Science. 1995;268:1358–62. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- 8.Mota MM, Thathy V, Nussenzweig RS, Nussenzweig V.Gene targeting in the rodent malaria parasite Plasmodium yoeliiMol Biochem Parasitol. 2001113271–8. [DOI] [PubMed] [Google Scholar]
- 9.Reece SE, Thompson J. Transformation of the rodent malaria parasite Plasmodium chabaudi and generation of a stable fluorescent line PcGFPCON. Malar J. 2008;7:183. doi: 10.1186/1475-2875-7-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner MJ, Hall N, Fung E, White OR, Berriman M, Hyman RWet al. Genome sequence of the human malaria parasite Plasmodium falciparumNature. 2002419498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlton JM, Angiuoli SV, Suh BB, Kooij TWA, Pertea M, Silva JCet al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoeliiNature. 2002419512–9. [DOI] [PubMed] [Google Scholar]
- 12.Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, Berriman M. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–6. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 13.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler Eet al. Comparative genomics of the neglected human malaria parasite Plasmodium vivaxNature. 2008455757–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pain A, Böhme U, Berry A, Mungall K, Finn RD, Jackson APet al. The genome of the simian and human malaria parasite Plasmodium knowlesiNature. 2008455799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto TD, Böhme U, Jackson AP, Hunt M, Franke-Fayard B, Hoeijmakers WAM et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moraes Barros RR, Straimer J, Sa JM, Salzman RE, Melendez-Muniz VA, Mu J et al. Editing the Plasmodium vivax genome, using zinc-finger nucleases. J Infect Dis. 2014;211(1)):125–9. doi: 10.1093/infdis/jiu423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udomsangpetch R, Kaneko O, Chotivanich K, Sattabongkot J.Cultivation of Plasmodium vivaxTrends Parasitol. 20082485–8. [DOI] [PubMed] [Google Scholar]
- 18.Limenitakis J, Soldati-Favre D.Functional genetics in Apicomplexa: potentials and limitsFEBS Lett. 20115851579–88. [DOI] [PubMed] [Google Scholar]
- 19.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio J-J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–21. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 20.Franke-Fayard B, Fonager J, Braks A, Khan SM, Janse C. Sequestration and tissue accumulation of human malaria parasites: can we learn anything from rodent models of malaria? PLoS Pathog. 2010;6:e1001032. doi: 10.1371/journal.ppat.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engwerda C, Belnoue E, Grüner AC, Rénia L. Experimental models of cerebral malaria. Curr Top Microbiol Immunol. 2005;297:103–43. [PubMed] [Google Scholar]
- 22.Hansen DS. Inflammatory responses associated with the induction of cerebral malaria: lessons from experimental murine models. PLoS Pathog. 2012;8:e1003045. doi: 10.1371/journal.ppat.1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafalla JC, Silvie O, Matuschewski K. Cell biology and immunology of malaria. Immunol Rev. 2011;240:297–316. doi: 10.1111/j.1600-065X.2010.00988.x. [DOI] [PubMed] [Google Scholar]
- 24.Jemmely NY, Niang M, Preiser PR. Small variant surface antigens and Plasmodium evasion of immunity. Future Microbiol. 2010;5:663–82. doi: 10.2217/fmb.10.21. [DOI] [PubMed] [Google Scholar]
- 25.Nacer A, Movila A, Baer K, Mikolajczak SA, Kappe SHI, Frevert U. Neuroimmunological blood brain barrier opening in experimental cerebral malaria. PLoS Pathog. 2012;8:e1002982. doi: 10.1371/journal.ppat.1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frevert U, Engelmann S, Zougbédé S, Stange J, Ng B, Matuschewski K et al. Intravital observation of Plasmodium berghei sporozoite infection of the liver. PLoS Biol. 2005;3:e192. doi: 10.1371/journal.pbio.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, Rennenberg A et al. Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science. 2006;313:1287–90. doi: 10.1126/science.1129720. [DOI] [PubMed] [Google Scholar]
- 28.Rug M, Maier AG. Transfection of Plasmodium falciparum. Methods Mol Biol. 2013;923:75–98. doi: 10.1007/978-1-62703-026-7_6. [DOI] [PubMed] [Google Scholar]
- 29.Mons B, van der Kaay HJ. The effect of cryopreservation on gametocytogenesis of Plasmodium berghei berghei: a preliminary report. Acta Leiden. 1982;48:9–16. [PubMed] [Google Scholar]
- 30.Mons B, Janse C, Boorsma EG, van der Kaay HJ. Synchronized erythrocytic schizogony and gametocytogenesis of Plasmodium bergheiin vivoin vitro. Parasitology. ;91(Pt. 1985;3:423–30. doi: 10.1017/s0031182000062673. [DOI] [PubMed] [Google Scholar]
- 31.Waters AP, Thomas AW, van Dijk MR, Janse C. Transfection of malaria parasites. Methods. 1997;13:134–47. doi: 10.1006/meth.1997.0506. [DOI] [PubMed] [Google Scholar]
- 32.Janse C, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 33.van Dijk MR, Vinkenoog R, Ramesar J, Vervenne RA, Waters AP, Janse C. Replication, expression and segregation of plasmid-borne DNA in genetically transformed malaria parasites. Mol Biochem Parasitol. 1997;86:155–62. doi: 10.1016/s0166-6851(97)02843-0. [DOI] [PubMed] [Google Scholar]
- 34.Andreadaki M, Morgan RN, Deligianni E, Kooij TWA, Santos JM, Spanos L et al. Genetic crosses and complementation reveal essential functions for the Plasmodium stage-specific actin2 in sporogonic development. Cell Microbiol. 2014;16:751–67. doi: 10.1111/cmi.12274. [DOI] [PubMed] [Google Scholar]
- 35.Iwanaga S, Khan SM, Kaneko I, Christodoulou Z, Newbold CI, Yuda M et al. Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host Microbe. 2010;7:245–55. doi: 10.1016/j.chom.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwanaga S, Kaneko I, Yuda M. A high-coverage artificial chromosome library for the genome-wide screening of drug-resistance genes in malaria parasites. Genome Res. 2012;22:985–92. doi: 10.1101/gr.124164.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunes A, Thathy V, Bruderer T, Sultan AA, Nussenzweig RS, Ménard R. Subtle mutagenesis by ends-in recombination in malaria parasites. Mol Cell Biol. 1999;19:2895–902. doi: 10.1128/mcb.19.4.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janse C, Kroeze H, van Wigcheren A, Mededovic S, Fonager J, Franke-Fayard B et al. A genotype and phenotype database of genetically modified malaria-parasites. Trends Parasitol. 2011;27:31–9. doi: 10.1016/j.pt.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 39.Kooij TWA, Franke-Fayard B, Renz J, Kroeze H, van Dooren MW, Ramesar J et al. Plasmodium berghei α-tubulin II: a role in both male gamete formation and asexual blood stages. Mol Biochem Parasitol. 2005;144:16–26. doi: 10.1016/j.molbiopara.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Pfander C, Anar B, Schwach F, Otto TD, Brochet M, Volkmann K et al. A scalable pipeline for highly effective genetic modification of a malaria parasite. Nat Methods. 2011;8:1078–82. doi: 10.1038/nmeth.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Spaendonk RM, Ramesar J, van Wigcheren A, Eling WMC, Beetsma AL, van Gemert G-J et al. Functional equivalence of structurally distinct ribosomes in the malaria parasite, Plasmodium berghei. J Biol Chem. 2001;276:22638–47. doi: 10.1074/jbc.M101234200. [DOI] [PubMed] [Google Scholar]
- 42.van Dijk MR, van Schaijk BCL, Khan SM, van Dooren MW, Ramesar J, Kaczanowski S et al. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 2010;6:e1000853. doi: 10.1371/journal.ppat.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs-Lorena VY, Mikolajczak SA, Labaied M, Vaughan AM, Kappe SHI. A dispensable Plasmodium locus for stable transgene expression. Mol Biochem Parasitol. 2010;17:40–4. doi: 10.1016/j.molbiopara.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kooij TWA, Rauch MM, Matuschewski K. Expansion of experimental genetics approaches for Plasmodium berghei with versatile transfection vectors. Mol Biochem Parasitol. 2012;185:19–26. doi: 10.1016/j.molbiopara.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Childs GE, Lambros C. Analogues of N-benzyloxydihydrotriazines: in vitro antimalarial activity against Plasmodium falciparum. Ann Trop Med Parasitol. 1986;80:177–81. doi: 10.1080/00034983.1986.11812002. [DOI] [PubMed] [Google Scholar]
- 46.Canfield CJ, Milhous WK, Ager AL, Rossan RN, Sweeney TR, Lewis NJ et al. PS-15: a potent, orally active antimalarial from a new class of folic acid antagonists. Am J Trop Med Hyg. 1993;49:121–6. doi: 10.4269/ajtmh.1993.49.121. [DOI] [PubMed] [Google Scholar]
- 47.Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, Vanichtanankul J, Sirawaraporn W, Taylor P et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol. 2003;10:357–65. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- 48.Ferone R, Burchall JJ, Hitchings GH. Plasmodium berghei dihydrofolate reductase. Isolation, properties, and inhibition by antifolates. Mol Pharmacol. 1969;5:49–59. [PubMed] [Google Scholar]
- 49.Ferone R. Dihydrofolate reductase from pyrimethamine-resistant Plasmodium berghei. J Biol Chem. 1970;245:850–4. [PubMed] [Google Scholar]
- 50.Donald RG, Roos DS. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc Natl Acad Sci U S A. 1993;90:11703–7. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Koning-Ward TF, Fidock DA, Thathy V, Ménard R, van Spaendonk RM, Waters AP et al. The selectable marker human dihydrofolate reductase enables sequential genetic manipulation of the Plasmodium berghei genome. Mol Biochem Parasitol. 2000;106:199–212. doi: 10.1016/s0166-6851(99)00189-9. [DOI] [PubMed] [Google Scholar]
- 52.Carvalho TG, Thiberge S, Sakamoto H, Ménard R. Conditional mutagenesis using site-specific recombination in Plasmodium berghei. Proc Natl Acad Sci U S A. 2004;101:14931–6. doi: 10.1073/pnas.0404416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braks JAM, Franke-Fayard B, Kroeze H, Janse C, Waters AP. Development and application of a positive–negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res. 2006;34:e39. doi: 10.1093/nar/gnj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ecker A, Moon R, Sinden RE, Billker O. Generation of gene targeting constructs for Plasmodium berghei by a PCR-based method amenable to high throughput applications. Mol Biochem Parasitol. 2006;145:265–8. doi: 10.1016/j.molbiopara.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 55.Lakshmanan V, Bray PG, Verdier-Pinard D, Johnson DJ, Horrocks P, Muhle RA et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picot S, Olliaro P, de Monbrison F, Bienvenu A-L, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J. 2009;8:89. doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sidhu ABS, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–3. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ecker A, Lakshmanan V, Sinnis P, Coppens I, Fidock DA. Evidence that mutant PfCRT facilitates the transmission to mosquitoes of chloroquine-treated Plasmodium gametocytes. J Infect Dis. 2011;203:228–36. doi: 10.1093/infdis/jiq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mamoun CB, Gluzman IY, Goyard S, Beverley SM, Goldberg DE. A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A. 1999;96:8716–20. doi: 10.1073/pnas.96.15.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganesan SM, Morrisey JM, Ke H, Painter HJ, Laroiya K, Phillips MA et al. Yeast dihydroorotate dehydrogenase as a new selectable marker for Plasmodium falciparum transfection. Mol Biochem Parasitol. 2011;177:29–34. doi: 10.1016/j.molbiopara.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franke-Fayard BMD, Trueman HE, Ramesar J, Mendoza J, van der Keur M, van der Linden R et al. Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol. 2004;137:23–33. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 62.Janse C, Franke-Fayard BMD, Waters AP. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat Protoc. 2006;1:614–23. doi: 10.1038/nprot.2006.88. [DOI] [PubMed] [Google Scholar]
- 63.Kenthirapalan S, Waters AP, Matuschewski K, Kooij TWA. Flow cytometry-assisted rapid isolation of recombinant Plasmodium berghei parasites exemplified by functional analysis of aquaglyceroporin. Int J Parasitol. 2012;42:1185–92. doi: 10.1016/j.ijpara.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matz JM, Matuschewski K, Kooij TWA. Two putative protein export regulators promote Plasmodium blood stage development in vivo. Mol Biochem Parasitol. 2013;191:44–52. doi: 10.1016/j.molbiopara.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Jobe O, Lumsden J, Mueller A-K, Williams J, Silva-Rivera H, Kappe SHI et al. Genetically attenuated Plasmodium berghei liver stages induce sterile protracted protection that is mediated by major histocompatibility complex class I-dependent interferon-gamma-producing CD8+T cells. J Infect Dis. 2007;196:599–607. doi: 10.1086/519743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Annoura T, van Schaijk BCL, Ploemen IHJ, Sajid M, Lin J-W, Vos MW et al. Two Plasmodium 6-Cys family-related proteins have distinct and critical roles in liver-stage development. FASEB J. 2014;28:2158–70. doi: 10.1096/fj.13-241570. [DOI] [PubMed] [Google Scholar]
- 67.Orr RY, Philip N, Waters AP. Improved negative selection protocol for Plasmodium berghei in the rodent malarial model. Malar J. 2012;11:103. doi: 10.1186/1475-2875-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manzoni G, Briquet S, Risco-Castillo V, Gaultier C, Topçu S, Ivănescu ML et al. A rapid and robust selection procedure for generating drug-selectable marker-free recombinant malaria parasites. Sci Rep. 2014;4:4760. doi: 10.1038/srep04760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin J-W, Annoura T, Sajid M, Chevalley-Maurel S, Ramesar J, Klop O et al. A novel “gene insertion/marker out” (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS One. 2011;6:e29289. doi: 10.1371/journal.pone.0029289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse C et al. A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog. 2009;5:e1000599. doi: 10.1371/journal.ppat.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tremp AZ, Dessens JT. Malaria IMC1 membrane skeleton proteins operate autonomously and participate in motility independently of cell shape. J Biol Chem. 2011;286:5383–91. doi: 10.1074/jbc.M110.187195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaughan AM, Wang R, Kappe SHI. Genetically engineered, attenuated whole-cell vaccine approaches for malaria. Hum Vaccin. 2010;6:107–13. doi: 10.4161/hv.6.1.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Borrmann S, Matuschewski K. Targeting Plasmodium liver stages: better late than never. Trends Mol Med. 2011;17:527–36. doi: 10.1016/j.molmed.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Nganou-Makamdop K, Sauerwein RW. Liver or blood-stage arrest during malaria sporozoite immunization: the later the better? Trends Parasitol. 2013;29:304–10. doi: 10.1016/j.pt.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 75.Ishino T, Boisson B, Orito Y, Lacroix C, Bischoff E, Loussert C et al. LISP1 is important for the egress of Plasmodium berghei parasites from liver cells. Cell Microbiol. 2009;11:1329–39. doi: 10.1111/j.1462-5822.2009.01333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haussig JM, Matuschewski K, Kooij TWA. Inactivation of a Plasmodium apicoplast protein attenuates formation of liver merozoites. Mol Microbiol. 2011;81:1511–25. doi: 10.1111/j.1365-2958.2011.07787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haussig JM, Burgold J, Hafalla JCR, Matuschewski K, Kooij TWA. Signatures of malaria vaccine efficacy in ageing murine immune memory. Parasite Immunol. 2014;36:199–206. doi: 10.1111/pim.12104. [DOI] [PubMed] [Google Scholar]
- 78.Sahu T, Boisson B, Lacroix C, Bischoff E, Richier Q, Formaglio P et al. ZIPCO, a putative metal ion transporter, is crucial for Plasmodium liver-stage development. EMBO Mol Med. 2014;6(11)):1387–97. doi: 10.15252/emmm.201403868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silvie O, Goetz K, Matuschewski K. A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PLoS Pathog. 2008;4:e1000086. doi: 10.1371/journal.ppat.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Schaijk BCL, Ploemen IHJ, Annoura T, Vos MW, Lander F, van Gemert G-J et al. A genetically attenuated malaria vaccine candidate based on P. falciparumb9/slarp gene-deficient sporozoites. Elife. 2014;3 doi: 10.7554/eLife.03582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baum J, Papenfuss AT, Mair GR, Janse C, Vlachou D, Waters AP et al. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009;37:3788–98. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci U S A. 2009;106:1554–9. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Combe A, Giovannini D, Carvalho TG, Späth S, Boisson B, Loussert C et al. Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe. 2009;5:386–96. doi: 10.1016/j.chom.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Lacroix C, Giovannini D, Combe A, Bargieri DY, Späth S, Panchal D et al. FLP/FRT-mediated conditional mutagenesis in pre-erythrocytic stages of Plasmodium berghei. Nat Protoc. 2011;6:1412–28. doi: 10.1038/nprot.2011.363. [DOI] [PubMed] [Google Scholar]
- 85.Siden-Kiamos I, Ganter M, Kunze A, Hliscs M, Steinbüchel M, Mendoza J et al. Stage-specific depletion of myosin A supports an essential role in motility of malarial ookinetes. Cell Microbiol. 2011;13:1996–2006. doi: 10.1111/j.1462-5822.2011.01686.x. [DOI] [PubMed] [Google Scholar]
- 86.Laurentino EC, Taylor S, Mair GR, Lasonder E, Bártfai R, Stunnenberg HG et al. Experimentally controlled downregulation of the histone chaperone FACT in Plasmodium berghei reveals that it is critical to male gamete fertility. Cell Microbiol. 2011;13:1956–74. doi: 10.1111/j.1462-5822.2011.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pino P, Sebastian S, Kim EA, Bush E, Brochet M, Volkmann K et al. A tetracycline-repressible transactivator system to study essential genes in malaria parasites. Cell Host Microbe. 2012;12:824–34. doi: 10.1016/j.chom.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elsworth B, Matthews K, Nie CQ, Kalanon M, Charnaud SC, Sanders PR et al. PTEX is an essential nexus for protein export in malaria parasites. Nature. 2014;511:587–91. doi: 10.1038/nature13555. [DOI] [PubMed] [Google Scholar]
- 89.Haussig JM, Matuschewski K, Kooij TWA. Identification of vital and dispensable sulfur utilization factors in the Plasmodium apicoplast. PLoS One. 2014;9:e89718. doi: 10.1371/journal.pone.0089718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tremp AZ, Khater EI, Dessens JT. IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes. J Biol Chem. 2008;283:27604–11. doi: 10.1074/jbc.M801302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carter V, Shimizu S, Arai M, Dessens JT. PbSR is synthesized in macrogametocytes and involved in formation of the malaria crystalloids. Mol Microbiol. 2008;68:1560–9. doi: 10.1111/j.1365-2958.2008.06254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tremp AZ, Al-Khattaf FS, Dessens JT. Distinct temporal recruitment of Plasmodium alveolins to the subpellicular network. Parasitol Res. 2014;113:4177–88. doi: 10.1007/s00436-014-4093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Franke-Fayard BMD, Waters AP, Janse C. Real-time in vivo imaging of transgenic bioluminescent blood stages of rodent malaria parasites in mice. Nat Protoc. 2006;1:476–85. doi: 10.1038/nprot.2006.69. [DOI] [PubMed] [Google Scholar]
- 94.Frischknecht F, Baldacci P, Martin B, Zimmer C, Thiberge S, Olivo-Marin J-C et al. Imaging movement of malaria parasites during transmission by Anopheles mosquitoes. Cell Microbiol. 2004;6:687–94. doi: 10.1111/j.1462-5822.2004.00395.x. [DOI] [PubMed] [Google Scholar]
- 95.Vanderberg JP, Frevert U. Intravital microscopy demonstrating antibody-mediated immobilisation of Plasmodium berghei sporozoites injected into skin by mosquitoes. Int J Parasitol. 2004;34:996. doi: 10.1016/j.ijpara.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 96.Amino R, Thiberge S, Martin B, Celli S, Shorte S, Frischknecht F et al. Quantitative imaging of Plasmodium transmission from mosquito to mammal. Nat Med. 2006;12:220–4. doi: 10.1038/nm1350. [DOI] [PubMed] [Google Scholar]
- 97.Tarun AS, Baer K, Dumpit RF, Gray S, Lejarcegui N, Frevert U et al. Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int J Parasitol. 2006;36:1283–93. doi: 10.1016/j.ijpara.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 98.Zhao H, Aoshi T, Kawai S, Mori Y, Konishi A, Ozkan M et al. Olfactory plays a key role in spatiotemporal pathogenesis of cerebral malaria. Cell Host Microbe. 2014;15:551–63. doi: 10.1016/j.chom.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 99.Stanway RR, Witt T, Zobiak B, Aepfelbacher M, Heussler VT. GFP-targeting allows visualization of the apicoplast throughout the life cycle of live malaria parasites. Biol Cell. 2009;101:415–30. doi: 10.1042/BC20080202. [DOI] [PubMed] [Google Scholar]
- 100.Stanway RR, Mueller N, Zobiak B, Graewe S, Froehlke U, Zessin PJM et al. Organelle segregation into Plasmodium liver stage merozoites. Cell Microbiol. 2011;13:1768–82. doi: 10.1111/j.1462-5822.2011.01657.x. [DOI] [PubMed] [Google Scholar]
- 101.Vanderberg JP. Imaging mosquito transmission of Plasmodium sporozoites into the mammalian host: immunological implications. Parasitol Int. 2014;63:150–64. doi: 10.1016/j.parint.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 102.Lawton JC, Benson RA, Garside P, Brewer JM. Using lymph node transplantation as an approach to image cellular interactions between the skin and draining lymph nodes during parasitic infections. Parasitol Int. 2014;63:165–70. doi: 10.1016/j.parint.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frevert U, Nacer A, Cabrera M, Movila A, Leberl M. Imaging Plasmodium immunobiology in the liver, brain, and lung. Parasitol Int. 2014;63:171–86. doi: 10.1016/j.parint.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Claser C, Malleret B, Peng K, Bakocevic N, Gun SY, Russell B et al. Rodent Plasmodium-infected red blood cells: imaging their fates and interactions within their hosts. Parasitol Int. 2014;63:187–94. doi: 10.1016/j.parint.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 105.Ferrer M, Martin-Jaular L, De Niz M, Khan SM, Janse C, Calvo M et al. Imaging of the spleen in malaria. Parasitol Int. 2014;63:195–205. doi: 10.1016/j.parint.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 106.Lima FA, Gómez-Conde I, Videira PA, Marinho CRF, Olivieri DN, Tadokoro CE. Intravital microscopy technique to study parasite dynamics in the labyrinth layer of the mouse placenta. Parasitol Int. 2014;63:254–9. doi: 10.1016/j.parint.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 107.Sakamoto H, Thiberge S, Akerman S, Janse C, Carvalho TG, Ménard R. Towards systematic identification of Plasmodium essential genes by transposon shuttle mutagenesis. Nucleic Acids Res. 2005;33:e174. doi: 10.1093/nar/gni173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balu B, Shoue DA, Fraser MJ, Adams JH. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyback. Proc Natl Acad Sci U S A. 2005;102:16391–6. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fonager J, Franke-Fayard BMD, Adams JH, Ramesar J, Klop O, Khan SM et al. Development of the piggyBac transposable system for Plasmodium berghei and its application for random mutagenesis in malaria parasites. BMC Genomics. 2011;12:155. doi: 10.1186/1471-2164-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ecker A, Bushell ESC, Tewari R, Sinden RE. Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol Microbiol. 2008;70:209–20. doi: 10.1111/j.1365-2958.2008.06407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tewari R, Straschil U, Bateman A, Böhme U, Cherevach I, Gong P et al. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–87. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guttery DS, Poulin B, Ramaprasad A, Wall RJ, Ferguson DJP, Brady D et al. Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell Host Microbe. 2014;16:128–40. doi: 10.1016/j.chom.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Ooij C, Tamez P, Bhattacharjee S, Hiller NL, Harrison T, Liolios K et al. The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathog. 2008;4:e1000084. doi: 10.1371/journal.ppat.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pasini EM, Braks JA, Fonager J, Klop O, Aime E, Spaccapelo R et al. Proteomic and genetic analyses demonstrate that Plasmodium berghei blood stages export a large and diverse repertoire of proteins. Mol Cell Proteomics. 2013;12:426–48. doi: 10.1074/mcp.M112.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matthews K, Kalanon M, Chisholm SA, Sturm A, Goodman CD, Dixon MWA et al. The Plasmodium translocon of exported proteins (PTEX) component thioredoxin-2 is important for maintaining normal blood-stage growth. Mol Microbiol. 2013;89:1167–86. doi: 10.1111/mmi.12334. [DOI] [PubMed] [Google Scholar]
- 116.Lin J-W, Meireles P, Prudencio M, Engelmann S, Annoura T, Sajid M et al. Loss-of-function analyses defines vital and redundant functions of the Plasmodium rhomboid protease family. Mol Microbiol. 2013;88:318–38. doi: 10.1111/mmi.12187. [DOI] [PubMed] [Google Scholar]
- 117.Frénal K, Tay CL, Mueller C, Bushell ES, Jia Y, Graindorge A et al. Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic. 2013;14:895–911. doi: 10.1111/tra.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.MacKellar DC, Vaughan AM, Aly ASI, DeLeon S, Kappe SHI. A systematic analysis of the early transcribed membrane protein family throughout the life cycle of Plasmodium yoelii. Cell Microbiol. 2011;13:1755–67. doi: 10.1111/j.1462-5822.2011.01656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mikolajczak SA, Aly ASI, Dumpit RF, Vaughan AM, Kappe SHI. An efficient strategy for gene targeting and phenotypic assessment in the Plasmodium yoelii rodent malaria model. Mol Biochem Parasitol. 2008;158:213–6. doi: 10.1016/j.molbiopara.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 120.Schwach F, Bushell E, Gomes AR, Anar B, Girling G, Herd C et al. PlasmoGEM, a database supporting a community resource for large-scale experimental genetics in malaria parasites. Nucleic Acids Res. 2015;43:D1176–82. doi: 10.1093/nar/gku1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gomes AR, Bushell ESC, Schwach F, Girling G, Anar B, Quail MA et al. A genome scale vector resource enables high throughput reverse genetic screening in a malaria parasite. Cell Host Microbe. 2015 doi: 10.1016/j.chom.2015.01.014. http://dx.doi.org/10.1016/j.chom.2015.01.014 [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang C, Xiao B, Jiang Y, Zhao Y, Li Z, Gao H et al. Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio. 2014;5:e01414. doi: 10.1128/mBio.01414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Franke-Fayard BMD, Djokovic D, Dooren MW, Ramesar J, Waters AP, Falade MO et al. Simple and sensitive antimalarial drug screening in vitroin vivo using transgenic luciferase expressing Plasmodium berghei parasites. Int J Parasitol. 2008;38:1651–62. doi: 10.1016/j.ijpara.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 124.Ploemen IHJ, Prudencio M, Douradinha BG, Ramesar J, Fonager J, van Gemert G-J et al. Visualisation and quantitative analysis of the rodent malaria liver stage by real time imaging. PLoS One. 2009;4:e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miller JL, Murray S, Vaughan AM, Harupa A, Sack B, Baldwin M et al. Quantitative bioluminescent imaging of pre-erythrocytic malaria parasite infection using luciferase-expressing Plasmodium yoelii. PLoS One. 2013;8:e60820. doi: 10.1371/journal.pone.0060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zuzarte-Luis V, Sales-Dias J, Mota MM. Simple, sensitive and quantitative bioluminescence assay for determination of malaria pre-patent period. Malar J. 2014;13:15. doi: 10.1186/1475-2875-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]