Background: CLEC18 is a novel C-type lectin not being characterized.
Results: The amino acid residue in the C-type lectin-like domain of CLEC18 contribute to the differential glycan-binding ability.
Conclusion: CLEC18 members are expressed in immune and non-immune cells, and preferentially bind to fucoidan, β-glucan, and galactan.
Significance: The biochemical features, tissue distribution, and the glycan-binding specificity suggest that CLEC18 may contribute to host immunity against pathogens.
Keywords: carbohydrate-binding protein, glycobiology, glycoconjugate, glycolipid, glycoprotein, lectin
Abstract
The human C-type lectin 18 (clec18) gene cluster, which contains three clec18a, clec18b, and clec18c loci, is located in human chromosome 16q22. Although the amino acid sequences of CLEC18A, CLEC18B, and CLEC18C are almost identical, several amino acid residues located in the C-type lectin-like domain (CTLD) and the sperm-coating protein/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS) domain, also known as the cysteine-rich secretory proteins/antigen 5/pathogenesis-related 1 proteins (CAP) domain, are distinct from each other. Genotyping by real-time PCR and sequencing further shows the presence of multiple alleles in clec18a/b/c loci. Flow cytometry analysis demonstrates that CLEC18 (CLEC18A, -B, and -C) are expressed abundantly in human peripheral blood cells. Moreover, CLEC18 expression is further up-regulated when monocytes differentiate into macrophages and dendritic cells. Immunofluorescence staining reveals that CLEC18 are localized in the endoplasmic reticulum, Golgi apparatus, and endosome. Interestingly, CLEC18 are also detectable in human sera and culture supernatants from primary cells and 293T cells overexpressing CLEC18. Moreover, CLEC18 bind polysaccharide in Ca2+-independent manner, and amino acid residues Ser/Arg339 and Asp/Asn421 in CTLD domain contribute to their differential binding abilities to polysaccharides isolated from Ganoderma lucidum (GLPS-F3). The Ser339 (CLEC18A) → Arg339 (CLEC18A-1) mutation completely abolishes CLEC18A-1 binding to GLPS-F3, and a sugar competition assay shows that CLEC18 preferentially binds to fucoidan, β-glucans, and galactans. Because proteins with the SCP/TAPS/CAP domain are able to bind sterol and acidic glycolipid, and are involved in sterol transport and β-amyloid aggregation, it would be interesting to investigate whether CLEC18 modulates host immunity via binding to glycolipids, and are also involved in glycolipid transportation and protein aggregation in the future.
Introduction
The superfamily of C-type lectin (CLEC)2 comprises a large group of glycoproteins with divergent functions, including host-pathogen interaction and cell-cell interaction (1, 2). The signature of CLEC is in the presence of a stretch of 115–130 amino acid residues known as the C-type lectin-like domain (CTLD), which is originally identified as carbohydrate-recognition domain in a family of Ca2+-dependent animal lectins (3). The CTLD-fold has a characteristic double-loop (“loop-in-a-loop”) stabilized by two highly conserved disulfide bridges (4). The long loop region contains a conserved “WND” or “ENC” motif for Ca2+ binding and dimerization in some C-type lectins (5). The CTLDs of CLEC members bind a variety of glycans, and it is now clear that even CTLDs with similar structures can bind ligands in distinct ways. Based on their ligand-binding properties, CTLD can be divided into two groups based on the presence of “glutamic acid-proline-asparagine” (EPN) and “glutamine-proline-aspartic acid” (QPD) motifs in a long loop region. CTLDs with the “EPN” motif usually bind mannose, N-acetylglucosamine, and fucose, whereas the CTLDs with the “QPD” motif usually bind galactose and N-acetylgalactosamine (5). However, whether this is a general rule to all the members of C-type lectins need to be further confirmed.
Recently, more and more evidence shows that not all proteins with CTLD interact with Ca2+ in the long loop region (6). The classical CLECs (such as selectins, collectins, and mannose-binding proteins) bind glycans in Ca2+-dependent manner, whereas members of Syk-coupled CLEC receptors (such as Dectin-1/CLEC7A and MDL1/CLEC5A) bind glycans in a Ca2+-independent manner (7, 8). Moreover, proteins with less closely related CTLDs (such as CD69, CD72, KLRF1, of NK receptor family) do not appear to have carbohydrate-binding activity.
In the human genome, there are at least 57 CTLD-containing proteins divided into XVI groups (9). Among these proteins, we are especially interested in group XV, the CLEC18 family, for following reasons: 1) the clec18 gene cluster contains three genes (clec18a, clec18b, and clec18c) located in human chromosome 16q22.1 (clec18a and clec18c), and 16q22.3 (clec18b). 2) Translation of clec18 cDNA predicted an N-linked polypeptide with a C-type lectin domain (CTLD) in the C terminus, and the SCP/TAPS (sperm-coating protein Tpx-1/Ag5/PR-1/Sc7) domain (also known as CAP (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1) domain), which is a conserved tertiary α-β-α structure stabilized by disulfide bonds. 3) Proteins of the SCP/TAPS/CAP domain have been proposed to play significant roles in host-pathogen interactions, reproduction, development, and immune function (10, 11). 4) Several amino acid residues located in the CTLD and SCP/TAPS/CAP domains are distinct among the sequences retrieved from the NCBI database. However, the biochemical features of CLEC18 have not been tested yet.
In this study, we developed a real-time reverse transcription PCR-based platform with fluorescent hybridization probe to detect the presence of CLEC18 in various cell lines and primary cells, followed by direct sequencing cDNAs to confirm the PCR-based typing method. We found that CLEC18 mRNAs and proteins were detectable in various cell lines and human peripheral blood cells. Furthermore, CLEC18 encode N-linked glycoproteins located in endoplasmic reticulum (ER), Golgi apparatus, and endosomes. Moreover, amino acid residues Ser/Arg339 and Asp/Asn421 located in the CTLD domain contribute to their differential glycan binding specificity. This observation suggests that CLEC18 may bind to various glycoconjugates with distinct affinity, and contribute to differential immune responses to glycoconjugates expressed on foreign antigens or altered self-antigen in ER, Golgi apparatus, and endosomes.
Experimental Procedures
Reverse Transcription and Cloning of CLEC18 by Polymerase Chain Reaction
Total RNA was extracted from cells using TRIzol according to the supplier's instructions, then subjected to reverse transcription using a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Burlington ON, Canada) as template for PCR amplification. The CLEC18 cDNAs were amplified by denaturing for 60 s at 95 °C, followed by 35 cycles of denaturation at 95 °C for 10 s, annealing at 70 °C for 45 s, and extension at 72 °C for 80 s. The amplified cDNA fragment was ligated into yTA vector (RBC TA cloning vector kit) before being subcloned into pCMV-Tag4A vector (Stratagene) for expression in mammalian cells. To sequence the CLEC18 cDNA fragment extending from the SCP/TAPS domain to the CTLD domain (nucleotides 211 to 1304), a cDNA fragment was performed by a thermocycler (GeneAmp® PCR System 9700, Applied Biosystem®). The PCR program (denature/annealing/extension) for amplification is as follows: 98 °C, 10 s at 63 °C, 30 s at 72 °C, and 70 s for 30 cycles.
Determination of CLEC18 Alleles by Real-time PCR
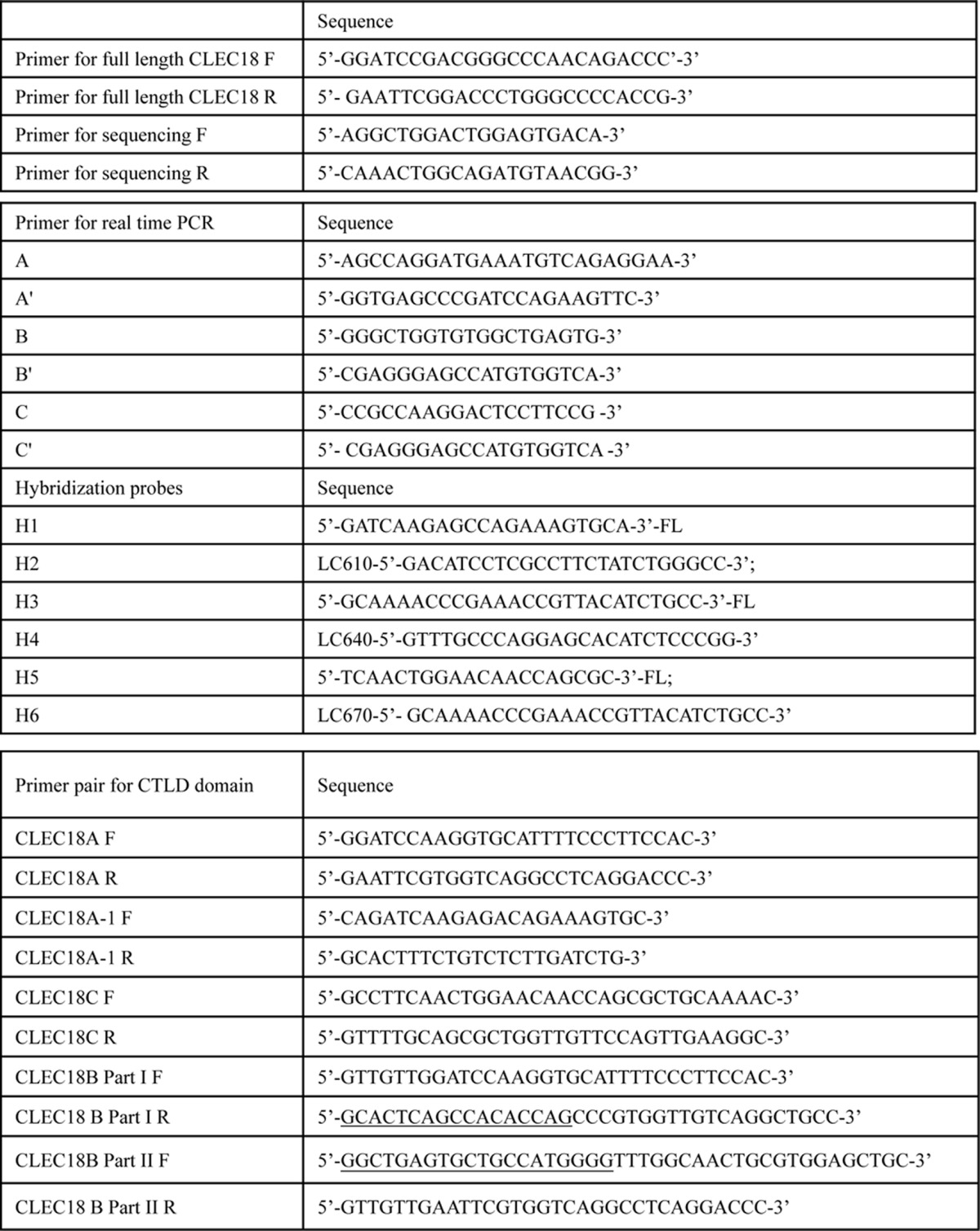
The polymorphism of CLEC18A/B/C was determined by real-time PCR with hybridization probes (Roche Life Sciences). The position of primers are as shown in Fig. 2, whereas the sequences of PCR primers and hybridization probes are listed in Table 1. PCR was performed by a thermocycler (LightCycler480® II, Roche), followed by heat denaturation at 95 °C for 60 s to determine CLEC18 polymorphism. The PCR program (denature/annealing/extension) for amplification of the CTLD domain and is as follows: 95 °C, 10 s at 66 °C, 10 s at 72 °C, and 10 s. The amplification cycles were followed by a melting cycle, in which DNA was denaturated at 95 °C for 60 s using a rate of 4.4 °C/s, cooled to 40 °C for 60 s using a rate of 1.5 °C/s and held for 30 s. The temperature was then raised to 75 °C with a transition rate of 0.03 °C/s. Fluorescence was continuously monitored during the melt.
FIGURE 2.
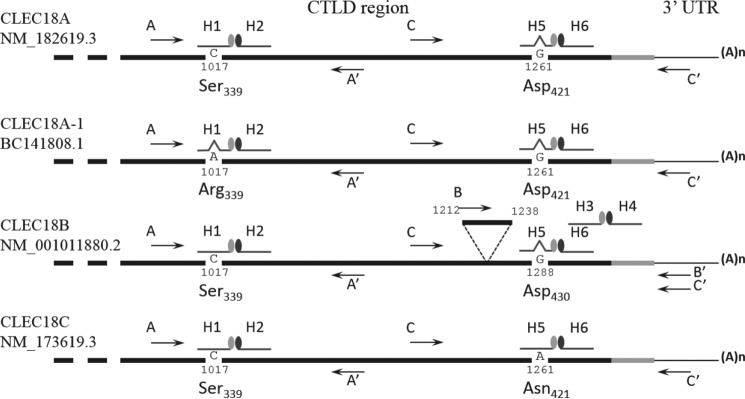
Typing of CLEC18 alleles by hybridization probes. Detection of CLEC18 alleles by real-time RT-PCR using hybridization probes. CLEC18 cDNAs from cell lines or primary cells are used as template, and amplified by primer pairs and hybridization probes to detect polymorphism in nucleotide 1017 (primers A and A′ with probes H1 and H2) nucleotide 1261 (primers C and C′ with probes H5 and H6). Exon B is detected by primers B and B′ with probes H3 and H4.
TABLE 1.
Primer lists
F indicated forward primer, R indicated reverser primer. Underlined nucleotides indicated complementary sequences of exon B.
Isolation of PBMC and Preparation of Human Macrophages and Dendritic Cells
Peripheral blood mononuclear cells (PBMCs) were isolated from the whole blood of healthy human donors by standard density-gradient centrifugation with Ficoll-Paque (Amersham Biosciences). To prepare primary macrophage, CD14+ cells were purified from PBMCs by high-gradient magnetic sorting, using the VarioMACS technique with anti-CD14 microbeads (Miltenyi Biotec GmbH). Cells were then incubated in complete RPMI 1640 medium (Hyclone) supplemented with human M-CSF or GM-CSF/IL-4 for 7 days as described previously (12, 13).
Western Blotting and Flow Cytometry Analysis
For Western blotting analysis, cells (1 × 106) were lysed by RIPA buffer, followed by fractionation on 12% SDS-PAGE, before blotting onto PVDF membrane. Lysates were probed with anti-CLEC18 mAb (clone 3A9E6), followed by incubation with peroxidase-conjugated goat anti-mouse polyclonal antibody (Millpore AP181P) and ImmobilonTM Western Chemiliminescent HRP Substrate (MilliporeTM). For flow cytometry analysis, cells were permeabilized with 0.1% saponin and incubated with the Alexa 647-conjugated anti-CLEC18 mAb (clone 3A9E6), then examined by flow cytometer (VerseTM, BD Biosciences). Data were analyzed by the FlowJowTM software.
Immunofluoresence Staining
Adherent cells were fixed with 4% paraformaldehyde for 1 h, then permeabilized with 0.5% Triton X-100 in PBS for 10 min before incubation in blocking buffer (10% BSA in PBS) for 60 min. Cells were then incubated with anti-CLEC18 mAb (40 μg/ml in 3% BSA/PBS, room temperature for 1 h), anti-calreticulin (dilution range 1:100 in 3% BSA/PBS, 4 °C, overnight), anti-GM130 (dilution range 1:150 in 3% BSA/PBS, 4 °C, overnight), and anti-EEA1 (dilution range 1:100 in 3% BSA/PBS, 4 °C, overnight), respectively. After washing, cells were incubated with Alexa 488-conjugated goat anti-mouse IgG (Jackson ImmunoResearch), TRITC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) for 1 h, and then Hoechst 33342 for 10 min. Samples were then mounted and observed under fluorescence confocal microscopy (Olympus FV10i). Alternatively, cells were incubated with MitoTracker Red (250 nm) or LysoTracker (100 nm) at 37 °C for 1 h to observe their colocalization with CLEC18.
Immunoprecipitation and Mass Spectrometry Analysis
To understand whether CLEC18A is a secretory protein, pCMV-Tag4A or pCMV-Tag4A-CLEC18A (2 μg) was transfected into 293T cells (2 × 105), respectively, followed by incubation at 37 °C for 48 h before harvesting. Cell lysates or supernatant were incubated with anti-FLAG M2 affinity gel (Sigma, A2220) to pulldown CLEC18A. The immunoprecipitates were fractionated on a 12% SDS-PAGE, then probed with anti-CLEC18 mAb (clone 3A9E6), and further incubated with peroxidase-conjugated goat anti-mouse polyclonal antibody (Millpore AP181P) and ImmobilonTM Western Chemiluminescent HRP Substrate (MilliporeTM) subsequently for detection. Alternatively, the immunoprecipitates were fractionated on 12% SDS-PAGE, then eluted from gel for mass spectrometry analysis. The eluted proteins were subjected to mass spectrometry analysis using the Thermo Finnigan LTQ FT Ultra High Performance Mass Spectrometer (Conquer scientific). The secretory CLEC18 in culture supernatants and sera was determined using a ELISA kit (CUSABIO Life Science).
Construction, Expression, and Purification of Fc-tagged Recombinant CLEC18.Fc Fusion Proteins
DNA fragments encoding the CTLD domain of CLEC18A/B/C were amplified by reverse transcriptase-PCR and subcloned into pcDNA3.1-hIgG1 Fc (mut) vector to generate CLEC18A/A-1/B/C Fc fusion proteins, respectively. PCR primers are listed in Table 1. The CLEC18B cDNA was generated by two steps of PCR to insert exon B using CLEC18A cDNA as template. The CLEC18.Fc fusion proteins were overexpressed by using the FreeStyle 293 Expression System (Invitrogen). Briefly, 3 × 107 293F cells in 28 ml of culture medium were transfected with a mixture of 30 μg of plasmid DNA and 60 μl of 293fectinTM in a total of 2 ml of Opti-MEM I (Invitrogen). The culture supernatants were harvested at days 3 and 5, and the recombinant CLEC18.Fc fusion proteins were purified by Protein A column (GE Healthcare).
Polysaccharide Binding Assay
Samples of GLPS-F3 were weighed, dissolved, and diluted with 100 mm Tris buffer (pH 9.5) to give 20 μg/ml as described previously (14). Briefly, the GLPS-F3 was immobilized in the wells of 96-well plates (50 μl/well, Corning) and incubated overnight at 4 °C, followed by incubation with 200 μl of blocking buffer (2% BSA/TBST) for 1 h at room temperature before washing with TBST (0.05% Tween 20/TBS) twice. Each well was incubated with 100 μl of CLEC.Fc fusion protein (2 μg/ml in 2 mm MgCl2, 2 mm CaCl2, 1% BSA/TBST) in the presence or absence of various glycans (Megazyme) as competitors for 1 h at room temperature. After washing with TBST, each well was incubated with 100 μl of peroxidase-conjugated goat anti-human IgG Ab (Jackson ImmunoResearch) in 1% BSA/TBST (1:5000) at room temperature for 30 min, followed by addition of 100 μl of tetramethylbenzidine substrate (BD Biosciences) for 20 min, and the reaction was stopped by 1 n H2SO4 before subjecting to analysis by an ELISA reader (TECAN). Alternatively, fusion protein was incubated with zymosans at 4 °C for 1 h and washed with PBS, followed by incubation with anti-hIgG1-FITC (1:200) at 4 °C for 30 min and then analyzed using a VerseTM flow cytometer (BD Biosciences), and data were analyzed using FlowJowTM software.
Results
Chromosomal Location and Structure of clec18 Gene Cluster
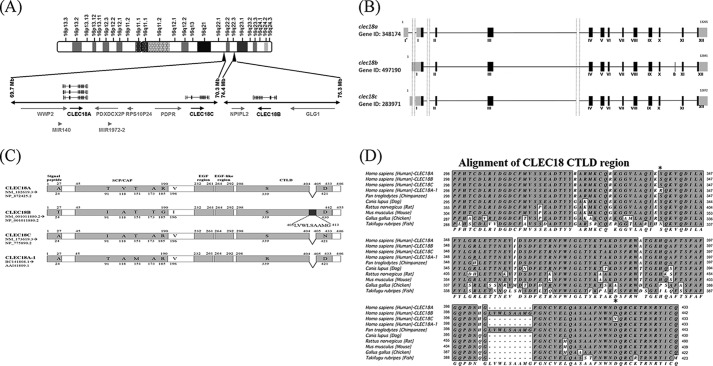
The clec18 gene cluster contains three loci: clec18a (16q22.1), clec18c (16q22.1), and clec18b (16q22.3). Both clec18a (Gene ID 348174) and clec18c (Gene ID 283971) comprise 12 exons, whereas a predicted extra exon (denoted as exon B) located between exons X and XI is found in clec18b (Gene ID 497190) (Fig. 1, A and B). clec18a (NM_182619.3) and clec18c (NM_173619.3) encode a 446-amino acid (aa) polypeptide, respectively, whereas clec18b is predicted to encode a 455-amino acid polypeptide with the extra nine amino acids (LVWLSAAMG) derived from exon B (AGG CUG GUG UGG CUG AGU GCU GCC AUG GGGU) within the CTLD (Fig. 1C). Compared with CLEC18A, CLEC18C is almost identical with CLEC18A except 2 amino acids (Thr91 → Ile91, Val118 → Als118) located in SCP/TAPS/CAP domain (aa 45–190) and 1 amino acid (Asp421 → Asn421) in WND motif located in CTLD. In addition, four amino acid residues (Thr91 → Ile91, Val118 → Ala118, Ala173 → Thr173, Arg185 → Gly185) located in the SCP/TAPS domain (aa 45–190), and one amino acid residue (Val196 → Ile196) located between the SCP/TAPS/CAP domain and EGF regions (aa 232–292) were noted among CLEC18A, CLEC18B, and CLEC18C. Moreover, CLEC18A-1 (AAI41809.1), which contains three distinct amino acid residues from CLEC18A (Val118 → Ala118 and Thr151 → Met151 in SCP/TAPS/CAP domain and Ser339 → Arg339 in CTLD) was found in the human genome database (Fig. 1C).
FIGURE 1.
Chromosomal location, structures, protein sequence, and alignment. A, the clelc18 gene family is located in human chromosomes 16q22.1 (clec18a and clec18c) and 16q22.3 (clec18b). B, the clec18a and clec18c comprise 12 exons, respectively, whereas an extra exon (exon B) is predicted in clec18b. C, clec18a and clec18c encode a polypeptide of 446 aa, respectively, whereas clec18b is predicted to encode a polypeptide of 455 aa due to the extra 9 amino acid residues translated from the exon B in clec18b. The polymorphic amino acid residues among members of the CLEC18 family are indicated. D, multiple sequence alignment of CTLD of human CLEC18 and other species. Only the human genome contains three genes (clec18a, clec18b, and clec18c), whereas other species only contain one gene (clec18a) in the genome. Only human and chimpanzee CLEC18 are predicted to contain exon B with 9-inserted amino acid residues (LVWLSAAMG) in the CTLD. *, the polymorphic amino acid residues (339 and 421) found in human CLEC18 family.
Alignment of human CLEC18 with CLEC18 of other species shows that CLEC18 is highly conserved in CTLD, but the predicted 9-amino acid insertion is only found in human CLEC18B and chimpanzee CLEC18 (Fig. 1D). The typical motifs of CTLDs, such as “WIGL” (aa 370–373), “QPD” (aa 399–401), and “WND” (aa 419–421), are found in both human and non-human CLEC18. It is interesting to note that the “Asp421/430” located in the “WND” motif of CLEC18A/B is replaced with “Asn421” in human CLEC18C. Moreover, the basic amino acid residues Arg322 and Arg329 found in human and chimpanzee are replaced with Gly322 and Gly/Glu329 in non-primates. Furthermore, the aliphatic amino acid residues Ile360, Ala385, and His403 found in human and chimpanzee are replaced with Thr/Met/Ser369, Thr/Asp385, and Gln403 in non-primate CLEC18.
Detection of CLEC18 Alleles by Real-time Reverse Transcription-PCR (RT-PCR)
Due to the highly conserved cDNA sequences among CLEC18, nucleotide residues (Cys/Ala1017 and Gly/Ala1261) located in CTLD were used to discriminate each member of the CLEC18 family by hybridization probe-based real-time RT-PCR assay. In addition, a probe hybridizing the extra exon B was used to detect CLEC18B (Fig. 2). We found that CLEC18A mRNA was expressed in almost all the cell lines tested, whereas CLEC18A-1 was detectable in CHME3, A549, and the human peripheral blood cells (PBCs) of three donors. CLEC18C mRNA was also detectable in some cell lines, but was less prevalent than CLEC18A. However, we cannot detect CLEC18B mRNA in all the cell lines by PCR using the probe hybridizing the exon B (Table 2, part A). We further investigated the expression of CLEC18 mRNAs in human PBMCs from healthy donors. Although CLEC18A mRNAs were detectable in all the samples tested, the expression of CLEC18A-1 and CLEC18C was less prevalent than CLEC18A. However, CLEC18B mRNA is not detectable in all the samples tested (Table 2, part B).
TABLE 2.
Typing of CLEC18 family in cell lines and human peripheral blood cell
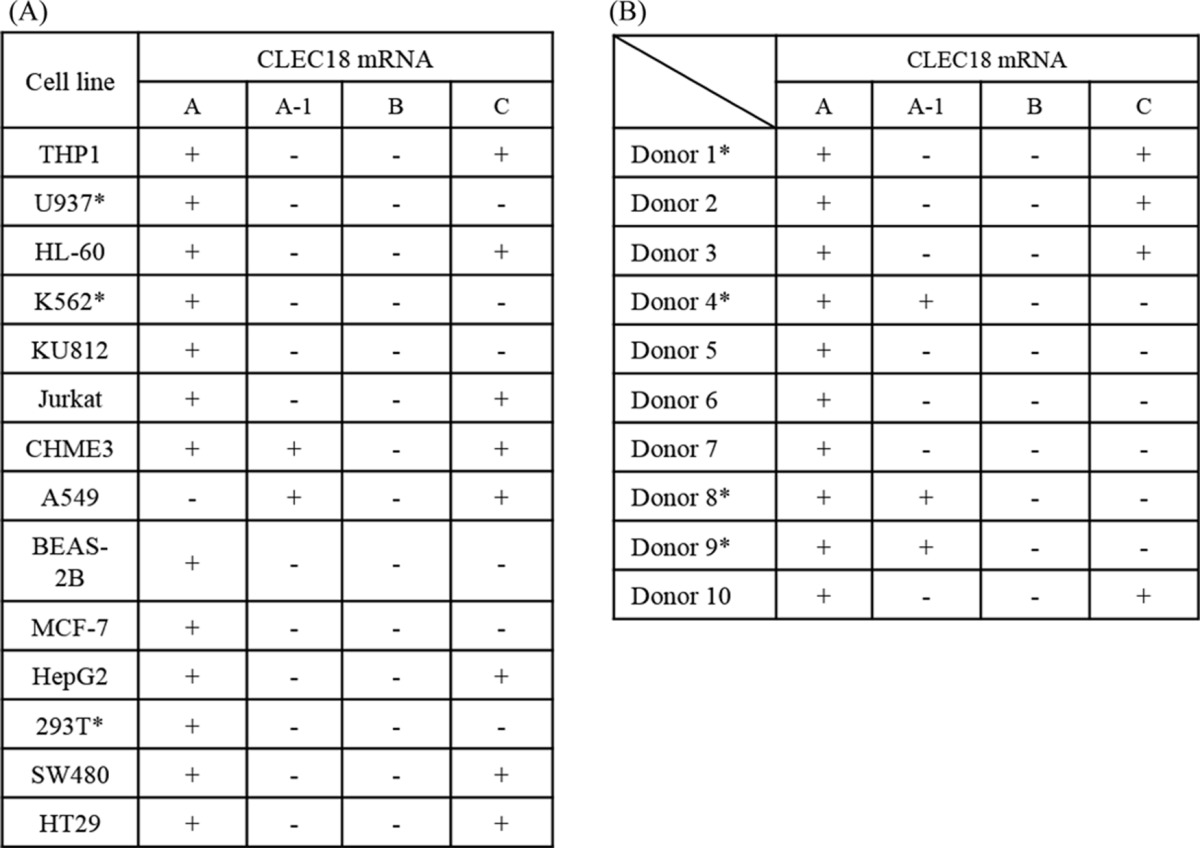
+, detectable; −:, undetectable; “∗” were confirmed by cDNA clone sequencing.
We further examined the presence of other CLEC18A/B/C alleles in cell lines and human PBCs by sequencing the CLEC18 cDNAs amplified by RT-PCR. Interestingly, two discrepancies with the reference sequences were found. 1) The dominant amino acid residues located in the SCP/TAPS/CAP domain of CLEC18A was Thr91-Ala118-Met151-Ala173-Arg185-Val196, not the Thr91-Val118-Thr151-Ala173-Arg185-Val196 (reference sequence: NP_872425.2) (Table 3, part A); 2) cDNA clones with the Ile91-Ala118-Thr151-Thr173-Gly185-Ile196 (characteristic of CLEC18B) in the SCP/TAPS/CAP domain do not contain the extra 9 amino acids (LVWLSAAMG) derived from exon B, among all the cell lines and human PBCs we sequenced (Table 3, B and C). This observation suggests that the putative exon B is absent in human CLEC18B, and multiple alleles are present in all the members of CLEC18 family.
TABLE 3.
Sequencing of CLEC18 family in cell lines and human peripheral blood cell
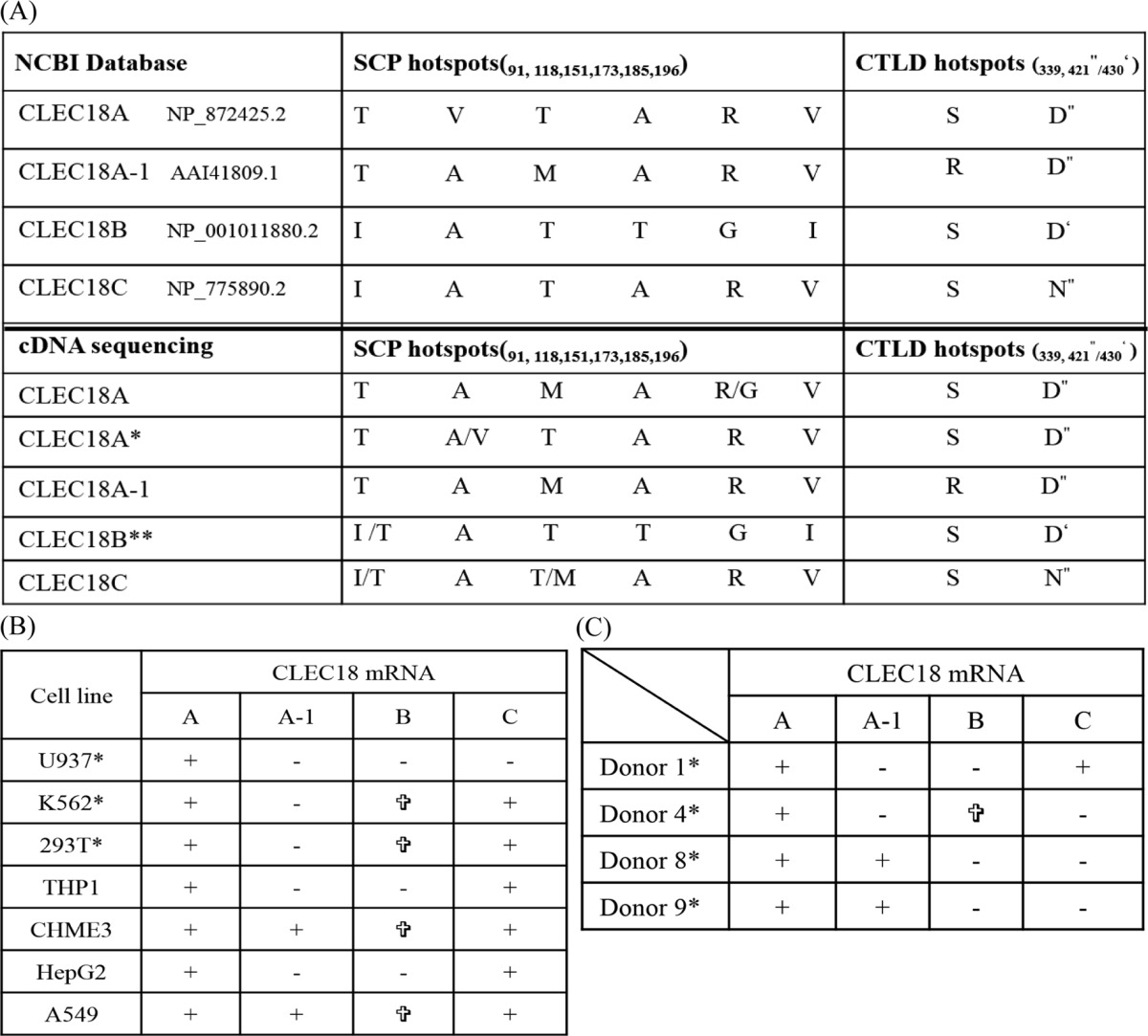
+, detectable; −, undetectable; “∗” were confirmed by cDNA clone sequencing; “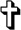 ”: real time PCR is negative, but sequencing is positive (but without the extra exon B).
”: real time PCR is negative, but sequencing is positive (but without the extra exon B).
CLEC18A∗, minor allele; CLEC18B∗∗, without exon B.
CLEC18 Are N-linked Glycoproteins Detectable in Cell Lysates and Culture Supernatant
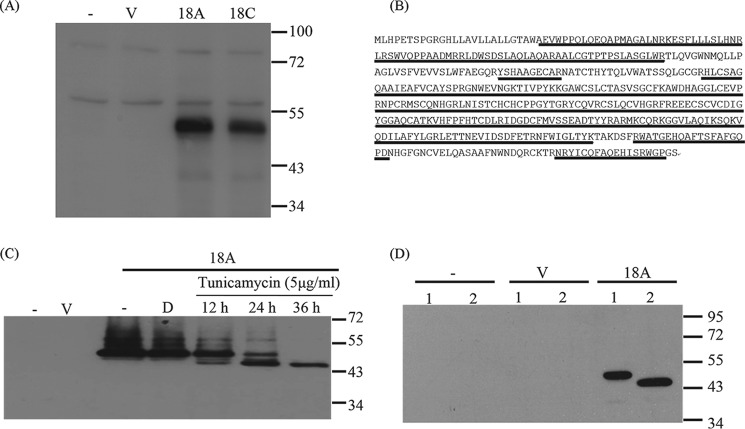
We further investigated the expression of CLEC18 by Western blotting and flow cytometry. We found that anti-CLEC18 mAb (clone 3A9E6) detects a specific band migrating at 50 kDa in 293T cells transfected with CLEC18A and CLEC18C cDNAs, respectively (Fig. 3A). The peptide was eluted for mass spectrometry analysis, and the sequences of peptides matched CLEC18 perfectly (Fig. 3B). Because two potential glycosylation sites (Asn144 and Asn243) were predicted when analyzing CLEC18 by Vector NTI software (version 11), we asked whether CLEC18 are N-linked glycosylated proteins. To address this question, FLAG-tagged CLEC18A-transfected 293T cells were incubated with tunicamycin to observe the change of Mr of CLEC18. We found the CLEC18A polypeptide migrated from 50 to 45 kDa in a time-dependent manner when probed with anti-FLAG mAb (Fig. 3C), and a similar result was observed after peptide:N-glycosidase F treatment (Fig. 3D). This observation indicates that CLEC18A is a 50-kDa N-linked glycoprotein.
FIGURE 3.
CLEC18 are an N-linked glycoprotein. A, the 293T cells were transfected with pCMV-Tag4A vector (V), pCMV-Tag4A-CLEC18A (18A), or pCMV-Tag4A-CLEC18C (18C), followed by Western blot analysis using anti-CLEC18 mAb (clone 3A9E6). B, mass spectrometry analysis of peptides recognized by anti-CLEC18 mAb (clone 3A9E6). The 50-kDa peptides were eluted for mass spectrometry analysis. The peptides (underlined) matched perfectly to CLEC18. C, CLEC18A-transfected 293T cells were incubated with tunicamycin for various time points, followed by Western blot analysis to determine the presence of N-linked glycosylation using anti-FLAG mAb. D, dimethyl sulfoxide. D, alternatively, 293T cells were transfected with vector (V) or CLEC18A (18A) for 48 h, and then the collected lysate (20 μg) was left untreated (lane 1) or treated with 500 units of peptide:N-glycosidase F (lane 2) for 37 °C for 2 h, followed by Western blot analysis using anti-FLAG mAb.
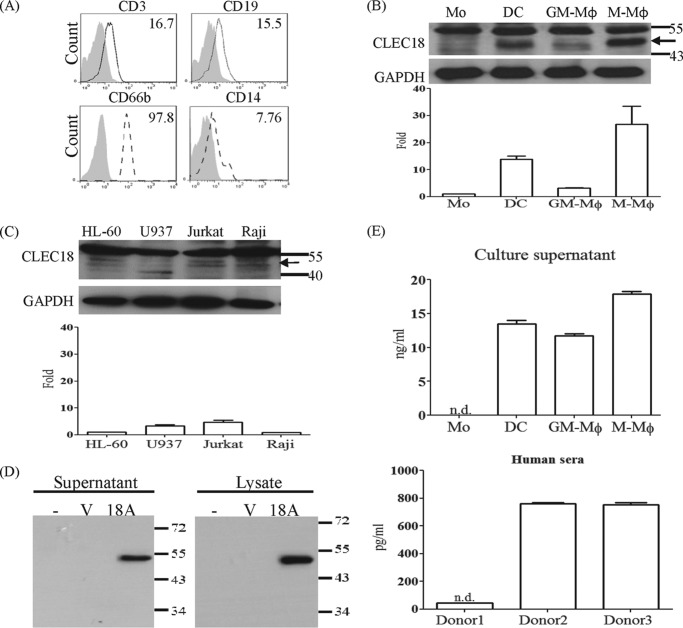
We further examined its expression in human PBCs and CD14+-derived macrophages and dendritic cells by flow cytometry and Western blotting. To address this question, CD14+-derived dendritic cells were incubated with GM-CSF + IL-4 (25 + 20 ng/ml), GM-CSF (10 ng/ml), and M-CSF (10 ng/ml) for 7 days, respectively, to differentiate into dendritic cells (DCs), GM-macrophages (GM-Mφ), and M-macrophages (M-Mφ). We found that CLEC18 are abundantly expressed in human PBCs, including T cells (CD3+), B cells (CD19+), granulocytes (CD66b+), and monocytes (CD14+) (Fig. 4A). Compared with monocytes, CLEC18 were further up-regulated in monocyte-derived DCs, GM-Mφ, and M-Mφ (Fig. 4B, upper), and the protein expression level in accord with CLEC18 mRNA levels was determined by real-time PCR (Fig. 4B, lower). This observation suggested the CLEC18 expression is up-regulated by cytokines. However, the expression of CLEC18 was low and barely detectable in several hematopoietic cell lines (Fig. 4C) and non-hematopoietic cell lines (data not shown).
FIGURE 4.
CLEC18 are a secretory protein. A, detection of CLEC18 by flow cytometry and real-time PCR in human peripheral blood cells. Shadow, isotype control; dash line, anti-CLEC18 mAb. B and C, detection of CLEC18 by Western blot and real-time PCR in DC/Mφ (B) and cell lines (C). Arrow, CLEC18. D, the 293T cells were transfected with vector or pCMV-Tag4A-CLEC18A for 48 h, and the supernatant or lysates were incubated with anti-FLAG mAb to pulldown CLEC18, followed by Western blot analysis. E, detection of secretory CLEC18 in culture media and human sera. Culture supernatants (upper) of monocyte, dendritic cells, and macrophages and human sera (lower) were determined by ELISA (CUSABIO Life Science). ND, not determined.
It has been shown that members of protein with the SCP/CAP domain are soluble proteins (11, 15), thus we asked whether CLEC18 are secretory proteins. To address this question, culture supernatants of pCMV-Flag-CLEC18A-transfected 293T cells were incubated with anti-FLAG mAb to pulldown soluble CLEC18A, followed by Western blot analysis using anti-CLEC18 mAb (clone 3A9E6). We found that CLEC18A is not only detectable in culture supernatant of pCMV-Flag-CLEC18A-transfected 293T cells (Fig. 4D), but also in the culture supernatant of DCs, GM-Mφ, and M-Mφ (upper, Fig. 4E), as well as in human sera (Fig. 4E, lower) as determined by ELISA. This observation suggests that CLEC18A is a secretory protein.
Subcellular Localization of CLEC18
Because CLEC18 were detectable in cell lysates, we further investigated its subcellular localization. To address this question, human CD14+-derived M-Mφ was incubated with anti-CLEC18A mAb, followed by incubation with Alexa 488-conjugated goat anti-mouse mAb. To determine its subcellular localization, M-Mφ was also co-incubated with anti-GM 130 mAb (rabbit), anti-calreticulin mAb (rabbit), and anti-EEA1 mAb (rabbit), respectively, followed by incubation with TRITC-conjugated goat anti-rabbit mAb. We found that CLEC18 were co-localized with GM 130 (marker for Golgi apparatus), calreticulin (marker for endoplasmic reticulum), and EEA1 (marker for early endosome), but not with MitoTracker (mitochondria dye) or LysoTracker (lysosome dye) under the same conditions (Fig. 5). This observation suggests that CLEC18 are located in the ER, Golgi apparatus, and early endosomes.
FIGURE 5.
CLEC18 are localized in the ER, Golgi apparatus, and early endosomes. CD14+-monocyte-derived macrophage (M-Mφ) were incubated with Alexa 488-conjugated anti-CLEC18 mAb (green) in the presence of TRITC-conjugated anti-GM130 mAb (red), TRITC-conjugated anti-calreticulin (red), TRITC-conjugated anti-EEA1 mAb (red), MitoTracker (red), or LysoTracker (red), respectively. Cells were examined under a confocal microscopy (Olympus FV10i). Scale bars, 10 μm.
Tissue Distribution of CLEC18
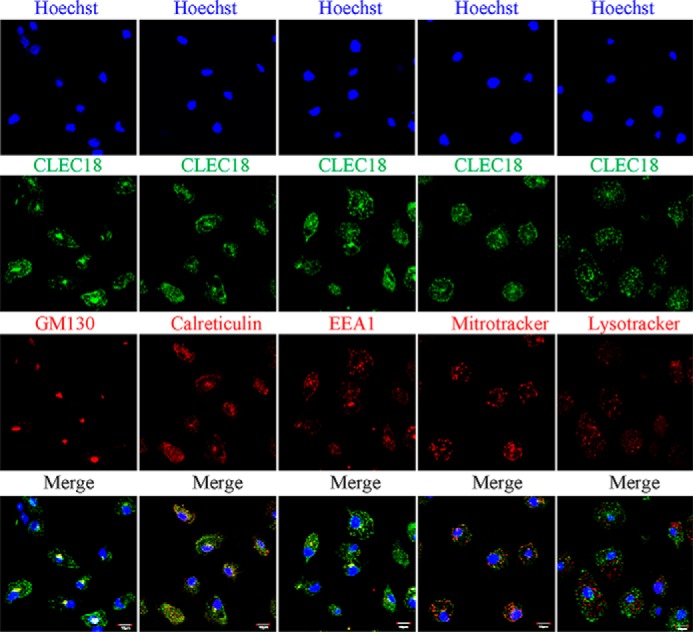
We further used a human tissue array to detect the expression of CLEC18 by immunohistochemistry staining. We found that CLEC18 were expressed in human spleen (leukocytes), brain (microglia), liver (hepatocytes), gall bladder (epithelial cells), fallopian tube (epithelial cells), and testis (epithelial cells) (Fig. 6). This observation suggests that CLEC18 are not only expressed in hematopoietic lineages, but also expressed in epithelial and parenchymal cells.
FIGURE 6.
Histochemical staining of human tissue array. The formalin-fixed human tissue array (prepared from normal human tissues in Taipei Veterans General Hospital) was incubated with anti-CLEC18 mAb (3A9E6) followed by the HISTOMOUSETM-MAX kit (Zymed Laboratories Inc.) according to the vendor's instructions. Arrowheads, leukocyte (spleen), microglia (brain), hepatocyte (liver), and epithelial cells (gall bladder, testis, fallopian tube).
Differential Glycan Binding Ability of CLEC18A, CLEC18A-1,and CLEC18C
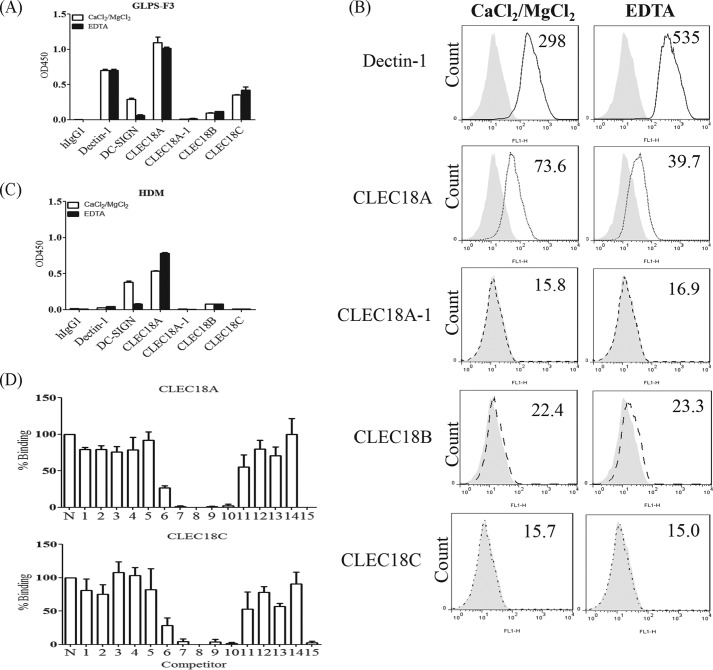
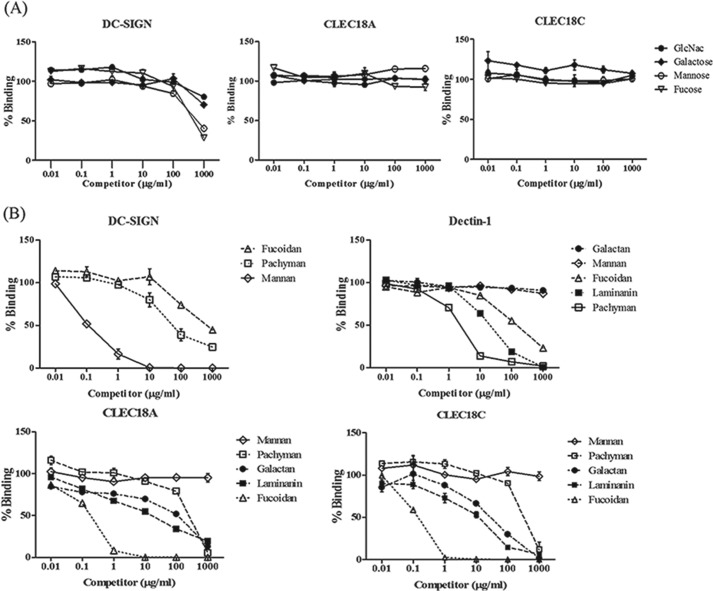
We further investigated the role of amino acid residues Ser/Arg339 and Asp/Asn421 in CTLD of the CLEC18 family in the binding ability to glycans. To address this question, Ser339 and Asp421 were located in CTLD of CLEC18A (Ser339, Asp421) were mutated into Arg339 and Asn421, respectively, to generate CLEC18A-1 (Arg339, Asp421) and CLEC18C (Ser339, Asn421). The CTLD domain of CLEC18A and mutants was ligated with pcDNA3.1-hIgG1 Fc (mut) to obtain pCLEC18A.Fc, pCLEC18A-1.Fc, and pCLEC18C.Fc, respectively, followed by transfection to 293F cells to obtain recombinant fusion proteins as described previously (8, 14, 16). To understand whether the presence of exon B (encoding “LVWLSAAMG”) affects the glycan-binding ability, we inserted the 9 amino acids into CLEC18A CTLD to generate pCLEC18B.Fc. The recombinant CLEC18A.Fc, CLEC18A-1.Fc, and CLEC18C.Fc fusion proteins were then used to hybridize with the synthetic oligosaccharide glycan arrays from CFG (Consortium for Functional Glycomics, slides 14508, 14506, and14507; PMT70 version 5.0 Alexa 48810/20/11 HJ). However, the fusion proteins did not bind any glycans spotted on the glass slides, because the signals are weak (relative fluorescence units < 10,000) and the coefficient variation is high (CV > 50%) (supplemental Data 1, A–C). Thus, we further tested their binding ability to F3 polysaccharides isolated from medicinal fungi Ganoderma lucidum (GLPS-F3), followed by a glycan competition assay to determine binding specificity as described previously (14, 16).
It has been shown that GLPS-F3 contains abundant polysaccharides comprising glucose, mannose, fucose, galactose, xylose, GlcNAc, and rhamnose (17), and GLPS-F3 is able to interact with several C-type lectins and Toll-like receptors (14). Thus, we examined the interaction between CLEC18 and GLPS-F3 using Dectin1.Fc and DC-SIGN.Fc as controls. Similar to Dectin1.Fc and DC-SIGN.Fc, we found that CLEC18A.Fc and CLEC18C.Fc also bound to F3 polysaccharides, whereas the Ser339 → Arg339 mutation (CLEC18A-1.Fc) and insertion of the 9-amino acid LVWLSAAMG in the CTLD domain (CLEC18B.Fc) abolished their binding ability to GLPS-F3 (Fig. 7A). Interestingly, EDTA was unable to abolish binding of Dectin1.Fc, CLEC18A.Fc, and CLEC18C.Fc to F3 polysaccharide, although EDTA inhibited DC-SIGN.Fc binding to F3 efficiently under the same condition (Fig. 7A). A similar observation was observed when incubating fusion proteins with zymosans (Fig. 7B) and house dust mite (Fig. 7C). Because Dectin-1.Fc binding to β-glucans is independent of Ca2+ (7), this observation suggests that CLEC18 binding to the F3 polysaccharides and zymosans is also Ca2+-independent, and amino acid residue Ser339 is critical for binding to polysaccharides.
FIGURE 7.
Determination of the sugar binding ability of CLEC18. A, polysaccharides isolated from the G. lucidum F3 fraction (GLPS-F3) were immobilized on an ELISA plate (20 μg/ml), followed by incubation with Dectin-1.Fc, DC-SIGN.Fc, and CLEC18.Fc family proteins (2 μg/ml), in the presence or absence of EDTA (5 mm). B, incubation of Dectin-1.Fc or CLEC18.Fc family proteins (10 μg/ml) with zymosan (10 mg/ml, Sigma), in the presence or absence of EDTA. The numbers represent the mean fluorescence intensity. C, house dust mite (5 μg/ml) were immobilized on a ELISA plate, followed by incubation with Dectin-1.Fc, DC-SIGN.Fc, and CLEC18.Fc family proteins (2 μg/ml), in the presence or absence of EDTA (5 mm). D, interaction of CLEC18A/C with various polysaccharides (as shown in Table 4) by ELISA. N, noncompetitor.
We further determined the glycan-binding specificity of CLEC18A and CLEC18C by a sugar competition assay (14) using monosaccharides (Fig. 8A) and polysaccharides (Fig. 8B) as competitors. Even though mannose and fucose competed with DC-SIGN.Fc binding to GLPS-F3 in a dose-dependent manner (Fig. 8A, left), none of the monosaccharides (GlcNAc, galactose, mannose, and fucose) were able to inhibit CLEC18A.Fc (Fig. 8A, middle) and CLEC18C.Fc (Fig. 8A, right most) binding to GLPS-F3 under the same condition. We further used polysaccharides to replace monosaccharides for competition assays. Among the 15 polysaccharides tested (Table 4 and Fig. 7D), fucoidan (sulfated fucose), laminarin (β-1,3-linked glucan with β-1,6-linked side chain), and galactan (β4-GlcNac) could inhibit CLEC18A.Fc and CLEC18C.Fc binding to GLPS-F3 (Fig. 8B, lower). Interestingly, pachyman (β-1,3-linked glucan) were less efficient than laminarin (β-1,3-linked glucan with β-1,6-linked side chain) to inhibit CLEC18A/C binding to GLPS-F3, although pachyman was more efficient than laminarin to inhibit Dectin-1 binding to GLPS-F3 (Fig. 8B, upper right). In contrast, mannan was unable to inhibit CLEC18A/C binding to GLPS-F3, even though mannan efficiently inhibited DC-SIGN binding to GLPS-F3 (Fig. 8B, upper left). Therefore, the CTLD domain of CLEC18A/C preferred binding to sulfated fucose, β-glucan, and galactan.
FIGURE 8.
Determination of sugar binding specify of CLEC18. Various monosaccharide (A) or polysaccharide (B) were used to compete DC-SIGN, Dectin-1, CLEC18A.Fc, and CLEC18C.Fc (2 μg/ml) binding to GLPS-F3 in the sugar competition assay.
TABLE 4.
Polysaccharides used for sugar competition assay
| Number | Name | Structure |
|---|---|---|
| 1 | α-Cyclodextrin | 6 α-1,4-Linked Glc ring molecule |
| 2 | d-(+)-Cellobiose | β-1,4-Linked Glc dissaccharides |
| 3 | Dextran | α-1,6-Linked glucan with 1–3-linked Glc side chains |
| 4 | Dextran | α-1,6-Linked glucan with 1–3-linked Glc side chains |
| 5 | CM-cellulose 4 m | β-1,4-Glc-linked glucan |
| 6 | Amylose | α-1,4-Glc-linked glucan |
| 7 | Laminarin | β-1,3-Linked glucan with β-1,6-linked side chains |
| 8 | Pachyman | β-1,3-Linked glucan |
| 9 | Fucoidan | Sulfated fucose polymer |
| 10 | Galactan | β-1,4-Linked galactan |
| 11 | Larch Arabinogalactan | β-1,4-Linked galactan |
| 12 | β-Gentiobiose | β-1,6-Linked Glc disaccharides |
| 13 | Galactosylmannotriose | β-1,4-Linked Mannan with α-1,6-Gal side chain |
| 14 | Di-galactosylmannopentaose | β-1,4-Linked Mannan with α-1,6-Gal side chain |
| 15 | F3 | Polysaccharide from G. lucidum |
Discussion
Several unusual features of CLEC18 are also noted in this study. 1) It is surprising to find that CLEC18A/C bind GLPS-F3 in a Ca2+-independent manner, even though the typical WND domain was present in CLEC18A. 2) Presence of the Gln399-Pro400-Asp401 tripeptide motif in CLEC18 CTLD predicts binding specificity to galactose and GalNAc. However, sugar competition assays showed that CLEC18A and CLEC18C displayed diverse binding specificity to various glycans, including galactan (β4-GlcNac), fucoidan, and β-glucan. In addition, amino acid residue Asp421/Asn421 has a mild effect on F3 binding, because laminarin and galactan have a better inhibitory effect to CLEC18C.Fc (Asn421) than CLEC18A.Fc (Asp421) in higher concentrations (100 μg/ml). Thus, amino acid residue Ser/Arg339 has a decisive role to determine CLEC18 binding to GLPS-F3, whereas Asp/Asn421 only has a mild effect to influence their binding to laminarin and galactan, and fucoidan (Fig. 8B). In addition, the stronger inhibitory effects of laminarin (β-1,3-linked glucan with β-1,6-linked side chain) than pachyman (β-1,3-linked glucan) suggests that CLEC18A.Fc and CLEC18C.Fc seem to be preferentially binding to the β-1,6-linked side chain of β-glucans. 3) CLEC18 are not only located in the ER, Golgi apparatus, and endosome of the primary macrophage (Fig. 5), but also detectable in the culture supernatant of 293T cells overexpressing CLEC18 (Fig. 4D). However, overexpression often results in a non-physiological cell response, thus the subcellular distribution of CLEC18 and the presence of soluble CLEC18 under physiological conditions needs to be verified further.
In contrast to L-type lectins calreticulin/calnextins, which are ER membrane proteins acting as chaperons to bind GlcM3 (Glc-α1→3-Man-α1→2-Man-α1→2-Man) of Glc1 Man9GlcNAc2 attached to glycoproteins in the ER for N-linked glycosylation (18), no significant binding to GlcM3 or other glycans spotted on CFG glycan arrays was noted by recombinant CLEC18A/A-1/C fusion proteins (supplemental Fig. S1, 92928 -247015 M A-C). Furthermore, CLEC18A.Fc and CLEC18C.Fc bind GLPS-F3 (Fig. 7A) and zymosans (Fig. 7B) in a Ca2+-independent manner, and display diverse binding specificity to various glycans (Fig. 7D and Table 4). Thus, CLEC18 does not seem to be involved in N-linked glycosylation in the ER and Golgi apparatus. This argument is further supported by the histochemical staining that CLEC18 are not ubiquitously expressed in all the cell types (Fig. 6).
Because CLEC18 does not contain the “H/KDEL” tetrapeptide motif critical for ER retention, we asked whether CLEC18 was retained in ER via association with other proteins. To address this question, FLAG-tagged CLEC18A was precipitated from CLEC18A-transfected 293T cells by anti-FLAG mAb, and the immunoprecipitates were subjected to mass spectrometry analysis. We found that CLEC18A was coimmunoprecipitated with the ER protein Grp78/Bip (data not shown). However, we were unable to co-precipitate CLEC18A and Grp78/Bip from cells with or without stress (data not shown), thus how CLEC18 were retained in the ER and Golgi apparatus needs to be further investigated in the future.
It has been shown that proteins with the SCP/TAPS/CAP domain, such as the pathogen-related yeast proteins and human CAP family member CRISP2, are necessary and sufficient for lipid export and sterol binding (15, 19). Because CLEC18 contains both CTLD and SCP/TAPS/CAP domains, it is reasonable to speculate that CLEC18 may bind glycolipids and are involved in the transport of glycolipids. The distinct amino acid residues in the SCP/TAPS/CAP domain may also contribute to binding specificity and affinity of CLEC18 to glycolipids and other glycoconjugates. Furthermore, it has been speculated that the plant pathogenesis-related 1 protein, a host-defense protein with the SCP/TAPS/CAP domain, may serve to inhibit pathogen proliferation by extracting sterols from the pathogen membrane (15). Therefore, CLEC18 may extract the sterol component of pathogen assembly in the ER and Golgi apparatus (such as members of flaviviruses) and attenuate their infectivity. This speculation is supported by the fact that CLEC18 are up-regulated in human macrophages and dendritic cells (Fig. 4B), which are the major targets of flaviviruses. Furthermore, the Golgi-associated plant pathogenesis related protein 1 (GAPR-1), a mammalian protein with SCP/TAPS/CAP domain, is shown to interact with acidic phospholipids and inhibits Aβ aggregation (20).
The above evidence suggests that CLEC18 may be involved in recognition and transportation of glycoconjugates, and it would be interesting to investigate how the polymorphic amino acid residues in SCP/TAPS/CAP and CTLD domains determine CLEC18 binding specificity and affinity to various glycoconjugates in the future.
Author Contributions
Y.-L. H., F.-S. P., Y.-T. T., and H.-C. M. designed, performed, and analyzed experiments; T.-L. H., C. Y. W., T. Y. C., W.-B. Y., C.-H. C., and C.-H. W. provided reagents and technique support; and S.-L. H. designed and analyzed experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the BioLegend® for their technique support and related antibody preparation. The glycan arrays were provided by the Consortium for the Functional Glycomics funded by National Institutes of Health NIGMS Grant GM62116.
This work was supported by the National Science Council Grants NSC 103-2321-B-001-044 and NSC 103-2320-B-001-010-MY3 and Summit and Thematic Research Project Grant AS-101-TP-B06-2 from Academia Sinica. The authors declare no competing financial interests.

This article contains supplemental Fig. S1.
- CLEC
- C-type lectin
- ER
- endoplasmic reticulum
- CTLD
- C-type lectin-like domain
- PBC
- peripheral blood cell
- aa
- amino acid(s)
- DC
- dendritic cell
- PBMC
- peripheral blood mononuclear cell
- TRITC
- tetramethylrhodamine isothiocyanate.
References
- 1. Weis W. I., Taylor M. E., Drickamer K. (1998) The C-type lectin superfamily in the immune system. Immunol. Rev. 163, 19–34 [DOI] [PubMed] [Google Scholar]
- 2. Cambi A., Figdor C. G. (2003) Dual function of C-type lectin-like receptors in the immune system. Curr. Opin. Cell Biol. 15, 539–546 [DOI] [PubMed] [Google Scholar]
- 3. Dodd R. B., Drickamer K. (2001) Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11, 71R–79R [DOI] [PubMed] [Google Scholar]
- 4. Zelensky A. N., Gready J. E. (2005) The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217 [DOI] [PubMed] [Google Scholar]
- 5. Drickamer K., Dodd R. B. (1999) C-Type lectin-like domains in Caenorhabditis elegans: predictions from the complete genome sequence. Glycobiology 9, 1357–1369 [DOI] [PubMed] [Google Scholar]
- 6. Poget S. F., Freund S. M., Howard M. J., Bycroft M. (2001) The ligand-binding loops in the tunicate C-type lectin TC14 are rigid. Biochemistry 40, 10966–10972 [DOI] [PubMed] [Google Scholar]
- 7. Brown G. D., Gordon S. (2001) Immune recognition. A new receptor for β-glucans. Nature 413, 36–37 [DOI] [PubMed] [Google Scholar]
- 8. Chen S. T., Lin Y. L., Huang M. T., Wu M. F., Cheng S. C., Lei H. Y., Lee C. K., Chiou T. W., Wong C. H., Hsieh S. L. (2008) CLEC5A is critical for dengue-virus-induced lethal disease. Nature 453, 672–676 [DOI] [PubMed] [Google Scholar]
- 9. Varki A., C. R., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) C-type Lectins. Essentials of Glycobiology. (Cummings R. D., McEver R. P., eds) 2nd Ed., Chapter 31, pp. 425–437, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 10. Cantacessi C., Campbell B. E., Visser A., Geldhof P., Nolan M. J., Nisbet A. J., Matthews J. B., Loukas A., Hofmann A., Otranto D., Sternberg P. W., Gasser R. B. (2009) A portrait of the ”SCP/TAPS“ proteins of eukaryotes: developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 27, 376–388 [DOI] [PubMed] [Google Scholar]
- 11. Gibbs G. M., Roelants K., O'Bryan M. K. (2008) The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897 [DOI] [PubMed] [Google Scholar]
- 12. Hsu T. L., Chang Y. C., Chen S. J., Liu Y. J., Chiu A. W., Chio C. C., Chen L., Hsieh S. L. (2002) Modulation of dendritic cell differentiation and maturation by decoy receptor 3. J. Immunol. 168, 4846–4853 [DOI] [PubMed] [Google Scholar]
- 13. Chang Y. C., Hsu T. L., Lin H. H., Chio C. C., Chiu A. W., Chen N. J., Lin C. H., Hsieh S. L. (2004) Modulation of macrophage differentiation and activation by decoy receptor 3. J. Leukoc. Biol. 75, 486–494 [DOI] [PubMed] [Google Scholar]
- 14. Hsu T. L., Cheng S. C., Yang W. B., Chin S. W., Chen B. H., Huang M. T., Hsieh S. L., Wong C. H. (2009) Profiling carbohydrate-receptor interaction with recombinant innate immunity receptor-Fc fusion proteins. J. Biol. Chem. 284, 34479–34489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneiter R., Di Pietro A. (2013) The CAP protein superfamily: function in sterol export and fungal virulence. Biomol. Concepts 4, 519–525 [DOI] [PubMed] [Google Scholar]
- 16. Yang C. Y., Chen J. B., Tsai T. F., Tsai Y. C., Tsai C. Y., Liang P. H., Hsu T. L., Wu C. Y., Netea M. G., Wong C. H., Hsieh S. L. (2013) CLEC4F is an inducible C-type lectin in F4/80-positive cells and is involved in α-galactosylceramide presentation in liver. PLoS ONE 8, e65070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y. Y., Khoo K. H., Chen S. T., Lin C. C., Wong C. H., Lin C. H. (2002) Studies on the immuno-modulating and antitumor activities of Ganoderma lucidum (Reishi) polysaccharides: functional and proteomic analyses of a fucose-containing glycoprotein fraction responsible for the activities. Bioorg. Med. Chem. 10, 1057–1062 [DOI] [PubMed] [Google Scholar]
- 18. Kozlov G., Pocanschi C. L., Rosenauer A., Bastos-Aristizabal S., Gorelik A., Williams D. B., Gehring K. (2010) Structural basis of carbohydrate recognition by calreticulin. J. Biol. Chem. 285, 38612–38620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choudhary V., Schneiter R. (2012) Pathogen-Related Yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 16882–16887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olrichs N. K., Mahalka A. K., Kaloyanova D., Kinnunen P. K., Bernd Helms J. (2014) Golgi-associated plant pathogenesis related protein 1 (GAPR-1) forms amyloid-like fibrils by interaction with acidic phospholipids and inhibits Aβ aggregation. Amyloid 21, 88–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.