Background: Telomerase elongation of chromosome ends is restricted within the cell cycle.
Results: A human telomerase holoenzyme subunit mediating Cajal body and telomere localization disassembles reversibly from the RNP catalytic core in each cell cycle.
Conclusion: Telomerase holoenzyme assembly and disassembly are favored at distinct stages of the cell cycle.
Significance: Human telomerase is relicensed for telomere synthesis in G1.
Keywords: cell cycle, mitosis, telomerase, telomerase reverse transcriptase (TERT), telomere, Cajal bodies, RNP assembly, RNP trafficking
Abstract
Human telomerase acts on telomeres during the genome synthesis phase of the cell cycle, accompanied by its concentration in Cajal bodies and transient colocalization with telomeres. Whether the regulation of human telomerase holoenzyme assembly contributes to the cell cycle restriction of telomerase function is unknown. We investigated the steady-state levels, assembly, and exchange dynamics of human telomerase subunits with quantitative in vivo cross-linking and other methods. We determined the physical association of telomerase subunits in cells blocked or progressing through the cell cycle as synchronized by multiple protocols. The total level of human telomerase RNA (hTR) was invariant across the cell cycle. In vivo snapshots of telomerase holoenzyme composition established that hTR remains bound to human telomerase reverse transcriptase (hTERT) throughout all phases of the cell cycle, and subunit competition assays suggested that hTERT-hTR interaction is not readily exchangeable. In contrast, the telomerase holoenzyme Cajal body-associated protein, TCAB1, was released from hTR in mitotic cells coincident with TCAB1 delocalization from Cajal bodies. This telomerase holoenzyme disassembly was reversible with cell cycle progression without any change in total TCAB1 protein level. Consistent with differential cell cycle regulation of hTERT-hTR and TCAB1-hTR protein-RNA interactions, overexpression of hTERT or TCAB1 had limited if any influence on hTR assembly of the other subunit. Overall, these findings revealed a cell cycle regulation that disables human telomerase association with telomeres while preserving the co-folded hTERT-hTR ribonucleoprotein catalytic core. Studies here, integrated with previous work, led to a unifying model for telomerase subunit assembly and trafficking in human cells.
Introduction
Safeguarding genomic stability requires a mechanism to maintain telomeres at the ends of chromosomes. The general eukaryotic solution to this obligation is the activity of telomerase, a reverse transcriptase specialized for the synthesis of telomeric repeats (1). By copying an RNA template within the active RNP,2 telomerase extends a 3′-overhang at the chromosome end. Telomerase is active in early human embryo cells but is repressed upon tissue differentiation (2). Without telomerase activity, human somatic cells count down cell divisions toward a proliferative limit due to progressive telomere shortening (3). To bypass this growth barrier, the majority of human cancers reactivate telomerase and balance the telomere attrition coupled to genome replication with new telomeric repeat synthesis (4). Knowledge about the regulation of human telomerase is therefore essential to understanding the changes in genome maintenance with human development and tumorigenesis.
Telomerase catalytic activity can be reconstituted by the combination of hTR and hTERT in a eukaryotic cell or cell extract (5, 6). In addition to these two RNP catalytic core subunits, many additional proteins are necessary for the in vivo assembly, subcellular trafficking, and telomere association of a functional telomerase holoenzyme (7, 8). Mature hTR biological stability requires precursor co-transcriptional assembly as an H/ACA small nucleolar RNP with dyskerin, NOP10, NHP2, and the chaperone NAF1, which is later replaced by GAR1. The crucial importance of this RNP biogenesis process is established by human gene mutations that cause telomerase deficiency diseases such as dyskeratosis congenita (9). After initial hTR H/ACA RNP biogenesis, a fraction of the biologically stable hTR RNP associates with hTERT through multiple direct protein-RNA interactions (10–12). Some or all of the hTR RNPs bind the telomerase Cajal body protein, TCAB1, via the Cajal body localization (CAB) motif in the hTR 3′-stem loop (13, 14). TCAB1 increases the steady-state Cajal body association of hTR and a subset of other H/ACA RNAs that also contain CAB boxes (15, 16). TCAB1 does not contribute to telomerase catalytic activation, but it is necessary for hTERT-hTR RNP recruitment to and extension of telomeres (16–18).
Cell cycle regulation imparts coordination to cellular processes such as chromosome replication and segregation that occur in ordered progression through a first gap phase (G1), DNA synthesis (S), a second gap phase (G2), and mitosis (M). As for many other DNA replication enzymes, telomerase action is under cell cycle control. Physical assays of 3′-overhang synthesis and processing in many organisms, including human cells (19, 20), support S/G2 as the interval for changes in telomeric DNA structure. Studies in budding and fission yeasts demonstrate that telomerase holoenzyme engagement of telomeres occurs only in S phase (8, 21–23). The telomere association of hTR detectable by in situ hybridization also occurs only in S phase (24, 25). Even in the ciliate Tetrahymena, which has thousands of chromosome ends per macronucleus, chromatin immunopurification (IP) assays show cell cycle restriction of telomerase-telomere engagement (26).
Cell cycle regulation of telomerase action at telomeres could derive in part from the regulation of telomerase holoenzyme. Yeast telomerase holoenzyme subunit interactions have cell cycle regulation, including the exchange of a shared DNA-binding subunit of mutually exclusive telomerase and telomere-capping complexes (27). In comparison, there has been relatively little investigation of human telomerase cell cycle regulation. Several studies have used lysates of cells harvested across synchronized cell cycle progression to survey for telomerase catalytic activity, with conclusions ranging from lack of regulation to S phase-maximal or S phase-specific activity (28–30). Subcellular localization studies suggest differential intranuclear distributions of hTERT and hTR until they coassemble in S phase in Cajal bodies or nucleoli (25, 30). Enrichment of hTR in Cajal bodies is regulated by the cell cycle, peaking in S phase when hTR foci colocalize with telomeres (15, 24, 25). However, conclusions differ about whether telomerase requires Cajal bodies for association with telomeres; coilin gene disruption does not affect telomerase function in telomere maintenance (31), whereas coilin depletion reduces endogenous hTR and hTERT recruitment to telomeres (18) or even recruitment of overexpressed hTR and hTERT (32). Curiously, no Cajal body localization has been detected for mouse telomerase RNA, which like hTR, becomes colocalized specifically in S phase with a few telomeres at a time (33), and overexpression of a CAB-box-mutant hTR lacking Cajal body association still elongates telomeres at least half as well as the wild type (34, 35). These findings leave open many possible hypotheses for cell cycle regulation of the human telomerase holoenzyme.
In this study, we investigated cell cycle regulation of the levels of hTR, hTERT, TCAB1, and their protein-RNA interactions. We also tested whether increasing the level of an individual telomerase holoenzyme or RNP biogenesis protein would affect hTERT-hTR or TCAB1-hTR interaction. We present a model in which hTERT and hTR assemble irreversibly, with synthesis of new hTR and active RNP maximal in G1. On the other hand, TCAB1 association with hTR undergoes cell cycle regulation. We observed a coordinate M-phase TCAB1 loss from hTR and from Cajal bodies, which was reversible with cell cycle progression. These and additional findings provide new understanding of human telomerase holoenzyme regulation.
Experimental Procedures
Cell Culture
Cell lines were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and 100 μg/ml Primocin (Invivogen). Cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C. Lipofectamine 2000 (Invitrogen) was used for transfection experiments.
Generation of Retrovirus
Retrovirus was produced by transfecting the 293 Phoenix packaging cell line with pBABE-HYGRO-ZZF-hTERT and pBABE-PURO-F-TCAB1, as well as empty vector, using the established protocol (36). HeLa S3 cells were plated on 10-cm dishes and infected in the presence of 5 μg/ml Polybrene (Sigma-Aldrich) the next day. The medium was replaced the following morning. Polyclonal stable cell lines were selected with either 300 μg/ml hygromycin B or 2 μg/ml puromycin.
Cell Staining
Cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were then washed with PBS three times and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Cells were then washed and blocked with 4% BSA in PBS for 1 h at room temperature before the addition of primary antibodies. Primary antibodies used were rabbit anti-TCAB1 (1:300, NB100-68252, Novus Biologicals), mouse anti-coilin (1:250, IH10, Abcam), and mouse anti-FLAG (1:2,000, F1804, Sigma-Aldrich). Following several PBS washes, cells were incubated for an additional hour with goat secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 568 (Invitrogen) and DAPI nuclear counterstain. Cells were then washed extensively with PBS and mounted using ProLong Gold (Invitrogen). Slides were dried overnight and imaged using a Zeiss LSM510 Meta confocal microscope with a ×100/1.49 Apo objective and 364-, 488-, 543-, and 647-nm laser excitation. Image processing was performed using NIH ImageJ. The collection of cells held in mitotic block was performed as above, except without Triton X-100 permeabilization.
Immunoblotting
After heating at 80 °C for 5 min, protein samples were cooled to room temperature and resolved by bis-Tris SDS-PAGE in MOPS buffer (250 mm MOPS, 250 mm Tris, 5 mm EDTA, 0.5% SDS, pH 7.0). Protein was then transferred to nitrocellulose membrane and subsequently blocked using 4% nonfat milk (Carnation) in TBS buffer (150 mm NaCl, 50 mm Tris, pH 7.5) for 1 h at room temperature. The membranes were then incubated with primary antibodies overnight at 4 °C. The primary antibodies used were mouse anti-tubulin (1:500, DM1A, Calbiochem), mouse anti-phosphoserine 10 histone H3 (1:2,000, sc-8656-R, Santa Cruz Biotechnology), mouse anti-FLAG (1:4,000, F1804, Sigma-Aldrich), rabbit anti-cyclin A (1:2,000, H-432, Santa Cruz Biotechnology), rabbit anti-TCAB1 (1:2,000, NB100-68252, Novus Biologicals), rabbit anti-Cdc2 pTyr-15 (1:1,000, 9111, Cell Signaling Technology), and rabbit IgG (1:10,000, Sigma-Aldrich). The membrane was washed in TBS and incubated with goat anti-mouse Alexa Fluor 680 (1:2,000, Life Technologies) and goat anti-rabbit IRDye 800CW (1:10,000, Rockland Immunochemicals) in 4% nonfat milk in TBS for 1 h at room temperature. After extensive washing with TBS, the membrane was visualized on a LI-COR Odyssey imager.
Cell Cycle Synchronization
Cells were plated at 30–40% confluency. For G1/S synchronization, cells were incubated with 2 mm thymidine for 24 h. Cells were then washed three times with PBS, and fresh medium was added. 8 h later, cells were reincubated with 2 mm thymidine for a second round of thymidine block. 16–18 h later, cells were washed with PBS three times, and fresh medium was added to continue synchronized cell cycle progression. For mitotic synchronization, cells underwent a single round of thymidine block as described above, and, three h after cells were washed of thymidine, 100 ng/ml nocodazole (Sigma-Aldrich) was added. Cells were held in nocodazole for 12 h. Cells were then collected, washed three times with PBS, and replated in fresh medium to continue the cell cycle progression. For synchronized progression into mitosis, cells were double thymidine block-synchronized as described above, released, and allowed to progress for 8 h before the addition of 9 nm S-trityl-l-cysteine (STLC) (Sigma-Aldrich). For fluorescence microscopy of mitotic cells, asynchronous cells were held in 9 nm STLC for 4 h to enrich the mitotic population before fixation. Synchronization was verified by cyclin A levels and the presence of histone H3 phosphoserine 10.
Cellular Cross-linking
The medium was aspirated, and cells were cross-linked on dishes in PBS containing 0.25% formaldehyde at 37 °C for 10 min. The formaldehyde was then quenched with 300 mm Tris, pH 8.0, for 10 min at room temperature. Cells were then washed three times with PBS, scraped into PBS, pelleted at 17,000 rcf for 5 min in Eppendorf tubes, and resuspended in radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 1 mm EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm DTT) before sonication. For cells held in mitotic block and for the collection of time points that included a mitotic synchronization step, cells were first scraped into suspension, centrifuged at 1000 rcf for 5 min in 15-ml conical tubes, and resuspended in PBS with 0.25% formaldehyde followed by the incubation and quenching steps as described above. Sonication was performed on ice with a Branson Sonifier 250 at 30% power, 50% duty cycle, and a duration of 15 s for three cycles. Samples were then centrifuged at 17,000 rcf for 5 min at 4 °C, and the supernatants were transferred to new tubes for processing.
RNP Purification
For non-denaturing IP, whole cell extract was generated via hypotonic cell lysis as before (37), except that the salt concentration never exceeded 200 mm NaCl. The whole cell extract was rotated with magnetic anti-FLAG M2 beads (Sigma-Aldrich) for 2 h at room temperature. The beads were then washed with HL150 buffer (20 mm HEPES, pH 8.0, 2.5 mm MgCl2, 0.25 EGTA, 10% glycerol, 1 mm DTT, 1 mm PMSF, and 150 mm NaCl) several times. After the final wash, a portion of the samples was eluted with 3×-FLAG peptide (Sigma-Aldrich) for protein SDS-PAGE, and the remainder was mixed with TRIzol (Life Technologies) for RNA purification. For denaturing IP of cross-linked whole cell extract, samples were rotated with magnetic anti-FLAG M2 beads, rabbit IgG-agarose resin (Sigma-Aldrich), or EZview Red anti-c-Myc affinity gel (Sigma-Aldrich) for 4 h at 4 °C. The samples were then washed with radioimmune precipitation assay buffer five times. After the final wash, beads were resuspended in 50 μl of radioimmune precipitation assay buffer containing 100 μg/ml proteinase K. Samples were then incubated at 50 °C for 1 h followed by 70 °C for 1 h for cross-linking reversal. TRIzol was then added directly to the samples.
Reverse Transcription and Quantitative PCR (RT-qPCR)
RNA was purified via TRIzol according to the manufacturer's instructions (Life Technologies) using linear polyacrylamide as carrier. RNA was resuspended in H2O and then treated with RQ1 DNase (Promega) in the presence of RNasin (Promega) for 30 min at 37 °C. DNase was then inhibited by the addition of RQ1 stop buffer and incubation at 65 °C for 10 min. cDNA generation from DNase-treated RNA samples was performed with target-specific oligonucleotides and Superscript III according to the manufacturer's instructions (Invitrogen) with slight modifications. Initial reverse transcription primer annealing was carried out with a 20-s incubation at 80 °C followed by 2 min at 65 °C and then 1 min at 50 °C before being cooled to 4 °C. Superscript III reverse transcriptase was added to the samples on ice, and then the samples were incubated on a prewarmed 55 °C block for 30 min. Reverse transcription was halted by incubation at 85 °C for 5 min. qPCR was performed on a CFX96 Touch real-time PCR detection system (Bio-Rad). 2 μl of cDNA was used per 20 μl of qPCR reaction with iTaq Universal SYBR Green Supermix and forward and reverse primers each at 300 μm. qPCR was carried out for 35 cycles at 95 °C for 15 s and 60 °C for 45 s after an initial 2-min 95 °C hot start. RT-qPCR data were quantified using the Δ-Δ Ct method. Statistical analysis was performed in GraphPad Prism 6 (GraphPad Software Inc.) using ANOVA with Tukey's multiple comparison test unless noted otherwise. The error bars represent ±S.E. The PCR amplification efficiencies were calculated using the LinRegPCR program (38). Reference gene quality was assessed by using the NormFinder Excel add-in (39). The following oligonucleotides were used (where the reverse transcription and the reverse primer for PCR are the same): hTR fwd (5′-CCCTAACTGAGAAGGGCGTA-3′) and hTR rev (5′-AGAATGAACGGTGGAAGGCG-3′); RNA POL II fwd (5′-ACGAGTTGGAGCGGGAATTT-3′) and RNA POL II rev (5′-TTCCTTGACTCCCTCCACCA-3′); U1 fwd (5′-TTTCCCAGGGCGAGGCTTAT-3′) and U1 rev (5′CCCCACTACCACAAATTATGCA-3′); precursor hTR fwd (5′-CTCGGCTCACACATGCAGTT-3′) and precursor hTR rev (5′-GCCCAGTCAGTCAGGTTTGG-3′); GAPDH fwd (5′-TGCACCACCAACTGCTTAGC-3′) and GAPDH rev (5′-GGCATGGACTGTGGTCATGAG-3′; and actin fwd (5′-CTGGAACGGTGAAGGTGACA-3′) and actin rev (5′-AAGGGACTTCCTGTAACAATGCA-3′).
Quantitative Telomeric Repeat Amplification Protocol (QTRAP)
QTRAP was performed on whole cell extract generated from hypotonic lysis similar to the previously published protocol (40). Protein concentration was determined by the BCA protein assay (Pierce). 2 μl of diluted whole cell extract was used per 20 μl of QTRAP reaction constituted of iTaq Universal SYBR Green Supermix (Bio-Rad), 0.1 μg of TS, and 0.02 μg of ACX primers (40). The samples were incubated at 30 °C for 30 min before a 2-min 95 °C hot start followed by 35 cycles at 95 °C for 15 s and 61 °C for 90 s. Relative telomerase activity was calculated by ΔCt to the reference sample. For RNase treatment, 20 μg/ml RNase A was added to whole cell extract.
Northern Blot
RNA was purified using TRIzol. Northern blot detection of hTR and the recombinant RNA recovery control for RNA precipitation was performed as described (41). U6 snRNA was detected using a 32P end-labeled probe (5′-AGTATATGTGCTGCCGAAGCG-3′) under conditions similar to hTR hybridization except at 37 °C.
Results
Quantitative Assays Detect Human Telomerase Subunit Assembly
We sought to evaluate cell cycle-dependent changes in hTR association with hTERT and TCAB1. Subunit interactions can be assayed under non-denaturing conditions after gentle cell lysis or under denaturing conditions after in vivo cross-linking and harsh cell lysis. The latter method is more discriminating for physical proximity but less sensitive, as a result of low cross-linking efficiency. However, nondenaturing cell extract can allow interactions to occur that differ from interactions in vivo. Previous studies have used eukaryotic cell extracts to assemble recombinant hTERT and hTR as a minimally active RNP, but on the other hand, the mixing of human 293T cell extracts containing overexpressed hTERT or overexpressed hTR shows little if any increase in active RNP (35, 42). Therefore, we first evaluated whether the production of cell extracts affects the determination of in vivo protein-RNA interactions.
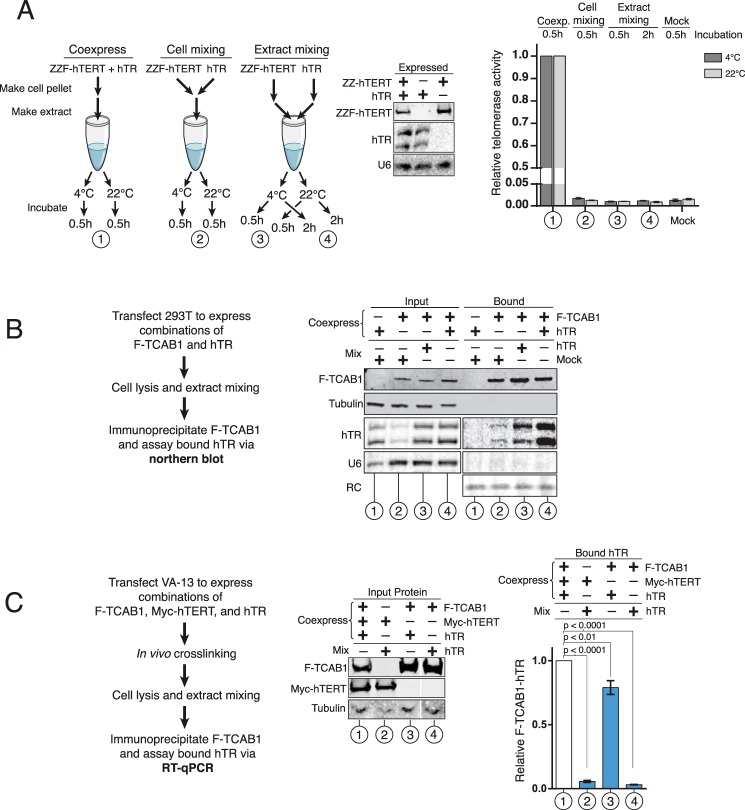
To test for whether telomerase subunit associations occurred de novo in extract, we transfected a telomerase-null immortalized human cell line, VA-13, to express a tandem protein A domain (ZZ) and 3×-FLAG-tagged (F) hTERT and hTR individually, combining the subunits after expression (Fig. 1A, left and middle panels). We used a SYBR Green quantitative PCR version of the telomeric repeat amplification protocol, QTRAP, to assay the amount of hTERT-hTR RNP product synthesis (40). Cell extracts were assayed using 200 ng of total protein, which gives a robust and specific signal well within in the quantifiable linear range and discriminates for small changes in the activity level (Fig. 2). Because we used VA-13 cells, there was no background of active RNP in the untransfected cell extract (Fig. 1A, Mock). As expected, robust assembly of active RNP was accomplished by coexpression of hTERT and hTR (Fig. 1A, set 1). In contrast, across a broad range of cell and extract mixing conditions including different times and temperatures, no active telomerase was assembled by mixing hTERT and hTR cell extracts after subunit synthesis (Fig. 1A, sets 2–4).
FIGURE 1.
Cross-linking preserves cellular TCAB1-hTR RNPs and prevents new RNP formation in cell extract. A, cell extract mixing using VA-13 cells expressing combinations of exogenous hTR and ZZF-hTERT. Mixing was performed at 4 and 22 °C for 0.5 or 2 h. Telomerase activity was detected by QTRAP. Values were normalized to VA-13 coexpressing hTR and ZZF-hTERT (n = 3). Note that mature hTR migrates as a doublet under the gel conditions used for Northern blot detection. The U6 snRNA is a control to demonstrate comparable amounts of input extract. B, cell extract mixing using 293T cells expressing combinations of exogenous hTR and F-TCAB1. hTR bound to F-TCAB1 was detected by FLAG IP and Northern blot of hTR. U6 snRNA and tubulin are controls to demonstrate comparable amounts of input extract and also IP specificity. RC, RNA precipitation recovery control. C, a similar experiment was carried out as described in B, except cells were formaldehyde cross-linked prior to cell lysis. hTR bound to F-TCAB1 was detected by FLAG IP followed by RT-qPCR. Input and F-TCAB1-bound hTR levels were each normalized to input RNA polymerase II mRNA and set relative to the triple coexpression sample (n = 3). All lanes were from the same blot; a gap indicates removal of extraneous samples.
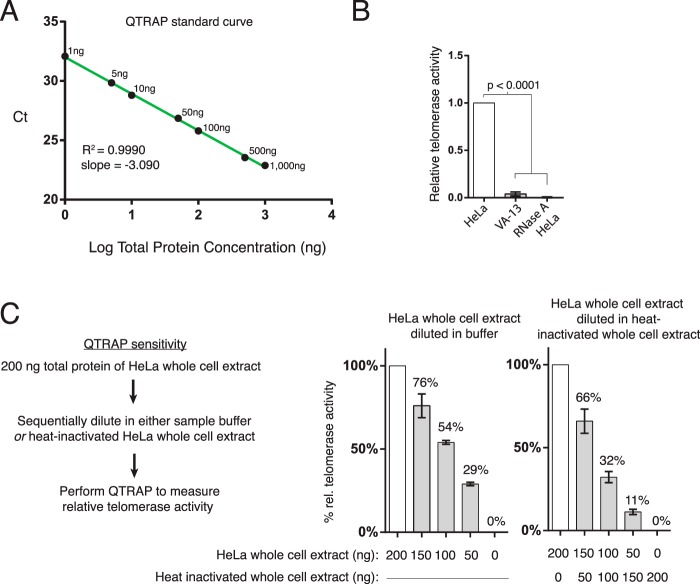
FIGURE 2.
Characterization of telomerase activity using QTRAP with HeLa cell extract. A, QTRAP standard curve using total protein titration of HeLa cell extract (n = 3). B and C, readout of QTRAP measurements with HeLa, VA-13, or RNase-treated HeLa cell extract (n = 6) and with sequentially diluted HeLa cell extract (n = 3).
We next evaluated native extract assembly of hTR and TCAB1. We transfected VA-13 (data not shown) or 293T (Fig. 1B) cells to express hTR and/or F-TCAB1 separately or together. We assayed protein-RNA interaction by FLAG antibody IP of F-TCAB1 followed by Northern blot for hTR. Maximal interaction was observed when the two subunits were coexpressed. TCAB1 IP of co-overexpressed hTR was much greater than TCAB1 IP of endogenous hTR (Fig. 1B, sets 2 and 4). Importantly, TCAB1 IP of hTR increased if extracts from cells overexpressing TCAB1 or hTR alone were mixed after cell lysis (Fig. 1B, sets 2 and 3). Therefore, unlike the hTERT-hTR interaction, the TCAB1-hTR interaction can occur in cell extract as well as in intact cells.
The binding of TCAB1 to hTR in lysed cell extract obscures the determination of RNP assembled in vivo. We therefore developed an in vivo cross-linking approach to detect biologically assembled RNP. We combined formaldehyde cross-linking, to capture snapshots of the cellular milieu, with hTR quantification by RT-qPCR, because cross-linked RNA detection required more sensitivity than provided by Northern blot hybridization. We designed RT-qPCR primers for hTR at the template/pseudoknot region and established their specificity for detecting hTR (Fig. 3, A and B). With the goal of reliably quantifying hTR-bound proteins across the cell cycle, we determined the best reference gene for relative quantification (Fig. 3C) based on expression constancy throughout the cell cycle (39). Using transient transfection of 293T cells to overexpress the subunits, we verified that formaldehyde cross-linking detected hTR interaction with proteins known to bind hTR directly such as hTERT and TCAB1 (see below). We also detected hTR cross-linking to dyskerin but not to the shelterin proteins TPP1 and TIN2 (data not shown).
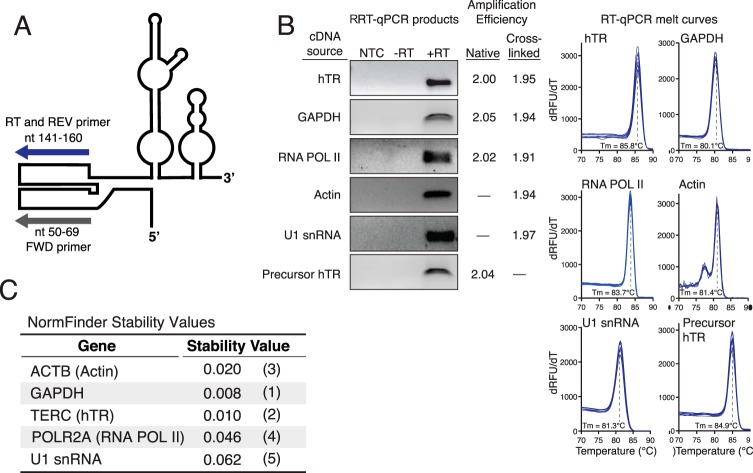
FIGURE 3.
Characterization of RT-PCR products for hTR and reference genes. A, schematic of hTR and RT-qPCR primers used for hTR cDNA generation and PCR amplification. B, RT-qPCR of hTR and normalization standards generates single products (+RT lane) and homogenous melt curves (n = 8) shown for RNA from native cell extract. PCR amplification efficiencies were measured using the LinReg program for samples from the relevant cell extract without or with in vivo cross-linking. No template control (NTC) samples lacked RNA template, and −RT lanes lacked reverse transcriptase. C, potential reference genes for cell cycle RT-qPCR measurements were compared using the NormFinder program. NormFinder stability values and rankings are shown in parentheses. GAPDH mRNA and hTR levels were the most consistent over the cell cycle.
We repeated the cell extract-mixing experiments using in vivo cross-linking and denaturing rather than native binding conditions. TCAB1 interaction with hTR was quantified using RT-qPCR and normalizing the bound to input hTR levels in each sample. TCAB1-hTR association was detected when the subunits were coexpressed by transfection of VA-13 cells, with or without coexpression of hTERT (Fig. 1C, sets 1–3). Unlike the case for native cell extracts, the mixing of extracts from cross-linked cells transfected to express only TCAB1 or hTR gave no detectable TCAB1-hTR interaction (Fig. 1C, set 4). The specificity of cross-linking and IP was confirmed by RT-qPCR for GAPDH mRNA and U1 snRNA, neither of which was detectable in association with F-TCAB1. These assays established in vivo cross-linking as a method of quantifying the biological assembly of hTR with TCAB1. Furthermore, the findings demonstrate that hTERT is not required for TCAB1-hTR interaction in vivo.
Endogenous Telomerase Subunit and Enzyme Catalytic Activity Levels Are Unchanged across the Cell Cycle
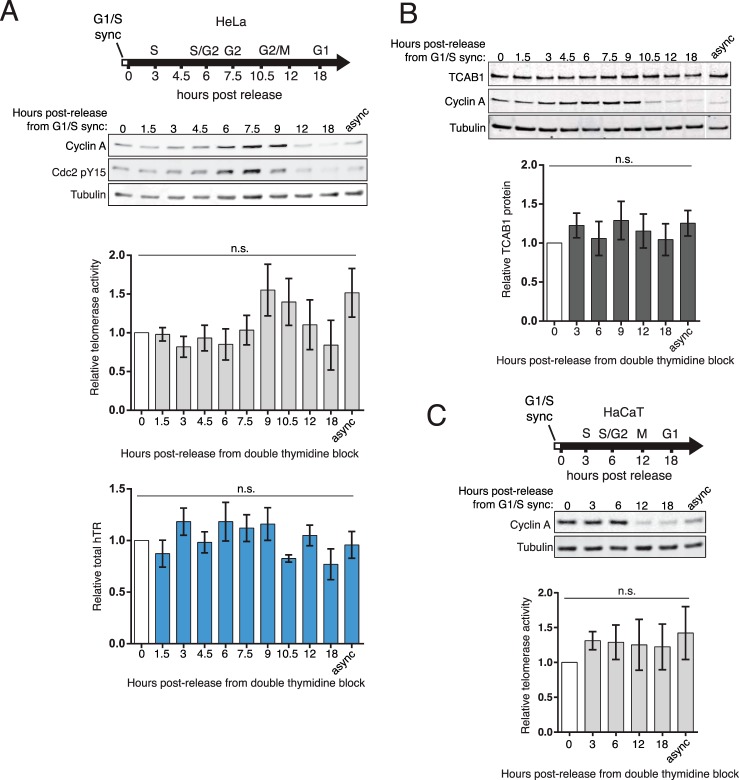
To begin a coordinated study of telomerase holoenzyme assembly dynamics, we first established total hTR level and hTERT-hTR RNP catalytic activity across a synchronized HeLa cell cycle. Although some findings differ (29, 30), several previous reports describe no substantial change, across the cell cycle, of hTR level (15, 43) or telomerase activity in cell extracts (28, 36). The endogenous hTERT protein level has not been possible to assay for cell cycle regulation because of low abundance. HeLa cells were synchronized at G1/S phase by double thymidine block, released to growth, and sampled with high temporal resolution of cell cycle progression. Cell cycle progression was followed by immunoblot for cyclin A (Fig. 4A, top). Cyclin A is an ideal marker for cell cycle progression because its expression steadily increases through S phase, peaks in G2, decreases as cells complete M, and remains low in G1 (44). For HeLa cells synchronized at the G1/S border, the cyclin A level peaked around 8 h post-release, indicating G2 (Fig. 4A, top). A dramatic decrease in the cyclin A level occurred around 12 h post-release from G1/S as cells completed M phase and entered G1. To verify that most cells recovered from the block to cell cycle progression and continued through S phase, we confirmed the loss of Cdc2 phosphorylation on tyrosine 15 (Fig. 4A).
FIGURE 4.
Telomerase activity, hTR, and TCAB1 levels do not change over the cell cycle. A, HeLa cells were released from a double thymidine induced G1/S block, and cells were harvested at time points as the cell cycle progressed. Telomerase activity was assayed by QTRAP, with values set relative to t = 0 at the G1/S block (n = 4). Total hTR was measured by RT-qPCR. Values were normalized to GAPDH mRNA and set relative to t = 0 (n = 3). B, total endogenous TCAB1 protein from HeLa cells synchronized by double thymidine block was measured by immunoblot and quantified relative to tubulin protein (n = 5). All lanes were from the same blot; a gap indicates removal of extraneous samples. C, HaCaT cells were synchronized at G1/S by double thymidine block. Telomerase activity was measured by QTRAP with values set relative to the t = 0 time point (n = 3). async, asynchronous cell cultures. n.s., not significant.
We used QTRAP to assay telomerase activity and RT-qPCR to assay total hTR. Consistent with previous studies, telomerase catalytic activity in extract and total hTR level were unchanged throughout the cell cycle as assayed here by time points taken every 1.5 or 2 h (Fig. 4A). The total TCAB1 protein level also did not fluctuate with cell cycle progression (Fig. 4B). In case the HeLa cells represented a unique case of telomerase cell cycle regulation, we measured telomerase activity across a synchronized cell cycle of the spontaneously immortalized HaCaT human keratinocyte cell line. As found for HeLa cells, the telomerase activity in HaCaT cell extracts remained at a stagnant level over the course of the cell cycle (Fig. 4C).
Increased hTERT or TCAB1 Level Does Not Stimulate Assembly of the Other Subunit with hTR
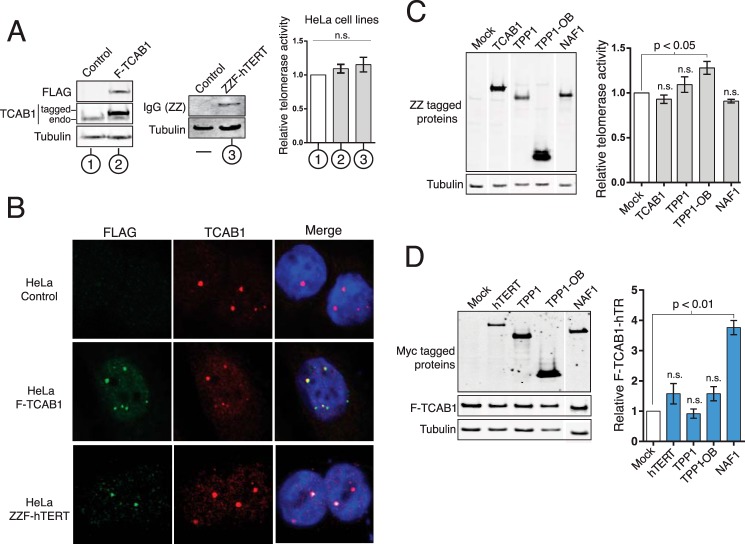
To investigate the cell cycle regulation of hTR physical interaction with hTERT and TCAB1, we generated HeLa cell lines stably expressing FLAG-tagged versions of the proteins. We verified the intended protein expression in HeLa ZZF-hTERT and HeLa F-TCAB1 cell lines by immunoblots (Fig. 5A). F-TCAB1 accumulated to a much higher level than ZZF-hTERT, as F-TCAB1 but not ZZF-hTERT was readily detected by a FLAG antibody immunoblot of whole cell extract. Telomerase activity assayed by QTRAP in the cell lines was similar to that of the parental HeLa cell line (Fig. 5A, right). In all cell lines, immunofluorescence (IF) localization of total TCAB1 showed protein concentration in discrete Cajal body foci colocalized with coilin (Fig. 5B; additional data not shown). In the F-TCAB1 HeLa cells, FLAG antibody IF strongly stained Cajal bodies as expected (Fig. 5B). A few cells expressing ZZF-hTERT also had FLAG antibody staining of a few Cajal body foci (Fig. 5B), but most cells in the population lacked FLAG antibody staining, consistent with the low expression level of ZZF-hTERT and heterogeneous hTERT localization dependent on factors including the cell cycle. None of the negative control parental HeLa cells stained with FLAG antibody (Fig. 5B).
FIGURE 5.
Overexpression of NAF1 affects TCAB1-hTR association. A, HeLa cells stably expressing ZZF-hTERT, F-TCAB1, or empty retroviral vector (Control) were confirmed by immunoblot, and relative telomerase activity was measured by QTRAP (n = 3). B, localization of ZZF-hTERT or F-TCAB1 was detected by IF for FLAG (green) and total TCAB1 (red) compared with DNA (blue). C, extract from ZZF-TERT HeLa cells transiently overexpressing the indicated individual protein was assayed via QTRAP, with values normalized to mock transfection (n = 3). Only some cells stained clearly for ZZF-hTERT, with one example shown. D, telomerase interactors were transiently overexpressed in the F-TCAB1 cell line. Effects on TCAB1-hTR RNP levels were assayed by RT-qPCR following F-TCAB1 IP. Bound hTR was normalized to input hTR and set relative to mock transfection (n = 4). All lanes were from the same blot; a gap indicates removal of extraneous samples.
We first used these cell lines to assay whether hTERT or TCAB1 reciprocally stimulated RNP assembly of the other subunit. Individual proteins were overexpressed in the ZZF-hTERT and F-TCAB1 cell lines by transient transfection, and the impact of subunit overexpression on hTERT-hTR or TCAB1-hTR interaction was determined by QTRAP of native cell extract for the hTERT-hTR interaction and in vivo cross-linking and RT-qPCR for the TCAB1-hTR interaction. In HeLa ZZF-hTERT cells, overexpression of TCAB1 did not stimulate an increase in the assembly of the active hTERT-hTR RNP (Fig. 5C). There was also a marginal, if any, change in hTERT-hTR RNP upon overexpression of the shelterin protein TPP1 or the TPP1-OB, which binds hTERT to assemble and activate telomerase at telomeres. Furthermore, none of several overexpressed H/ACA RNP biogenesis factors had an impact on telomerase activity level, including overexpression of the H/ACA RNP assembly chaperone, NAF1 (Fig. 5C; additional data not shown).
We next assayed for factors affecting the level of hTR association with TCAB1. In HeLa F-TCAB1 cells, overexpression of hTERT did not substantially increase the amount of TCAB1-hTR RNP in vivo (Fig. 5D), paralleling the comparable TCAB1-hTR interaction in transfected VA-13 cells with or without coexpressed hTERT (Fig. 1C). Also, despite sharing a role with TCAB1 in telomerase recruitment to telomeres, neither TPP1 nor the TPP1-OB stimulated TCAB1-hTR interaction (Fig. 5D). Furthermore, overexpression of many H/ACA RNP biogenesis factors did not affect TCAB1-hTR interaction (data not shown). In contrast, overexpression of NAF1 increased TCAB1-hTR association by ∼3.5-fold (Fig. 5D). This could reflect a role for NAF1 in targeting immature H/ACA RNPs to Cajal bodies for the initial steps of RNA modification and RNP subunit exchange. Alternately, NAF1 overexpression could increase TCAB1 availability for hTR interaction indirectly.
The Amount of hTERT-hTR Interaction Is Independent of Cell Cycle Progression
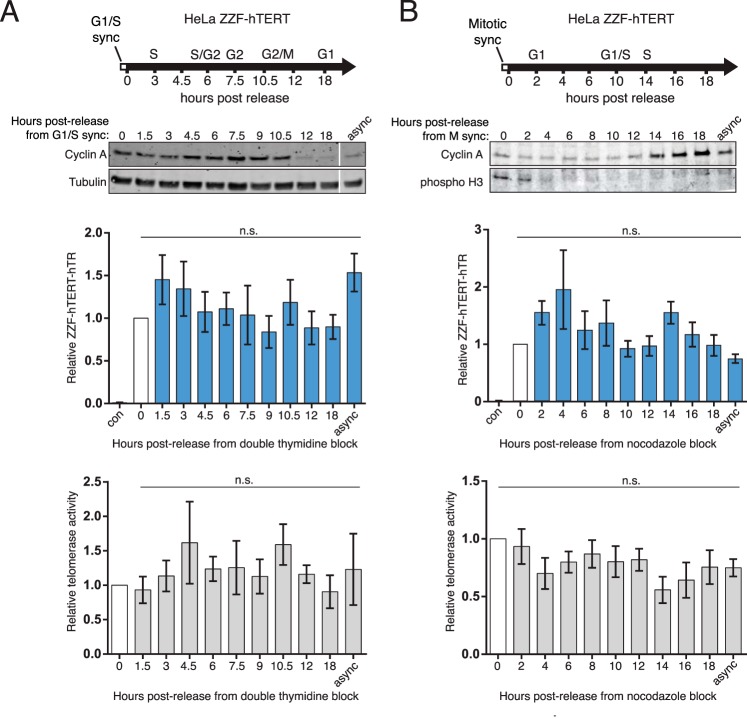
To capture the assembly states of hTR across the cell cycle, we assayed hTERT-hTR and TCAB1-hTR interactions by cross-linking. The ZZF-hTERT HeLa cell line was synchronized at G1/S by double thymidine block before being released to cell cycle progression, as monitored by the cyclin A level (Fig. 6A, top). Post-release time points extensively covered S and G2 phases (0–10.5 h post-release) and also M/G1 and G1 (12 and 18 h post-release). We measured input hTR levels, GAPDH mRNA for normalization, and hTR bound to hTERT. The association of hTERT and hTR remained constant over the sampled 18 h of the cell cycle (Fig. 6A, middle). This result is consistent with the lack of change in RNP catalytic activity monitored by QTRAP of native ZZF-hTERT cell extracts (Fig. 6A, bottom), recapitulating the results for the HeLa parental cell line (Fig. 4A).
FIGURE 6.
hTERT-hTR RNP and telomerase activity do not significantly change over the cell cycle. A, HeLa ZZF-hTERT cells were synchronized at G1/S phase by double thymidine block. The amount of hTR cross-linked to ZZF-hTERT was assayed via RT-qPCR. Bound hTR values were normalized to input GAPDH mRNA and set relative to t = 0 (n = 3). Input hTR normalized to input GAPDH was also invariant (not shown). Telomerase activity was measured by QTRAP, with values set relative to t = 0 (n = 3). B, HeLa ZZF-hTERT cells were synchronized at prometaphase using nocodazole following a single round of thymidine block and release. The amounts of hTERT-hTR RNP and telomerase activity were assayed as described in A (n = 3). The negative control sample (con) was the asynchronously dividing parental HeLa cells. async, asynchronous cell cultures; n.s., not significant.
To complement cell cycle synchronization by release from G1/S block, we synchronized ZZF-hTERT cells at prometaphase by a single round of thymidine block followed by the addition of the spindle checkpoint activator nocodazole. Mitotic block was confirmed by immunoblot for phosphorylation of histone H3 serine 10 (Fig. 6B, top) and the ability of adherent cell cultures to easily lift from the dish bottom. Release from mitotic block into G1 and then S phase was evidenced by the loss of histone H3 serine 10 phosphorylation (Fig. 6B, top). The cyclin A level was minimal in post-release cells completing M phase and began increasing after 12–14 h as cells entered S phase (Fig. 6B, top). Cell cycle progression occurred without change in hTERT-hTR interaction as assayed by cross-linking and RT-qPCR (Fig. 6B, middle) and QTRAP (Fig. 6B, bottom), corroborating the results using synchronization by double thymidine block (Fig. 6A). Of relevance to the TCAB1 results described below, the prolonged mitotic block had no impact on the amount hTERT-hTR RNP (Fig. 6B, compare t = 0 of release from mitotic block with that of the asynchronous cell culture).
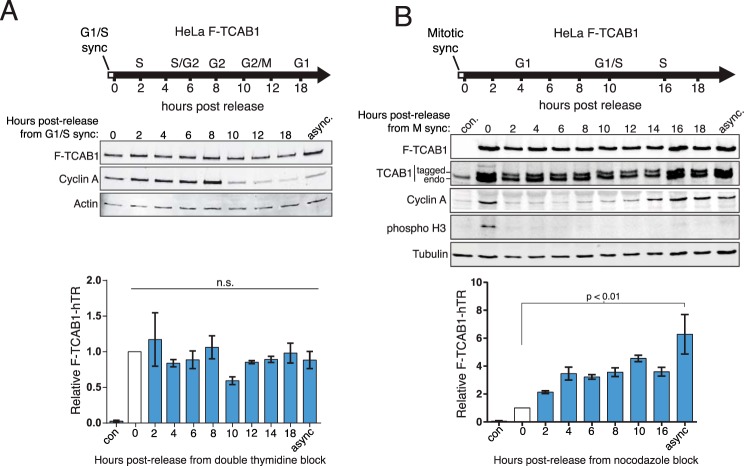
The Amount of TCAB1-hTR Interaction Varies with Cell Cycle Progression
We next interrogated the cell cycle progression changes in TCAB1-hTR interaction using in vivo cross-linking and quantification by RT-qPCR. No change was observed in the amount of hTR bound to TCAB1 in F-TCAB1 cells held at G1/S or released from G1/S block (Fig. 7A). However, in stark contrast to hTERT-hTR interaction, cells in nocodazole-induced mitotic block showed an hTR loss of interaction with TCAB1 (Fig. 7B). TCAB1-hTR interaction was regained with 4–10 h of cell cycle progression post-release (Fig. 7B). Importantly, the TCAB1 level was not affected by the mitotic block (Fig. 7B, top). Therefore, the TCAB1-hTR association was lost, whereas TCAB1 and hTR subunit levels and hTERT-hTR RNP level all remained unaffected.
FIGURE 7.
TCAB1-hTR RNP is altered by mitotic block. A, HeLa F-TCAB1 cells were synchronized at G1/S using double thymidine block. hTR cross-linked to F-TCAB1 was quantified by RT-qPCR, with values normalized to input hTR and set relative to t = 0 (n = 3). B, HeLa F-TCAB1 cells were synchronized at prometaphase using nocodazole after a single round of thymidine block and then released. F-TCAB1-hTR levels were assayed by RT-qPCR with normalization to input hTR and set relative to t = 0 (n = 2). The negative control sample (con) was the asynchronously dividing parental HeLa cells. async, asynchronous cell cultures; n.s., not significant.
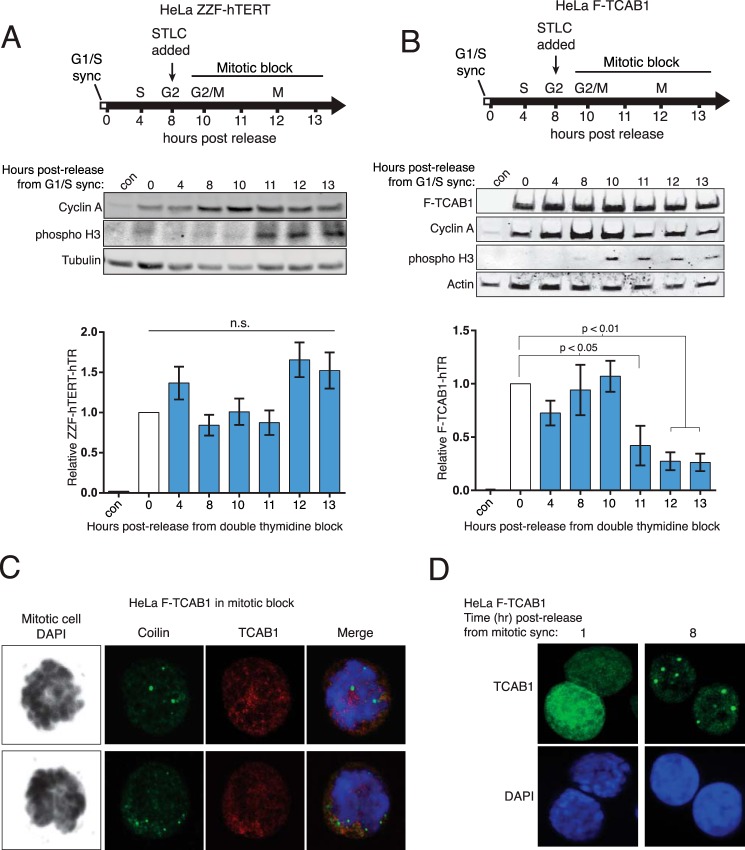
To investigate the M-phase dynamics of telomerase holoenzyme composition, we released G1/S-synchronized cells into mitosis. Cells were synchronized at G1/S using a double thymidine block and released to progress toward S phase, and then at 8 h post-release from double thymidine block, we added the mitotic kinesin inhibitor STLC (Fig. 8, A and B, top). The cell population became enriched in M-phase cells, as evidenced by phosphorylation of histone H3 serine 10 and morphological changes, at 11–13 h post-release from the initial G1/S block (Fig. 8, A and B, top). The amount of hTERT associated with hTR did not decrease as cells approached and entered mitotic block (Fig. 8A). In contrast, TCAB1 association with hTR decreased ∼5-fold in the M phase-enriched cell population (Fig. 8B).
FIGURE 8.
TCAB1 transiently changes nuclear localization and association with hTR. A, HeLa ZZF-hTERT cells were synchronized at G1/S using double thymidine treatment and subsequently released into mitotic block using STLC. ZZF-TCAB1-hTR RNP levels were quantified by RT-qPCR, normalized to input hTR, and set relative to t = 0 (n = 3). B, HeLa F-TCAB1 cells were synchronized at G1/S and led into mitotic block as above. F-TCAB1-hTR RNP was assayed as described in A (n = 3). C, IF of coilin (green), TCAB1 (red), and DNA (black/blue) in HeLa F-TCAB1 cells under STLC induced mitotic block. D, IF of TCAB1 (green) and DNA (blue) in HeLa F-TCAB1 cells released from STLC mitotic block. The negative control sample (con) was the asynchronously dividing parental HeLa cells. n.s., not significant.
The decrease in TCAB1-hTR interaction prompted the question of whether Cajal bodies were still present in cells experiencing STLC-induced mitotic block. IF for coilin revealed that Cajal bodies remained present during mitotic block (Fig. 8C), consistent with many previous visualizations of Cajal bodies in cells throughout the cell cycle. However, during mitotic block, TCAB1 lost colocalization with coilin. Instead of concentrating in Cajal bodies, TCAB1 was diffuse throughout the nucleus (Fig. 8C). This diffuse localization was reversed with restored prominent foci of TCAB1 by 8 h post-release (Fig. 8D). This timing coincides with the restoration of TCAB1-hTR interaction following release from nocodazole-induced mitotic block (Fig. 7B). We concluded that an M-phase regulation of TCAB1 affects its association with both RNA and protein partners.
Holoenzyme Exchanges TCAB1 but Not hTERT
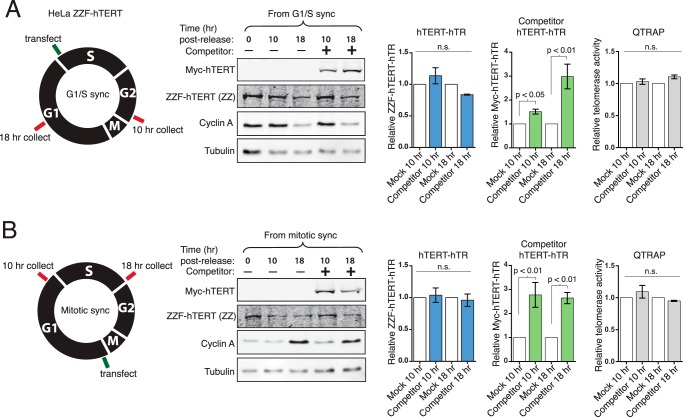
We next investigated holoenzyme subunit exchange. We synchronized the ZZF-hTERT HeLa line at the G1/S border, using a double thymidine block, or at prometaphase, using STLC after a single round of thymidine block. Cells were then released into cell cycle progression for 1 h before being transfected to express a competitor Myc-tagged hTERT followed by continued growth for a total of 10 or 18 h post-release (Fig. 9, A and B, left). Expression of ZZF-hTERT remained constant, and expression of the competitor Myc-hTERT was detected only in cells transfected with this construct versus empty vector (Fig. 9, A and B). We could not estimate a specific fold overexpression of the competitor Myc-hTERT relative to ZZF-hTERT, because only the overexpressed competitor was detectable by immunoblot with hTERT antibody (data not shown). Cyclin A levels indicate progression through the cell cycle following the release from synchronization and transfection. For the G1/S-synchronized cells, cyclin A levels were elevated at 10 h post-release, indicating S/G2, and subsequently decreased 18 h post-release, when cells completed M and entered G1 phase (Fig. 9A). Conversely, in M phase-synchronized cells, cyclin A levels were low at 10 h post-release, indicating that cells were still in G1, and were heightened 18 h post-release as cells were passing through S/G2 (Fig. 9B).
FIGURE 9.
The hTERT-hTR RNP does not undergo subunit exchange. A, HeLa ZZF-hTERT cells were synchronized at G1/S using double thymidine block. Cells were released from block and after 1 h were transfected with empty vector (no competitor, mock) or competitor Myc-hTERT plasmid. Levels of hTR bound to ZZF-hTERT and the competing Myc-hTERT were assayed using RT-qPCR at 10 and 18 h post-release. hTR values were normalized to input hTR and set relative to the corresponding mock transfection sample (n = 3). QTRAP used total cell extract (n = 3). B, HeLa ZZF-hTERT cells were synchronized at prometaphase using STLC following a single round of thymidine block. One hour post-release, cells were transfected with empty vector or competitor Myc-hTERT plasmid. Bound hTR was assayed as above (n = 3). n.s., not significant.
We quantified the amount of hTR bound to ZZF-hTERT and the transiently overexpressed competitor Myc-hTERT using in vivo cross-linking, IP, and RT-qPCR. The bound hTR was normalized to input hTR and set relative to control samples transfected with empty vector. The amount of hTR bound to stably expressed ZZF-hTERT remained unchanged at 10 or 18 h post-release from G1/S phase (Fig. 9A) or prometaphase (Fig. 9B). This suggests that there was little or no exchange of previously assembled ZZF-hTERT with competitor Myc-hTERT. We detected the assembly of competitor Myc-hTERT RNP at all of the time points post-release, but the major increase in Myc-hTERT RNP correlated with cells progressing through G1 either between 10 and 18 h post-release from G1/S phase (Fig. 9A) or within 10 h post-release from M phase (Fig. 9B). There appeared to be no gain in Myc-hTERT RNP between 10 and 18 h post-release from M-phase block (Fig. 9B), consistent with preferential assembly of new hTERT-hTR RNP in G1. Interestingly, the total telomerase activity was not substantially increased by competitor hTERT expression (Fig. 9, A and B, right), suggesting that overexpression of hTERT without hTR overexpression does not increase the amount of active RNP above its normal steady-state level.
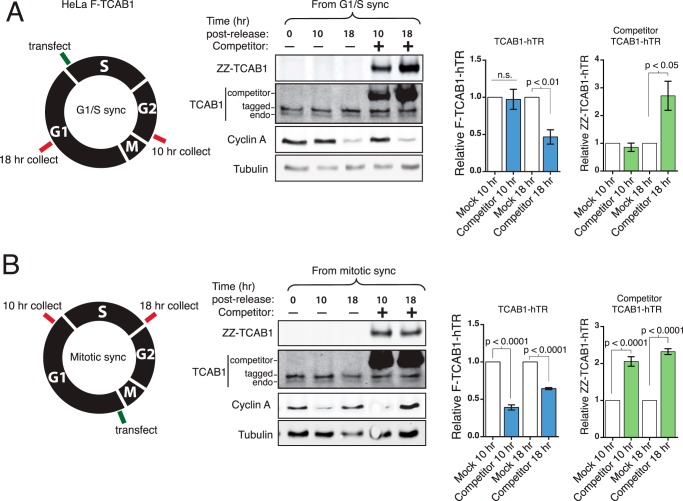
We then tested the dynamics of hTR exchange of TCAB1. Following release of HeLa F-TCAB1 cells from synchronization at G1/S or M phase, cells were transfected to express competitor ZZ-TCAB1 (Fig. 10, A and B, left). New expression of ZZ-TCAB1 and the continued presence of F-TCAB1 were verified by immunoblot (Fig. 10). The amount of hTR bound to each tagged TCAB1 was assayed at 10 and 18 h post-release from synchronization by cross-linking, IP, and RT-qPCR. In cells blocked at G1/S and released into S/G2, there was no redistribution of hTR between the preassembled and competitor TCAB1 by 10 h post-release (Fig. 10A), suggesting that there had been no exchange and limited if any new TCAB1-hTR RNP assembly during this time. However, between 10 and 18 h post-release from G1/S, after cells completed M phase and entered G1, as indicated by the decrease in the cyclin A level, hTR was exchanged between the F-TCAB1 and the competitor ZZ-TCAB1 populations (Fig. 10A). The hTR bound to F-TCAB1 decreased, whereas hTR bound to the competitor ZZ-TCAB1 increased. These results suggest that TCAB1 exchanges preferentially in cells cycling through M phase. Strengthening this conclusion, in cells released from mitotic block immediately prior to ZZ-TCAB1 expression (Fig. 10B), the hTR bound to F-TCAB1 decreased, whereas hTR bound to the competitor ZZ-TCAB1 increased within 10 h post-release into G1. There was minimal change in the distribution of hTR between F-TCAB1 and ZZ-TCAB1 from 10 to 18 h post-release from M-phase block (Fig. 10B), suggesting G1 as the period of maximal TCAB1-hTR RNP assembly.
FIGURE 10.
The TCAB1-hTR RNP dissociates and reassembles with progression through mitosis. A, HeLa F-TCAB1 cells were synchronized at G1/S using double thymidine block. Cells were released from block and after 1 h were transfected with empty vector (no competitor, mock) or competitor ZZ-TCAB1 plasmid. Levels of hTR bound to F-TCAB1 and the competing ZZ-TCAB1 were assayed by RT-qPCR at 10 and 18 h post-release. hTR values were normalized to input hTR and set relative to the corresponding mock transfection sample (n = 6 at 10 h, n = 3 at 18 h; unpaired t test). B, HeLa F-TCAB1 cells were synchronized at prometaphase using STLC following a single round of thymidine block. One hour post-release, cells were transfected with empty vector or competitor ZZ-TCAB1 plasmid. Bound hTR was assayed as above (n = 6 at 10 h, n = 3 at 18 h; unpaired t test). n.s., not significant.
New hTR RNP Biogenesis Has Cell Cycle Coordination
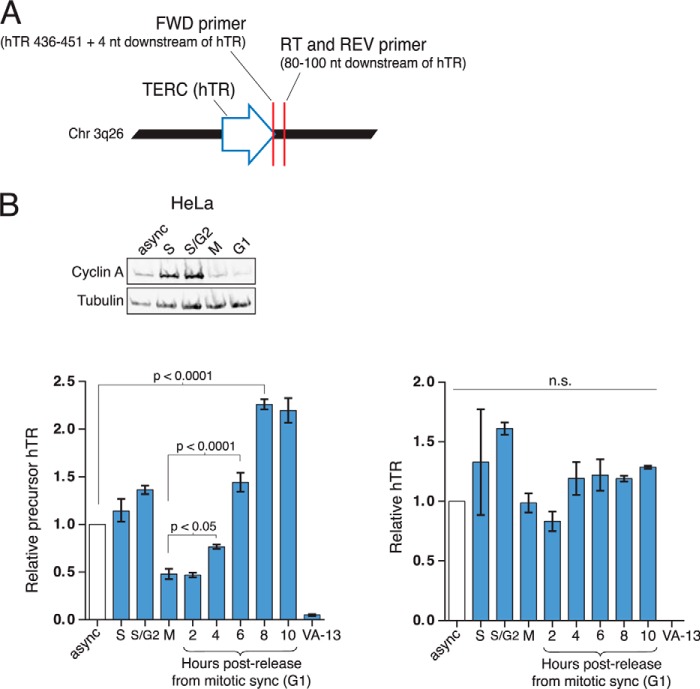
The long half-life of hTR (45) suggests that most cellular hTR RNP is retained through multiple rounds of cell division. However, the initial biogenesis and folding of active RNP could occur preferentially at certain stage(s) of the cell cycle, as does transcription of budding yeast TLC1 (46). To investigate the cell cycle timing of initial hTR RNP biogenesis, we designed RT-qPCR primers to detect 3′-extended hTR precursor (Fig. 11A). Although not every 3′-extended transcript is necessarily an intermediate to functional RNP, 3′-extended hTR provides a reasonable surrogate for mature hTR precursor because transcripts not protected by H/ACA RNP assembly are rapidly degraded (47, 48). Precursor hTR RT-qPCR was benchmarked in parallel to mature hTR RT-qPCR (Fig. 3B) and validated for specificity in parallel using VA-13 cell extract as a negative control (Fig. 11B, left side of each set of assays). Curiously, the hTR precursor level was lowest in M phase and highest in G1 (Fig. 11B, left) in comparison with the constant level of mature hTR across the cell cycle (Fig. 11B, right). This hTR precursor expression profile is consistent with maximal endogenous de novo hTR RNP biogenesis in G1. An integrated model of telomerase holoenzyme assembly coupled to cell cycle progression unifies the observations reported in this work and other relevant studies (Fig. 12, and see “Discussion” below).
FIGURE 11.
New hTERT-hTR RNP may be assembled predominantly in G1. A, RT-qPCR primers were designed to detect the immediate downstream region of the TERC (hTR) gene present only on the hTR precursor before 3′-end processing to mature hTR. B, precursor hTR detection in HeLa cells over the cell cycle. HeLa cells were held in S phase by a single round of thymidine block (S-phase sample) and released. At 3 h post-release (S/G2 sample), STLC was added for mitotic block. At 9 h after release, an M-phase sample was collected. Mitotic block was subsequently released, and the time points were collected after the hours of release indicated. The G1 immunoblot sample is 4 h post-release from mitotic block. Precursor hTR and total hTR levels were measured by RT-qPCR. Values were normalized to GAPDH mRNA and set relative to the sample of cells dividing asynchronously (async) (n = 3). n.s., not significant.
FIGURE 12.
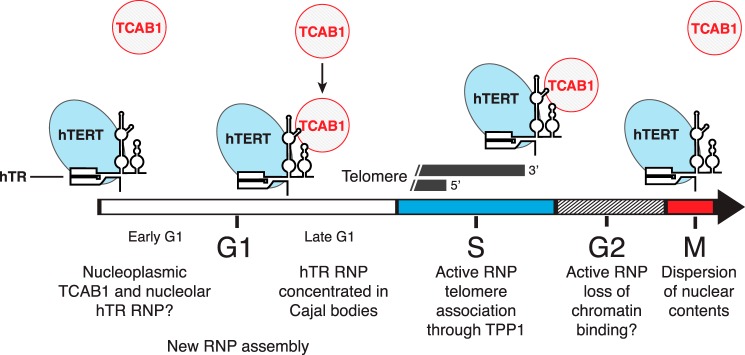
Model for telomerase holoenzyme subunit assembly and disassembly. Although the catalytically active core of telomerase RNP remains intact, TCAB1 dissociates and is reassembled with progression through M and G1 phases, respectively. New hTR RNP biogenesis may peak in G1, accompanied by RNP assembly with hTERT.
Discussion
Like many chromosome replication factors, telomerase action is restricted within the cell cycle. Some of this restriction arises from S/G2-specific remodeling of telomeric chromatin, which can sequester chromosome 3′-ends from access by telomerase or DNA repair proteins (49, 50). How much cell cycle regulation is imposed directly on telomerase remains an open question. Yeast telomerase subunits have been relatively well characterized for changes in expression, interaction, and post-translational modification across the cell cycle, with numerous holoenzyme subunits implicated as having some type of regulation (23, 51, 52). Also, although steady-state levels of TLC1 and telomerase activity in cell extract do not vary with the progression of the cell cycle, TLC1 precursor transcription is maximal in G1 phase (46). These observations provide a context for this work, but the divergence of telomerase and telomere biology between yeast and human cells precludes a direct comparison. In particular, telomerase RNP assembly and nuclear trafficking are phylogenetically variable; vertebrate telomerase is an H/ACA RNP assembled in the nucleus, whereas yeast telomerases are Sm RNPs trafficked between nucleus and cytoplasm (7, 53, 54). A further difference is the nuclear envelope breakdown that occurs prior to chromosome segregation in human but not yeast cells, which obliges hTR RNP relocalization in the nucleus with each G1 phase. Control of telomerase recruitment to telomeres differs as well, as telomerase extends every HeLa cell telomere during every cell cycle, whereas only a small minority of budding yeast telomeres is elongated in each cell cycle (55, 56).
This study establishes a fundamental description of human telomerase holoenzyme subunit levels and interactions across the cell cycle. We developed and combined quantitative assays for hTR, hTERT-hTR, and TCAB1-hTR interactions. The use of in vivo cross-linking captured biological interactions without RNP rearrangement in the cell extract. We cross-compared the results from different cell cycle synchronization methods with a cell cycle progression sampled at high temporal resolution. Our findings establish that total hTR and telomerase catalytic activity remained remarkably consistent across the cell cycle, and total TCAB1 protein level also remained constant with cell cycle progression. Against this context of invariant steady-state levels of the holoenzyme subunits and apparently non-exchanging physical interaction between hTR and hTERT, the cell cycle regulation of hTR association to TCAB1 is dramatic. TCAB1 dissociated from hTR in M-phase cells synchronized by nocodazole block. Dissociation of the TCAB1-hTR interaction also was observed in cells allowed to progress into M phase after release from synchronization at G1/S phase. Because this regulation of holoenzyme composition was captured by in vivo cross-linking prior to cell lysis, it does not have the caveat of sampling dependent on cell extract. A recent study suggested that hTR assembly with hTERT and TCAB1 occurs de novo in each S phase (30), which differs from the conclusions of this work in proposed dynamics of hTERT-hTR assembly but mirrors this work in suggesting cell cycle regulation of TCAB1.
The cell cycle dynamic of TCAB1-hTR interaction can account for endogenous hTR localization changes described by in situ hybridization, which in landmark studies have revealed hTR concentration in Cajal bodies peaking in S phase (24, 25). The progressive concentration of hTR in Cajal bodies over G1 would result from restoration of TCAB1-hTR interaction. Although one consequence of TCAB1-hTR interaction is hTR Cajal body localization, we suggest that the cell cycle regulation of TCAB1-hTR interaction serves a more general role in licensing catalytically active hTERT-hTR RNP for telomere association. This suggestion arises from the observation that Cajal body disruption does not necessarily impede telomerase function; HeLa cells without coilin and thus without Cajal bodies can support telomere recruitment of telomerase and telomere length maintenance (31). As the essential role of TCAB1 appears not to be telomerase localization to Cajal bodies per se, TCAB1 role in telomere maintenance remains to be determined. The significance of M-phase removal of TCAB1 from hTR also remains to be elucidated, although an obvious possibility is that this disassembly ensures the release of telomerase from telomeric chromatin after each round of genome replication. Curiously, of all of the telomerase RNP biogenesis and trafficking factors tested, only NAF1 overexpression led to an increase in TCAB1-hTR interaction. This could reflect a physiological targeting of NAF1-bound RNPs to Cajal bodies, for example to accomplish the early RNP biogenesis step of hTR 5′-trimethylguanosine cap modification (57). However the nonphysiological consequences of NAF1 overexpression are not excluded.
The hTERT association to hTR remains at a consistent level regardless of whether cells are blocked in or progressing though different stages of the cell cycle, as conclusively demonstrated here by in vivo cross-linking. This conclusion was anticipated by the detection of telomerase activity in extracts of cells lysed after harvesting at different stages of the cell cycle (28, 36). That the cellular level of assembled active RNP is not cell cycle-regulated is somewhat surprising given the apparent cell cycle dependence of subunit colocalization; endogenous hTERT and hTR appear to colocalize only at telomeres. Thus, although the level of hTERT-hTR RNP is constant, it is not detected as concentrated at a particular nuclear site until TCAB1 and other interaction partners cluster the RNP at Cajal bodies and telomeric chromatin. Other than at telomeres, hTERT-hTR RNP localization is speculative. Individual subunit localizations detected for endogenous or exogenous expression reflect a combination of coassembled RNP and either hTERT-free hTR or hTR-free hTERT.
Surprisingly, in cells not in G1, a minimal redistribution of hTR between preassembled and competitor hTERT or TCAB1 was detected. These findings suggest that there could be a non-exchangeable subunit content of telomerase RNPs, except as accomplished by cell cycle regulation of TCAB1. It is possible that the substantial steady-state pool of hTERT-free hTR RNP (43) is not competent for active RNP assembly. This suggestion is consistent with poor assembly of active RNP by mixing hTERT and hTR cell extracts or separately folded subunits in vitro (58). It will be interesting to apply the in vivo cross-linking approach in combination with competition assays to examine the cell cycle dynamics of telomerase holoenzyme subunit interactions in budding yeast, which is suggested to include disassembly of the catalytic core (52).
At least in cultured cell lines, because hTR has a long biological half-life of many cell cycles (45), only a limited amount of new active RNP assembly needs to occur. Although speculative, based on the preferential G1 assembly of new hTERT-hTR RNP in the subunit competition experiments and the putative increased G1 level of hTR precursor, we suggest that the biogenesis of active RNP could occur predominantly in G1 phase. Contemporary with this work, hTERT mRNA was reported to increase ∼3-fold in HeLa cell M phase (43). Combining that observation with the findings presented here, our prediction is that hTERT mRNA translation would prestock newly synthesized hTERT prior to new hTR RNP biogenesis. Because TCAB1 is maximally dissociated from hTR in M and early G1 phases, and because hTR without TCAB1 and hTERT is nucleolar (15, 16, 59), hTR synthesized in early G1 may be trafficked to the nucleoli (Fig. 12). This could account for the nucleolar association of ∼7% of hTR in asynchronously cycling HeLa cells, as quantified by subcellular fractionation (53). Curiously, the fraction of hTERT associated to nucleoli also appears maximal in G1 compared with its predominantly nucleoplasmic distribution in late S/G2 phase in IMR90 cells stably expressing a low level of GFP-hTERT (36). Thus, as originally proposed by others many years ago (60), the nucleoli are candidate sites of initial hTERT-hTR interaction.
As hTR RNPs gain TCAB1 association and TCAB1 returns to Cajal bodies with the elapse of G1, hTR RNPs would shift to a more nucleoplasmic and Cajal body distribution (Fig. 12). Because hTERT increases hTR Cajal body concentration (61) and hTERT localization is affected by DNA damage and other signaling pathways (36, 62), there could be many inputs governing what fraction of hTERT-hTR RNP is in Cajal bodies prior to S phase. We suggest that partially dependent and partially independent of association to Cajal bodies per se, TCAB1 assembly with hTR licenses the catalytically active hTERT-hTR RNP for recruitment to telomeres. This licensing allows telomere extension in S phase, coupled to genome replication, and potentially also in G2 phase, when telomeres can be accessible as substrates for DNA repair (50).
Author Contributions
J. M. V. and K. C. conceived and interpreted the experiments and wrote the paper. J. M. V. performed all experiments. Both authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank George Katibah, Michael Rape, Dirk Hockemeyer, Heather Upton, Alec Sexton, and other members of the Collins/Hockemeyer group for thoughtful discussion and/or comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 HL079585 (to K. C.) and T32 GM007232 (to J. M.V.). The authors declare that they have no conflicts of interest with the contents of this article.
- RNP
- ribonucleoprotein
- hTR
- human telomerase RNA
- hTERT
- human telomerase reverse transcriptase
- CAB
- Cajal body localization
- G1
- gap phase 1
- S
- DNA synthesis phase
- G2
- gap phase 2
- M
- mitosis
- IP
- immunopurification
- STLC
- S-trityl-l-cysteine
- RT-qPCR
- reverse transcription and quantitative PCR
- QTRAP
- quantitative telomeric repeat amplification protocol
- IF
- immunofluorescence
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- rcf
- relative centrifugal force
- OB
- oligonucleotide/oligosaccharide fold.
References
- 1. Blackburn E. H., Greider C. W., Szostak J. W. (2006) Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 2. Forsyth N. R., Wright W. E., Shay J. W. (2002) Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation 69, 188–197 [DOI] [PubMed] [Google Scholar]
- 3. Aubert G. (2014) Telomere dynamics and aging. Prog. Mol. Biol. Transl. Sci. 125, 89–111 [DOI] [PubMed] [Google Scholar]
- 4. Shay J. W., Wright W. E. (2011) Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 21, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weinrich S. L., Pruzan R., Ma L., Ouellette M., Tesmer V. M., Holt S. E., Bodnar A. G., Lichtsteiner S., Kim N. W., Trager J. B., Taylor R. D., Carlos R., Andrews W. H., Wright W. E., Shay J. W., Harley C. B., Morin G. B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17, 498–502 [DOI] [PubMed] [Google Scholar]
- 6. Podlevsky J. D., Chen J. J. (2012) It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat. Res. 730, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Egan E. D., Collins K. (2012) Biogenesis of telomerase ribonucleoproteins. RNA 18, 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nandakumar J., Cech T. R. (2013) Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Armanios M., Blackburn E. H. (2012) The telomere syndromes. Nat. Rev. Genet. 13, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitchell J. R., Collins K. (2000) Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol. Cell 6, 361–371 [DOI] [PubMed] [Google Scholar]
- 11. Blackburn E. H., Collins K. (2011) Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 3, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang J., Brown A. F., Wu J., Xue J., Bley C. J., Rand D. P., Wu L., Zhang R., Chen J. J., Lei M. (2014) Structural basis for protein-RNA recognition in telomerase. Nat. Struct. Mol. Biol. 21, 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tycowski K. T., Shu M. D., Kukoyi A., Steitz J. A. (2009) A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell 34, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009) A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jády B. E., Bertrand E., Kiss T. (2004) Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhong F., Savage S. A., Shkreli M., Giri N., Jessop L., Myers T., Chen R., Alter B. P., Artandi S. E. (2011) Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 25, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Batista L. F., Pech M. F., Zhong F. L., Nguyen H. N., Xie K. T., Zaug A. J., Crary S. M., Choi J., Sebastiano V., Cherry A., Giri N., Wernig M., Alter B. P., Cech T. R., Savage S. A., Reijo Pera R. A., Artandi S. E. (2011) Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 474, 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stern J. L., Zyner K. G., Pickett H. A., Cohen S. B., Bryan T. M. (2012) Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol. Cell. Biol. 32, 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao Y., Sfeir A. J., Zou Y., Buseman C. M., Chow T. T., Shay J. W., Wright W. E. (2009) Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chow T. T., Zhao Y., Mak S. S., Shay J. W., Wright W. E. (2012) Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 26, 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moser B. A., Nakamura T. M. (2009) Protection and replication of telomeres in fission yeast. Biochem. Cell Biol. 87, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallardo F., Laterreur N., Wellinger R. J., Chartrand P. (2012) Telomerase caught in the act: united we stand, divided we fall. RNA Biol. 9, 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ribeyre C., Shore D. (2013) Regulation of telomere addition at DNA double-strand breaks. Chromosoma 122, 159–173 [DOI] [PubMed] [Google Scholar]
- 24. Jády B. E., Richard P., Bertrand E., Kiss T. (2006) Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 17, 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tomlinson R. L., Ziegler T. D., Supakorndej T., Terns R. M., Terns M. P. (2006) Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Upton H. E., Hong K., Collins K. (2014) Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol. Cell. Biol. 34, 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li S., Makovets S., Matsuguchi T., Blethrow J. D., Shokat K. M., Blackburn E. H. (2009) Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holt S. E., Aisner D. L., Shay J. W., Wright W. E. (1997) Lack of cell cycle regulation of telomerase activity in human cells. Proc. Natl. Acad. Sci. U.S.A. 94, 10687–10692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu X., Kumar R., Mandal M., Sharma N., Sharma H. W., Dhingra U., Sokoloski J. A., Hsiao R., Narayanan R. (1996) Cell-cycle dependent modulation of telomerase activity in tumor cells. Proc. Natl. Acad. Sci. U.S.A. 93, 5091–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J. H., Lee Y. S., Jeong S. A., Khadka P., Roth J., Chung I. K. (2014) Catalytically active telomerase holoenzyme is assembled in the dense fibrillar component of the nucleolus during S phase. Histochem. Cell Biol. 141, 137–152 [DOI] [PubMed] [Google Scholar]
- 31. Chen Y., Deng Z., Jiang S., Hu Q., Liu H., Songyang Z., Ma W., Chen S., Zhao Y. (2015) Human cells lacking coilin and Cajal bodies are proficient in telomerase assembly, trafficking and telomere maintenance. Nucleic Acids Res. 43, 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong F. L., Batista L. F., Freund A., Pech M. F., Venteicher A. S., Artandi S. E. (2012) TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tomlinson R. L., Li J., Culp B. R., Terns R. M., Terns M. P. (2010) A Cajal body-independent pathway for telomerase trafficking in mice. Exp. Cell Res. 316, 2797–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fu D., Collins K. (2007) Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cristofari G., Adolf E., Reichenbach P., Sikora K., Terns R. M., Terns M. P., Lingner J. (2007) Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell 27, 882–889 [DOI] [PubMed] [Google Scholar]
- 36. Wong J. M., Kusdra L., Collins K. (2002) Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4, 731–736 [DOI] [PubMed] [Google Scholar]
- 37. Mitchell J. R., Wood E., Collins K. (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 [DOI] [PubMed] [Google Scholar]
- 38. Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66 [DOI] [PubMed] [Google Scholar]
- 39. Andersen C. L., Jensen J. L., Ørntoft T. F. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 [DOI] [PubMed] [Google Scholar]
- 40. Wege H., Chui M. S., Le H. T., Tran J. M., Zern M. A. (2003) SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res. 31, E3–E3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu D., Collins K. (2003) Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell 11, 1361–1372 [DOI] [PubMed] [Google Scholar]
- 42. Wenz C., Enenkel B., Amacker M., Kelleher C., Damm K., Lingner J. (2001) Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 20, 3526–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xi L., Cech T. R. (2014) Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 42, 8565–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Donjerkovic D., Scott D. W. (2000) Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 10, 1–16 [DOI] [PubMed] [Google Scholar]
- 45. Yi X., Tesmer V. M., Savre-Train I., Shay J. W., Wright W. E. (1999) Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 19, 3989–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dionne I., Larose S., Dandjinou A. T., Abou Elela S., Wellinger R. J. (2013) Cell cycle-dependent transcription factors control the expression of yeast telomerase RNA. RNA 19, 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Richard P., Kiss A. M., Darzacq X., Kiss T. (2006) Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 26, 2540–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Egan E. D., Collins K. (2012) An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol. Cell. Biol. 32, 2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Lange T. (2010) How shelterin solves the telomere end-protection problem. Cold Spring Harb. Symp. Quant. Biol. 75, 167–177 [DOI] [PubMed] [Google Scholar]
- 50. Cesare A. J., Karlseder J. (2012) A three-state model of telomere control over human proliferative boundaries. Curr. Opin. Cell Biol. 24, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wellinger R. J., Zakian V. A. (2012) Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191, 1073–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tucey T. M., Lundblad V. (2014) Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev. 28, 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitchell J. R., Cheng J., Collins K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seto A. G., Zaug A. J., Sobel S. G., Wolin S. L., Cech T. R. (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401, 177–180 [DOI] [PubMed] [Google Scholar]
- 55. Zhao Y., Abreu E., Kim J., Stadler G., Eskiocak U., Terns M. P., Terns R. M., Shay J. W., Wright W. E. (2011) Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol. Cell 42, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Teixeira M. T., Arneric M., Sperisen P., Lingner J. (2004) Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117, 323–335 [DOI] [PubMed] [Google Scholar]
- 57. Fu D., Collins K. (2006) Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 20, 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu R. A., Collins K. (2014) Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 33, 921–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Theimer C. A., Jády B. E., Chim N., Richard P., Breece K. E., Kiss T., Feigon J. (2007) Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol. Cell 27, 869–881 [DOI] [PubMed] [Google Scholar]
- 60. Etheridge K. T., Banik S. S., Armbruster B. N., Zhu Y., Terns R. M., Terns M. P., Counter C. M. (2002) The nucleolar localization domain of the catalytic subunit of human telomerase. J. Biol. Chem. 277, 24764–24770 [DOI] [PubMed] [Google Scholar]
- 61. Tomlinson R. L., Abreu E. B., Ziegler T., Ly H., Counter C. M., Terns R. M., Terns M. P. (2008) Telomerase reverse transcriptase is required for the localization of telomerase RNA to Cajal bodies and telomeres in human cancer cells. Mol. Biol. Cell 19, 3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Collins K. (2008) Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 129, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]