Background: hsp90β binds to heme-free sGCβ1 subunit to enable heme insertion during maturation, but the mechanism is unclear.
Results: Their interaction involves the hsp90β M domain and the Per-Arnt-Sim (PAS) domain of apo-sGCβ1.
Conclusion: hsp90β may modulate the apo-sGCβ1 through its PAS domain, suggesting a means to aid heme insertion.
Significance: This work expands our knowledge of how hsp90 regulates PAS-containing client proteins like sGC.
Keywords: cyclic GMP (cGMP), fluorescence anisotropy, heat shock protein 90 (hsp90), hydrogen exchange mass spectrometry, protein-protein interaction, signal transduction, H-NOX domain, PAS domain, heme insertion, structural modeling
Abstract
Heat shock protein 90 (hsp90) drives heme insertion into the β1 subunit of soluble guanylate cyclase (sGC) β1, which enables it to associate with a partner sGCα1 subunit and mature into a nitric oxide (NO)-responsive active form. We utilized fluorescence polarization measurements and hydrogen-deuterium exchange mass spectrometry to define molecular interactions between the specific human isoforms hsp90β and apo-sGCβ1. hsp90β and its isolated M domain, but not its isolated N and C domains, bind with low micromolar affinity to a heme-free, truncated version of sGCβ1 (sGCβ1(1–359)-H105F). Surprisingly, hsp90β and its M domain bound to the Per-Arnt-Sim (PAS) domain of apo-sGC-β1(1–359), which lies adjacent to its heme-binding (H-NOX) domain. The interaction specifically involved solvent-exposed regions in the hsp90β M domain that are largely distinct from sites utilized by other hsp90 clients. The interaction strongly protected two regions of the sGCβ1 PAS domain and caused local structural relaxation in other regions, including a PAS dimerization interface and a segment in the H-NOX domain. Our results suggest a means by which the hsp90β interaction could prevent apo-sGCβ1 from associating with its partner sGCα1 subunit while enabling structural changes to assist heme insertion into the H-NOX domain. This mechanism would parallel that in other clients like the aryl hydrocarbon receptor and HIF1α, which also interact with hsp90 through their PAS domains to control protein partner and small ligand binding interactions.
Introduction
Heat shock protein 90 (hsp90)4 is an ATP-dependent molecular chaperone that plays important roles in cell proliferation, signaling, and survival (1, 2). hsp90 stabilizes or activates numerous protein clients by binding to their unfolded, partly folded, or native-like forms (3, 4). hsp90 functions as a homodimer, with each monomer consisting of a C-terminal dimerization domain (C), a middle domain (M) that is often involved in client binding, and an N-terminal domain (N) that contains the majority of the ATP-binding site and that participates in some client-binding interactions (5, 6). The hsp90 dimer functions in client maturation by undergoing large conformational rearrangements that are governed by ATP site occupancy and hydrolysis, co-chaperone proteins, and post-translational modifications (7–9). The molecular details of hsp90-client protein interactions, and their associated structural and functional impacts, are under active investigation (6, 10–14).
Proteins that bind iron protoporphyrin IX (hemeproteins) are ubiquitous in nature and perform important roles in metabolism, energy transduction, communication, and host defense (15, 16). hsp90 interacts with a number of hemeproteins, including the nitric-oxide synthases (17–19), eIF2a kinase (20), HAP1 (21), NADPH oxidases (22), and the intracellular signaling enzyme soluble guanylate cyclase (sGC) (23). Association with hsp90 has been shown to increase sGC lifetime and enzymatic activity in cells and tissues (23, 24), but the mechanisms involved are unclear. In its active form, sGC is a heterodimer composed of similar α and β subunits, but only the β subunit contains a binding site for heme (25, 26). In recent studies, we found that hsp90 specifically associates with the heme-free form of the sGCβ1 subunit (apo-sGCβ1) in cells (27, 28), consistent with hsp90's recognized role of stabilizing immature clients. Moreover, we found that hsp90 promotes heme insertion into apo-sGCβ1 in an ATP-dependent process. hsp90 subsequently dissociates from heme-loaded sGCβ1, which concurrently associates with an sGCα1 partner subunit to form the mature sGC heterodimer (27, 28).
Only heme-containing sGCβ1 can form heterodimers that bind and respond to nitric oxide (NO), the native sGC-activating molecule in signaling cascades (29). Thus, elucidation of the molecular details of the hsp90-sGCβ1 interaction is needed to understand hsp90's role in sGC maturation. To address this, we utilized fluorescence polarization measurements and hydrogen-deuterium exchange mass spectrometry (HxDMS) to characterize the interaction between hsp90β1 and apo-sGCβ1, representative isoforms of the two proteins. We determined that hsp90β interacts not with the heme-binding H-NOX domain of heme-free sGCβ1 but rather with the adjacent Per-Arnt-Sim (PAS) domain, also termed the H-NOXA (H-NOX-associated) domain. We found that the M domain of hsp90β primarily mediates this interaction; moreover, we identified specific regions on the hsp90β M domain and sGCβ1 PAS domain that participate in or respond to the interaction. Our findings (i) provide unifying concepts regarding hsp90 interactions with PAS domain client proteins and (ii) suggest a possible mechanism whereby hsp90β anchors apo-sGCβ1 through its PAS domain for heme insertion during client maturation.
Experimental Procedures
hsp90β Plasmid Constructs, Protein Expression, and Purification
Dr. Tomasz Religa (Case Western Reserve University) kindly provided pet15MHL plasmids containing the coding sequences of full-length human hsp90β or the hsp90β N domain (residues 1–227) and a pet28MHL plasmid encoding the hsp90β M domain (residues 284–543). These constructs add a TEV-cleavable hexa-histidine tag at the N or C termini of the respective constructs. We cloned the human hsp90β C domain (residues 621–724) into pGEX-4T1 plasmid to generate a construct with an N-terminal thrombin-cleavable GST tag. All constructs were sequenced for verification.
Plasmids coding for full-length hsp90β, the N domain, or the M domain were transformed into Escherichia coli BL21-codon+. Overnight cultures grown at 37 °C in Luria Broth (LB) were used to inoculate 2 liters of Terrific broth. Expression cultures were grown at 37 °C until they reached an A600 of 0.8–1.0, induced with 1 mm isopropyl β-d-1-thiogalactopyranoside, and shaken for 20 h at 25 °C. Cells were harvested by centrifugation, and all subsequent steps were carried out at 4 °C. Cell pellets were suspended in 50 ml of phosphate-buffered saline (PBS, 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4) supplemented with phenylmethanesulfonyl fluoride (PMSF), protease inhibitor mixture (aprotinin, leupeptin, pepstatin, and 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride), and lysozyme and disrupted by sonication on ice. Supernatants were clarified by centrifugation and applied onto a gravity column of 20 ml of Ni-NTA-agarose resin (Qiagen) pre-equilibrated with PBS. The resin was washed with 100 ml of PBS followed by 100 ml of PBS + 80 mm imidazole. Bound protein was eluted with 75 ml of PBS + 200 mm imidazole and concentrated by centrifugal ultrafiltration. Concentrated protein was dialyzed into PBS + 1 mm DTT, flash-frozen, and stored at −70 °C.
The hsp90β C domain was expressed in E. coli BL21. Cells were grown, induced, and processed as described above. Clarified cell lysate was bound to 10–40 ml of 50% glutathione-Sepharose-4B in PBS. The resin was washed five times in 30 ml of PBS per wash, and bound protein was eluted with 20 ml of fresh 10 mm glutathione, 10 mm Tris, pH 8.0. This solution was concentrated by ultrafiltration and dialyzed twice in 1 liter of PBS + 1 mm DTT. GST-free hsp90β C domain was prepared by incubating the sample with 10 units of thrombin/mg of protein for several hours at room temperature, followed by tumbling for 30 min with 10–40 ml of 50% GST-Sepharose-4B resin, centrifugation (250 rpm for 5 min), and collection of the supernatant. The resin was washed three times with 20 ml of PBS to recover the remaining C domain, and this solution was combined with the supernatant for concentration, flash-freezing, and storage at −70 °C.
sGCβ1 Plasmid Constructs, Protein Expression, and Purification
sGCβ1 constructs were obtained from pET21 expression plasmids containing bovine sGCβ1(1–197) (the heme-binding H-NOX domain) or sGCβ1(1–359) (H-NOX domain, PAS domain, and N-terminal portion of the coiled-coil domain), which were described previously (30). A H105F substitution was introduced into sGCβ1(1–359) using the QuikChange mutagenesis kit (Agilent Technologies). A construct of the sGCβ1 PAS domain (residues 202–342) plusa C-terminal TEV cleavage site was generated from the sGCβ1(1–359) DNA by PCR and subcloned into a pET20b+ plasmid, adding a C-terminal hexa-histidine affinity tag.
Plasmids coding for sGCβ1(1–197), sGCβ1(1–359), sGCβ1(1–359)-H105F, or sGCβ1-PAS were transformed into BL21Codon+ cells. Overnight cultures were grown at 37 °C in Luria Broth and used to inoculate 2-liter expression cultures grown in Terrific broth. Expression cultures were grown until the A600 = 2.0–3.0 and subsequently cold-shocked for 1 h in an ice bath. Protein expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside for 20 h at 25 °C at 250 rpm. Cells were pelleted by centrifugation and resuspended in 20 ml of buffer A (20 mm Tris, 50 mm NaCl, 10% glycerol, pH 7.5) supplemented with protease inhibitors, benzonase, and lysozyme. Resuspended cells were lysed by sonication and clarified by centrifugation.
The sGCβ1(1–197), sGCβ1(1–359), and sGCβ1(1–359)-H105F proteins were purified by anion exchange chromatography on a 25-ml Q-Sepharose column pre-equilibrated with buffer A. The column was washed with 3 column volumes of buffer A, and the sGCβ1 protein was eluted with a linear gradient of 50 mm to 1 m NaCl in buffer A. Fractions containing sGCβ1 protein were pooled, concentrated to 2 ml, and dialyzed into buffer A. The Q-Sepharose purification was then repeated using buffers adjusted to pH 8.5, and fractions containing the sGCβ1 proteins were pooled, concentrated, flash-frozen, and stored at −70 °C.
The sGCβ1-PAS construct was purified on a gravity column containing 20 ml of Ni-NTA-agarose resin (Qiagen) pre-equilibrated with buffer A. The resin was washed with 100 ml of buffer A and 100 ml of buffer A + 40 mm imidazole. Bound protein was eluted with 50 ml of buffer A + 160 mm imidazole and concentrated to 2 ml by centrifugal ultrafiltration. Concentrated protein was dialyzed into buffer A + 1 mm DTT, and the His6 tag was removed by incubation with 1000 units of TEV protease (Invitrogen) overnight at 4 °C. To remove uncut protein, the TEV digest was redialyzed twice into buffer A (without DTT) and applied to a gravity column containing 5 ml of Ni-NTA-agarose resin pre-equilibrated with buffer A. The flow-through was retained, and 20 ml of buffer A + 10 mm imidazole was used to elute residual sGCβ1-PAS from the beads. Residual sGCβ1-PAS was combined with the original flow-through, concentrated to 2 ml, dialyzed against buffer A + 1 mm DTT, and flash-frozen and stored at −70 °C. Protein molecular weight estimations were performed using a gel filtration column and protein standards.
Fluorescence Polarization Measurements
For labeling, selected proteins were dialyzed into 0.1 m sodium bicarbonate, pH 9.3. FITC, dissolved in the bicarbonate buffer, was added to the proteins at a ratio of 20 μg of FITC to 1 mg of protein. After incubation for 1 h at room temperature in the dark, the reaction mixture was passed through a PD-10 column to capture excess FITC and to exchange the protein sample into PBS buffer. FITC labeling efficiency was calculated based on 495 nm based on a FITC extinction coefficient of 63,000 m−1 cm−1 at 495 nm.
FITC-labeled proteins were diluted in PBS in a fluorescence cuvette to a final concentration of 0.5 or 1 μm, and unlabeled prospective binding partners were titrated into the cuvette. The excitation wavelength was 495 nm, and the emission was monitored at 520 nm. The fluorescence intensity was recorded multiple times after each addition, with polarization filters set in the parallel and perpendicular positions, and then used to calculate the average polarized parallel intensity (I1) and average polarized perpendicular intensity (I2). The fluorescence polarization at each titration point was calculated by the formula: (I1 − I2)/(I1 + I2). In addition, selected titrations were performed in triplicate using 0.2 μm FITC-labeled sGCβ1(1–359)-H105F or sGCβ1-PAS in 100-μl volumes in a 96-well plate, using a Flexstation 3 multimode microplate reader (Molecular Devices). The resulting binding curves were fit in Origin Lab 8.0 (Origin Lab Corp.) to calculate binding affinities. In some cases, the Flexstation titrations also included 1 mm ATP, ADP, or AMP-PNP. All titration experiments were repeated independently 2 or 3 times.
Peptide Mapping by HPLC-Tandem Mass Spectrometry
hsp90β M domain (8 μg) or sGCβ1(1–359)-H105F (10 μg) in 100 μl of 40 mm EPPS, pH 7.6, 150 mm NaCl was mixed with 10 μl of 1 mm HCl to lower the pH to 2.4. Samples were digested by incubation with 10 μg of porcine pepsin (1 mg/ml), dissolved in 0.05% TFA, for 5 min on ice. The final hsp90β M and sGCβ1(1–359)-H105F concentrations were 2 μm. The digested sample was injected into a micropeptide trap (Michrom Bioresources) connected to a C18 HPLC column (5 cm × 1 mm, Alltech) coupled to a ThermoElectron LTQ ion-trap mass spectrometer (ThermoElectron). Peptic fragments were eluted using a gradient of acetonitrile (Burdick and Jackson) at a flow rate of 50 μl/min for a tandem mass spectrometry experiment to sequence each of the peptic fragments. Peptic fragments were identified using the search algorithm SEQUEST (ThermoElectron) and by manual inspection.
Hydrogen/Deuterium Exchange Reactions of Individual Proteins and Proteins in Complex
Purified hsp90β M domain and sGCβ1(1–359)-H105F were utilized singly or were incubated together (20 μm each) for 45 min at room temperature in 40 mm EPPS buffer, pH 7.6, 150 mm NaCl to form a complex. Each protein sample was rapidly diluted 10-fold with 40 mm EPPS buffer, pD 7.6, 150 mm NaCl dissolved in D2O (Cambridge Isotope Laboratories) at 25 °C to initiate deuterium exchange. Reactions were quenched at different time points by addition of 1 mm HCl to lower the pH to 2.4, followed by quick-freezing in a dry ice-ethanol bath. Samples were stored at −80 °C before further analysis.
Isotope Analysis by HPLC-Electrospray Ionization Mass Spectrometry
Frozen hydrogen/deuterium exchange samples were quickly thawed and digested with 10 μg of pepsin (1 mg/ml, dissolved in 0.05% TFA, and H2O) for 5 min on ice immediately followed by injection into a micropeptide trap connected to a C18 HPLC column (5 cm ×1 mm, Alltech) coupled to a ThermoElectron LTQ ion-trap mass spectrometer. Peptic peptides were eluted in 15 min using a gradient of 5–45% acetonitrile at a flow rate of 50 μl/min. The micropeptide trap and C18 HPLC column were immersed in a water/ethanol bath at 0.2 °C during the entire run to minimize back exchange. Peaks were identified manually, and centroid masses were calculated using the software MagTran (Amgen, Thousand Oaks, CA). A fully deuterated reference sample for each protein was used to correct for deuterium loss during sample handling. Deuterium incorporation for each peptide fragment was calculated and is presented as normalized percentage of deuterium uptake (% deuteration or % deuterium uptake) using Equation 1,
where mt is the centroid mass of the deuterated peptic fragment at incubation time t, and m0% and m100% are the centroid masses of the unlabeled and fully deuterated peptic fragments, respectively. All deuterium uptake experiments were performed in triplicate for individual proteins and their complexes.
Structural Modeling
Homology models of sGCβ1 domains and hsp90β (full-length or domains) were generated using I-Tasser (31), either without explicitly specified templates or using homologous structures/models from the protein model portal (32) as inputs. The best I-Tasser outputs were manually adjusted in Coot (33) and refined using ModRefiner (34). We generated all molecular graphics using PyMOL 1.7 (Schrödinger, LLC).
The human sGCβ1 H-NOX model was generated based on the equivalent domain of a Nostoc sp. PC7120 H-NOX protein (35, 36). The sGCβ1 PAS domain was modeled as a monomer and dimer using PAS domains from Manduca sexta sGCα and signal transduction histidine kinase of Nostoc punctiforme sp. PCC73102 as templates (37, 38). We modeled a short segment of the coiled-coil domain of human sGCβ1 based on the nearly identical equivalent in rat sGCβ1 (39). The crystal structures of the human hsp90β N and M domains were solved previously ((40) and Protein Data Bank code 3PRY). We generated a model of the hsp90β C domain based on the crystal structure of the M and C domains of human hsp90α (41). The I-tasser Z-scores of all domain models were greater than −0.25, indicating a high likelihood that the domains had correct folds and correct overall tertiary structure. We further estimated the accuracy of the individual domain models using ModEval (42). For each model, the predicted root mean square deviation from the true structure was less than 3 Å, whereas the estimated Native Overlap 3.5 score (the fraction of model Cα atoms within 3.5 Å from their true positions) was above 85%. These scores suggest that the models have correct overall folds and realistically represent the individual domain structures.
A multidomain model of sGCβ1(1–359) was generated using a previously described model of truncated M. sexta sGCα that encompasses the H-NOX, PAS, and coiled-coil regions (43). The M. sexta model was based on restraints derived from small angle x-ray scattering and chemical cross-linking experiments, and our current model satisfies the equivalent distance restraints. We superimposed individual human sGCβ1 domain models onto their counterparts in the M. sexta model, added missing or linker residues using Coot (33), and refined the structure using FG-MD (44) and ModRefiner (34). We generated models of the full-length hsp90β dimer in a similar manner. Experimental structures or models of the hsp90β N, M, or C domains were superimposed onto their counterparts in a closed structure of the Saccharomyces cerevisiae hsp90 dimer (45) or onto an open dimer of the E. coli hsp90 orthologue HtpG (46).
To generate models of sGCβ1(1–359) complexes with hsp90β, we docked the former onto one full-length monomer of each hsp90β dimer model using HADDOCK (Haddock 2.2 software web portal) (47). We designated residues in regions of sGCβ1(1–359) and hsp90β M domain that exhibited substantial decreases in H/D exchange upon complexation and had solvent-exposed side chains, as “active” residues in the Haddock inputs (48, 49). These included sGCβ1 residues 266, 269–271, 273–275, 333, 338–340, 343–345, and 348–349 as well as hsp90β residues 371–373, 399, 402–403, 405–406, 409–410, 413–414, 473–475, 477–478, 501, 504–505, and 507–508. “Passive” residues were automatically defined by the software. A random fraction of 50% of ambiguous restraints was allowed to be excluded during docking. We also allowed the sGCβ1 HNOX-PAS linker segment (residues 182–207) and the PAS-coiled-coil linker (residues 324–351) to be fully flexible during docking.
Results
Properties of the sGCβ1 and hsp90β Proteins
Fig. 1 illustrates the protein constructs we used in the study. The sGCβ1(1–359) construct is similar to others that have been extensively studied (26) and is composed of the N-terminal regulatory domain (H-NOX or heme domain), the PAS domain, and an N-terminal portion of the coiled-coil domain. This construct lacks most of the coiled-coil domain and the entire catalytic domain. Unlike the full-length sGCβ1 protein, however, it can be expressed well in E. coli (26). The sGCβ1(1–197) and sGCβ1(202–342) (sGCβ1-PAS) constructs, respectively, correspond to the individual H-NOX and PAS domains, including inter-domain linker sequences. All proteins were expressed in E. coli. The sGCβ1(1–359) purified as a minor fraction of heme-enriched and a major fraction of heme-free (apo) protein; the two forms were separated using an anion exchange column. The sGCβ1(1–359)-H105F variant was at least 95% heme-free and was predominantly dimeric with some higher oligomers as purified (data not shown), confirming that the Phe substitution resulted in the intended heme-binding defect (25, 26). The sGCβ1-PAS protein purified as a mixture of apparent dimeric and trimeric forms. The sGCβ1(1–197) protein has also been extensively studied (25). In our hands, this construct was predominantly heme-free after purification even without the H105F mutation (data not shown).
FIGURE 1.
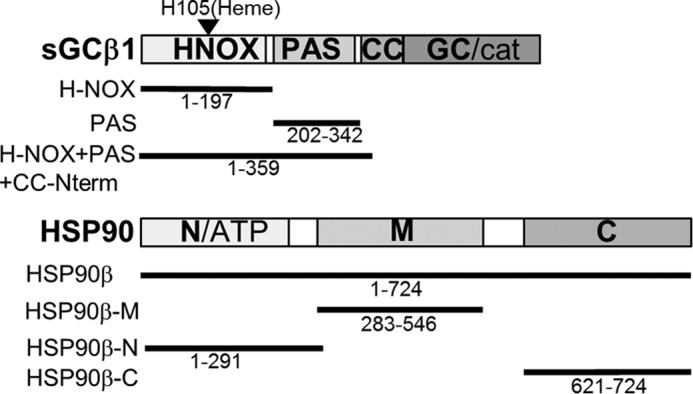
Protein constructs used in the study. The domain arrangements of sGCβ and hsp90β are shown. The solid lines and residue ranges show the corresponding constructs of each protein utilized in our study.
Analysis of Binding Interactions between hsp90β and Apo-sGCβ1
Previous studies that used affinity resins or antibody capture established that hsp90 associates with apo-sGCβ1 in cells, primarily through the hsp90 M domain (23, 24). To understand the interaction in greater detail, we utilized fluorescence polarization measurements on recombinant purified forms sGCβ1 and hsp90β. These measurements detect binding between a fluorophore-labeled protein and an unlabeled partner protein, and they have been used previously to characterize binding between hsp90 and clients or hsp90-cochaperones (10, 50, 51).
We measured the residual fluorescence polarization of FITC-labeled sGCβ1(1–359)-H105F titrated with hsp90β or with individual hsp90β N, M, or C domains. We observed a concentration-dependent, saturable increase in the residual polarized fluorescence emission in titrations with full-length hsp90β or its M domain. In contrast, we observed only weaker or no binding between sGCβ1(1–359)-H105F and the hsp90β N domain, and no apparent binding to the hsp90β C domain (Fig. 2A). We concluded that sGCβ1(1–359)-H105F interacted directly with hsp90β, primarily through the hsp90β M domain. We obtained similar results using a purified heme-depleted form of wild-type sGCβ(1–359) instead of the H105F mutant (data not shown). In a complementary experiment, we titrated unlabeled sGCβ1(1–359)-H105F against FITC-labeled hsp90β M domain (Fig. 2B). The results confirmed that the binding interaction did not depend on which protein was labeled with FITC.
FIGURE 2.
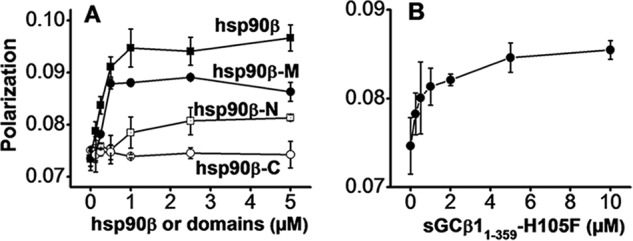
Binding interaction of hsp90β or its N, M, and C domains with sGCβ1. A, residual polarized fluorescence of FITC-labeled sGCβ1(1–359)-H105F (0.9 FITC per mol, 0.5 μm) titrated with full-length hsp90β (solid squares) or with the hsp90β N domain (open squares), hsp90β M domain (solid circles), or hsp90β C domain (open circles). B, residual polarized fluorescence of FITC-labeled hsp90β M domain (0.95 FITC per mol, 1.0 μm) titrated with sGCβ1(1–359)-H105F. The points depict mean ± S.D. data collected from three consecutive titrations and are representative of three independent experiments.
We utilized FITC-labeled apo-sGCβ1(1–197) or sGCβ1 PAS proteins to further define which sGCβ1 domains interact with hsp90β. Titration of FITC-labeled PAS domain with either hsp90β or the M domain led to saturable increases in the residual polarized emission (Fig. 3A). In contrast, we observed no increase when labeled apo-sGCβ1(1–197) was titrated with full-length hsp90β or with the hsp90β M domain (Fig. 3B). We carried out similar titrations to determine dissociation constants for the relevant interactions. We measured the binding of either sGCβ1(1–359)-H105F or sGCβ1 PAS to hsp90β or its M domain (Fig. 4, A–D). The calculated dissociation constants ranged between 1 and 1.5 μm, showing that hsp90β and its M domain display similar affinities toward both sGCβ1(1–359)-H105F and the sGCβ1 PAS domain. Inclusion of 1 mm ATP, ADP, or AMP-PNP in the experiment did not significantly alter the binding of hsp90β with sGCβ1(1–359)-H105F (data not shown). Together, the results show that the hsp90β interaction with sGCβ1(1–359)-H105F is primarily mediated by their respective M- and PAS domains. The hsp90β N and C domains and the sGCβ1 H-NOX domain appear to contribute little to the interaction.
FIGURE 3.
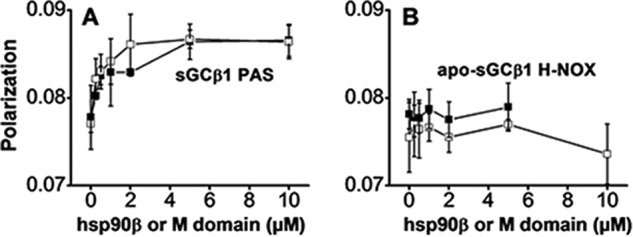
Binding interaction of hsp90β or the M domain with the sGCβ1 PAS or H-NOX domains. A, FITC-labeled sGCβ1 PAS domain (1.0 FITC per mol, 1 μm). B, FITC-labeled apo-sGC β1(1–197) (0.9 FITC per mol, H-NOX domain, 1 μm) was titrated with full-length hsp90β (open squares) or its M domain (solid squares), and the residual polarized fluorescence was measured. The graphs depict mean ± S.D. data collected from three consecutive replica titrations and are representative of two independent experiments.
FIGURE 4.
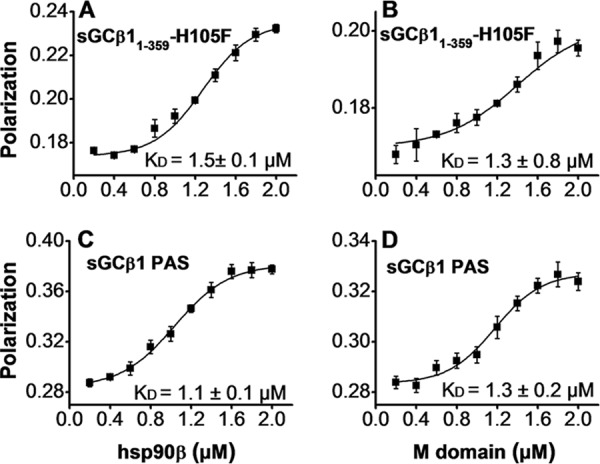
Titrations to determine binding affinities of the four principal interactions. A–D, each indicated FITC-labeled protein (0.2 μm) was titrated with the indicated concentrations of hsp90β or the M domain in a Flexstation instrument, and the residual polarized fluorescence was plotted and fit to determine the apparent KD value of each interaction (mean ± S.D., n = 3, written in each panel).
HxDMS Studies
We utilized HxDMS to identify regions within the hsp90β M domain and in apo-sGCβ1(1–359)-H105F that mediate or respond to their interaction. This approach was used previously to identify regions in hsp90 that bind cochaperones or small molecule inhibitors (10, 11, 52, 53). HxDMS was similarly used to characterize conformational changes in sGCβ1 induced by NO binding to its heme (54). We mapped 14 and 17 peptide sequences within sGCβ1(1–359)-H105F and the hsp90β M domain, respectively (Figs. 5A and 6A). We measured the kinetics of deuterium exchange for each mapping peptide by analyzing samples taken at 10 time points (10–3000 s) from exchange reactions that contained sGCβ1(1–359)-H105F, hsp90β M domain, or both proteins in a pre-equilibrated complex (Figs. 7 and 8).
FIGURE 5.
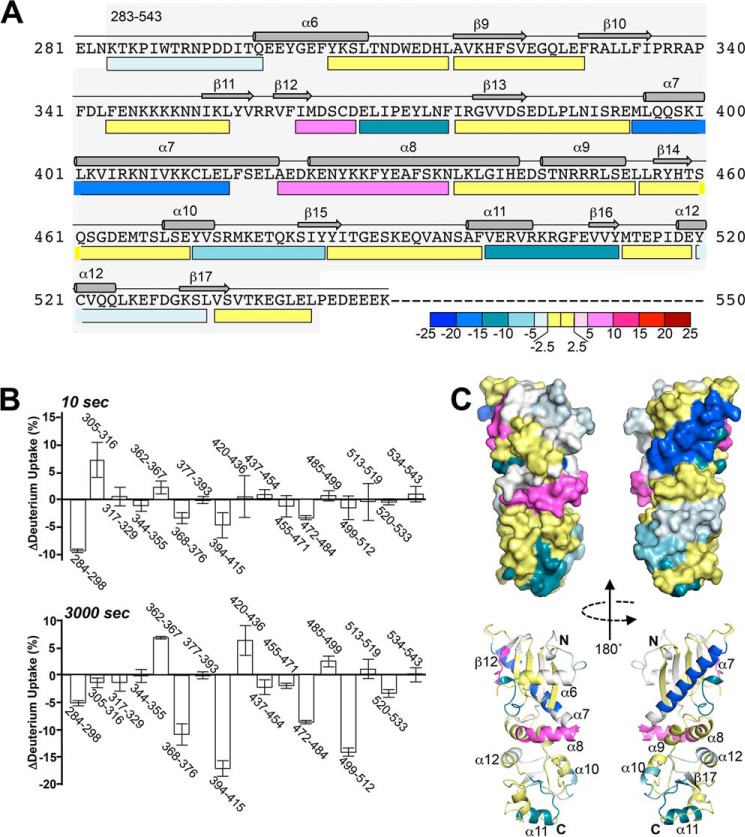
Change in hsp90β M domain deuterium exchange pattern upon binding to sGCβ1(1–359)-H105F. A, M domain mapping peptides are represented by bars below the protein primary sequence, with secondary structural features shown above. Bar colors indicate the difference in the percent of exchange (after 3000 s) observed in the reactions of the complexed versus free M domain. Decreased, increased, or equivalent exchanges in the bound hsp90β M domain relative to the free form are indicated by blue, red, and yellow bars, respectively. B, histogram of exchange differences for the reporter peptides after 10 and 3000 s in exchange reactions (n = 3, mean ± S.D.). Negative and positive values respectively indicate reduced exchange (protection) and increased exchange upon complexation of the M domain with sGCβ1(1–359)-H105F. C, surface and schematic representations of the hsp90β M domain (Protein Data Bank code 3PRY) colored according to deuterium exchange differences at 3000 s. Regions lacking HxDMS coverage are colored white.
FIGURE 6.
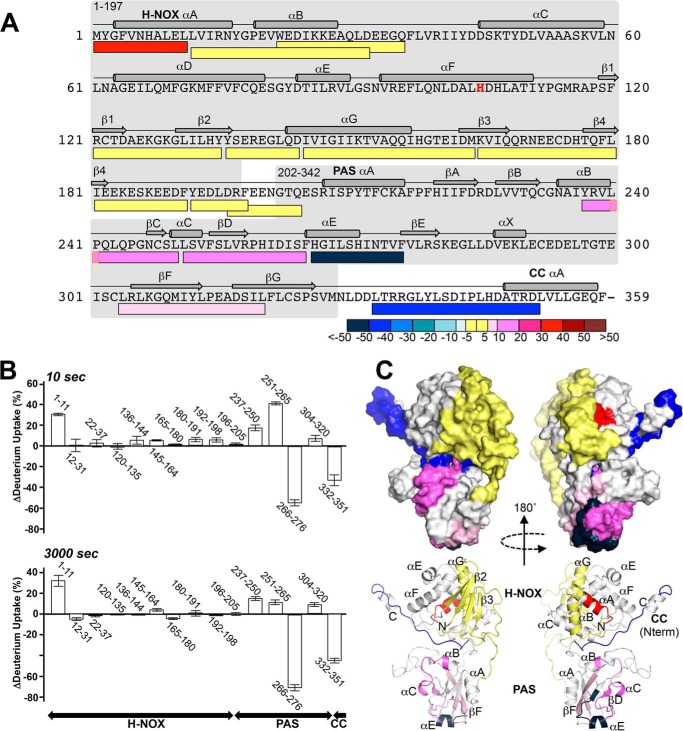
Change in sGCβ1(1–359)-H105F deuterium exchange pattern upon binding to hsp90βM domain. A, mapping peptides for sGCβ1(1–359)-H105F are represented by bars below the protein primary sequence, with secondary structural features shown above. Bar colors indicate the difference in the percent of exchange observed (after 3000 s) in the reactions of complexed versus free sGCβ1(1–359)-H105F. Decreased, increased, or equivalent exchanges in bound sGCβ1(1–359)-H105F relative to the free form are indicated by blue, red, and yellow bars, respectively. B, histogram of the exchange differences for reporter peptides after 10 and 3000 s in the exchange reactions (n = 3, mean ± S.D.). Negative and positive values respectively indicate decreased exchange (protection) and increased exchange upon complexation of sGCβ1(1–359)-H105F with hsp90β M domain. C, surface and schematic representations of sGCβ1(1–359) (generated as described under “Experimental Procedures”) colored according to deuterium exchange differences at 3000 s. Regions lacking HxDMS coverage are colored white.
FIGURE 7.
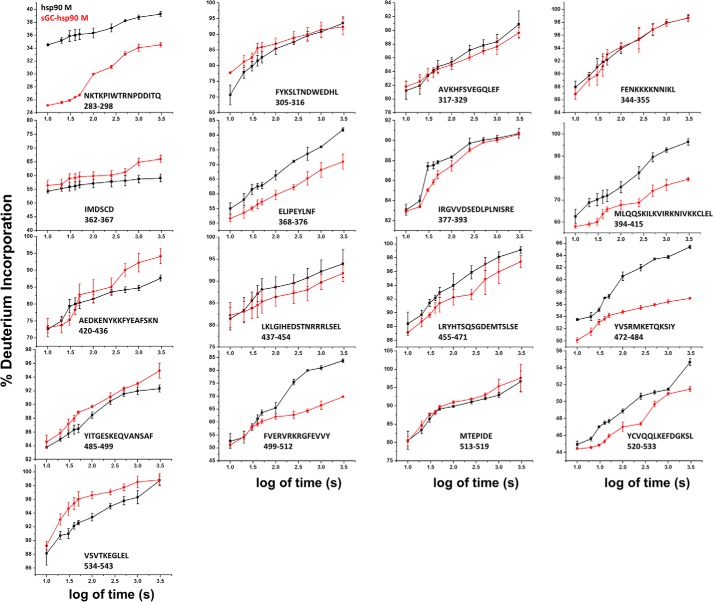
Kinetic traces of deuterium exchange on hsp90β M domain. Plots show the time dependence of deuterium incorporation (as a percentage of full deuterium incorporation) in the hsp90β M domain alone (black) or preincubated with sGCβ1(1–359)-H105F (red). Values represent the mean ± S.D. for three experiments.
FIGURE 8.
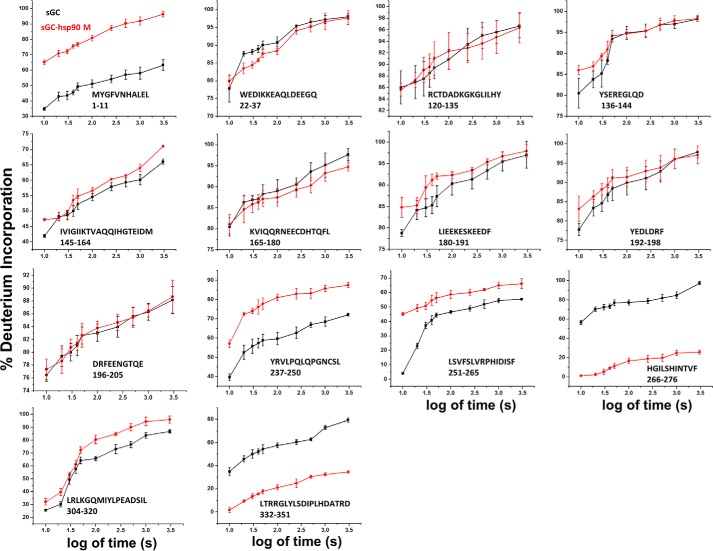
Kinetic traces of deuterium exchange on sGCβ1(1–359)-H105F. Plots show the time dependence of deuterium incorporation (as a percentage of full deuterium incorporation) in sGCβ1(1–359)-H105F alone (black) or preincubated with hsp90β M domain (red). Values represent the mean ± S.D. for three experiments.
Impact of Complex Formation on hsp90β M Domain
Binding to sGCβ1(1–359)-H105F altered the exchange behaviors of several reporter peptides within the hsp90β M domain (Figs. 5, A and B, and 7). Two peptides near the N terminus showed the largest percentage change in rapidly exchanging protons due to complex formation (Fig. 5B, 10 s histogram). Several reporter peptides located in the middle and C-terminal regions of the M domain displayed differences in the extent of exchange that accrued over time (Fig. 5B, 3000 s histogram), which reflected reduced or increasing exchange progression as a result of complex formation (Fig. 7). Four M domain peptides exhibited 7–18% decreases in exchange, while two peptides exhibited 6–8% increases (Fig. 5B, 3000 s histogram). These results identify specific sequences within the hsp90β M domain that may interact directly with sGCβ1(1–359)-H105F and/or respond structurally upon complexation. Fig. 5C maps these sequences onto a crystal structure of the hsp90 M domain (55, 56). The sequences that exhibited reduced exchange upon complexation at 3000 s form two discrete noncontiguous surfaces, one each on the N-and C-terminal halves of the hsp90β M domain. Surface-exposed helices on each of these surfaces (α7 and α11, respectively) manifest the greatest decreases in exchange. Interestingly, the two surfaces are flanked and separated by elements that are less affected or even undergo an increase in exchange, indicating a local relaxation of structure (for example, at the edge of β12 residues 362–367 and the first half of helix α8 residues 420–436).
Impact of Complex Formation on sGCβ1(1–359)-H105F
Complex formation with the hsp90β M domain increased the extent of rapid proton exchange for one mapping peptide located at the N terminus of the H-NOX domain, but it did not affect exchange in any other segment of the H-NOX (Figs. 6, A and B, and 8). In conjunction with our fluorescence polarization data, these HxDMS data suggest that the H-NOX domain does not interact directly with the hsp90β M domain, but it still dynamically responds to complex formation.
In contrast, complex formation with the hsp90β M domain resulted in more extensive differences in exchange within the PAS domain (Figs. 6, A and B, and 8). The percentage of rapidly exchanging protons increased in two PAS peptides from 5 to 45% (residues 251–65) and from 40 to 57% (residues 237–50) of total exchangeable protons (Fig. 6B, 10 s histogram, and 8). The exchange accrued over time also increased in these two peptides and in a third PAS peptide (residues 304–20). Together, these peptides correspond to portions of the central β-sheet (βC, βD, βF, and βG) of the PAS domain and two helical segments (αB and αC), representing a substantial increase in dynamics over approximately one-third of the domain (Fig. 6B, 3000 s histogram). At the same time, complex formation strongly reduced rapid exchange within two peptides (residues 266–276 and 332–351) that otherwise rapidly exchanged 35 and 58% of their total exchangeable protons (Figs. 6B, 10 s histogram, and 8). These sequences respectively correspond to helix αE, which flanks one side of the PAS domain, as well as the C-terminal linker sequence (residues 332–351) that connects the PAS domain to the coiled-coil domain of sGCβ1. Complex formation also reduced slower exchange kinetics in these two segments by 60 and 45% (Fig. 6B, 3000 s histogram). These findings are consistent with our fluorescence polarization experiments, which showed that the PAS domain interacts directly with the hsp90β M domain. The above-mentioned sequences correspond to regions within the PAS domain that respond to the complex formation and possibly interact directly with hsp90β. Fig. 6C illustrates our exchange results in the context of a structural model for sGCβ1(1–359)-H105F that is based on existing structural and biophysical studies (37, 38, 43, 57).
Modeling Complexes of sGCβ1(1–359) with hsp90β
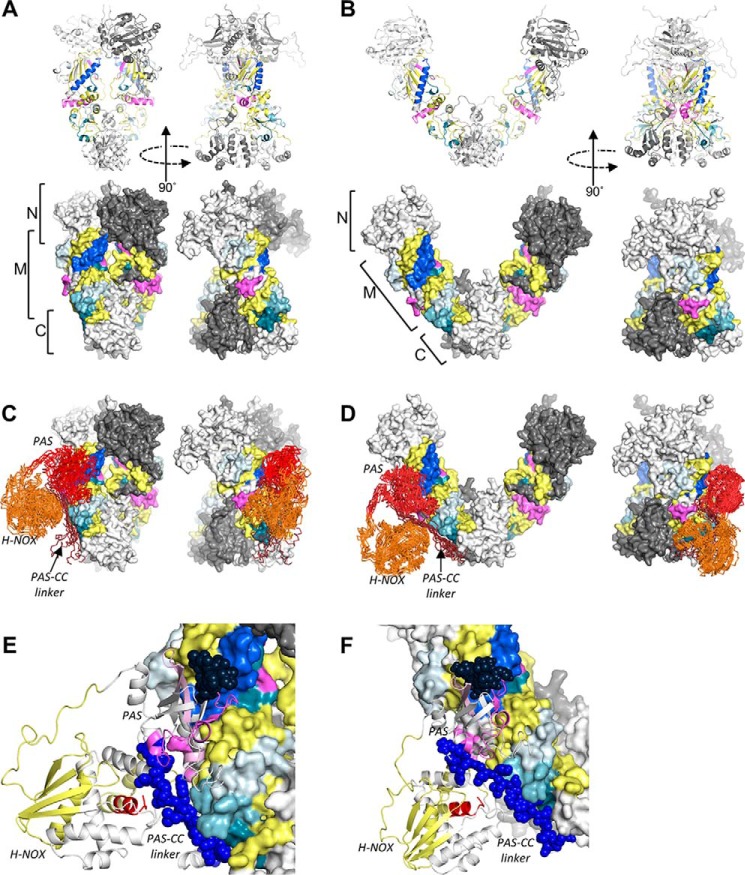
The segments of the hsp90β M domain that were protected by sGCβ1(1–359) define essentially two surface-exposed interfaces. Full-length hsp90β dimers undergo conformational changes between open and closed forms in nucleotide- and cochaperone-driven reaction cycles (5, 6). We built homology models of full-length hsp90β dimers based on available crystal structures and determined that the protected surfaces are solvent-exposed in both closed and open conformations (Fig. 9, A and B). These data support the notion that these surfaces correspond to direct binding sites for the sGCβ1 PAS domain. Correspondingly, two distinct segments on sGCβ1 also exhibited decreased exchange upon complex formation. One of these, the linker segment that connects the PAS and coiled-coil domains of sGCβ1 is structurally flexible and could adopt different orientations relative to the PAS domain. We therefore carried out computational docking of sGCβ1(1–359) onto open and closed hsp90β dimers using HADDOCK (47–49), specifying the two most strongly protected segments of each binding partner as potential interaction sites. As illustrated in Fig. 9, C–F, the lowest-energy ensembles of models readily accommodate such interactions. All low energy models involved packing of the PAS domain onto hsp90β helix α7 and nearby surfaces. The flexible linker segment of sGCβ1 extends toward and packed against surfaces formed by helices α10 and α11 of the M domain. Although no explicit interprotein restraints were specified during the docking, no low-energy models from the docking exhibited the opposite arrangement.
FIGURE 9.
Full-length hsp90β dimers and docked models of hsp90β-sGCβ1(1–359) complexes. A, HxDMS results from hsp90β M domain mapped onto hsp90β dimer in a closed conformation showing both ribbon and surface representations. Coloring is identical to that shown in Fig. 5. Regions lacking HxDMS coverage are colored white and dark gray in the respective monomers. B, HxDMS results from hsp90β M domain mapped onto hsp90β dimer in an open conformation. C, results of HADDOCK docking of sGCβ1(1–359) onto the closed hsp90β dimer. The 10 lowest energy sGCβ1(1–359) structures from the highest scoring HADDOCK output cluster are shown in ribbon (Cα-trace) representation. The sGCβ1 H-NOX, PAS, and C-terminal linker regions are colored orange, red, and dark red-brown, respectively. D, HADDOCK docking of sGCβ1(1–359) onto the open hsp90β dimer. The 10 lowest energy sGCβ1(1–359) structures from the highest scoring HADDOCK output cluster are shown. E, close-up view of a representative low energy docked configuration of sGCβ1(1–359) bound to the closed hsp90β dimer colored as in Fig. 6 and shown as a schematic. Spheres designate the two segments of sGCβ1(1–359) that undergo a significant decrease in exchange upon binding to the hsp90β M domain. F, close-up view of a representative low energy configuration of sGCβ1(1–359) docked onto the open hsp90β dimer.
Discussion
hsp90 binds to apo-sGCβ1 and drives its heme insertion in an ATP-dependent process (27). We utilized purified hsp90β, apo-sGCβ1(1–359), a heme-deficient sGCβ1(1–359) mutant, and individual domains to investigate how hsp90β and sGCβ1 interact with each other. We determined that the hsp90β M domain and the PAS domain of sGCβ1 are the primary mediators of interaction between the two proteins.
Our data place sGCβ1 in context relative to other selected hsp90 clients that contain PAS domains, including the aryl hydrocarbon receptor (AhR) and the hypoxia-inducing factor-1α (HIF1α) (58–61). There are notable parallels and differences in how the hsp90-PAS interaction regulates the biological functions of apo-sGCβ1, AhR, and HIF1α. For example, the hsp90-bound form of all three proteins is inactive, because hsp90 replaces a partner subunit (the sGCα1 subunit for apo-sGCβ1) or partner protein (ARNT for AhR; HIF1β, P300, or RACK1 for HIF1α) that is otherwise essential for biological function (27, 58–62). The hsp90-PAS interaction influences the binding of a small molecule ligand, the heme cofactor, to apo-sGCβ1 (28); similarly, hsp90 regulates binding of aromatic hydrocarbon ligands to AhR (60, 61, 63). Both ligand-binding events also require hsp90 ATPase activity. However, in the PAS domain of AhR, the binding sites for hsp90 and aromatic hydrocarbons overlap, and it has been suggested that hsp90 binding and ligand binding directly regulate each other (59, 63, 64). A similar mechanism may hold for ligand-binding domains of other hsp90 clients, including the glucocorticoid hormone receptor (10, 11) and multiple hsp90-chaperoned protein kinases (14, 65). In these cases, the hsp90-binding interface overlaps with the steroid ligand-binding site and with the ATP-binding site of the respective clients.
The PAS domain of sGCβ1 has a more densely packed hydrophobic core than typical PAS domains (such as that of AhR). The sGCβ1 PAS domain does not contain a discernible small molecule- or ligand-binding pocket (modeling data not shown). Rather, the heme-binding site of sGCβ1 is located in the adjacent H-NOX domain and therefore is separated from the PAS. For this client, therefore, hsp90 binds to a PAS domain and thereby may modulate a distinct ligand-binding domain. These considerations suggest that hsp90 operates by a blend of similar and distinct mechanisms to modulate sGCβ1 maturation and function, as compared with other PAS-containing clients or even other small molecule ligand-binding clients.
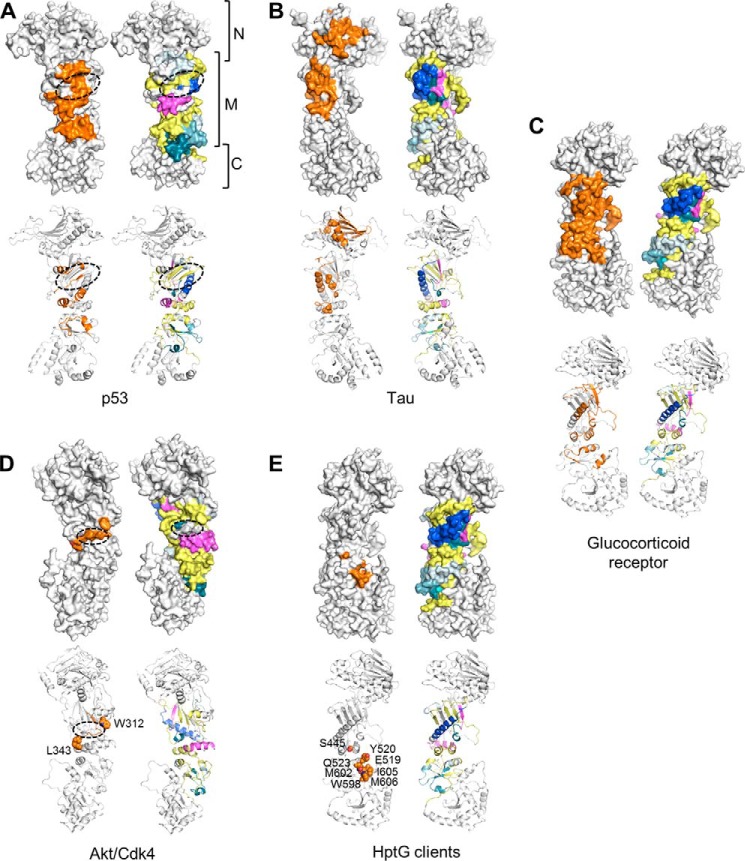
Our data show that sGCβ1(1–359)-H105F, and presumably full-length apo-sGCβ1, primarily interact with the hsp90β M domain. For the two M domain regions that displayed increased protection at 3000 s primarily due to their having a slower exchange after initial complex formation, the protection could arise from distant allosteric effects or could also result from a slow binding event that directly involves these protein regions. In either case, these regions of the M domain only partly coincide with binding sites proposed for other client proteins. For example, the binding sites for the DNA-binding domain of p53 (50) and sGCβ1-PAS overlap relatively little (Fig. 10A), and p53 binding involves an M domain groove formed by one side of β9 and elements from helices α6, α7, and α8 that are not protected upon sGCβ1-PAS binding. Binding sites for Tau (an intrinsically disordered protein) and sGCβ1 partly overlap within the M domain (66). However, Tau also interacts with the hsp90β N domain, and, unlike Tau, sGCβ1 engages the C-terminal portion of the M domain (Fig. 10B). The putative sGCβ1-PAS binding interface of hsp90β also differs from those of Akt/PKB (65), kinases such as CDK4 that are co-chaperoned by cdc37 (55, 56), and the glucocorticoid receptor ligand binding domain (Fig. 10, C and D) (10, 11). In particular, a flexible basic loop that connects strands β10 and β11 of the hsp90 M domain has been suggested to interact directly with kinases and ligand-binding domains (small molecule-binding PAS domains and steroid hormone receptor ligand-binding domains), but it is unaffected by the sGCβ1 PAS domain. Finally, the sGCβ1-PAS-binding site also differs from the M domain and C-terminal domain regions identified as binding sites for model client proteins (fragments of staphylococcal nuclease or ribosomal protein L2) of the E. coli hsp90 orthologue HtpG (Fig. 10E)(13, 53).
FIGURE 10.
Comparison with other hsp90-client protein interactions. Surface and schematic representations of hsp90 monomers are shown for each comparison. Residues implicated in client binding are colored orange. For comparison, an hsp90 monomer colored as described in Fig. 5 (depicting areas affected by complexation with sGCβ1(1–359)-H105F) is shown for each client interaction. A, putative hsp90-interaction surface of p53 DNA-binding domain. The dashed oval indicates a putative binding groove for several loops of the p53 DNA-binding domain (50) formed by one side of β9 and elements from helices α6, α7, and α8. B, distributed interaction surface of Tau protein on N and M domains of hsp90 identified by mutagenesis, NMR, and small angle x-ray scattering data (66). C, putative interaction surface of glucocorticoid receptor ligand-binding domain identified by NMR, mutagenesis, and cryo-EM data (10, 11). D, putative kinase-interacting epitopes of hsp90. Dashed oval indicates location of an Akt/PKB-interacting loop (65) connecting strands β10 and β11. Residues Trp-312 and Leu-343, shown as spheres, are implicated in binding of kinases such as CDK4, which are cochaperoned by cdc37 (55). E, residues on C-terminal half of M domain and C domain corresponding to client-binding residues on E. coli HptG (13, 53). Residues are shown as spheres.
The respective PAS domains of sGCα1 and sGCβ1 interact with each other in a stable heterodimeric sGC (37, 38). The sGCβ1 PAS domain also homodimerizes, and several representative crystal structures have been solved, including a homodimer of the PAS domain of M. sexta sGCα (38). However, a homology model of a human sGCβ1 PAS homodimer shows that the two regions we found to be heavily protected against proton exchange by the hsp90β M domain (sGCβ1 residues 266–276 and 332–351) lie outside the putative PAS dimer interface (Fig. 11, A and B). In addition, among the PAS domain peptides that became deprotected (showed increased exchange) due to complex formation with the hsp90β M domain, one peptide (residues 204–244) lies within the PAS homodimer interface in a region that contains residues that are essential for sGC heterodimerization (Fig. 11) (37). Thus, the binding of the hsp90β M domain at specific C-terminal regions in the sGCβ1 PAS domain appears to destabilize upstream regions in the PAS domain, including one needed in sGC heterodimer stabilization. This might represent a mechanism by which hsp90β prevents apo-sGCβ1 binding to its sGCα1 partner, an important outcome of chaperoning that would prevent formation of futile sGC heterodimers that do not respond to NO.
FIGURE 11.
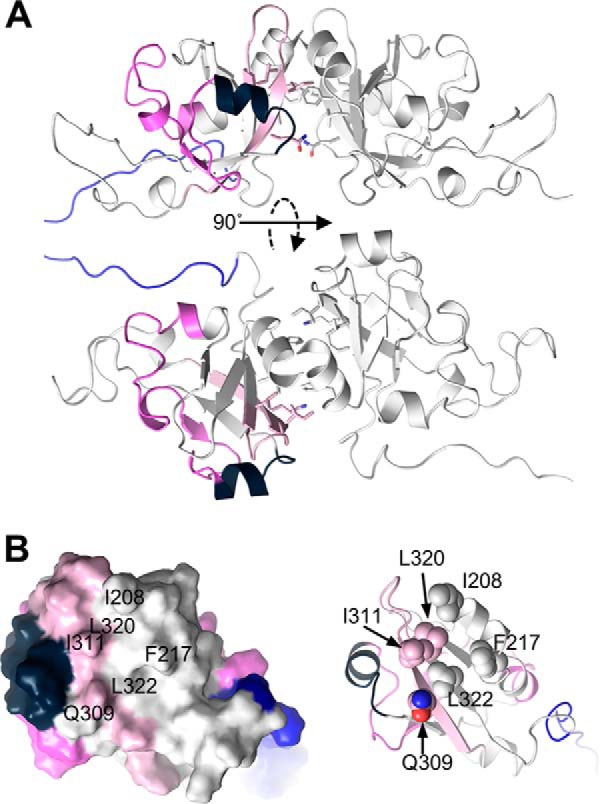
Model of sGCβ1 PAS domain dimer, highlighting the proton exchange differences induced by hsp90β M domain. The PAS domain and dimer structure were generated as described under “Experimental Procedures.” A, PAS dimer in two orientations, with one monomer colored as in Fig. 5. B, view directly into the dimer interface of the PAS domain monomer, shown in surface and schematic representations. Residues previously implicated as being important for dimerization are labeled (surface representation) and shown as spheres (schematic representation). Peptides and residues undergoing increased or decreased deuterium exchange upon complexation with the hsp90β M domain are colored as in Fig. 5.
Regarding binding stoichiometry, our hsp90β M domain is monomeric, whereas our full-length hsp90β is dimeric (by gel filtration analysis). Because we observed no significant difference in the apparent binding affinity between hsp90β and the M domain, the hsp90β dimeric structure and the included N- and C-terminal domains do not appear to be too important. This result is consistent with our other data indicating that little or no sGCβ1 binding takes place with the individual hsp90β N- or C-terminal domains, and the binding interaction appears to involve the solvent-facing regions of the M domain. For our sGCβ1(1–359)-H105F protein, we utilized the longest construct that can be successfully expressed in bacteria, which in our hands was primarily dimeric (with some higher oligomers also present). It is presently unclear whether complex formation with hsp90β or with the M domain weakened or dissociated the sGCβ1(1–359)-H105F homodimeric structure, but as noted such weakening may not be necessary for complex formation to occur because the putative interacting surfaces are solvent-exposed. In any case, our current results can now guide investigation into the hsp90β interactions that take place within cells during the course of sGCβ1 maturation.
A prior HxDMS study provided strong evidence that the sGCβ1 PAS domain may also engage in intramolecular interactions with the (heme-loaded) H-NOX domain. These results were obtained using an sGCβ1 construct spanning residues 1–385 (67), which is similar to our sGCβ1(1–359) construct. The PAS domain interacts with an H-NOX interface defined by helices αE, αF, and αG, which flank the heme-binding pocket and, in the case of αF, respond conformationally when NO binds to the heme. The prior study did not identify which elements of the PAS domain are involved in the interaction, and we did not obtain HxDMS data from the H-NOX peptides located in the putative interface. However, it is conceivable that the segments of the sGCβ1 PAS domain that become deprotected upon hsp90β M binding (and are outside the PAS dimerization interface) may be PAS surfaces that interact with the H-NOX domain. Such surfaces could thus become exposed upon binding to hsp90β. Our modeling data (Figs. 7 and 11) also suggest that interactions between the hsp90β M domain and the sGCβ1 PAS-coiled-coil linker segment could modulate the tertiary arrangement of the sGCβ1 domains (Figs. 7 and 11). Such interaction could displace intramolecular contacts between the linker segment and the H-NOX or PAS domains, indirectly affecting their interactions with each other as well as their stability and dynamics. By abstracting intramolecular interaction elements of the PAS domain and PAS-coiled-coil linker, hsp90 may indirectly drive a heme-loading competent state of the H-NOX domain.
Intriguingly, the two structural elements in sGCβ1 PAS that were indicated by our study to interact with hsp90β M domain may also help to orchestrate sGC activation by NO and to enable sGC heterodimer formation. A recent study (54) showed that NO binding to heme-containing sGCβ1 increased the degree of exchange, and thus deprotected the same two regions in the PAS domain (residues 266–276 and 332–351) that become strongly protected upon binding of the hsp90β M domain. One of these regions (residues 332–351) was shown by deletion analysis to be essential for the sGCα/β heterodimerization (68). Likewise, NO binding to the sGCβ1 heme was reported to stabilize the N-terminal helices of the H-NOX domain (54), including the same H-NOX segment that we find becomes deprotected by complex formation with hsp90β (helix αA). Because this segment lies adjacent to the heme pocket in H-NOX, it conceivably might influence the kinetics or energetics of heme binding. That the same few structural elements in sGCβ1 might provide a basis for it to alternate between an inactive heme-free form that is associated with hsp90β and an active heme-containing form that is associated with sGCα1 (27, 28) is an interesting concept emerging from our data, and it deserves further study.
A current model for hsp90 function in heme insertion is based on its role in steroid or aromatic hydrocarbon binding (18). In these cases, hsp90 may directly regulate the conformation of an “open cleft” in a client protein domain to help it accept a small ligand. Thus, one might have predicted that hsp90β would predominantly interact with the H-NOX domain of apo-sGCβ1 to stabilize and shape it for heme insertion, but our results suggest that the regulation is not so straightforward. Instead, hsp90β interacts through its M domain with the apo-sGCβ1 PAS domain, and this appears to cause some specific H-NOX structural relaxation and at the same time helps to block premature association with sGCα1. Further studies may now investigate how its anchoring of the PAS domain and accompanying structural perturbations enable hsp90β to direct heme insertion into the H-NOX domain of apo-sGCβ1.
Author Contributions
A. S. and Y. D. designed and performed the experiments, analyzed data, and helped with manuscript preparation. M. M. H. designed and made the sGC and hsp90 variants and helped with manuscript preparation. E. D. provided parent sGC constructs and purification protocols and helped with manuscript preparation. F. S. generated the parent sGC constructs. A. G. analyzed the data and prepared Figs. 7 and 8. W. R. M. provided structural models and helped analyze the data. S. L. H. provided HxDMS instrumentation and advice. S. M. helped design and interpret experiments, helped with manuscript preparation, and performed computer modeling and protein docking simulations. D. J. S. directed the research, analyzed the data, and helped in manuscript preparation. All authors performed a critical reading of the paper prior to submission and approved the final version of the paper.
Acknowledgments
We thank Drs. Tom McIntyre and Jonathan Smith (Cleveland Clinic) for advice and use of their fluorescence spectrometers, Belinda Willard of the Cleveland Clinic proteomics core for protein mass spectroscopy, Drs. Gary Gerstenecker and Valentin Gogonea of the Hazen laboratory for advice and assistance in the HxDMS instrumentation, and Deb Durra for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM097041 and GM051491 (to D. J. S.) and HL076491 (to S. L. H.). This work was also supported by American Heart Association Awards 10SDG2600345 (to E. D. G.) and 13PRE17000045 (to F. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- hsp90
- heat shock protein 90
- NO
- nitric oxide
- sGC
- soluble guanylate cyclase
- PAS domain
- Per-Arnt-Sim domain
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- HxDMS
- hydrogen-deuterium exchange mass spectrometry
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid
- AhR
- aryl hydrocarbon receptor
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1. Miyata Y., Nakamoto H., Neckers L. (2013) The therapeutic target Hsp90 and cancer hallmarks. Curr. Pharm. Des 19, 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Whitesell L., Santagata S., Mendillo M. L., Lin N. U., Proia D. A., Lindquist S. (2014) HSP90 empowers evolution of resistance to hormonal therapy in human breast cancer models. Proc. Natl. Acad. Sci. U.S.A. 111, 18297–18302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. da Silva V. C., Ramos C. H. (2012) The network interaction of the human cytosolic 90 kDa heat shock protein Hsp90: a target for cancer therapeutics. J. Proteomics 75, 2790–2802 [DOI] [PubMed] [Google Scholar]
- 4. Stankiewicz M., Mayer M. P. (2012) The universe of Hsp90. Biomol. Concepts 3, 79–97 [DOI] [PubMed] [Google Scholar]
- 5. Jackson S. E. (2013) Hsp90: structure and function. Top. Curr. Chem. 328, 155–240 [DOI] [PubMed] [Google Scholar]
- 6. Li J., Buchner J. (2013) Structure, function and regulation of the hsp90 machinery. Biomed. J. 36, 106–117 [DOI] [PubMed] [Google Scholar]
- 7. Mollapour M., Neckers L. (2012) Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta 1823, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ratzke C., Hellenkamp B., Hugel T. (2014) Four-colour FRET reveals directionality in the Hsp90 multicomponent machinery. Nat. Commun. 5, 4192. [DOI] [PubMed] [Google Scholar]
- 9. Southworth D. R., Agard D. A. (2011) Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-EM structure of the human Hsp90:Hop complex. Mol. Cell 42, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirschke E., Goswami D., Southworth D., Griffin P. R., Agard D. A. (2014) Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorenz O. R., Freiburger L., Rutz D. A., Krause M., Zierer B. K., Alvira S., Cuéllar J., Valpuesta J. M., Madl T., Sattler M., Buchner J. (2014) Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol. Cell 53, 941–953 [DOI] [PubMed] [Google Scholar]
- 12. Blacklock K., Verkhivker G. M. (2014) Allosteric regulation of the Hsp90 dynamics and stability by client recruiter cochaperones: protein structure network modeling. PLoS One 9, e86547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Genest O., Reidy M., Street T. O., Hoskins J. R., Camberg J. L., Agard D. A., Masison D. C., Wickner S. (2013) Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol. Cell 49, 464–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D., Karras G. I., Lindquist S. (2012) Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider S., Marles-Wright J., Sharp K. H., Paoli M. (2007) Diversity and conservation of interactions for binding heme in b-type heme proteins. Nat. Prod. Rep. 24, 621–630 [DOI] [PubMed] [Google Scholar]
- 16. Tsiftsoglou A. S., Tsamadou A. I., Papadopoulou L. C. (2006) Heme as key regulator of major mammalian cellular functions: molecular, cellular, and pharmacological aspects. Pharmacol. Ther. 111, 327–345 [DOI] [PubMed] [Google Scholar]
- 17. Ghosh A., Chawla-Sarkar M., Stuehr D. J. (2011) Hsp90 interacts with inducible NO synthase client protein in its heme-free state and then drives heme insertion by an ATP-dependent process. FASEB J. 25, 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng H. M., Morishima Y., Pratt W. B., Osawa Y. (2012) Modulation of heme/substrate binding cleft of neuronal nitric-oxide synthase (nNOS) regulates binding of Hsp90 and Hsp70 proteins and nNOS ubiquitination. J. Biol. Chem. 287, 1556–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pritchard K. A., Jr., Ackerman A. W., Gross E. R., Stepp D. W., Shi Y., Fontana J. T., Baker J. E., Sessa W. C. (2001) Heat shock protein 90 mediates the balance of nitric oxide and superoxide anion from endothelial nitric-oxide synthase. J. Biol. Chem. 276, 17621–17624 [DOI] [PubMed] [Google Scholar]
- 20. Mukai K., Shimizu T., Igarashi J. (2011) Phosphorylation of a heme-regulated eukaryotic initiation factor 2α kinase enhances the interaction with heat-shock protein 90 and substantially upregulates kinase activity. Protein Pept. Lett. 18, 1251–1257 [DOI] [PubMed] [Google Scholar]
- 21. Lee H. C., Zhang L. (2009) A unique mechanism of chaperone action: heme regulation of Hap1 activity involves separate control of repression and activation. Protein Pept. Lett. 16, 642–649 [DOI] [PubMed] [Google Scholar]
- 22. Chen F., Pandey D., Chadli A., Catravas J. D., Chen T., Fulton D. J. (2011) Hsp90 regulates NADPH oxidase activity and is necessary for superoxide but not hydrogen peroxide production. Antioxid. Redox Signal. 14, 2107–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nedvetsky P. I., Meurer S., Opitz N., Nedvetskaya T. Y., Müller H., Schmidt H. H. (2008) Heat shock protein 90 regulates stabilization rather than activation of soluble guanylate cyclase. FEBS Lett. 582, 327–331 [DOI] [PubMed] [Google Scholar]
- 24. Papapetropoulos A., Zhou Z., Gerassimou C., Yetik G., Venema R. C., Roussos C., Sessa W. C., Catravas J. D. (2005) Interaction between the 90-kDa heat shock protein and soluble guanylyl cyclase: physiological significance and mapping of the domains mediating binding. Mol. Pharmacol. 68, 1133–1141 [DOI] [PubMed] [Google Scholar]
- 25. Derbyshire E. R., Marletta M. A. (2012) Structure and regulation of soluble guanylate cyclase. Annu. Rev. Biochem. 81, 533–559 [DOI] [PubMed] [Google Scholar]
- 26. Friebe A., Koesling D. (2009) The function of NO-sensitive guanylyl cyclase: what we can learn from genetic mouse models. Nitric Oxide 21, 149–156 [DOI] [PubMed] [Google Scholar]
- 27. Ghosh A., Stuehr D. J. (2012) Soluble guanylyl cyclase requires heat shock protein 90 for heme insertion during maturation of the NO-active enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghosh A., Stasch J. P., Papapetropoulos A., Stuehr D. J. (2014) Nitric oxide and heat shock protein 90 activate soluble guanylate cyclase by driving rapid change in its subunit interactions and heme content. J. Biol. Chem. 289, 15259–15271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ignarro L. J., Buga G. M., Wood K. S., Byrns R. E., Chaudhuri G. (1987) Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 84, 9265–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Purohit R., Fritz B. G., The J., Issaian A., Weichsel A., David C. L., Campbell E., Hausrath A. C., Rassouli-Taylor L., Garcin E. D., Gage M. J., Montfort W. R. (2014) YC-1 binding to the β subunit of soluble guanylyl cyclase overcomes allosteric inhibition by the α subunit. Biochemistry 53, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnold K., Kiefer F., Kopp J., Battey J. N., Podvinec M., Westbrook J. D., Berman H. M., Bordoli L., Schwede T. (2009) The protein model portal. J. Struct. Funct. Genomics 10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu D., Zhang Y. (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 101, 2525–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winter M. B., Herzik M. A., Jr., Kuriyan J., Marletta M. A. (2011) Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain. Proc. Natl. Acad. Sci. U.S.A. 108, E881–E889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma X., Sayed N., Beuve A., van den Akker F. (2007) NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 26, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma X., Sayed N., Baskaran P., Beuve A., van den Akker F. (2008) PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J. Biol. Chem. 283, 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Purohit R., Weichsel A., Montfort W. R. (2013) Crystal structure of the α subunit PAS domain from soluble guanylyl cyclase. Protein Sci. 22, 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma X., Beuve A., van den Akker F. (2010) Crystal structure of the signaling helix coiled-coil domain of the β1 subunit of the soluble guanylyl cyclase. BMC Struct. Biol. 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wright L., Barril X., Dymock B., Sheridan L., Surgenor A., Beswick M., Drysdale M., Collier A., Massey A., Davies N., Fink A., Fromont C., Aherne W., Boxall K., Sharp S., et al. (2004) Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chem. Biol. 11, 775–785 [DOI] [PubMed] [Google Scholar]
- 41. Lee C. C., Lin T. W., Ko T. P., Wang A. H. (2011) The hexameric structures of human heat shock protein 90. PLoS One 6, e19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eramian D., Eswar N., Shen M. Y., Sali A. (2008) How well can the accuracy of comparative protein structure models be predicted? Protein Sci. 17, 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fritz B. G., Roberts S. A., Ahmed A., Breci L., Li W., Weichsel A., Brailey J. L., Wysocki V. H., Tama F., Montfort W. R. (2013) Molecular model of a soluble guanylyl cyclase fragment determined by small-angle x-ray scattering and chemical cross-linking. Biochemistry 52, 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang J., Liang Y., Zhang Y. (2011) Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shiau A. K., Harris S. F., Southworth D. R., Agard D. A. (2006) Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127, 329–340 [DOI] [PubMed] [Google Scholar]
- 47. de Vries S. J., van Dijk M., Bonvin A. M. (2010) The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 48. de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 49. Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 50. Hagn F., Lagleder S., Retzlaff M., Rohrberg J., Demmer O., Richter K., Buchner J., Kessler H. (2011) Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nat. Struct. Mol. Biol. 18, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 51. Schmid A. B., Lagleder S., Gräwert M. A., Röhl A., Hagn F., Wandinger S. K., Cox M. B., Demmer O., Richter K., Groll M., Kessler H., Buchner J. (2012) The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31, 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Phillips J. J., Yao Z. P., Zhang W., McLaughlin S., Laue E. D., Robinson C. V., Jackson S. E. (2007) Conformational dynamics of the molecular chaperone Hsp90 in complexes with a co-chaperone and anticancer drugs. J. Mol. Biol. 372, 1189–1203 [DOI] [PubMed] [Google Scholar]
- 53. Street T. O., Lavery L. A., Verba K. A., Lee C. T., Mayer M. P., Agard D. A. (2012) Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. J. Mol. Biol. 415, 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Underbakke E. S., Iavarone A. T., Chalmers M. J., Pascal B. D., Novick S., Griffin P. R., Marletta M. A. (2014) Nitric oxide-induced conformational changes in soluble guanylate cyclase. Structure 22, 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. (2003) Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11, 647–658 [DOI] [PubMed] [Google Scholar]
- 56. Vaughan C. K., Gohlke U., Sobott F., Good V. M., Ali M. M., Prodromou C., Robinson C. V., Saibil H. R., Pearl L. H. (2006) Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell 23, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Campbell M. G., Underbakke E. S., Potter C. S., Carragher B., Marletta M. A. (2014) Single-particle EM reveals the higher-order domain architecture of soluble guanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 111, 2960–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Minet E., Mottet D., Michel G., Roland I., Raes M., Remacle J., Michiels C. (1999) Hypoxia-induced activation of HIF-1: role of HIF-1α-Hsp90 interaction. FEBS Lett. 460, 251–256 [DOI] [PubMed] [Google Scholar]
- 59. Perdew G. H., Bradfield C. A. (1996) Mapping the 90 kDa heat shock protein binding region of the Ah receptor. Biochem. Mol. Biol. Int. 39, 589–593 [DOI] [PubMed] [Google Scholar]
- 60. Heid S. E., Pollenz R. S., Swanson H. I. (2000) Role of heat shock protein 90 dissociation in mediating agonist-induced activation of the aryl hydrocarbon receptor. Mol. Pharmacol. 57, 82–92 [PubMed] [Google Scholar]
- 61. Tsuji N., Fukuda K., Nagata Y., Okada H., Haga A., Hatakeyama S., Yoshida S., Okamoto T., Hosaka M., Sekine K., Ohtaka K., Yamamoto S., Otaka M., Grave E., Itoh H. (2014) The activation mechanism of the aryl hydrocarbon receptor (AhR) by molecular chaperone HSP90. FEBS Open. Bio. 4, 796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soshilov A., Denison M. S. (2011) Ligand displaces heat shock protein 90 from overlapping binding sites within the aryl hydrocarbon receptor ligand-binding domain. J. Biol. Chem. 286, 35275–35282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Coumailleau P., Poellinger L., Gustafsson J. A., Whitelaw M. L. (1995) Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J. Biol. Chem. 270, 25291–25300 [DOI] [PubMed] [Google Scholar]
- 65. Sato S., Fujita N., Tsuruo T. (2000) Modulation of Akt kinase activity by binding to Hsp90. Proc. Natl. Acad. Sci. U.S.A. 97, 10832–10837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Karagöz G. E., Duarte A. M., Akoury E., Ippel H., Biernat J., Morán Luengo T., Radli M., Didenko T., Nordhues B. A., Veprintsev D. B., Dickey C. A., Mandelkow E., Zweckstetter M., Boelens R., Madl T., Rüdiger S. G. (2014) Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 156, 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Underbakke E. S., Iavarone A. T., Marletta M. A. (2013) Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 110, 6777–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rothkegel C., Schmidt P. M., Atkins D. J., Hoffmann L. S., Schmidt H. H., Schröder H., Stasch J. P. (2007) Dimerization region of soluble guanylate cyclase characterized by bimolecular fluorescence complementation in vivo. Mol. Pharmacol. 72, 1181–1190 [DOI] [PubMed] [Google Scholar]