Background: RME-8 is involved in clathrin removal at early endosomes.
Results: RME-8 possesses a novel PI(3)P-binding motif within its N terminus.
Conclusion: Association of RME-8 with PI(3)P is required for endosomal clathrin removal and alters the steady state localization of retrograde transport cargo CI-MPR.
Significance: Regulation of PI(3)P association or PI(3)P levels is likely critical for controlling early endosomal organizational activities.
Keywords: 70-kilodalton heat shock protein (Hsp70), clathrin, endocytosis, protein motif, protein sorting, protein-protein interaction, PI(3)P, RME-8, SNX1, retrograde transport
Abstract
Receptor-mediated endocytosis 8 (RME-8) is a DnaJ domain containing protein implicated in translocation of Hsc70 to early endosomes for clathrin removal during retrograde transport. Previously, we have demonstrated that RME-8 associates with early endosomes in a phosphatidylinositol 3-phosphate (PI(3)P)-dependent fashion. In this study, we have now identified amino acid determinants required for PI(3)P binding within a region predicted to adopt a pleckstrin homology-like fold in the N terminus of RME-8. The ability of RME-8 to associate with PI(3)P and early endosomes is largely abolished when residues Lys17, Trp20, Tyr24, or Arg26 are mutated resulting in diffuse cytoplasmic localization of RME-8 while maintaining the ability to interact with Hsc70. We also provide evidence that RME-8 PI(3)P binding regulates early endosomal clathrin dynamics and alters the steady state localization of the cation-independent mannose 6-phosphate receptor. Interestingly, RME-8 endosomal association is also regulated by the PI(3)P-binding protein SNX1, a member of the retromer complex. Wild type SNX1 restores endosomal localization of RME-8 W20A, whereas a SNX1 variant deficient in PI(3)P binding disrupts endosomal localization of wild type RME-8. These results further highlight the critical role for PI(3)P in the RME-8-mediated organizational control of various endosomal activities, including retrograde transport.
Introduction
Endocytosis is a highly regulated and diverse process of plasma membrane and extracellular matrix internalization that participates in a variety of cellular processes (1–3). Recent work in the field has revealed that endocytic vesicles are heterogeneous macromolecular complexes, making it critical to further identify and fully characterize factors that associate and regulate endocytic events such as endosomal signaling, endosomal maturation, cargo sorting, cargo recycling, and cargo degradation.
Receptor-mediated endocytosis 8 (RME-8)3 was first discovered utilizing genetic screens in Caenorhabditis elegans, where rme-8 mutants exhibited defects in receptor-mediated yolk endocytosis in the oocyte and fluid phase endocytosis in the coelomocyte (4). Likewise, studies of rme-8 mutants in Drosophila displayed blockage in the internalization of the Bride of Sevenless receptor causing the formation of the rough eye phenotype (5). RME-8 has not only been shown to be highly conserved in the animal kingdom (6, 7), but it is also present in plants, where studies in Arabidopsis thaliana have demonstrated that RME-8 mutants exhibit gravitropism defects and associate with endosomal structures (8). Finally, a recent study has determined that a gain of function mutation in RME-8 correlates with Parkinson disease (9), highlighting the importance of studying RME-8 biology in more detail.
RME-8 is a large protein composed of more than 2000 amino acids. It contains four IWN repeats of unknown function and a DnaJ binding domain located between the second and the third IWN repeat that has been shown to associate with heat shock protein Hsc70 in a variety of species (5–7, 10). DnaJ protein family members act as coupling factors to stimulate ATP hydrolysis by its partner heat shock protein and thus they function as co-chaperones (11). The pleiotropic Hsc70 has a well established role in the disassembly of clathrin (12). Clathrin is crucial for vesicle formation at the plasma membrane during clathrin-mediated endocytosis and for protein sorting from early endosomes (13). In the case of endocytosis, the DnaJ domain protein auxillin recruits Hsc70 to release clathrin coats from clathrin-coated vesicles by binding to the terminal domain of clathrin heavy chain (12, 14, 15). Clathrin-coated vesicles are also featured on early endosomes and are the target of the RME-8·Hsc70 complex where they are employed to sort cargo from early endosomes to the trans-Golgi network (TGN) during retrograde transport (5, 10, 16, 17).
In addition to binding Hsc70, RME-8 has also been shown to associate and co-localize with the endosome membrane remodeling component SNX1 (10, 18). SNX1 when complexed with SNX2 recruits the Vps26-Vps29-Vps35 retromer trimer to form the heteropentameric coat, also known as retromer (19). SNX1 contains a phox (PX) domain that binds specifically to phosphatidylinositol 3-phosphate (PI(3)P). Subsequently, the Vps26-Vps29-Vps35 trimer is recruited and recognizes the transmembrane cargo to be sorted from early endosomes tothe trans-Golgi network during retrograde transport (19–21). In mammals, a well characterized transmembrane protein that is sorted by a retromer is the cation-independent mannose 6-phosphate receptor (CI-MPR) (19, 20). Generally, newly synthesized acid hydrolase precursor proteins are recognized by the CI-MPRs at the TGN membranes and are later sorted at early endosomes. After acid hydrolases reach the lumen of early endosomes, they dissociate from the receptor due to the acidic environment. They are then directed to lysosomes to degrade biological material, whereas the acid hydrolase receptors escape the early-to-late endosomal degradation pathway and are transported to the TGN through the retrograde transport pathway (19, 22, 23).
A prominent mechanism during endosomal processing events is the recruitment of target effector proteins through association with PI(3)P on the surface of early endosomes. We have recently discovered that RME-8 associates with PI(3)P-containing early endosomes in a myotubularin-related-2-dependent manner (24). Here, we have now identified critical residues mediating PI(3)P binding within the N terminus of RME-8. We have characterized this PI(3)P binding region in terms of its biochemical properties and examined its requirement for RME-8 activities at the early endosome.
Experimental Procedures
Plasmid Constructs
The plasmids encoding GFP-RME-8 were a kind gift from Drs. Fujibayashi and Sekiguchi from Osaka University, Osaka, Japan (6). Site-directed mutagenesis was performed to generate RME-8 point mutants. The forward and reverse primers for the GFP-RME8 K8A mutant are 5′-TAATTAGGGAAAATGCGGATCTGGCATGTT-3′ and 5′-AACATGCCAGATCCGCATTTTCCCTAATTA-3′. The forward and reverse primers for generation of the GFP-RME-8 K17A mutant are 5′-TTCTACACAACAGCACATTCATGGAG-3′ and 5′-CTCCATGAATGTGCTGTTGTGTAGAA-3′. To generate the GFP-RME-8 W20A mutant, the forward primer used was 5′-CAACAAAACATTCAGCGAGGGGGAAGTATA-3′, whereas the reverse primer was 5′-TATACTTCCCCCTCGCTGAATGTTTTGTTG-3′. The forward primer for GFP-RME-8 Y24A was 5′-CATGGAGGGGGAAGGCTAAGCGTGTCTTTT-3′, and the reverse was 5′-AAAAGACACGCTTAGCCTTCCCCCTCCATG-3′. The forward primer for the generation of GFP-RME-8 K25A was 5′-GGGAAGTATGCGCGTGTCTTTTC-3′, and the reverse was 5′-AAGACACGCGCATACTTCCCCCT-3′. The forward primer for the generation of GFP-RME-8 R26A was 5′-GGGAAGTATAAGGCTGTCTTTTCAGTT-3′, and the reverse was 5′-AACTGAAAAGACAGCCTTATACTTCCC-3′. All the constructs generated were verified by automated DNA sequencing (ACGT Corp). FLAG-Hsc70 and mRFP-SNX1 constructs were the kind gifts from Dr. Frank R. Sharp (University of California at Davis) and Dr. Peter J. Cullen (University of Bristol), respectively. Site-directed mutagenesis was performed to generate the mRFP-SNX1 K214A point mutant. The forward and reverse primers for mRFP-SNX1 K214A are 5′- CCGCCCCCGGAGGCAAGCCTCATAGGGA-3′ and 5′-TCCCTATGAGGCTTGCCTCCGGGGGCGG-3′.
Bioinformatics Analysis
RME-8 sequences were obtained by NCBI (www.ncbi.nlm.nih.gov), and orthologue sequences were aligned utilizing ClustalW multiple sequence alignment program (25). The three-dimensional structure prediction model of RME-8 was obtained by submitting the sequence of the first 100 amino acids of RME-8 to Protein Homology/AnalogY Recognition Engine Version 2.0 (Phyre2) (26, 27). The predicted three-dimensional structure of RME-8 generated from Phyre2 was superimposed onto the structure of the FERM domain of talin (Protein Data Bank code d1mixa2) using the Swiss-PDB Viewer (Deep View) (28, 29).
Cell Culture, Transfection, and Cell Lysis
HeLa (ATCC) and HEK293 (ATCC) cells were maintained in DMEM/F-12 with 10% FBS, 2 mm l-glutamine and supplemented with 1% penicillin/streptomycin antibiotics at 37 °C and 5% CO2. Cells were seeded 24 h before transfection. Following transient transfection with 5 μg of DNA using FuGENE HD (Roche Applied Science) using the manufacturer's protocol, cells were lysed with lysis buffer (50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 76 mm NaCl, 2 mm EGTA, 10% glycerol) supplemented with protease inhibitors 1 mm phenylmethylsulfonyl fluoride and 1 mm aprotinin.
Knockdown of endogenous RME-8 and SNX1 protein expression was performed using ON-TARGET siRNA SMARTpools (Dharmacon). HeLa cells were transfected with RME-8 and SNX1 siRNAs to a final concentration of 25 nm using DharmaFECT 1 transfection reagent following the manufacturer's protocols. Cells were then incubated with antibiotic-free media for 48 h before analysis.
Phosphatidylinositol Pulldown
HEK293 were transiently transfected with GFP-RME-8 variants and lysed as described above. Cellular lysates were incubated overnight at 4 °C with various phosphatidylinositol (PI) phosphates conjugated to resin (Echelon Biosciences), including control PI beads, PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3. Following three washing steps with washing/binding buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.25% Nonidet P-40), protein samples were subjected to SDS-PAGE analysis. Afterward, the samples were transferred onto a PVDF membrane and incubated with goat anti-GFP (Rockland) and anti-goat HRP (Rockland) antibodies at room temperature for 1 h. Proteins were visualized using SuperSignal West Femto Reagent (Thermo Scientific).
Liposome Flotation Assay
40 and 30% Opti-Prep density gradient medium (Sigma) was prepared in washing/binding buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.25% Nonidet P-40). HEK293 cells were lysed with 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 76 mm NaCl, 2 mm EGTA, 10% glycerol lysis buffer. Cellular lysates were incubated with control PI or PI(3)P-polymerized liposomes (PolyPIPosomes, Echelon Biosciences) for 2 h at room temperature. Following incubation, 60% Opti-Prep density gradient medium (Sigma) was added to obtain a final 40% Opti-Prep density gradient. The protein/liposome mixtures were added to thick-walled polycarbonate tubes (Beckman Coulter). 30% Opti-Prep density gradient medium was then overlaid on top of the 40% layer. Finally, wash/binding buffer was added as the top layer. Samples were spun in an ultracentrifuge at 50,000 rpm for 3 h at room temperature. After centrifugation, SDS-PAGE loading dye was added, and protein samples were analyzed by immunoblotting.
Co-immunoprecipitation
For the RME-8/Hsc70 co-immunoprecipitation, we co-transfected HEK293 cells with GFP-RME-8 variants and FLAG-Hsc70 for 24 h, and cells were lysed as described above. Cellular lysates were incubated with FLAG-protein A-agarose beads (Sigma) overnight at 4 °C. Immunoprecipitates were washed three times with 50 mm Tris-HCl, pH 7.4, 0.1% Triton X-100, 150 mm NaCl, 0.1% SDS and analyzed by immunoblotting using anti-FLAG (Sigma), goat anti-GFP (Rockland), rabbit anti-actin (Sigma), and secondary antibodies anti-mouse HRP (Promega), anti-goat HRP (Rockland), and anti-rabbit HRP (Vector Laboratories). For RME-8/SNX1 co-immunoprecipitations, we transfected cells with GFP-RME-8 variants for 24 h. We pre-bound the protein A-agarose beads with 1 μg of goat anti-GFP antibody for 45 min at room temperature. Following washing steps, we incubated cellular lysates with the conjugated beads overnight at 4 °C. Samples were washed three times as described above and analyzed by immunoblotting. Anti-SNX1 (BD Transduction Laboratories) was used to detect endogenous SNX1.
Immunofluorescence and Image Analysis
HeLa cells were seeded on four-chamber slides (BD Biosciences) 24 h prior to transient transfection with 0.5 μg of DNA using FuGENE HD. Slides were fixed for 15 min with 3.7% paraformaldehyde at room temperature. Cells were then permeabilized with 0.15% Triton X-100 for 2 min and blocked with 5% BSA for 1 h at room temperature. Primary antibodies, including mouse anti EEA-1 (BD Biosciences), rabbit anti-EEA-1 (Sigma), goat anti-clathrin (Santa Cruz Biotechnology), mouse anti-CI-MPR (Abcam), and rabbit anti-SNX1 were incubated for 1 h. Rabbit anti-RME-8 antibody was a kind gift from Dr. Peter McPherson (McGill University, Canada). Following washes, cells were incubated with the appropriate Alexa secondary antibodies (Life Technologies, Inc.) for 1 h at room temperature. Cells were washed, stained with Hoechst stain (0.5 mg/ml) (Invitrogen) for 2 min, and mounted on the slide. Confocal fluorescence microscopy was utilized to capture the images using a ×40 oil and a ×60 water objective. To determine the extent of co-localization between EEA1-positive endosomes and RME-8, Pearson's correlation coefficients were measured. Pearson's correlation coefficient values were calculated using ImageJ software via JACoP plugin (National Institutes of Health) (30). Data in the bar graphs represent the average of 30 randomly selected cells examined for co-localization.
Results and Discussion
Identification of the RME-8 PI(3)P-binding Motif
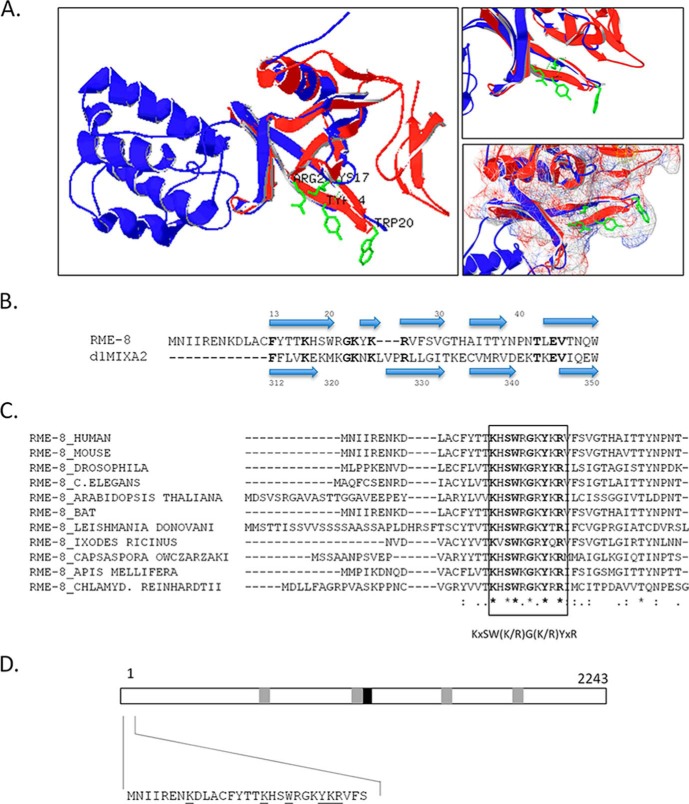
Earlier studies have shown that RME-8 associates with early endosomes (6, 7, 18), and recently our laboratory has discovered RME-8 to be a novel PI(3)P-binding protein that is regulated by the PI(3)P phosphatase MTMR2 (24). Because RME-8 does not contain a classical PI(3)P binding domain (e.g. FYVE domain, PX domain) we sought to localize the PI(3)P-binding motif in RME-8. To guide this process, the primary sequence of RME-8 was analyzed by the Phyre2 protein folding prediction program (26, 27). The result of this analysis was a high confidence score predicting the presence of a pleckstrin homology (PH)-like binding fold present in the first 100 amino acids of RME-8. Along with predicting homology with PH domains from a variety of proteins, the highest homology was with the PH domain of the PI(4,5)P2-binding protein Talin) (94% confidence) (Fig. 1, A and B) (31, 32. Further homology modeling using the Swiss-PDB Viewer and the three-dimensional structure of the PH domain of Talin (Protein Data Bank code d1mixa2) was performed (Fig. 1A). The resulting structural model revealed strong alignment with a portion of the PH fold of Talin, complementing the Phyre2 prediction program result. Generally, most PH domain binding interfaces reside on loop structures that link β-strands (33). Examining these regions on the RME-8/Talin model revealed residues in the extreme N terminus of RME-8 predicted to reside on such loop structures (Fig. 1A). Moreover, when examining this N-terminal region in RME-8, it was determined that many of these residues are highly conserved among orthologues (Fig. 1C), leading us to hypothesize their involvement in PI(3)P binding (Fig. 1, C and D). This region contains many positively charged residues and several bulky hydrophobic residues conserved among orthologues. Such residues have been shown to be functionally critical in PH domains (33) as well as other PI(3)P binding domains such as PX and FYVE domains (34, 35).
FIGURE 1.
RME-8 contains a putative phosphoinositide-binding motif. A, structural homology model depicting RME-8 (red) aligned with the solved structure of Talin's FERM third subdomain (blue) (27). The first 100 amino acids of RME-8 sequence were first submitted to the Phyre2 server for homology detection followed by Swiss-PDB Viewer to produce the structural model (see “Experimental Procedures”). Solvent-exposed amino acids of interest (Lys17, Trp20, Tyr24, and Arg26) in RME-8 that potentially represent a phosphoinositide-binding interface are shown in green. B, alignment of RME-8 and FERM's PH-like subdomain (d1mixa2). Sequences are aligned based on the secondary structure homology predicted by the Phyre2 server. Blue arrows represent β-strands, and the amino acids in bold represent amino acid identity. C, multiple sequence alignment of the N-terminal region of RME-8 orthologues. The RefSeq accession numbers used were the following: Homo sapiens, AAV41096.1; Mus musculus, NP_001156498.1; Drosophila melanogaster, NP_610467.1; C. elegans, AF372457_1; A. thaliana, AEC07904.1; Desmodus rotundus, JAA50105.1; Leishmania donovani, CBZ36243.1; Ixodes ricinus, JAB76328.1; Capsaspora owczarzaki, EFW44108.1; Apis mellifera, XP_394533.4; and Chlamydomonas reinhardtii, EDP05338.1. Based on the ClustalW alignment, the putative signature phosphoinositide-binding motif of RME-8 to is KXSW(K/R)G(K/R)YXR. D, schematic presentation of the human RME-8 protein with its IWN repeats (gray boxes) and its DnaJ domain (black box) shown. The proposed RME-8 PI(3)P-binding motif targeted for subsequent mutagenesis studies is highlighted.
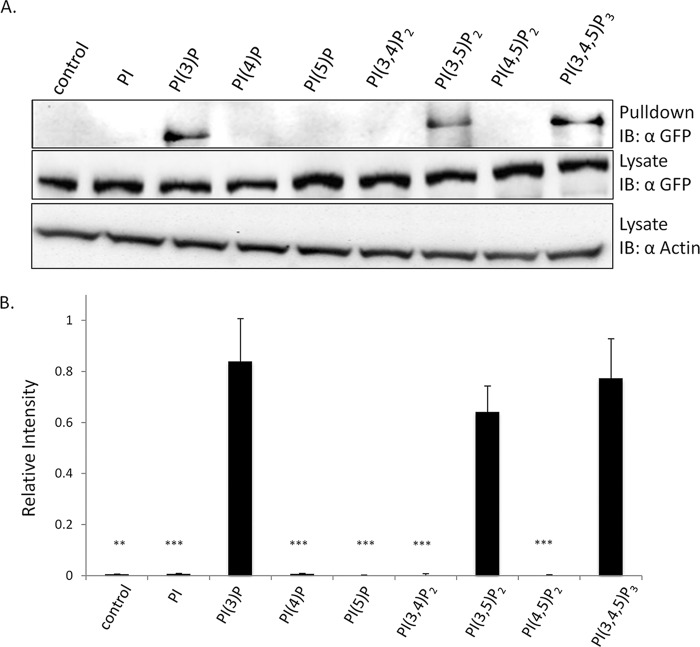
PH domains have been shown to bind to a variety of phosphoinositide isoforms, in particular isoforms with vicinal phosphates such as PI(3,4)P2 and PI(4,5)P2 (33). Therefore, it was important to examine the phosphoinositide specificity of RME-8. To accomplish this, HEK293 cells expressing wild type RME-8 were lysed and incubated with each of the seven phosphatidylinositol phosphates (PIP) conjugated to resin, and binding specificity was assessed via immunoblotting (Fig. 2). Consistent with earlier studies (24), RME-8 associates with PI(3)P and PI(3,5)P2 while failing to associate with the products of MTMR2, PI or PI(5)P. Furthermore, RME-8 shows no appreciable association with PI(4)P, PI(3,4)P2, or PI(4,5)P2. Interestingly, RME-8 did associate with PI(3,4,5)P3 to a similar extent as PI(3)P and PI(3,5)P2. Although the finding that RME-8 can associate with resin functionalized with PI(3,5)P2 and PI(3,4,5)P3 is potentially biologically relevant (see discussion below), the focus of this study was regulation of RME-8 at early endosomes. Thus, subsequent analysis focused on delineating residues important for association with PI(3)P.
FIGURE 2.
RME-8 binds PI(3)P, PI(3,5)P2, and PI(3,4,5)P3in vitro. A, HEK293 cells were transiently transfected with wild type RME-8 for 24 h. Cells were then lysed as described under “Experimental Procedures” and subjected to pulldowns with the following lipid groups: PI, PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3. Following elution, immunoblots (IB) were probed with anti-GFP antibody to detect GFP-RME-8. Whole cell lysates were analyzed by GFP and actin immunoblotting to confirm equal GFP RME-8 and total protein levels. B, lipid pulldown assay was quantified by densitometry using ImageJ software and normalized to actin levels. Means ± S.D. of three independent experiments are shown (n = 3). p values were calculated by the Student's t test comparing each sample to GFP-RME-8. **, p < 0.01; *** p < 0.001.
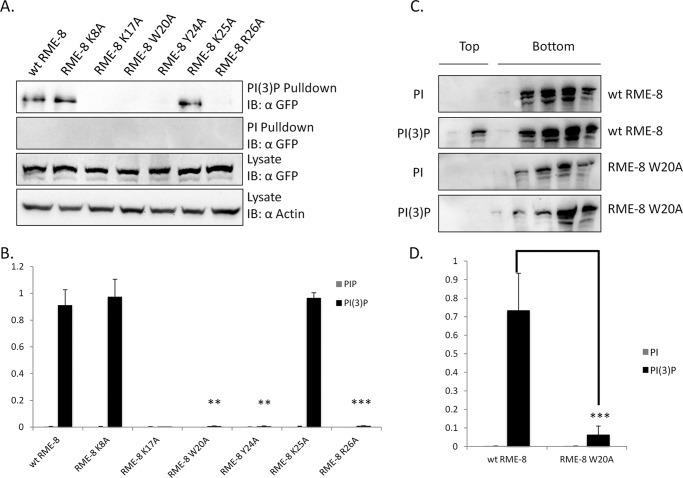
As noted above, the predicted PH-like fold region in the N terminus of RME-8 contains several highly conserved residues with the potential to mediate PI(3)P binding. Therefore, we examined the effect of substituting a variety of positively charged and bulky hydrophobic residues within this region for PI(3)P binding using the PIP pulldown assay (Fig. 3). Lysates generated from HEK293 cells expressing RME-8 variants were incubated with resin containing PI or PI(3)P. Following washing and SDS-PAGE separation, the PI(3)P binding activity of RME-8 variants was determined by immunoblotting. As expected, wild type RME-8 associates with PI(3)P. Moreover, RME-8 K8A and RME-8 K25A variants also display no significant difference in their ability to interact with PI(3)P. In contrast, the RME-8 variants K17A, W20A, Y24A, and R26A show markedly reduced capacity to associate with PI(3)P in this in vitro pulldown assay (Fig. 3, A and B). These results demonstrate that Lys17, Trp20, Tyr24, and Arg26 represent possible determinants for an N-terminal PI(3)P-binding motif within RME-8.
FIGURE 3.
Lys17, Trp20, Tyr24, and Arg26 are involved in RME-8 PI(3)P binding in vitro. A, HEK293 cells were transiently transfected with wild type RME-8, RME-8 K8A, RME-8 K17A, RME-8 W20A, RME-8 Y24A, RME-8 K25A, and RME-8 R26A constructs for 24 h. Cells were lysed as described under “Experimental Procedures” and subjected to PI and PI(3)P lipid pulldowns. Subsequent immunoblots were probed with anti-GFP to detect GFP-RME-8 variants. Whole cell lysates were analyzed by GFP immunoblotting (IB) to confirm equal GFP-RME-8 expression levels. A representative blot from three independent experiments is shown (n = 3). B, lipid pulldown assays quantified by densitometry using ImageJ software and normalized to actin levels. Means ± S.D. of three independent trials are shown. p values were calculated by the Student's t test and compared with wild type RME-8, **, p < 0.01; ***, p < 0.001. C, HEK293 cells expressing wild type RME-8 (wt) and RME-8 W20A were subjected to liposomal flotation assays. Lysates incubated with PI liposomes and PI(3)P liposomes were separated using an Opti-Prep gradient and ultracentrifugation. Gradient fractions were then analyzed by immunoblot analysis. Representative blots from three independent experiments are shown (n = 3). D, liposome flotation assay quantified by densitometry using ImageJ software. Means ± S.D. of three independent trials are shown. p values were calculated by the Student's t test and compared with wild type RME-8. ***, p < 0.001.
To complement the bead-based pulldown assay, we also examined the ability of wild type RME-8 and a representative binding mutant RME-8 W20A to associate with liposomes containing PI(3)P using liposome flotation assays (36). HEK293 cells expressing wild type RME-8 or RME-8 W20A were incubated with PI- or PI(3)P-containing liposomes followed by the addition of an Opti-Prep density gradient solution. Samples were subjected to ultracentrifugation upon which the PI(3)P liposomes (along with any bound RME-8) migrate to the top portion of the gradient. Gradients were then fractionated and analyzed by immunoblotting (Fig. 3, C and D). We reproducibly observed a significant portion of wild type RME-8 fractionate with the top PI(3)P liposome containing fraction but not with liposomes loaded with PI. Moreover, RME-8 W20A was defective at fractionating with the PI(3)P liposomes suggesting that RME-8 can associate with PI(3)P liposomes and that Trp20 is required for PI(3P) binding activity.
PI(3)P-binding Mutants Attenuate RME-8 Association with PI(3)P-rich Early Endosomes
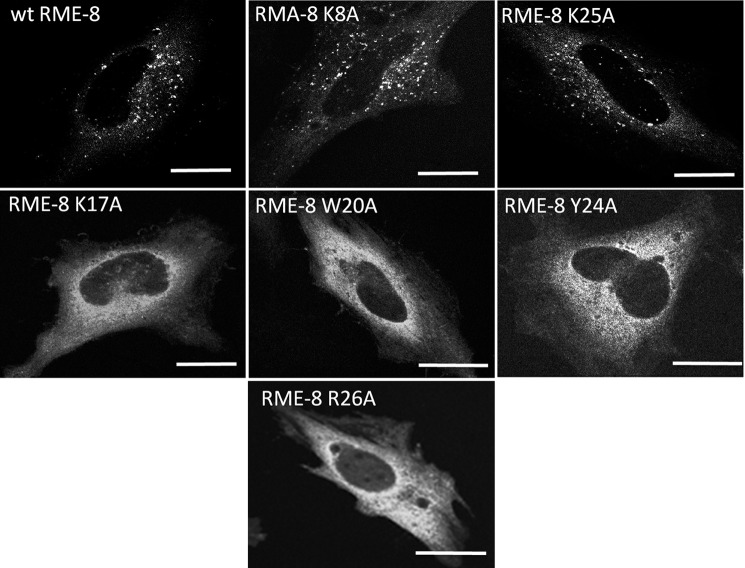
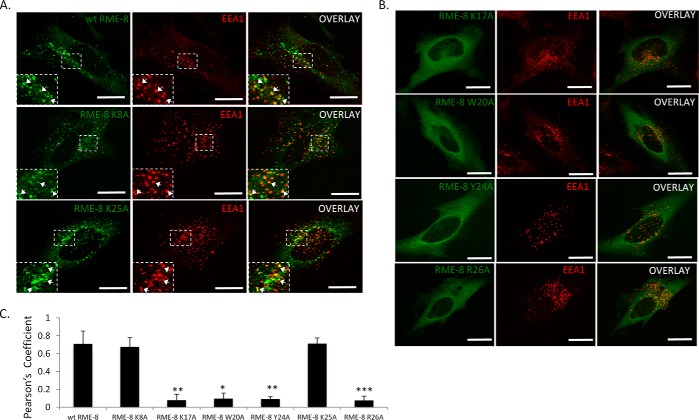
It has been demonstrated that RME-8 displays a punctate localization pattern due to its association with early endosomal structures (4, 6, 7, 18, 24, 37) that can be disrupted by the PI(3)-kinase inhibitor wortmannin and expression of the PI(3)P phosphatase MTMR2 (24). Therefore, to complement the above biochemical assays, we screened the various RME-8 mutants for subcellular localization changes from punctate to a diffused cytoplasmic pattern. As shown in Fig. 4, wild type RME-8, RME-8 K8A, and RME-8 K25A variants display a punctate pattern consistent with functional endosomal localization. In contrast RME-8 K17A, RME-8 W20A, RME-8 Y24A, and RME-8 R26A significantly lost the RME-8 punctate pattern, instead displaying largely diffuse cytoplasmic localization.
FIGURE 4.
RME-8 K17A, W20A, Y24A, and R26A alter the subcellular localization of RME-8. HeLa cells were transiently transfected with wild type RME-8, RME-8 K8A, RME-8 K17A, RME-8 W20A, RME-8 Y24A, RME-8 K25A, and RME-8 R26A and analyzed by confocal microscopy as described under “Experimental Procedures.” Images were collected using a ×40 objective. Scale bars, 15 μm.
To test whether these residues would be involved in dictating RME-8 PI(3)P-dependent early endosomal localization in vivo, we utilized immunofluorescence microscopy to examine co-localization of RME-8 variants with endogenous EEA-1, which is a well characterized PI(3)P early endosome marker (Fig. 5) (35, 38, 39). Consistent with our pulldown experiments, RME-8 K8A and RME-8 K25A co-localize with EEA1 to a similar extent as wild type RME-8 (Fig. 4, A and C), indicating that these Lys residues likely are not critical for PI(3)P binding. On the contrary, RME-8 K17A, RME-8 W20A, RME-8 Y24A, and RME-8 R26A failed to associate with PI(3)P-rich EEA1 early endosomes as indicated by loss of co-localization with EEA-1 along with diffuse cytoplasmic localization (Fig. 5, B and C).
FIGURE 5.
RME-8 early endosome localization requires Lys17, Trp20, Tyr24, and Arg26. A and B, HeLa cells were transfected with wild type RME-8, RME-8 K8A, RME-8 K17A, RME-8 W20A, RME-8 Y24A, RME-8 K25A, and RME-8 R26A (green) for 24 h and stained for EEA1 (red). Co-localization was analyzed by confocal microscopy. A, wild type RME-8, RME-8 K8A, and RME-8 K25A co-localize with EEA-1-positive early endosomes. B, absence of co-localization between RME-8 K17A, RME-8 W20A, RME-8 Y24A, and RME-8 R26A with EEA1 at early endosomes. Solid white arrows show regions of co-localization. Expanded boxes represent regions of interest. Images were collected using a ×40 objective. Scale bars, 15 μm. C, Pearson's correlation coefficient (PC) was utilized to quantify the extent of co-localization between RME-8 and EEA-1-positive endosomes. Means ± S.D. of three independent experiments (n = 30 cells) are shown. p values were calculated by the Student's t test and compared with wild type RME-8, *, p < 0.1; **, p < 0.01; ***, p < 0.001.
Taken together, our data indicate that RME-8 binds PI(3)P biochemically in vitro and on early endosomes in vivo through a novel N-terminal binding motif that is predicted to reside within a PH-like folded domain. Moreover, residues Lys17, Trp20, Tyr24, and Arg26 were identified as key determinants for competent PI(3)P association. Akin to other PI(3)P-binding proteins, RME-8 utilizes a combination of hydrophobic and positive charge functional groups for association with PI(3)P. In particular, the PX domain also possesses invariant Lys and Tyr residues to drive PI(3)P binding (40, 41). It is interesting to note that analogous to RME-8, PX-containing proteins have also been shown to associate with PI(3,5)P2 (22, 42, 43). This is in contrast to the FYVE domain, whose structurally distinct zinc coordination and small positively charged pocket is postulated to be too small to accommodate PI(3,5)P2 (44). Another distinguishing feature of RME-8's PIP specificity is the interaction with PI(3,4,5)P3 in the PIP pulldown assay (Fig. 2). Further structure-function analysis will be required to experimentally determine whether the N-terminal region of RME-8 truly adopts a PH-like fold and to examine whether RME-8's association with PI(3,5)P2 and PI(3,4,5)P3 is biologically relevant. However, it is interesting to note that in addition to early endosomes, RME-8 has been reported to localize to late endosomes and recycling endosomes, which are known to contain PI(3,5)P2 and PI(3,4,5)P3, respectively (4, 7, 45). Thus, it will be interesting to assess whether these PIP isoforms localized on distinct endosome subtypes regulate subcellular localization of RME-8.
RME-8 Does Not Require its PI(3)P-binding Motif for Association with Hsc70
Hsc70 binds DnaJ domain-containing cofactor proteins and is involved in ATP-dependent clathrin dissociation from clathrin-coated vesicles (46). Studies in Drosophila, C. elegans, and humans have shown that RME-8 binds the ATPase domain of Hsc70 through its DnaJ domain (5, 7, 10, 18, 46). Intriguingly, RME-8 is the only known PI(3)P-binding protein that recruits Hsc70 to early endosomes via its DnaJ domain. Because the mechanism by which these two molecules regulate clathrin is not well characterized, it was of interest to explore whether the RME-8/Hsc70 interaction was maintained in RME-8 PI(3)P-binding mutants. Co-immunoprecipitation experiments were conducted using HEK293 cells expressing GFP-RME-8 variants and FLAG-Hsc70 followed by GFP immunoprecipitation and immunoblot analysis (Fig. 6A). All of the RME-8-binding mutants that were found to disrupt PI(3)P association were able to co-immunoprecipitate Hsc70 to a similar extent when compared with wild type RME-8. The GFP-RME-8 ΔC1165 truncation mutant, which is devoid of its DnaJ domain (6), was also analyzed as a negative control, and as expected it was largely defective at co-immunoprecipitating Hsc70. The data strongly indicate that the RME-8/Hsc70 interaction is not perturbed by the ability of RME-8 to bind PI(3)P. The notion that the PI(3)P binding activity of RME-8 is segmented from its protein-protein binding activity was confirmed using another interacting partner, SNX1. As shown in Fig. 6B, we observed RME-8 W20A to co-immunoprecipitate endogenous SNX1 to a similar extent as wild type RME-8. In addition to indicating that mutating residues in the N-terminal PI(3)P binding region does not disrupt the functional fold of other RME-8 domains, this finding also signifies that RME-8 may potentially associate with Hsc70 or SNX1 away from endosomal structures.
FIGURE 6.
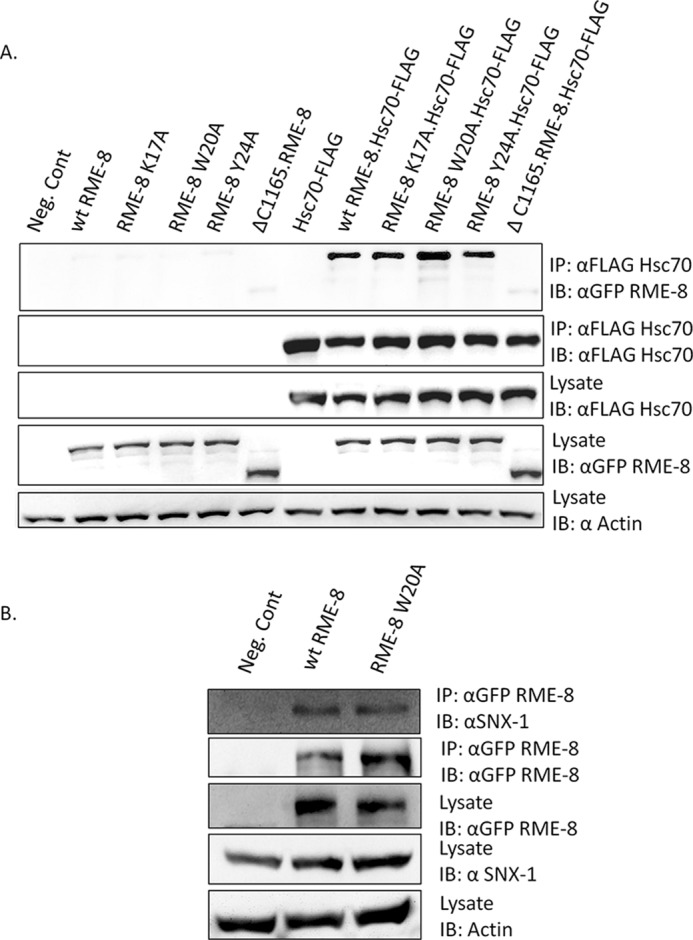
Wild type RME-8 and PI(3)-binding mutants associate with Hsc70 and SNX1. A, HEK293 cells were transiently co-transfected with GFP-RME-8 and FLAG-Hsc70 constructs for 24 h. Cellular lysates were then incubated with anti-FLAG protein-A-agarose beads. Following extensive washing steps, RME-8 bound to FLAG-Hsc70 was analyzed via immunoblotting (IB) using an anti-GFP antibody. IP, immunoprecipitation. Actin served as a cell number control. B, HEK293 cells were transiently transfected with wild type GFP-RME-8 or GFP-RME-8 W20A for 24 h. Lysates were subjected to anti-GFP immunoprecipitation and probed for endogenous SNX1 using immunoblot analysis. Actin served as a cell number control (Neg. Cont). A representative blot from three independent experiments is shown (n = 3).
It remains unclear how Hsc70 is distributed between auxillin and RME-8 during endocytosis and endosome maturation, respectively. One possibility is that due to the relative high concentration of Hsc70 in the cell, there are sufficient amounts of Hsc70 protein to partition within the two complex pools independently. Alternatively, PI(3)P levels may play a role in signaling the cell toward forming the RME-8·Hsc70 complex for sorting processing. In either case, our results signify that the endosomal clathrin uncoating activity of Hsc70 may potentially be indirectly regulated by PI(3)P levels via its interaction with RME-8.
Functional PI(3)P Binding Is Required for RME-8-mediated Early Endosomal Clathrin Regulation
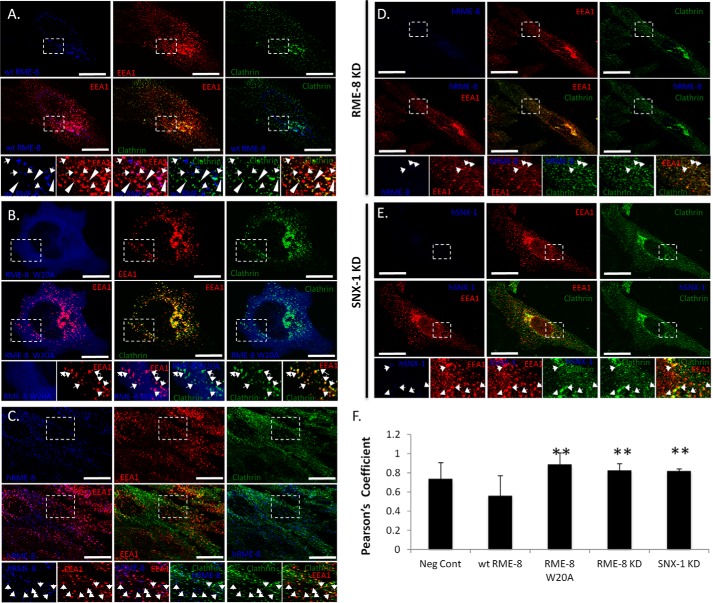
One of the hallmarks of membrane trafficking is cargo recognition and sorting from one cellular compartment to the other. Following cargo concentration into patches, the membrane undergoes deformation into smaller coated vesicles containing the cargo of interest (47). One of these coats is clathrin, which has been shown to regulate the trafficking of retrograde transport receptors at early endosomes (10, 18, 47, 48) in addition to its role in the endocytosis of receptors at the plasma membrane. Recent studies have revealed that the retromer complex controls endosomal clathrin levels through its incorporation of the RME-8·Hsc70 tandem (10, 18). Interestingly, the loss of either RME-8 or Hsc70 results in early endosomal clathrin accumulation (5, 10). Having established that the RME-8/Hsc70 association is preserved in RME-8 PI(3)P-binding mutants, we hypothesized that early endosomal clathrin levels would be perturbed in RME-8 PI(3)P-binding mutants due to the inability of Hsc70 to access the endosomal clathrin structures. To examine this hypothesis, HeLa cells were transiently transfected with wild type or the RME-8 W20A variant. We chose the RME-8 W20A variant as the representative binding mutant for this and all subsequent experiments due to its extensive conservation among RME-8 orthologues and based on the structural model that predicts Trp20 of residing on an exposed loop that could potentially serve as a strong binding interface (Fig. 1). Following fixation, cells were triple-labeled for endogenous EEA-1 (Fig. 7, red), endogenous clathrin heavy chain (green), and GFP-RME-8 (blue). The extent of co-localization between clathrin (green) and EEA-1 (red) was calculated as a measure of co-localization between the two channels (Fig. 7E). Of interest were RME-8-decorated early endosomes that were positive for PI(3)P (EEA-1). Analysis of these structures revealed a clear pattern where vesicles that were positive for RME-8 and PI(3)P (EEA-1) were largely devoid of clathrin (Fig. 7A). Interestingly, early endosomes that were lacking RME-8 (shown by arrowheads, Fig. 7A) displayed strong co-localization between PI(3)P (EEA-1) and clathrin. These results emphasize the function that RME-8 plays at the early endosomes and its involvement in clathrin regulation. Conversely, HeLa cells transfected with siRNA targeting RME-8 expression resulted in an endosomal clathrin phenotype opposite of overexpressing wild type RME-8 with increased co-localization between clathrin and the PI(3)P marker EEA-1 (Fig. 7D). We observed an analogous phenotype by knocking down the protein expression of SNX1 by siRNA (Fig. 7E). Interestingly, similar to the siRNA-treated samples, cells expressing RME-8 W20A displayed a statistically significant increase in co-localization of early endosomal clathrin on EEA-1-positive early endosomes compared with wild type RME-8-expressing cells (Fig. 7, B and F). In concert with RME-8 W20A failing to associate with PI(3)P-positive endosomes, the clear increased accumulation of clathrin at EEA1-positive structures compared with wild type and control cells points to RME-8 W20A possibly behaving as a dominant negative mutant. RME-8 W20A can still associate with Hsc70 (Fig. 6A) and thus likely competes with endogenous RME-8 for Hsc70, resulting in clathrin remaining at early endosomes. Thus, our results suggest that RME-8 binding to PI(3)P through its PI(3)P-binding motif plays a critical mechanistic role in regulating early endosomal clathrin levels in collaboration with its binding partner Hsc70.
FIGURE 7.
RME-8 PI(3)P binding affects early endosomal clathrin dynamics. HeLa cells were transiently transfected with wild type RME-8 (blue) (A) and RME-8 W20A (blue) (B) and for 24 h. Cells were triple-stained to observe endogenous EEA1 (red) and endogenous endosomal clathrin (green). A, wild type RME-8 at EEA1 early endosomes is displayed as magenta colored puncta, and clathrin at EEA1 early endosomes is displayed as yellow puncta. EEA1-positive and RME-8-positive endosomes display reduced clathrin levels indicated by arrows, and early endosomes devoid of wild type RME-8 (shown by arrowheads) display strong co-localization between EEA1 and clathrin. B, cells expressing RME-8 W20A display high co-localization between clathrin and EEA1. C, control cells are stained for endogenous RME-8 (blue), EEA1 (red), and clathrin (green). D and E, HeLa cells were transfected with siRNA targeting RME-8 (D) and SNX1 (E). Cells were stained for RME-8 (blue), EEA1 (red), and clathrin (green) to observe clathrin accumulation at EEA1 positive endosomes. F, Pearson's correlation coefficient (PC) was utilized to quantify the extent of co-localization between clathrin and EEA-1 in control, wild type RME-8, RME-8 W20A variant, RME-8 KD, and SNX1 KD samples. Means ± S.D. from three independent experiments (n = 30 cells) are shown. p values were calculated by the Student's t test and compared with wild type RME-8. **, p < 0.01. The values of Pearson's correlation coefficients of all the experiments were related and compared with RME-8 W20A set at 100% due to its largest statistical value. Solid white arrows, arrowheads, and expanded boxes represent regions of interest. Images were collected using a ×40 objective. Scale bars, 15 μm.
Perturbation of Mannose 6-Phosphate Receptor Localization by the RME-8 W20A PI(3)P-binding Mutant
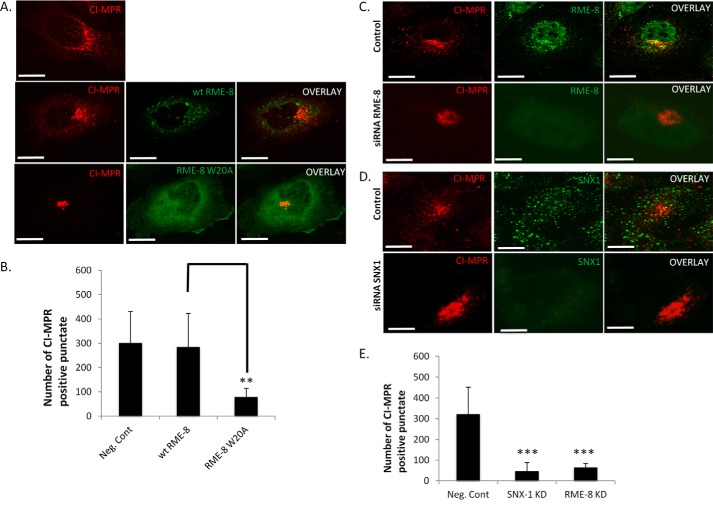
Clathrin and retromer coats have been implemented in the retrograde transport of transmembrane proteins from early endosomes to the trans-Golgi network (TGN) (48). The action of these two coats working in a coordinated sequential fashion is facilitated by the RME-8/Hsc70-mediated clathrin uncoating process at early endosomes (10, 17, 18, 48). RME-8 is a component of the retromer coat, and it has been shown to be important for the retrograde transport of transmembrane proteins such as the CI-MPR (10, 18, 19). The sorting of the acid hydrolase precursors starts at the TGN, and their delivery to early endosomes is mediated by their binding to CI-MPR embedded in the membrane followed by retrograde recovery of CI-MPR back to the TGN (19, 20, 49). Proper functioning of the retrograde transport is regulated by RME-8. Knockdown of RME-8 expression has been shown to mis-sort retrograde transport receptors such as MIG-14 in C. elegans and CI-MPR in mammalian cells (7, 10, 18). Because RME-8 regulates CI-MPR transport through clathrin disassembly (10), we investigated whether CI-MPR sorting would be affected in cells expressing RME-8 PI(3)P-binding mutants. Therefore, HeLa cells expressing wild type RME-8 and RME-8 W20A were stained for endogenous CI-MPR. In control cells as well as in cells expressing wild type RME-8, CI-MPR was observed to display mainly a perinuclear localization pattern with moderate staining at dispersed cytoplasmic punctate consistent with previous reports on the steady state localization of CI-MPR to TGN and endosomal structures (49–51). However, in RME-8 W20A-expressing cells, CI-MPR localization on cytoplasmic punctate structures was significantly diminished and was predominantly found in the perinuclear region (Fig. 8, A and B). The observed localization shift of CI-MPR was similarly reported in studies where RME-8 expression was knocked down using siRNA (7, 18). Analogous results have also been reported when other members of retromer components were knocked down in mammalian cells (20, 49). In these studies, it was concluded that disruption of retrograde transport of CI-MPR from endosomes to TGN results in rapid degradation of CI-MPR due to re-routing of CI-MPR from the endosome to the lysosome. This results in the loss of CI-MPR localization on endocytic punctate structures and the observed perinuclear localization pattern (7, 18, 20, 49). Thus, as a control, we examined HeLa cells treated with siRNA targeting RME-8 and SNX1 (Fig. 8, C–E). Under these conditions, we also observed a significant decrease in CI-MPR localized to punctate structures, confirming previous studies and supporting the RME-8 W20A result. Overall, our results demonstrate that PI(3)P association is critical for RME-8 cellular activities, as disrupting the PI(3)P binding ability of RME-8 yields a phenotype resembling knocking down RME-8 or SNX1 protein levels.
FIGURE 8.
Alteration of CI-MPR cellular localization in RME-8 W20A-expressing cells. A, HeLa cells expressing wild type RME-8 and RME-8 W20A were stained for endogenous CI-MPR. Cells were analyzed by confocal microscopy. Images were collected using a ×40 objective. Scale bars, 15 μm. ImageJ was utilized to count the number of total punctate in negative control, wild type RME-8, and RME-8 W20A-transfected cells. HeLa cells expressing RME-8 W20A exhibit a large decrease in the peripheral number of CI-MPR positive punctate displaying perinuclear localization. B, means ± S.D. of three independent experiments (n = 30 cells) are shown. p values were compared with wild type RME-8. **, p < 0.01. C and D, both RME-8- and SNX1-depleted HeLa cells have been labeled with CI-MPR antibody and also displayed a decrease in the total number of CI-MPR-positive punctate. E, bar graph representing the effect that RME-8 KD and SNX1 KD p values were compared with WT RME-8 in A and to negative control in C and D. All cells were analyzed by confocal microscopy. Means ± S.D. of three independent experiments (n = 30 cells) are shown. p values were calculated by Student's t test and compared with wild type RME-8 and SNX1. ***, p < 0.001.
SNX1 Rescues RME-8 W20A Dispersed Cellular Localization
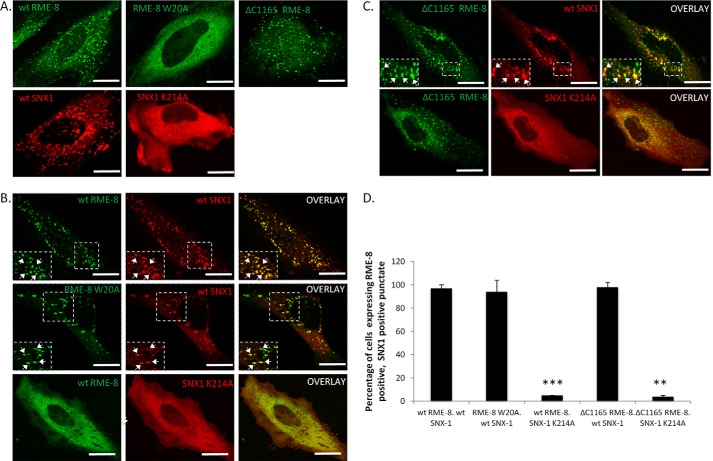
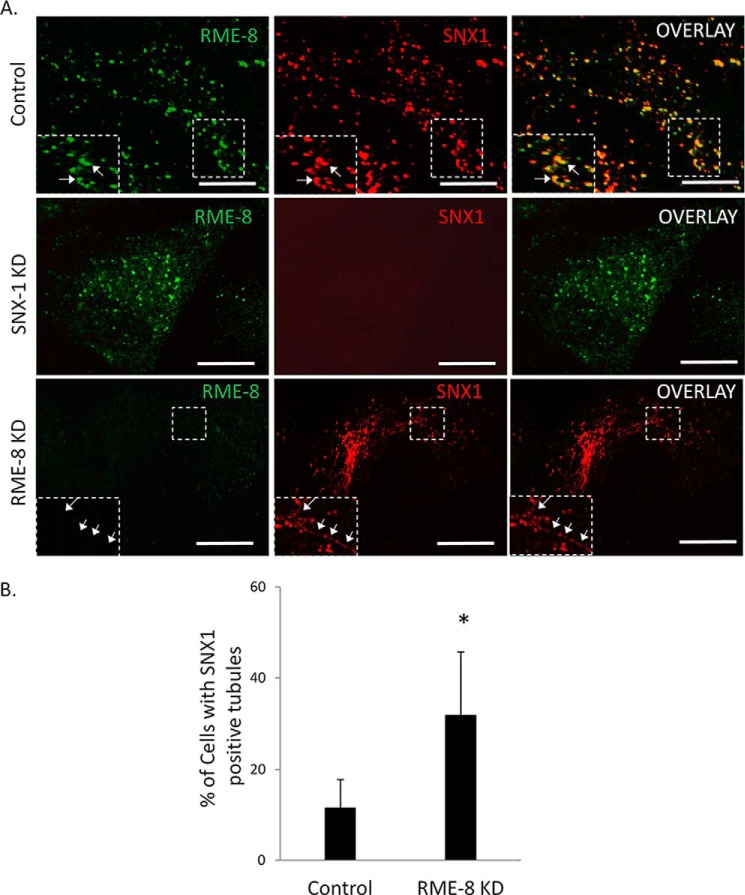
The retromer complex is important in recognizing the cytosolic tail of MPRs for efficient transport from endocytic structures (49). The retromer constituent SNX1 is a PX domain containing PI(3)P-binding protein (17, 42) that has been shown to play a role in sensing and driving membrane curvature (10, 17, 18). RME-8 associates with SNX1 through a region C-terminal to its DnaJ domain, containing the third IWN repeat and the linker region between the third and the fourth IWN repeat (10). To examine the relationship between the PI(3)P binding ability of RME-8 and its association with SNX1, we co-transfected HeLa cells with wild type SNX1/RME-8 and variants of both that are defective in PI(3)P association. As expected, wild type RME-8 and SNX1 strongly co-localize on peripheral structures consistent with endocytic vesicles (Fig. 9B). Surprisingly, cells co-expressing both RME-8 W20A and wild type SNX1 exhibited a drastic reversal in the RME-8 W20A subcellular localization pattern from diffused cytoplasmic to punctate (Fig. 9, A and B). Moreover, the rescued RME-8 W20A mutant strongly co-localizes with wild type SNX1. We next examined whether wild type RME-8 could rescue a well characterized PX domain point mutant of SNX1 (K214A) that disrupts PI(3)P binding (43, 52). As shown in Fig. 9B, SNX1 K214A remains diffusely localized throughout the cytoplasm when co-expressed with wild type RME-8, indicating that RME-8 is not able to recover the PI(3)P-mediated localization of SNX1 to endocytic structures when its PX domain is compromised. More striking, however, was the observation that when co-expressed, SNX K124A induced significant loss of endocytic localization of wild type RME-8 (Fig. 9B). One possible interpretation of this result is that RME-8 requires SNX1 to localize to endocytic structures. To test this hypothesis, we utilized a C-terminal deletion mutant of RME-8 (RME-8 ΔC1165) that lacks the SNX1 binding region and thus is unable to associate with SNX1 (6). However, this deletion construct possesses the N-terminal PI(3)P-binding motif. When analyzed for localization, RME-8 ΔC1165 was fully competent at targeting to peripheral punctate structures (Fig. 9, A and C). Moreover, overexpression of SNX1 K124A failed to disrupt localization of RME-8 ΔC1165 indicating that the PI(3)P-binding motif is sufficient for localization to endocytic vesicles. Interestingly, RME-8 ΔC1165 and wild type SNX1 strongly co-localized to these punctate structures suggesting that their interaction is not required for targeting to these vesicles. To test this hypothesis without the use of overexpressed proteins, we examined endogenous RME-8 localization in cells treated with SNX1 siRNA (Fig. 10). We observed no significant alteration in the RME-8 localization pattern in SNX1-depleted cells, confirming that RME-8 does not require SNX1 for endosomal localization. We also investigated the effect of knocking down RME-8 protein levels on SNX1 subcellular localization. As shown in Fig. 10, although SNX1 was still observed on endocytic structures, there was a significant increase in SNX1 tubulation in RME-8-depleted cells. An increase in SNX1 tubulation in response to RME-8 depletion has been reported by others and is thought to represent stalled cargo sorting (18, 53), indicating the importance of RME-8 to retromer-mediated trafficking events.
FIGURE 9.
SNX1 expression affects RME-8 early endosome targeting. A–C, HeLa cells were transfected with the indicated constructs and analyzed by confocal microscopy. Co-localizations between RME-8 and SNX1 variants are shown with arrows depicting regions of co-localization. In the middle panel, HeLa cells were co-transfected with RME-8 W20A and wild type SNX1, and arrows display regions of co-localization. Images were collected using a ×40 objective. Expanded boxes represent regions of interest. Scale bars, 15 μm. D, quantitation of co-localization between RME-8 and SNX1. Means ± S.D. of three independent experiments (n = 30 cells) are shown. p values were calculated by Student's t test and compared with wild type RME-8 and SNX1, **, p < 0.01; ***, p < 0.001.
FIGURE 10.
RME-8 expression regulates SNX1 tubulation. A, HeLa cells were transfected with RME-8 siRNA and SNX1 siRNA for 48 h. Knockdown of SNX1 protein expression does not affect RME-8 endosomal localization (middle panel). Knockdown of RME-8 protein expression results in the formation of SNX1-positive tubules (bottom panel). Images were collected using a ×60 objective. Expanded boxes represent regions of interest. Scale bars, 15 μm. B, bar graph representation of the percentage of cells altering from punctate to tubules in response to knockdown of RME-8 expression. Means ± S.D. of three independent experiments (n = 30 cells). p values were calculated by Student's t test and compared with control cells. *, p < 0.1.
The above results are intriguing as they reveal that although RME-8 endosomal targeting is mediated through its PI(3)P-binding motif, its interaction with SNX1 can also affect its endosomal association. A possible explanation for these observations is that the SNX1/RME-8 interaction could be simply stronger than the PI(3)P/RME-8 association, which could explain why the PX mutant of SNX1 was able to remove wild type RME-8 from endocytic structures, whereas wild type SNX1 was able to maintain RME-8 W20A at these vesicles (Fig. 9B). Additionally, the SNX1 interaction may stabilize RME-8 at the early endosome or perhaps serve an allosteric role during clathrin disassembly. Although further studies are required, our results indicate the intriguing possibility that regulatory mechanisms acting on SNX1 PI(3)P association (post-translational modifications and protein/protein interactions) will also potentially regulate RME-8 and subsequently Hsc-70-mediated clathrin disassembly.
In conclusion, we have shown that RME-8 associates with PI(3)P-positive endosomes through an N-terminal PI(3)P-binding motif predicted to adopt a PH-like fold and characterized by the presence of conserved basic and aromatic residues. The data support the notion that RME-8 at PI(3)P-rich early endosomes facilitates the process of clathrin removal, and the failure to do so may alter retrograde transport of cargo such as CI-MPR. Our results are consistent with a previous study reporting that RME-8 is located in SNX1-positive, clathrin-negative subdomains (17). This study by McGough and Cullen (17) elegantly showed that clathrin is not required for SNX1-BAR tubulation during retrograde transport as SNX1-BAR tubules start forming at RME-8-positive, SNX1-positive, and clathrin-negative subdomains of the early endosomes. Because RME-8 has not been found to be present in the tubules themselves, it indicates that it likely plays a proximal role mainly at the endosomal level when clathrin is present. Collectively, our data extend this model by which RME-8 utilizes PI(3)P binding in its role in shedding off clathrin before SNX1-BAR tubulation takes place in the retrograde transport pathway.
Author Contributions
B. X. and P. O. V. designed and conceived the study. B. X. performed all experimental work. P. O. V. coordinated the study. B. X. and P. O. V. wrote and edited the manuscript. Both of the authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Christopher Bonham for helpful discussions and Jasloveleen Sohi for technical assistance.
This work was supported in part by Operating Grant 123241 from Canadian Institutes of Health Research (to P. O. V.) and by a University of Windsor Golden Jubilee Health Research Award (to P. O. V.). The authors declare that they have no conflicts of interest with the contents of this article.
- RME-8
- receptor-mediated endocytosis 8
- PI(3)P
- phosphatidylinositol 3-phosphate
- PIP
- phosphatidylinositol phosphate
- PI
- phosphatidylinositol
- FYVE domain
- Fab-1, YGL023, Vps27, and EEA1
- PX
- Phox domain
- CI-MPR
- cation-independent mannose 6-phosphate receptor
- TGN
- trans-Golgi network
- CCV
- clathrin-coated vesicle
- PH
- pleckstrin homology.
References
- 1. Mellman I. (1996) Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575–625 [DOI] [PubMed] [Google Scholar]
- 2. Mukherjee S., Ghosh R. N., Maxfield F. R. (1997) Endocytosis. Physiol. Rev. 77, 759–803 [DOI] [PubMed] [Google Scholar]
- 3. Conner S. D., Schmid S. L. (2003) Regulated portals of entry into the cell. Nature 422, 37–44 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y., Grant B., Hirsh D. (2001) RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell 12, 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang H. C., Hull M., Mellman I. (2004) The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J. Cell Biol. 164, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujibayashi A., Taguchi T., Misaki R., Ohtani M., Dohmae N., Takio K., Yamada M., Gu J., Yamakami M., Fukuda M., Waguri S., Uchiyama Y., Yoshimori T., Sekiguchi K. (2008) Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct. Funct. 33, 35–50 [DOI] [PubMed] [Google Scholar]
- 7. Girard M., Poupon V., Blondeau F., McPherson P. S. (2005) The DnaJ-domain protein RME-8 functions in endosomal trafficking. J. Biol. Chem. 280, 40135–40143 [DOI] [PubMed] [Google Scholar]
- 8. Silady R. A., Kato T., Lukowitz W., Sieber P., Tasaka M., Somerville C. R. (2004) The gravitropism defective 2 mutants of Arabidopsis are deficient in a protein implicated in endocytosis in Caenorhabditis elegans. Plant Physiol. 136, 3095–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vilariño-Güell C., Rajput A., Milnerwood A. J., Shah B., Szu-Tu C., Trinh J., Yu I., Encarnacion M., Munsie L. N., Tapia L., Gustavsson E. K., Chou P., Tatarnikov I., Evans D. M., Pishotta F. T., et al. (2014) DNAJC13 mutations in Parkinson disease. Hum. Mol. Genet. 23, 1794–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi A., Sun L., Banerjee R., Tobin M., Zhang Y., Grant B. D. (2009) Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 28, 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walsh P., Bursać D., Law Y. C., Cyr D., Lithgow T. (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lemmon S. K. (2001) Clathrin uncoating: auxilin comes to life. Curr. Biol. 11, R49–R52 [DOI] [PubMed] [Google Scholar]
- 13. Brodin L., Löw P., Shupliakov O. (2000) Sequential steps in clathrin-mediated synaptic vesicle endocytosis. Curr. Opin. Neurobiol. 10, 312–320 [DOI] [PubMed] [Google Scholar]
- 14. Ungewickell E., Ungewickell H., Holstein S. E., Lindner R., Prasad K., Barouch W., Martin B., Greene L. E., Eisenberg E. (1995) Role of auxilin in uncoating clathrin-coated vesicles. Nature 378, 632–635 [DOI] [PubMed] [Google Scholar]
- 15. Scheele U., Alves J., Frank R., Duwel M., Kalthoff C., Ungewickell E. (2003) Molecular and functional characterization of clathrin- and AP-2-binding determinants within a disordered domain of auxilin. J. Biol. Chem. 278, 25357–25368 [DOI] [PubMed] [Google Scholar]
- 16. Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C., Wakeham D. E. (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17, 517–568 [DOI] [PubMed] [Google Scholar]
- 17. McGough I. J., Cullen P. J. (2013) Clathrin is not required for SNX-BAR-retromer-mediated carrier formation. J. Cell Sci. 126, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Popoff V., Mardones G. A., Bai S. K., Chambon V., Tenza D., Burgos P. V., Shi A., Benaroch P., Urbé S., Lamaze C., Grant B. D., Raposo G., Johannes L. (2009) Analysis of articulation between clathrin and retromer in retrograde sorting on early endosomes. Traffic 10, 1868–1880 [DOI] [PubMed] [Google Scholar]
- 19. Bonifacino J. S., Hurley J. H. (2008) Retromer. Curr. Opin. Cell Biol. 20, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seaman M. N. (2004) Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verges M. (2007) Retromer and sorting nexins in development. Front. Biosci. 12, 3825–3851 [DOI] [PubMed] [Google Scholar]
- 22. Johannes L., Popoff V. (2008) Tracing the retrograde route in protein trafficking. Cell 135, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 23. Braulke T., Bonifacino J. S. (2009) Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793, 605–614 [DOI] [PubMed] [Google Scholar]
- 24. Xhabija B., Taylor G. S., Fujibayashi A., Sekiguchi K., Vacratsis P. O. (2011) Receptor-mediated endocytosis 8 is a novel PI(3)P-binding protein regulated by myotubularin-related 2. FEBS Lett. 585, 1722–1728 [DOI] [PubMed] [Google Scholar]
- 25. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 29. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolte S., Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 31. Saltel F., Mortier E., Hytönen V. P., Jacquier M. C., Zimmermann P., Vogel V., Liu W., Wehrle-Haller B. (2009) New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control β3-integrin clustering. J. Cell Biol. 187, 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martel V., Racaud-Sultan C., Dupe S., Marie C., Paulhe F., Galmiche A., Block M. R., Albiges-Rizo C. (2001) Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 276, 21217–21227 [DOI] [PubMed] [Google Scholar]
- 33. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 34. Wurmser A. E., Emr S. D. (2002) Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 158, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kutateladze T. G., Ogburn K. D., Watson W. T., de Beer T., Emr S. D., Burd C. G., Overduin M. (1999) Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell 3, 805–811 [DOI] [PubMed] [Google Scholar]
- 36. Busse R. A., Scacioc A., Hernandez J. M., Krick R., Stephan M., Janshoff A., Thumm M., Kühnel K. (2013) Qualitative and quantitative characterization of protein-phosphoinositide interactions with liposome-based methods. Autophagy 9, 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang F. N., Navickas I. J., Au C., Budzilowicz C. (1978) Identification of the methylated ribosomal proteins in HeLa cells and the fluctuation of methylation during the cell cycle. Biochim. Biophys. Acta 518, 89–94 [DOI] [PubMed] [Google Scholar]
- 38. Lee S. A., Eyeson R., Cheever M. L., Geng J., Verkhusha V. V., Burd C., Overduin M., Kutateladze T. G. (2005) Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. U.S.A. 102, 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gillooly D. J., Raiborg C., Stenmark H. (2003) Phosphatidylinositol 3-phosphate is found in microdomains of early endosomes. Histochem. Cell Biol. 120, 445–453 [DOI] [PubMed] [Google Scholar]
- 40. Bravo J., Karathanassis D., Pacold C. M., Pacold M. E., Ellson C. D., Anderson K. E., Butler P. J., Lavenir I., Perisic O., Hawkins P. T., Stephens L., Williams R. L. (2001) The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell 8, 829–839 [DOI] [PubMed] [Google Scholar]
- 41. Ellson C. D., Andrews S., Stephens L. R., Hawkins P. T. (2002) The PX domain: a new phosphoinositide-binding module. J. Cell Sci. 115, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 42. Gallop J. L., McMahon H. T. (2005) BAR domains and membrane curvature: bringing your curves to the BAR. Biochem. Soc. Symp. 72, 223–231 [DOI] [PubMed] [Google Scholar]
- 43. Cozier G. E., Carlton J., McGregor A. H., Gleeson P. A., Teasdale R. D., Mellor H., Cullen P. J. (2002) The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 277, 48730–48736 [DOI] [PubMed] [Google Scholar]
- 44. Misra S., Hurley J. H. (1999) Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell 97, 657–666 [DOI] [PubMed] [Google Scholar]
- 45. Koumandou V. L., Boehm C., Horder K. A., Field M. C. (2013) Evidence for recycling of invariant surface transmembrane domain proteins in African trypanosomes. Eukaryot. Cell 12, 330–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eisenberg E., Greene L. E. (2007) Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic 8, 640–646 [DOI] [PubMed] [Google Scholar]
- 47. McGough I. J., Cullen P. J. (2011) Recent advances in retromer biology. Traffic 12, 963–971 [DOI] [PubMed] [Google Scholar]
- 48. Popoff V., Mardones G. A., Tenza D., Rojas R., Lamaze C., Bonifacino J. S., Raposo G., Johannes L. (2007) The retromer complex and clathrin define an early endosomal retrograde exit site. J. Cell Sci. 120, 2022–2031 [DOI] [PubMed] [Google Scholar]
- 49. Arighi C. N., Hartnell L. M., Aguilar R. C., Haft C. R., Bonifacino J. S. (2004) Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klumperman J., Hille A., Veenendaal T., Oorschot V., Stoorvogel W., von Figura K., Geuze H. J. (1993) Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J. Cell Biol. 121, 997–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geuze H. J., Stoorvogel W., Strous G. J., Slot J. W., Bleekemolen J. E., Mellman I. (1988) Sorting of mannose 6-phosphate receptors and lysosomal membrane proteins in endocytic vesicles. J. Cell Biol. 107, 2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong Q., Lazar C. S., Tronchère H., Sato T., Meerloo T., Yeo M., Songyang Z., Emr S. D., Gill G. N. (2002) Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. U.S.A. 99, 6767–6772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seaman M., Freeman C. L. (2014) Analysis of the Retromer complex-WASH complex interaction illuminates new avenues to explore in Parkinson disease. Commun. Integr. Biol. 7, e29483. [DOI] [PMC free article] [PubMed] [Google Scholar]