Background: The androgen receptor (AR) plays a key role in prostate cancer progression.
Results: USP7 is required for binding of the AR to chromatin.
Conclusion: USP7 is a novel AR co-regulator that mediates AR activity.
Significance: Pharmacological inhibition of USP7 represents a compelling therapeutic strategy for the treatment of prostate cancer.
Keywords: androgen receptor, gene transcription, prostate cancer, transcriptional coactivator, ubiquitin-dependent protease
Abstract
The androgen receptor (AR), a nuclear receptor superfamily transcription factor, plays a key role in prostate cancer. AR signaling is the principal target for prostate cancer treatment, but current androgen-deprivation therapies cannot completely abolish AR signaling because of the heterogeneity of prostate cancers. Therefore, unraveling the mechanism of AR reactivation in androgen-depleted conditions can identify effective prostate cancer therapeutic targets. Increasing evidence indicates that AR activity is mediated by the interplay of modifying/demodifying enzymatic co-regulators. To better understand the mechanism of AR transcriptional activity regulation, we used antibodies against AR for affinity purification and identified the deubiquitinating enzyme ubiquitin-specific protease 7, USP7 as a novel AR co-regulator in prostate cancer cells. We showed that USP7 associates with AR in an androgen-dependent manner and mediates AR deubiquitination. Sequential ChIP assays indicated that USP7 forms a complex with AR on androgen-responsive elements of target genes upon stimulation with the androgen 5α-dihydrotestosterone. Further investigation indicated that USP7 is necessary to facilitate androgen-activated AR binding to chromatin. Transcriptome profile analysis of USP7-knockdown LNCaP cells also revealed the essential role of USP7 in the expression of a subset of androgen-responsive genes. Hence, inhibition of USP7 represents a compelling therapeutic strategy for the treatment of prostate cancer.
Introduction
The male hormone androgens are critical for normal biological function of the prostate and for prostate cancer initiation and progression. The cellular effects of androgens are mediated by AR,2 a nuclear receptor superfamily transcription factor (1–3). AR consists of an N-terminal domain, a DNA-binding domain, a hinge region, and a C-terminal ligand-binding domain (LBD). The transactivation domain, AF-1 and AF-2 domains located within the N-terminal, and LBD domains all have a role in mediating AR transcriptional activity (3).
AR signaling begins with circulating androgens diffusing across the cell membrane of target cells and binding directly to AR in the cytosol. Upon androgen binding, AR undergoes a conformational change, homodimerization, and unmasking of the nuclear localization signal. The ligand-activated AR then translocates to the nucleus, where it binds to specific DNA sequences known as androgen-responsive elements (AREs) on the target genes. The basic transcriptional machinery and a series of co-regulators that exhibit distinct functions are recruited to support ligand-dependent AR transcriptional regulation (1–3).
Because of the central role of AR in the development of prostate cancer, androgen-deprivation therapy is the most widely used treatment for advanced prostate cancer. Androgen-deprivation therapy targets AR actions by reducing the circulating androgens with the administration of anti-androgens that compete for binding to the AR. However, as lethal castration-resistant prostate cancer (CRPC) develops, androgen-deprivation therapy is no longer effective at abolishing AR signaling. Given that AR signaling remains functional in CRPC, unraveling the mechanism of AR reactivation under androgen-depleted conditions will help to identify new therapeutic targets for prostate cancer (4–6).
Several mechanisms are proposed to cause AR signaling reactivation during CRPC progression, including AR gene amplification, AR mutation, which results in ligand promiscuity, and the alteration of AR co-regulator expression and activity (5–7). AR co-regulators modulate AR transcriptional activity and specificity through AR post-translational modifications such as phosphorylation, acetylation, methylation, and ubiquitination (8–13).
Increasing evidence indicates the importance of ubiquitination in AR activity regulation. Numerous E3 ubiquitin ligases have been identified as AR co-regulators implicated in prostate cancer progression. For example, MDM2, a well established AR E3 ubiquitin ligase (14–17), and STUB1 control AR stability and activity (17). Similarly, SIAH2 targets a select pool of transcriptionally inactive ARs for ubiquitin-dependent degradation, thereby regulating AR transcriptional activity. This regulation is required for CRPC development (18). In contrast to the effects of MDM2 and SIAH2 on AR degradation, RNF6 generates Lys6- and Lys27-linked ubiquitin chains that serve as scaffolding for ARA54 co-activator recruitment and regulate gene expression (19). Deubiquitination of AR is also critical for its activity and stability. The USP12, which is a deubiquitinating enzyme, is co-localized with AR in the cytoplasm and also can promote AR transcriptional activity by opposing the ubiquitin-dependent degradation of ARs (20). USP26 also can counteract AR activation and degradation by reversing MDM2-mediated ubiquitination (21).
In this study, we identified USP7 as a novel AR co-regulator in prostate cancer cells using affinity purification with antibodies against AR. USP7, also known as herpesvirus-associated ubiquitin-specific protease, has varied substrates including the tumor suppressor protein TP53 and its regulator MDM2 as well as the transcription factor FOXO4 (22–27). Additionally, USP7 is overexpressed in prostate cancer cells and is directly correlated with tumor aggressiveness (28). Here we present evidence that USP7 associates with the AR in an androgen-dependent manner and mediates its deubiquitination. The USP7-AR interaction is essential to facilitate AR binding to chromatin. Analysis of the transcriptome of LNCaP cells, a human prostate cancer cell line, also suggested that USP7 cooperates with the AR to promote expression of a subset of genes that are required for proliferation of prostate cancer cells.
Experimental Procedures
Antibodies
Antibodies for Western blotting were: anti-AR (N20, Santa Cruz Biotechnology, sc-816, 1:3000), anti-USP7 (Abcam, ab4080, 1:3000), anti-ubiquitin (P4D1, Santa Cruz Biotechnology, sc-8017, 1:1000), anti-FLAG M2 (Sigma, F1804, 1:5000), anti-α-Tubulin (Sigma, T6074, 1:5000), and anti-β-Actin (Abcam, ab8224, 1:5000). Antibodies for IP and ChIP were: anti-AR (441, Santa Cruz Biotechnology, sc-7305, 2 μg/sample), anti-USP7 (Bethyl Laboratory, A300–033A, 2 μg/sample), and anti-FLAG M2 affinity gel (Sigma, A2220).
Reagents
The androgen 5α-dihydrotestosterone (DHT) (Sigma) was dissolved in ethanol (EtOH) to make a 10−3 m stock and then a 10−5 m stock, which was diluted to 10−8 m prior to use. Cycloheximide (Sigma) was dissolved in dimethyl sulfoxide to make a 50 mg ml−1 stock. The proteasome inhibitor MG132 (Z-Leu-Leu-Leu-H, PEPTIDE INSTITUTE, 3175-v) was dissolved in dimethyl sulfoxide to make a 10 mm stock. The AR antagonist bicalutamide (Sigma) was dissolved in dimethyl sulfoxide to make a 10−3 m stock.
Cell Culture
LNCaP and CWR22Rv1 cells were grown at 37 °C under 5% CO2 in 10-cm plastic dishes containing 10 ml of RPMI 1640 medium with 10% FBS, 0.2 mm l-glutamine, 100 units ml−1 of penicillin, and 100 μg ml−1 of streptomycin. HEK293T cells were cultured in DMEM supplemented with 0.2 mm l-glutamine, 100 units ml−1 of penicillin, and 100 μg ml−1 of streptomycin. Before treatment with DHT, cells were incubated in phenol red-free medium with 5% charcoal-stripped FBS for 3 days.
Immunoprecipitation (IP) and Western Blotting
For IP and Western blots, cell lysates were prepared using lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5% (w/v) Nonidet P-40, cOmplete Protease Inhibitor Mixture (Roche Applied Science), 1 mm Na3VO4, 2 mm NaF, 1 mm PMSF, 10 mm N-ethylmaleimide). Antibodies were added, and the antibodies/lysate mixtures were rotated at 4 °C for 16 h. Immunocomplexes were collected using protein G or A Dynabeads (Invitrogen) that were washed with lysis buffer. To detect ubiquitinated proteins, lysis was performed under denaturing conditions (1% SDS and heating at 95 °C for 10 min). The lysates were then diluted with lysis buffer to reduce the SDS concentration to 0.1% and incubated at 4 °C for 30 min before antibodies were added. For Western blotting, cell lysates were separated by SDS-PAGE (7–15% polyacrylamide gradient), and the proteins were transferred to a PVDF membrane. Blots were incubated with primary antibodies followed by detection with HRP-conjugated secondary antibodies.
Purification and Identification of USP7 Using an Anti-AR-conjugated Column
Anti-AR (441) or negative control mouse IgG was cross-linked to protein G Dynabeads using dimethyl pimelimidate. The lysates from CWR22Rv1 prostate cancer cells were incubated with the antibody cross-linked beads at 4 °C overnight. The beads were then extensively washed with lysis buffer containing 150 mm NaCl, and the bound proteins were eluted with 0.05% TFA and collected by centrifugation. The eluates were subjected to TCA precipitation, separated by SDS-PAGE, visualized with silver staining, and excised for LC-MS/MS. Equivalent regions of a gel lane in which IgG had been electrophoresed were similarly analyzed to serve as negative controls. For LC-MS/MS analysis, gel slices were destained in 50% acetonitrile in 100 mm NH4HCO3, lyophilized, and rehydrated in 50 μl of 10 mm DTT in 100 mm NH4HCO3. After incubation at 56 °C for 1 h, the rehydration solution was removed and 50 μl of 55 mm iodoacetamide solution was added, followed by incubation in the dark for 45 min. After the supernatant was removed, the gel slices were lyophilized and digested with trypsin at 37 °C overnight. The tryptic peptides were extracted from the gel slices using 5% formic acid in 50% acetonitrile. This solution was lyophilized and the peptides reconstituted in 0.1% formic acid prior to LC-MS/MS analysis.
Plasmid Construction and Transfection
USP7 cDNA fragments were generated by PCR and subcloned into the vector FLAG-pcIneo (Invitrogen) at the BamHI and EcoRV sites. The primers used are shown in Table 1. The vector sequence was confirmed by DNA sequencing. The expression plasmid for wild-type AR (WT-AR) was kindly provided by Dr. Eriko Matsuura-Suzuki (Institute of Molecular and Cellular Biosciences, University of Tokyo). Cells were transiently transfected with plasmids using X-treme GENE HP DNA Transfection Reagent (Roche Applied Science).
TABLE 1.
Primers used in this study
| Target | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Cloning | ||
| FLAG-USP7 | GGAACTCGAGATGAACCACCAGCAGCAGCAGCAGC | GGAACCCGGGTCAGTTATGGATTTTAATGGCC |
| siRNA | ||
| USP7 | CCGGGACCUGUUAGAAGAAUGUAAA | UUUACAUUCUUCUAACAGGUCCCGG |
| ChlP-qPCR | ||
| FKBP5 | CTTCACGCCTGTGTTGCTTTTA | AGGGTGCAGGACGTTCCA (38) |
| PDE9A-ARE (−77,800) | GCCTCCCCCGTGCAG | TGCAAGGCACGTCTCAATTC (40) |
| PSA-ARE (−4366 to −3874) | TGGGACAACTTGCAAACCTG | CCAGAGTAGGTCTGTTTTCAATCCA (39) |
| RT-PCR | ||
| ACTB | TGGCACCCAGCACAATGAA | CTAAGTCATAGTCCGCCTAGAAGCA |
| AR | CCTGGCTTCCGCAACTTACAC | GGACTTGTGCATGCGGTACTCA |
| CLDN8 | CGGCTGGAATCATCTTCATCA | TTGGCAACCCAGCTCACAG |
| FKBP5 | GCGGAGAGTGACGGAGTC | TGGGGCTTTCTTCATTGTTC |
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
| NDRG1 | GTGGAGAAAGGGGAGACCAT | ACAGCGTGACGTGAACAGAG |
| PDE9A | GATCCCAATGTTTGAAACAGTGAC | TCCCAAAGTGGCTGCAGC |
| PSA | TGTGTGCTGGACGCTGGA | CACTGCCCCATGACGTGAT |
| TMPRSS2 | GGACAGTGTGCACCTCAAAGAC | TCCCACGAGGAAGGTCCC |
RNA Isolation and RT-PCR
Total RNA was isolated using TRIzol (Invitrogen) and the Nucleospin RNA II kit (Macherey-Nagel). cDNA was synthesized using the ReverTra qPCR RT Master Mix (TOYOBO), and the cDNA was amplified with the KAPA SYBR FAST qPCR kit (Kapa Biosystems) on an ABI 7500 real-time PCR system (Applied Biosystems). Primers used in these experiments are listed in Table 1.
RNAi
The siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen). The USP7 siRNA oligonucleotide sequences used in this study are shown in Table 1.
Chromatin Immunoprecipitation (ChIP) and Quantitative PCR (qPCR)
LNCaP cells were cross-linked with 1% formaldehyde for 10 min, quenched with 125 mm glycine, and then sonicated. ChIP experiments were carried out as described (29). Soluble chromatin was immunoprecipitated using anti-AR (441), anti-USP7 (A300–033A), or control antibody cross-linked to protein G or A Dynabeads overnight at 4 °C. Immunoprecipitates were washed and eluted with elution buffer (50 mm Tris-HCl, 10 mm EDTA, 1% SDS) for 20 min at 65 °C. Eluates were incubated at 65 °C overnight to reverse the cross-links and then treated with RNase A followed by proteinase K. The samples were purified using a PCR purification kit (Qiagen). qPCR was performed using the KAPA SYBR FAST qPCR kit (Kapa Biosystems) on the ABI 7500 real-time PCR system. For sequential ChIP, the first-round antibody cross-linked beads were added to chromatin extracts and incubated overnight at 4 °C. The bound protein complexes were eluted with 10 mm DTT at 37 °C for 30 min, and the eluate was diluted 10-fold and subsequently re-immunoprecipitated with the specific antibodies or control IgG overnight at 4 °C. The results are presented as fold-enrichment over control IgG or as the percentage of input chromatin that was precipitated.
RNA Sequencing (RNA-seq)
After transfection with USP7 siRNA for 72 h, LNCaP cells were exposed to 10 nm DHT or EtOH for 16 h and subjected to RNA extraction. Total RNA was isolated using TRIzol and the Nucleospin RNA II kit. RNA (1 μg) was used for library preparation with the TruSeq RNA Sample Preparation version 2 kit (Illumina), and the samples were then subjected to RNA-seq (Solexa HiSeq2000, Illumina). Sequenced RNA reads were aligned to the National Center for Biotechnology Information (NCBI) RefSeq RNA database (mRNA accession numbers only) using Bowtie, with three mismatches in the first 28 bases allowed per read (−n3 option) (30). Aligned reads were merged for a single gene. To accommodate for transcriptional variants, we used both uniquely and multiply aligned reads. We used the best match for multiply aligned reads (−k option). The gene expression level was calculated in reads per kilobase per million reads and normalized by the trimmed mean of M values (31). Using a threshold of one read per kilobase per million reads yielded 11,460 expressed genes. Data graphics were produced with R (32). Gene Ontology analysis was performed using the web tool Database for Annotation, Visualization and Integrated Discovery (DAVID) (33). These sequence results have been deposited in the NCBI Sequence Read Archive (SRP049426).
Cell Proliferation Assay
LNCaP and CWR22Rv1 cells maintained in 10-cm dishes were transfected with USP7 siRNA for 24 h before cells were seeded in 12-well plates and incubated in RPMI 1640 containing 5% charcoal-stripped FBS. The cell numbers were counted using a Neubauer chamber at the indicated times.
Statistical Analysis
Statistical differences were determined by two-tailed Student's t test. Statistical significance is displayed as: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Results
USP7 Is a Novel Androgen-dependent AR-associated Protein in Prostate Cancer Cell Lines
To better understand the regulation of AR activity in the progression of prostate cancer to a castration-resistant state, endogenous AR-associated proteins were evaluated in CRPC CWR22Rv1 cells. CWR22Rv1 cells express full-length AR and AR splice variants lacking the AR LBD (AR-V7) and serve as a model of the transition between hormone-sensitive and castration-resistant prostate cancer (34, 35). In addition, because CWR22Rv1 cells possess easily detectable and stable levels of AR expression, they are a valuable material from which to purify endogenous AR-associated proteins.
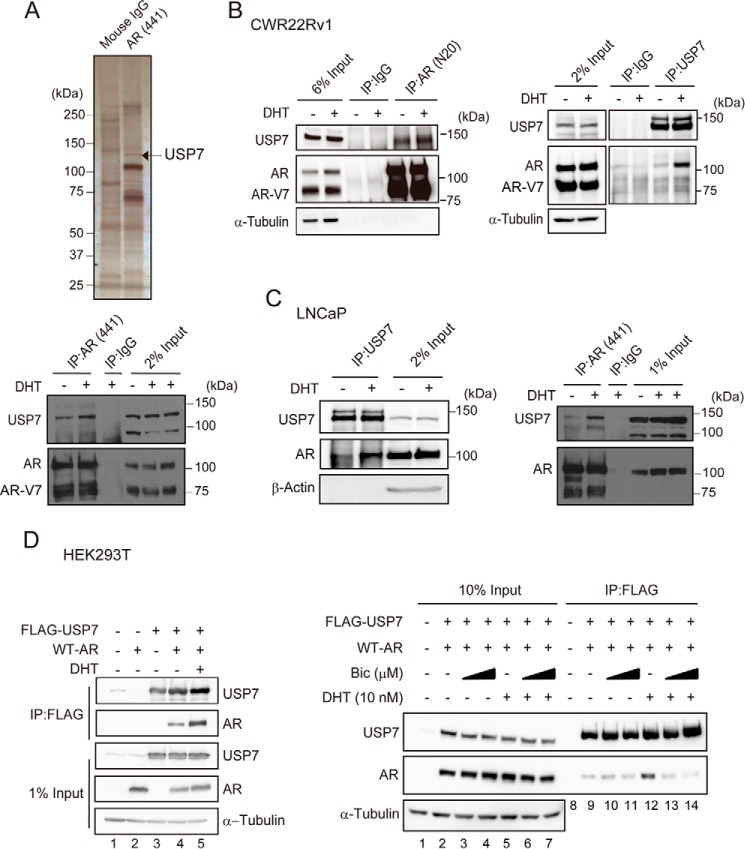
AR-associated proteins were obtained by affinity purification using the AR antibody anti-AR (441), which is specific to the central region of the AR transactivation domain (amino acids 299–315). The AR-associated proteins were separated by SDS-PAGE and visualized with silver staining. The proteins in the gel were excised for LC-MS/MS analysis. Among the proteins that specifically bound to AR but not to the IgG control, 128-kDa USP7, a deubiquitinating enzyme, was one of the top scoring proteins identified by LC-MS/MS (with a threshold false discovery rate of <1% and peptide number of >10) (Fig. 1A, upper panel). The LC-MS/MS identification results are shown in supplemental Table 1. The USP7-AR interaction was also observed using Western blotting (Fig. 1A, bottom panel).
FIGURE 1.
USP7 is a novel androgen-dependent AR-associated protein in prostate cancer cells. A, upper panel: the lysates from DHT-treated CWR22Rv1 cells were purified using anti-AR (441)- or control IgG-conjugated beads. Bound proteins were eluted, subjected to SDS-PAGE, and visualized by silver staining. The band identified as USP7 by LC-MS/MS is indicated. Lower panel: cell lysates from CWR22Rv1 cells were subjected to IP using anti-AR (441). The DHT-stimulated USP7-AR association was confirmed by Western blotting. Before DHT stimulation, cells were cultured in phenol red-free medium with 5% charcoal-stripped FBS for 3 days. B and C, the endogenous USP7-AR interaction was enhanced in a ligand-dependent manner in CWR22Rv1 (B) and LNCaP (C) cells. Cells were cultured in phenol red-free medium with 5% charcoal-stripped FBS for 3 days before 10 nm DHT treatment, and the cell lysates were subjected to IP using the indicated antibodies and analyzed by Western blotting. D, left panel, HEK293T cells were transfected with wild-type AR (WT-AR) and FLAG-USP7 expression vectors. At 24 h post-transfection, cells were treated with 10 nm DHT for 30 min before harvesting. The cell lysates were subjected to IP using anti-FLAG M2 affinity gel, and proteins were visualized by Western blotting. Right panel, HEK293T cells transfected with WT-AR and FLAG-USP7 were treated with 10 or 40 μm bicalutamide (Bic) for 2 h before stimulation with DHT or EtOH for 30 min. Cells were harvested and subjected to IP using anti-FLAG M2 affinity gel.
To validate the USP7-AR interaction, we performed reciprocal co-IP assays using another antibody against AR, anti-AR (N20), and anti-USP7 (A300–033A) in CWR22Rv1 cells. As shown in Fig. 1B (left panel), the USP7-AR interaction was observed using anti-AR (N20) but not the IgG control, confirming the identification result. Additionally, USP7 was associated with full-length AR rather than AR-V7 in an androgen-dependent manner (Fig. 1B, right panel).
Given that USP7 associated with AR in a DHT-dependent manner, we used the androgen-sensitive prostate cancer cell line LNCaP to assess this androgen-sensitive interaction between USP7 and AR. As shown in Fig. 1C, the USP7-AR association was clearly increased in DHT-treated LNCaP cells regardless of whether anti-AR or anti-USP7 was used.
To further investigate whether USP7 binds specifically to androgen-liganded AR, we tested the impact of bicalutamide on the exogenously expressed USP7-AR interaction. Bicalutamide is an AR antagonist that inhibits AR activation by competitively inhibiting the binding of endogenous androgens to AR (36). The androgen-dependent USP7-AR association was confirmed in USP7- and WT-AR-overexpressing HEK293T cells (Fig. 1D, left panel, lanes 4 and 5). Whereas DHT obviously enhanced the USP7-AR interaction (Fig. 1D, right panel, compare lane 9 with 12), bicalutamide had no noticeable effect on the USP7-AR interaction even upon high concentration stimulation (Fig. 1D, compare lane 9 with 10 and 11). Also, the DHT-enhanced USP7-AR association was attenuated by bicalutamide treatment (Fig. 1D, compare lane 12 with lanes 13 and 14). This result demonstrated that USP7 preferentially associates with androgen-liganded AR.
USP7 Mediates AR Deubiquitination in an Androgen-dependent Manner and Controls Its Stability
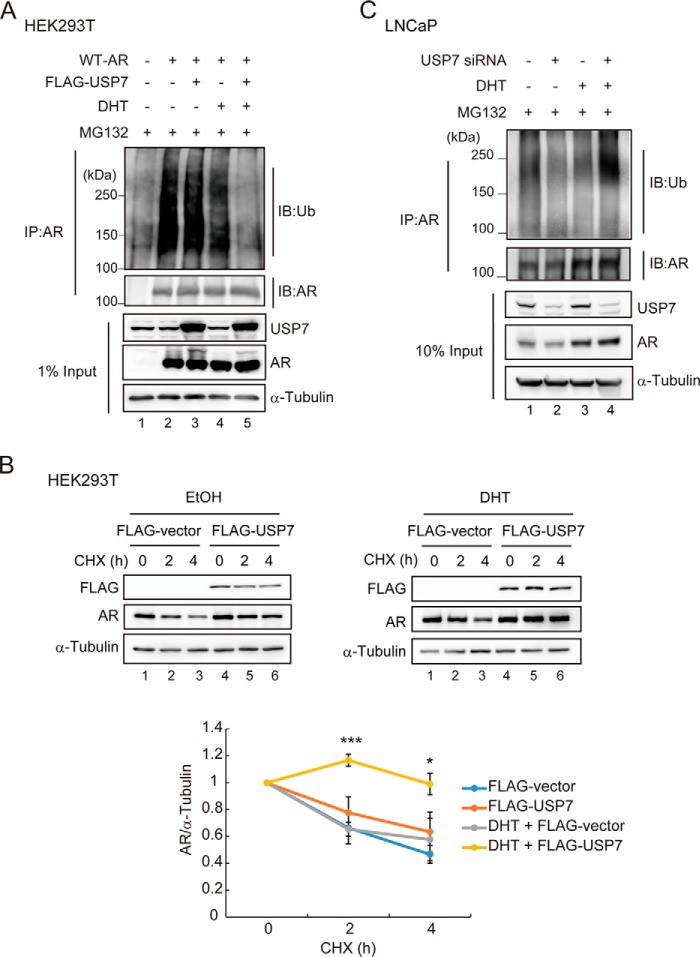
Because USP7 associates with AR in an androgen-dependent manner, we next examined whether the USP7-AR interaction affects the AR ubiquitination status. HEK293T cells were transfected with WT-AR either alone or with FLAG-USP7 in the presence or absence of DHT and were treated with the proteasome inhibitor MG132. The cell lysates were denatured and the exogenously expressed AR was immunoprecipitated using anti-AR (441). The ubiquitination status of exogenously expressed AR in HEK293T cells was detected by Western blotting using anti-ubiquitin. The AR ubiquitination was markedly decreased by DHT stimulation (Fig. 2A, compare lane 2 with 4). We also found that overexpression of USP7 notably attenuated ubiquitinated AR upon DHT stimulation (Fig. 2A, compare lane 4 with 5).
FIGURE 2.
USP7 mediates AR deubiquitination in an androgen-dependent manner. A, HEK293T cells were transfected with protein expression vectors as indicated for 48 h followed by treatment with 10 nm DHT or EtOH and 10 μm MG132 as indicated for 6 h. Cells were harvested, and lysates were denatured and subjected to IP using anti-AR (441). The protein ubiquitination status was visualized by Western blotting. B, HEK293T cells were transfected with expression vectors as indicated. At 24 h post-transfection, 10 nm DHT or EtOH was added for 2 h followed by 150 μg/ml of cycloheximide (CHX) addition for the indicated times before cells were harvested. AR was quantified by densitometry and normalized to α-Tubulin. Values are the mean ± S.D. of three independent experiments. C, USP7 siRNA-transfected LNCaP cells were treated with 10 nm DHT and 2 μm MG132 for 16 h before cells were harvested. The lysates were denatured and subjected to IP using anti-AR (441). The protein ubiquitination status was visualized by Western blotting.
To determine whether the deubiquitination effect of USP7 on AR alters AR protein stability, we analyzed the exogenous WT-AR protein degradation rate in USP7-overexpressing HEK293T cells. It had been reported that the protein half-life of exogenously expressed AR is ∼1–1.5 h in the absence of androgens (37); hence, we observed AR protein degradation 2 and 4 h after cycloheximide treatment (Fig. 2B, upper panel). The results showed that overexpression of USP7 had only a small effect on AR protein half-life without DHT stimulation. However, after DHT stimulation, the AR protein half-life was significantly prolonged by USP7. The results from three independent experiments were quantified by densitometry (Fig. 2B, bottom panel).
The effect of USP7 on endogenous AR protein ubiquitination status was further assessed in LNCaP cells. To determine this, USP7 in LNCaP cells was knocked down by siRNA, and cells were harvested after DHT and MG132 treatments. The endogenous AR in denatured lysates was immunoprecipitated by anti-AR (441), and the AR ubiquitination status was evaluated. Consistent with Fig. 2A, knockdown of USP7 increased the endogenously ubiquitinated AR in an androgen-dependent manner (Fig. 2C, lanes 3 and 4). These results demonstrated that the USP7-AR interaction is important for deubiquitination and stabilization of androgen-liganded AR.
USP7 Associates with AR on AREs in an Androgen-dependent Manner and Modulates Its Binding to Chromatin
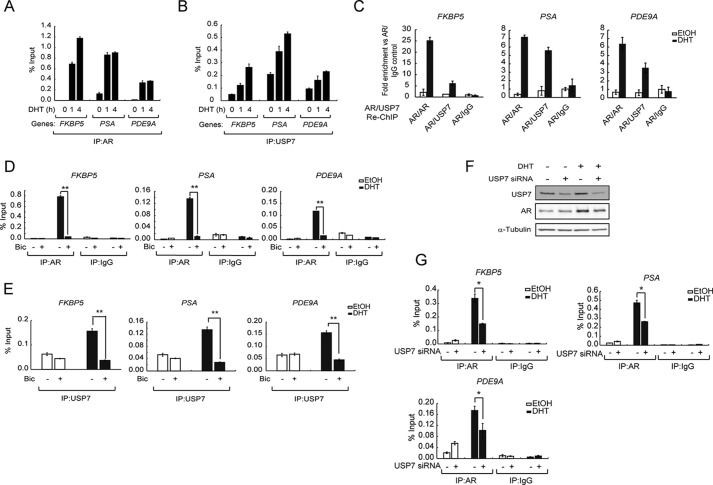
Our results showed that USP7 preferentially associates with and deubiquitinates androgen-liganded AR, suggesting a role for USP7 in AR activity regulation. To assess whether USP7 and AR are colocalized on target gene AREs to regulate gene transcription, we performed a ChIP-qPCR assay to examine the recruitment of USP7 to AREs of target genes upon DHT stimulation. LNCaP cells were stimulated with DHT for the indicated times, and the binding of AR and USP7 to AREs of the well characterized AR target genes FKBP5, PSA, and PDE9A (38–40) was measured. As expected, DHT induced the recruitment of AR to AREs of FKBP5, PSA, and PDE9A (Fig. 3A). Also, we found that the binding of USP7 to the same AREs showed a 2–3-fold increase upon DHT stimulation (Fig. 3B). These data suggested that USP7 associates with AR on AREs of target genes upon DHT stimulation.
FIGURE 3.
USP7 associates with AR on AREs in an androgen-dependent manner and is essential for AR binding to chromatin. A and B, LNCaP cells were treated with 10 nm DHT for the indicated times and subjected to ChIP-qPCR using anti-AR (441) (A) or anti-USP7 (B). Antibody-bound DNA was assessed by qPCR. C, LNCaP cells were treated with 10 nm DHT or EtOH for 1 h and subjected to sequential ChIP (Re-ChIP) using anti-AR as the first antibody and anti-AR, anti-USP7, or anti-IgG as the second antibody, as indicated. Antibody-bound DNA was assessed by qPCR. D and E, LNCaP cells were treated with 10 μm bicalutamide (Bic) for 2 h before induction with DHT for 4 h. ChIP-qPCR using anti-AR (D) or anti-USP7 (E) was performed to observe AR (D) and USP7 (E) recruitment to the AREs of FKBP5, PSA, and PDE9A as indicated. F, LNCaP cells were transfected with USP7 siRNA for 72 h followed by 10 nm DHT stimulation for 1 h before cells were harvested. Protein levels were assessed using Western blotting with the indicated antibodies. G, LNCaP cells were transfected with USP7 siRNA for 72 h before stimulation with DHT or EtOH for 1 h. The binding of AR to AREs of FKBP5, PSA, and PDE9A was visualized using ChIP-qPCR. Values are the mean ± S.D. of triplicate samples from one experiment.
To further determine the association between USP7 and AR on AREs of target genes, a sequential ChIP assay was performed after DHT stimulation using anti-AR (441) as the first-round antibody followed by anti-USP7 (A300–033A) as the second-round antibody. Anti-AR and anti-IgG were also included in the second round as positive and negative controls, respectively. Consistent with DHT-stimulated AR and USP7 binding to FKBP5, PSA, and PDE9A AREs, USP7 was detected in the AR-containing protein complex assembled on the AREs of FKBP5, PSA, and PDE9A upon DHT stimulation (Fig. 3C). This confirmed our assumption that USP7 and AR form a complex on target gene AREs upon DHT stimulation and might regulate gene transcription.
To further assess whether the binding of USP7 to AREs is through an interaction with androgen-liganded AR, we observed the binding of USP7 to AREs of target genes in bicalutamide-treated LNCaP cells. As shown in Fig. 3D, the DHT-stimulated AR binding to FKBP5, PSA, and PDE9A AREs was blocked by bicalutamide. In parallel with inhibition of AR recruitment by bicalutamide, the binding of USP7 to the AREs of these genes was also decreased to background levels (Fig. 3E). Considering that bicalutamide blocked the association between USP7 and androgen-liganded AR (Fig. 1D, right panel), these data indicated that USP7 binds to AREs through its association with androgen-liganded AR. After confirming the association between USP7 and AR on AREs of target genes, we further assessed whether the USP7-AR interaction is essential for AR transcriptional activity.
The effect of USP7 knockdown on the recruitment of AR to chromatin in LNCaP cells was determined. The level of AR protein in USP7-knockdown LNCaP cells was first assessed by Western blotting and shown to be slightly attenuated (Fig. 3F). We further investigated the impact of USP7 knockdown on DHT-stimulated AR binding to FKBP5, PSA, and PDE9A AREs. The USP7-knockdown LNCaP cells were stimulated with DHT and subjected to ChIP-qPCR using anti-AR. Consistent with the decrease in AR protein by USP7 knockdown, the DHT-stimulated recruitment of AR to chromatin AREs of FKBP5, PSA, and PDE9A was significantly reduced (Fig. 3G), indicating the USP7-AR interaction is important for regulation of AR-chromatin binding.
USP7 Cooperates with AR to Regulate Androgen-responsive Gene Expression in Prostate Cancer Cells
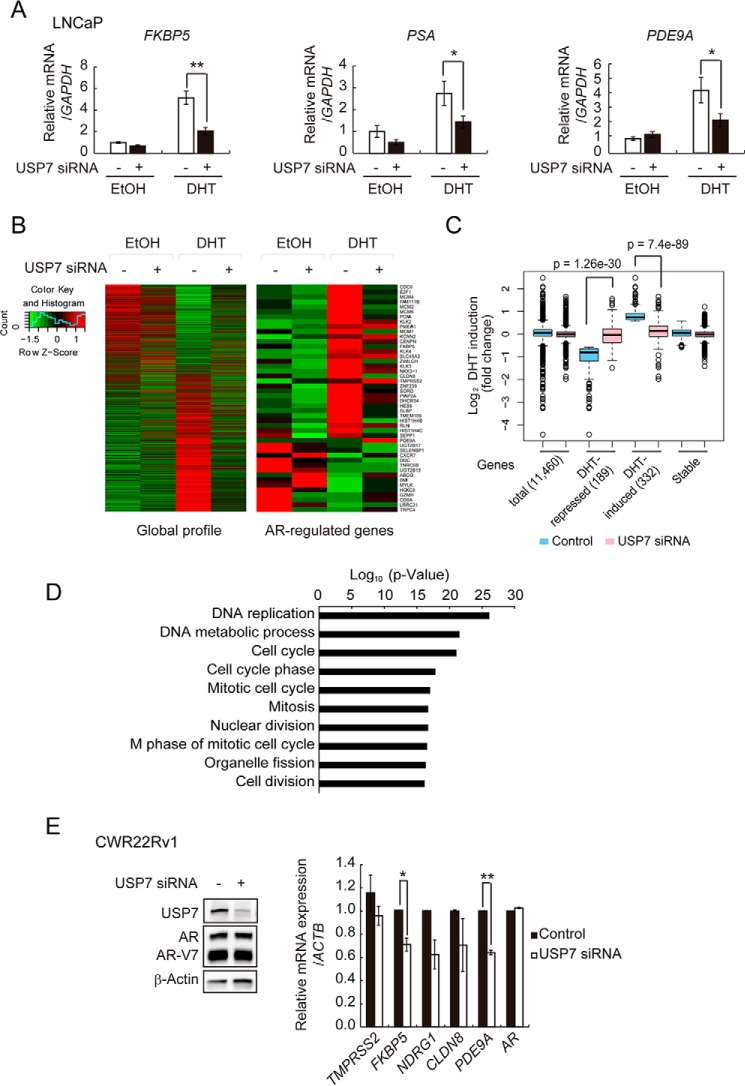
To assess whether USP7-modulated AR-chromatin binding alters its transcriptional output, we assessed the effect of USP7 knockdown on FKBP5, PSA, and PDE9A target gene expression. USP7 siRNA-transfected LNCaP cells were stimulated with DHT, and mRNA expression of these genes was investigated. As expected, mRNA expression of FKBP5, PSA, and PDE9A was attenuated by USP7 knockdown (Fig. 4A).
FIGURE 4.
USP7 is essential in androgen-responsive gene regulation. A, LNCaP cells were transfected with USP7 siRNA for 72 h before incubation with DHT or EtOH for 8 h. Cells were harvested for RNA isolation, and FKBP5, PSA, and PDE9A mRNA expression was determined by qPCR and normalized against expression of GAPDH. B, USP7 siRNA-transfected LNCaP cells were incubated with 10 nm DHT or EtOH for 16 h before cells were harvested. The isolated RNAs were subjected to RNA-seq analysis. Heat maps of all 11,460 transcripts (left) and several androgen-responsive genes (right) that were quantified by RNA-seq are shown. Up-regulated genes are in red; down-regulated genes are in green. C, DHT induction (fold) among four groups of genes is visualized by box plots. The number of genes is shown in parentheses. D, DAVID functional annotation of 179 DHT-induced genes that were differentially expressed upon USP7 knockdown. The top 10 overrepresented (p < 0.05) Gene Ontology biological process categories are shown. E, CWR22Rv1 cells were transfected with USP7 siRNA for 72 h before RNA isolation. Left, USP7 and AR levels in USP7-knockdown CWR22Rv1 cells were assessed by Western blotting. β-Actin was included as a loading control. Right, expression of several AR target genes was determined by qPCR and normalized against expression of ACTB. Values are the mean ± S.D. of three independent experiments.
To further evaluate the global effect of USP7 on AR transcriptional output, we used RNA-seq to analyze the effects of USP7 knockdown on the transcriptome of LNCaP cells. The USP7-knockdown cells were incubated with or without 10 nm DHT for 16 h before harvesting, and RNA-seq analysis was performed to determine altered gene profiling. We identified 11,460 transcripts that were expressed in the cells, and the quantitative results were visualized with a heat map (Fig. 4B, left). USP7 knockdown profoundly altered global transcription; the primary genes affected were DHT-responsive genes, illustrating the essential role of USP7 in regulating the expression of AR target genes. The expression levels of several well known AR target genes implicated in prostate cancer progression are shown in Fig. 4B (right). USP7 knockdown significantly decreased the expression of DHT-induced genes, including FAM111B, KLK2, PSA, and FKBP5. Among the DHT-repressed genes, USP7 knockdown derepressed BMF, CXCR7, and TRPC4 (Fig. 4B, right panel). We also found that E2F1 and HES6, which were recently identified as crucial regulators in CRPC development (41), were down-regulated by USP7 knockdown (Fig. 4B, right panel). The relative expression levels of the top 10 DHT-induced and -repressed genes in USP7-depleted cells are shown in Tables 2 and 3.
TABLE 2.
Top 10 DHT-induced genes
| Gene symbol | Description | Relative expression levels |
|||
|---|---|---|---|---|---|
| Control | USP7 siRNA | DHT | DHT + USP7 siRNA | ||
| KLK2 | Kallikrein-related peptidase 2 | 1.00 | 1.12 | 5.61 | 3.54 |
| FAM111B | Family with sequence similarity 111, member B | 1.00 | 0.48 | 4.27 | 0.91 |
| KLK3 | Kallikrein-related peptidase 3 | 1.00 | 0.83 | 4.13 | 2.22 |
| CDC6 | Cell division cycle 6 | 1.00 | 0.45 | 3.54 | 0.99 |
| TMEM 160 | Transmembrane protein 160 | 1.00 | 2.46 | 3.45 | 2.77 |
| LOC391322 | d-Dopachrome tautomerase-like | 1.00 | 1.94 | 3.37 | 2.67 |
| C4orf48 | Chromosome 4 open reading frame 48 | 1.00 | 2.15 | 3.33 | 3.05 |
| E2F1 | E2F transcription factor 1 | 1.00 | 0.47 | 3.19 | 1.17 |
| CCDC85B | Coiled-coil domain containing 85B | 1.00 | 1.89 | 3.18 | 1.46 |
| GNMT | Glycine N-methyltransferase | 1.00 | 0.76 | 3.07 | 1.42 |
TABLE 3.
Top 10 DHT-repressed genes
| Gene symbol | Description | Relative expression levels |
|||
|---|---|---|---|---|---|
| Control | USP7 siRNA | DHT | DHT + USP7 siRNA | ||
| ZCWPW2 | Zinc finger, CW type with PWWP domain 2 | 1.00 | 0.08 | 0.10 | 0.15 |
| TRPC4 | Transient receptor potential cation channel, subfamily C, member 4 | 1.00 | 0.12 | 0.11 | 0.29 |
| GZMH | Granzyme H (cathepsin G-like 2, protein h-CCPX) | 1.00 | 0.11 | 0.11 | 0.33 |
| CD8A | CD8a molecule | 1.00 | 0.15 | 0.13 | 0.30 |
| MT1A | Metallothionein 1A | 1.00 | 0.14 | 0.14 | 0.30 |
| CHODL | Chondrolectin | 1.00 | 0.11 | 0.14 | 0.29 |
| SPIC | Spi-C transcription factor (Spi-1/PU.1 related) | 1.00 | 0.12 | 0.15 | 0.25 |
| UPB1 | Ureidopropionase, β | 1.00 | 0.18 | 0.18 | 0.18 |
| FAM71F2 | Family with sequence similarity 71, member F2 | 1.00 | 0.16 | 0.19 | 0.42 |
| CKM | Creatine kinase, muscle | 1.00 | 0.12 | 0.20 | 0.22 |
To validate this finding using qPCR, we chose several well characterized AR target genes including PSA, FKBP5, and PDE9A and the DNA replication factors MCM2, MCM6, and CDC6 as androgen-induced genes. BMF and SLC44A1 were chosen as DHT-repressed genes. The qPCR results correlated very well with our global RNA-seq data (data not show).
Because the RNA-seq result was validated, we identified the USP7-regulated androgen-responsive genes. Genes exhibiting a ≥1.5-fold change in expression after DHT stimulation (relative to vehicle) were scored as DHT-responsive genes. Based on this threshold, we defined 332 genes as DHT induced and 189 genes as DHT repressed. To identify which of these DHT-responsive genes were USP7 dependent, we similarly defined genes exhibiting ≥1.5-fold change in expression upon USP7 knockdown (relative to levels in control transfected cells) as USP7-regulated genes. We then examined the global effects of USP7 knockdown on DHT-stimulated gene expression compared with total and stable groups of genes. As shown in Fig. 4C, USP7 knockdown significantly reduced the effect of DHT on both repressed and induced genes, suggesting an essential role for USP7 in AR transcriptional output.
We further investigated the cooperation of USP7 and AR to regulate gene expression. Knockdown of USP7 decreased 54% of DHT-induced genes and derepressed 46% of DHT-repressed genes (data not shown). These data suggested that, in the presence of DHT, USP7 regulates the expression of DHT-responsive genes in the same direction as AR.
To determine the biological function of USP7 in androgen signaling, the subset of DHT-responsive genes that were regulated by USP7 was subjected to functional analysis. Gene Ontology analysis of 179 DHT-induced genes revealed that they are highly associated with DNA replication (Fig. 4D), suggesting that USP7 plays a critical role in DHT-regulated prostate cancer cell DNA replication.
We also examined the effect of USP7 knockdown in CRPC CWR22Rv1 cells. USP7 siRNA reduced the level of USP7 in these cells (Fig. 4E, left) and also decreased AR target gene expression (Fig. 4E, right).
USP7 Is Required for Prostate Cancer Cell Proliferation
AR plays a fundamental role in prostate cancer progression by promoting cell proliferation. Because our data suggested that USP7 cooperates with AR to regulate the expression of a subset of DHT-responsive genes, we further evaluated the physiological relevance of USP7 in prostate cancer progression. For this purpose, we investigated the effect of USP7 knockdown on LNCaP and CWR22Rv1 cell proliferation with or without DHT stimulation. USP7 knockdown attenuated the proliferation of LNCaP cells, particularly in the presence of DHT (Fig. 5A), demonstrating the key role of USP7 in androgen-sensitive prostate cancer cell proliferation. In addition, knockdown of USP7 also impaired CWR22Rv1 cell proliferation in the absence and presence of DHT (Fig. 5B).
FIGURE 5.
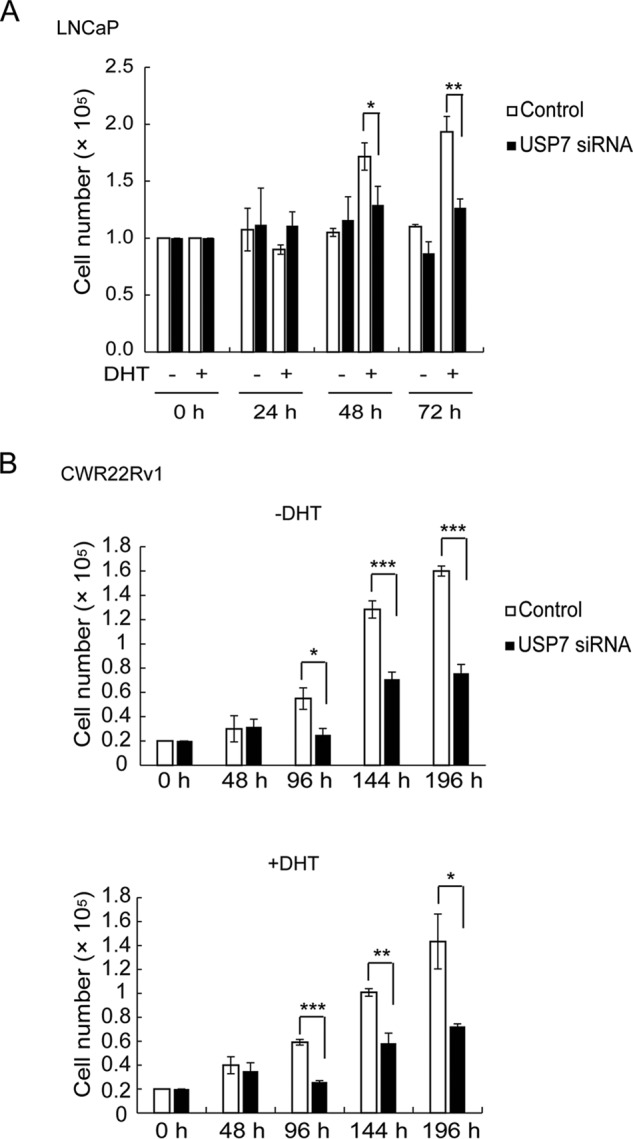
USP7 is required for prostate cancer cell proliferation. A, LNCaP cells were transfected with USP7 siRNA, and cells were counted at the indicated times after transfection in the presence or absence of DHT. B, CWR22Rv1 cells were transfected with USP7 siRNA, and cells were counted at the indicated times after transfection in the absence (upper) or presence (lower) of DHT. Values are the mean ± S.D. of three independent experiments.
Discussion
In this study, we identified USP7 as a novel AR co-regulator that mediates AR deubiquitination and stabilization. The USP7-AR interaction is also essential to androgen-activated AR binding to chromatin. The ability of USP7 to modulate the binding of AR to chromatin might contribute to appropriate regulation of AR transcriptional output.
We first demonstrated that USP7 mediates AR protein ubiquitination and protein stability in an androgen-dependent manner. The association of USP7 with androgen-activated AR on target gene regulatory elements was also confirmed. This effect of USP7 on androgen-liganded AR might have an important role in facilitating the transcriptional-activating binding of AR to target gene regulatory elements.
Many AR E3 ubiquitin ligase co-regulators have been identified in prostate cancer cells that modulate AR transcriptional activity and specificity, including MDM2 and SIAH2 (14–18). It is possible that USP7 coordinates with E3 ubiquitin ligase co-regulators to modulate the AR ubiquitination status, thereby fine-tuning AR transcriptional activity. This hypothesis is supported by the action of USP7 on another transcription factor, NF-κB, the central transcription factor of inflammation and immunity genes. USP7 controls the ubiquitination/deubiquitination status of DNA-bound NF-κB, which modulates its effects on transcription (42). These observations suggest that USP7-mediated deubiquitination and stabilization of transcription factors, including NF-κB as well as AR, might be a common mechanism for regulating transcriptional output.
In addition to AR itself, USP7 might also target other substrates that are involved in AR-chromatin binding and transcriptional activity regulation. Recently, USP10 was reported to deubiquitinate histone variant H2A.Z, which is required for AR transcriptional activation (43, 44). Because USP7 is proposed to modulate H2A and H2B ubiquitination via direct or indirect pathways (45, 46), the effect of USP7 on AR-chromatin binding and transcriptional activity could be a combinatorial consequence of both direct deubiquitination of AR and indirect deubiquitination of histones.
This study also revealed that USP7 associates with full-length AR in an androgen-dependent manner but not with AR-V7, which lacks the LBD, indicating that the AR LBD is critical for USP7 binding to AR. In addition, USP7 preferentially associated with androgen-liganded AR but not with bicalutamide-liganded AR. An analysis of the AR LBD crystal structural indicates that different ligands (e.g. agonists, antagonists) induce different conformational changes in the LBD (47), implying that androgen-induced repositioning of the AR LBD is critical for USP7 binding. This explains why USP7 deubiquitination and stabilization of AR are androgen dependent.
Many AR point mutations that allow AR to be activated by endogenous non-androgenic steroids as well as synthetic AR antagonists are found in CRPC (5, 6). Given that USP7 preferentially interacts with androgen-bound, transcriptionally active AR, USP7 may also bind to these antagonist-bound, transcriptionally active mutant forms of AR and regulate their activities in CRPC.
The function of USP7 in CRPC cells is still unknown, although this study showed that E2F1 and HES6, which have recently been identified as crucial regulators in CRPC development (41), were down-regulated by USP7 knockdown. Additionally, USP7 depletion affected AR target gene expression and the proliferation of CRPC CWR22Rv1 cells. These data suggest that USP7 also functions in CRPC development.
Given the multiple biological functions of USP7, it is also likely to promote prostate cancer progression via an AR-independent pathway (23, 48–50). Indeed, it has been reported that overexpression of USP7 in prostate cancer results in predominantly nuclear-excluded PTEN, which contributes to cancer progression (28). Here, we demonstrated that USP7 associates with AR in an androgen-dependent manner and forms a complex on AREs of target genes. The transcriptome profiling of LNCaP cells also showed strong overlap of the USP7-regulated genes with androgen-responsive genes only upon DHT stimulation. These data demonstrated that USP7 cooperates with AR to regulate gene expression in androgen signaling. Hence, USP7 might promote prostate cancer progression via both AR-dependent and AR-independent pathways.
Although the mechanism of USP7 gene overexpression or mutation in prostate cancer and its correlation with tumor aggressiveness have not been established, our study indicates that inhibition of USP7 can counteract androgen-regulated AR activation in prostate cancer cell lines. Therefore, pharmacological inhibition of USP7 represents a compelling therapeutic strategy for prostate cancer therapy.
Author Contributions
S. C., M. B., and K. S. designed experiments; S. C. and M. O. performed biochemical analyses; S. C., R. N., and M. B. carried out the RNA sequencing analysis; S. C., M. B., M. O., R. N., and K. S. interpreted the data; S. C., M. B., M. O., K. I., and K. S. drafted the manuscript.
Supplementary Material
Acknowledgments
We thank K. Nakagawa, N. Yokota, and S. Watanabe for technical assistance.
This work was supported by Grants-in-Aid for Scientific Research from the Research Program of Innovative Cell Biology by Innovative Technology, Grant-in-Aid for Scientific Research (to K. S.) (category S), Grant-in-Aid for Scientific Research (to M. B.) (category B) and Japan Society for the Promotion of Science KAKENHI Grant Number 24-5985. The authors declare no competing financial interests.

This article contains supplemental Table S1.
- AR
- androgen receptor
- ARE
- androgen-responsive element
- CRPC
- castration-resistant prostate cancer
- DHT
- 5α-dihydrotestosterone
- IP
- immunoprecipitation
- qPCR
- quantitative PCR
- RNA-seq
- RNA sequencing
- LBD
- ligand-binding domain.
References
- 1. Wilson J. D., Griffin J. E., George F. W., Leshin M. (1983) The endocrine control of male phenotypic development. Aust. J. Biol. Sci. 36, 101–128 [DOI] [PubMed] [Google Scholar]
- 2. Whitfield G. K., Jurutka P. W., Haussler C. A., Haussler M. R. (1999) Steroid hormone receptors: evolution, ligands, and molecular basis of biologic function. J. Cell Biochem. 32, 110–122 [DOI] [PubMed] [Google Scholar]
- 3. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) The nuclear receptor superfamily: the second decade. Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nelson W. G., De Marzo A. M., Isaacs W. B. (2003) Prostate Cancer. N. Engl. J. Med. 349, 366–381 [DOI] [PubMed] [Google Scholar]
- 5. Feldman B. J., Feldman D. (2001) The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1, 34–45 [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto T., Sakari M., Okada M., Yokoyama A., Takahashi S., Kouzmenko A., Kato S. (2013) The androgen receptor in health and disease. Annu. Rev. Physiol. 75, 201–224 [DOI] [PubMed] [Google Scholar]
- 7. Lee H. J., Chang C. (2003) Recent advances in androgen receptor action. Cell Mol. Life Sci. 60, 1613–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heemers H. V., Tindall D. J. (2009) Unraveling the complexities of androgen receptor signaling in prostate cancer cells. Cancer Cell 15, 245–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKenna N. J., O'Malley B. W. (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108, 465–474 [DOI] [PubMed] [Google Scholar]
- 10. van der Steen T., Tindall D. J., Huang H. (2013) Posttranslational modification of the androgen receptor in prostate cancer. Int. J. Mol. Sci. 14, 14833–14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heinlein C. A., Chang C. (2002) Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 23, 175–200 [DOI] [PubMed] [Google Scholar]
- 12. Heemers H. V., Tindall D. J. (2007) Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr. Rev. 28, 778–808 [DOI] [PubMed] [Google Scholar]
- 13. Coffey K., Robson C. N. (2012) Regulation of the androgen receptor by post-translational modifications. J. Endocrinol. 215, 221–237 [DOI] [PubMed] [Google Scholar]
- 14. Lin H. K., Wang L., Hu Y. C., Altuwaijri S., Chang C. (2002) Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21, 4037–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaughan L., Logan I. R., Neal D. E., Robson C. N. (2005) Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 33, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Felten A., Brinckmann D., Landsberg G., Scheidtmann K. H. (2013) Zipper-interacting protein kinase is involved in regulation of ubiquitination of the androgen receptor, thereby contributing to dynamic transcription complex assembly. Oncogene 32, 4981–4988 [DOI] [PubMed] [Google Scholar]
- 17. Chymkowitch P., Le May N., Charneau P., Compe E., Egly J. M. (2011) The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 30, 468–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi J., Tripathi M., Mishra R., Sahgal N., Fazli L., Fazil L., Ettinger S., Placzek W. J., Claps G., Chung L. W., Bowtell D., Gleave M., Bhowmick N., Ronai Z. A. (2013) The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell 23, 332–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu K., Shimelis H., Linn D. E., Jiang R., Yang X., Sun F., Guo Z., Chen H., Li W., Chen H., Kong X., Melamed J., Fang S., Xiao Z., Veenstra T. D., Qiu Y. (2009) Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 15, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burska U. L., Harle V. J., Coffey K., Darby S., Ramsey H., O'Neill D., Logan I. R., Gaughan L., Robson C. N. (2013) Deubiquitinating enzyme Usp12 is a novel co-activator of the androgen receptor. J. Biol. Chem. 288, 32641–32650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dirac A. M., Bernards R. (2010) The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Mol. Cancer Res. 8, 844–854 [DOI] [PubMed] [Google Scholar]
- 22. Everett R. D., Meredith M., Orr A., Cross A., Kathoria M., Parkinson J. (1997) A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16, 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nicholson B., Suresh Kumar K. G. (2011) The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 60, 61–68 [DOI] [PubMed] [Google Scholar]
- 24. Li M., Chen D., Shiloh A., Luo J., Nikolaev A. Y., Qin J., Gu W. (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 [DOI] [PubMed] [Google Scholar]
- 25. Kon N., Kobayashi Y., Li M., Brooks C. L., Ludwig T., Gu W. (2010) Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29, 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M., Brooks C. L., Kon N., Gu W. (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 [DOI] [PubMed] [Google Scholar]
- 27. van der Horst A., de Vries-Smits A. M., Brenkman A. B., van Triest M. H., van den Broek N., Colland F., Maurice M. M., Burgering B. M. (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell. Biol. 8, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 28. Song M. S., Salmena L., Carracedo A., Egia A., Lo-Coco F., Teruya-Feldstein J., Pandolfi P. P. (2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deardorff M. A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D., Cole K. E., De Baere E., Decroos C., Di Donato N., Ernst S., Francey L. J., Gyftodimou Y., Hirashima K., Hullings M., Ishikawa Y., Jaulin C., Kaur M., Kiyono T., Lombardi P. M., Magnaghi-Jaulin L., et al. (2012) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson M. D., Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 33. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 34. Sramkosk R. M., Ii T. G. P., Giaconia J. M., Pretlow T. P., Schwartz S., Sy M., Marengo S. R., Rhim J. S., Zhang D., Jacobbergew J. W. (1999) A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev. Biol. Anim. 145, 403–409 [DOI] [PubMed] [Google Scholar]
- 35. Tepper C. G., Boucher D. L., Ryan P. E., Ma A.-H., Xia L., Lee L.-F., Pretlow T. G., Kung H. J. (2002) Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 62, 6606–6614 [PubMed] [Google Scholar]
- 36. Taplin M. E. (2007) Drug insight: role of the androgen receptor in the development and progression of prostate cancer. Nat. Clin. Pract. Oncol. 4, 236–244 [DOI] [PubMed] [Google Scholar]
- 37. Kemppainen J. A., Lane M. V., Sar M., Wilson E. M. (1992) Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation: specificity for steroids and antihormones. J. Biol. Chem. 267, 968–974 [PubMed] [Google Scholar]
- 38. Chng K. R., Chang C. W., Tan S. K., Yang C., Hong S. Z., Sng N. Y., Cheung E. (2012) A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 31, 2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Q., Carroll J. S., Brown M. (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19, 631–642 [DOI] [PubMed] [Google Scholar]
- 40. Wang Q., Li W., Liu X. S., Carroll J. S., Jänne O. A., Keeton E. K., Chinnaiyan A. M., Pienta K. J., Brown M. (2007) A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramos-Montoya A., Lamb A. D., Russell R., Carroll T., Jurmeister S., Galeano-Dalmau N., Massie C. E., Boren J., Bon H., Theodorou V., Vias M., Shaw G. L., Sharma N. L., Ross-Adams H., Scott H. E., Vowler S. L., Howat W. J., Warren A. Y., Wooster R. F., Mills I. G., Neal D. E. (2014) HES6 drives a critical AR transcriptional programme to induce castration-resistant prostate cancer through activation of an E2F1-mediated cell cycle network. EMBO Mol. Med. 6, 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colleran A., Collins P. E., O'Carroll C., Ahmed A., Mao X., McManus B., Kiely P. A., Burstein E., Carmody R. J. (2013) Deubiquitination of NF-κB by ubiquitin-specific protease-7 promotes transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faus H., Meyer H. A., Huber M., Bahr I., Haendler B. (2005) The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol. Cell Endocrinol. 245, 138–146 [DOI] [PubMed] [Google Scholar]
- 44. Draker R., Sarcinella E., Cheung P. (2011) USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 39, 3529–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Knaap J. A., Kumar B. R., Moshkin Y. M., Langenberg K., Krijgsveld J., Heck A. J., Karch F., Verrijzer C. P. (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695–707 [DOI] [PubMed] [Google Scholar]
- 46. Lecona E., Narendra V., Reinberg D. (2015) Usp7 cooperates with Scml2 to regulate the activity of Prc1. Mol. Cell Biol. 35, 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Helsen C., Claessens F. (2014) Looking at nuclear receptors from a new angle. Mol. Cell. Endocrinol. 382, 97–106 [DOI] [PubMed] [Google Scholar]
- 48. Becker K., Marchenko N. D., Maurice M., Moll U. M. (2007) Hyperubiquitylation of wild-type p53 contributes to cytoplasmic sequestration in neuroblastoma. Cell Death Differ. 14, 1350–1360 [DOI] [PubMed] [Google Scholar]
- 49. Cheng C., Niu C., Yang Y., Wang Y., Lu M. (2013) Expression of HAUSP in gliomas correlates with disease progression and survival of patients. Oncol. Rep. 29, 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Masuya D., Huang C., Liu D., Nakashima T., Yokomise H., Ueno M., Nakashima N., Sumitomo S. (2006) The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J. Pathol. 208, 724–732 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.