Background: The bacterial glycyl radical enzyme CutC converts choline to trimethylamine, a metabolite involved in pathogenesis of several diseases.
Results: The structures of substrate-bound and substrate-free CutC revealed significant differences.
Conclusion: Choline binding to the active site triggers a conformational change from the open to closed form.
Significance: A novel substrate-driven conformational mechanism and a potential target for drug design have been identified.
Keywords: choline, conformational change, enzyme catalysis, enzyme structure, radical
Abstract
CutC choline trimethylamine-lyase is an anaerobic bacterial glycyl radical enzyme (GRE) that cleaves choline to produce trimethylamine (TMA) and acetaldehyde. In humans, TMA is produced exclusively by the intestinal microbiota, and its metabolite, trimethylamine oxide, has been associated with a higher risk of cardiovascular diseases. Therefore, information about the three-dimensional structures of TMA-producing enzymes is important for microbiota-targeted drug discovery. We have cloned, expressed, and purified the CutC GRE and the activating enzyme CutD from Klebsiella pneumoniae, a representative of the human microbiota. We have determined the first crystal structures of both the choline-bound and choline-free forms of CutC and have discovered that binding of choline at the ligand-binding site triggers conformational changes in the enzyme structure, a feature that has not been observed for any other characterized GRE.
Introduction
Trimethylamine (TMA)2 is a tertiary amine that is produced by bacteria from the quaternary amine compounds choline (1, 2) and carnitine (3), which are present in food. The human organism is incapable of synthesizing TMA, and therefore, the bacteria of the microbiota are the only source of this metabolite. After absorption in the bloodstream, TMA is further metabolized to trimethylamine oxide (TMAO) by FMO3 (flavin-containing monooxygenase 3) in the liver. Defective or insufficient amounts of FMO3 cause TMA accumulation in the blood, which manifests as trimethylaminuria, or fish odor syndrome (4). Recent studies have linked the increased availability of tertiary amines with a higher risk of cardiovascular disease via an intestinal microbiota-dependent pathway (5). High levels of TMAO, which are produced from choline (6) and carnitine (3) with the assistance of the intestinal microflora, have been shown to be a cause of atherosclerosis in mice. Studies with human volunteers (3, 7) have confirmed TMA production from carnitine and choline by the intestinal microflora, and clinical data cross-examinations have associated high levels of TMAO with an elevated risk of atherosclerosis (7), severe heart failure (8), and renal failure (9). Lowering TMA and TMAO levels in humans could therefore have potentially preventive and therapeutic effects on trimethylaminuria and cardiovascular diseases. TMAO also increased glucose tolerance and adipose tissue inflammation in mice fed a high-fat diet (10), suggesting involvement in the development of diabetes. Structural biology studies of microbiota enzymes are important to guide the discovery of novel inhibitor compounds.
CutC choline lyase, a TMA-producing glycyl radical enzyme (GRE), has recently been identified and characterized (11, 12). A common feature of all GREs is a glycyl radical in the active site. The GRE-activating enzyme, a 4Fe-4S cluster protein, cleaves S-adenosylmethionine (SAM) into a radical (13) that further generates a glycyl radical on the active site glycine. The structures of numerous GREs that catalyze different reactions, such as pyruvate formate-lyase (PFL) (14), ribonucleotide reductase (15), glycerol dehydratase (GD) (16), and benzylsuccinate synthase (BSS) (17), have been determined. Along with the activating enzyme CutD, CutC cleaves choline, forming TMA and acetaldehyde under anaerobic conditions. Under aerobic conditions, catalysis is impossible because the glycyl radical readily reacts with oxygen, cleaves the main chain, and irreversibly inactivates the enzyme (18).
The cutC/cutD gene cluster is possessed by several representatives of the human intestinal microbiota, including bacteria in the genera Klebsiella, Escherichia, Streptococcus, and Enterobacter (11). The mean abundance of representatives of these genera in the stool microbiota is low, but in certain individuals, it can rise to 0.56% for Klebsiella pneumoniae and to 96% for Escherichia coli (19). Not all bacteria of the microbiota are capable of producing TMA, including Bacteroides, the most abundant genus in stool (19, 20). Therefore, even a small increase in the amount of TMA-producing microorganisms could substantially affect the amounts of TMA and TMAO in an individual. These findings suggest that the CutC/CutD metabolic pathway could be accountable for a significant portion of TMA production in human intestines.
It has been shown that the production of TMAO can be suppressed by shifting the microbial degradation pattern of supplemental/dietary tertiary amines (21). In addition, the cardioprotective drug meldonium, an inhibitor of l-carnitine biosynthesis and transport (22), has been shown to lower TMAO levels in both rats and humans (21, 23) through inhibition of TMA production by the intestinal microbiota.
CutC/CutD enzymatic activity in Desulfovibrio alaskensis has previously been demonstrated both in vivo and in vitro (11, 12). Our model organism, K. pneumoniae, has previously been shown to be capable of producing TMA from both choline and carnitine (21), but the activity of the CutC/CutD enzymes in this organism has not been demonstrated. To our knowledge, no crystal structure of CutC has been determined until now. Here, we report the crystal structure of the CutC enzyme from K. pneumoniae in both its choline-free and choline-bound forms.
Experimental Procedures
Plasmid Construction and Protein Expression
Genomic DNA was extracted from K. pneumoniae culture (The Microbial Strain Collection of Latvia) by proteinase K treatment and precipitation with ethanol. CutC (GenBankTM accession number EPO20241.1) and CutD (accession number EPO20361.1) sequences from the GenBankTM accession number ARRZ01000032.1 entry were used for primer design (CutC, ATATTCATGACGGCACACTACAACTTAACGCCGC (forward) and AATTAAGCTTTTAGAACTTCTCAATCACCGTACGGC (reverse); and CutD, TATAGGCCTCATCGCAAAACAAGAATTAACGGG (forward) and AATTCCATGGTTAATGGCGGACTAAGCGAATATC (reverse)). The cutC and cutD genes were amplified by PCR and cloned into pRSFDuet and pET1 vectors containing N-terminal His6 tags with tobacco etch virus protease cleavage sites. The constructs were sequenced for confirmation. The proteins were expressed in E. coli BL21(DE3) cells. Upon reaching A540 > 0.3, induction was performed with 1 mm isopropyl β-d-thiogalactopyranoside in 2× Tryptone/yeast extract medium for 3 h at +37 °C.
CutC Purification
Frozen cell paste was suspended in lysis buffer (100 mm Tris-HCl (pH 8.0), 200 mm NaCl, 1% Triton X-100, 20 mm MgSO4, 0.1 mg/ml DNase, 1 mg/ml lysozyme, 1 mm PMSF, and 2 mm DTT). Cells were lysed by ultrasound, and the lysate was centrifuged at 14,000 × g for 40 min. CutC protein was purified by nickel affinity chromatography using a 1-ml HisTrap column (GE Healthcare). For this step, 20 mm imidazole in 40 mm Tris-HCl (pH 8.0) and 300 mm NaCl was used as a washing buffer, and 300 mm imidazole in 40 mm Tris-HCl (pH 8.0) and 300 mm NaCl was used as an elution buffer.
Limited Proteolysis
For preparative purposes, the 124-kDa CutC protein obtained in HisTrap elution buffer was immediately digested with chymotrypsin using 1 mg of chymotrypsin/100 mg of CutC protein for 30 min at room temperature. The 90–92-kDa CutC fragment was separated from the digestion products and chymotrypsin by gel filtration on a Superdex 200 column in 20 mm Tris-HCl (pH 8.0). The protein was concentrated by ultrafiltration and stored in 50% glycerol at −20 °C.
For analytic purposes, after purification from the HisTrap column, the 124-kDa CutC protein was transferred to 20 mm Bistris propane buffer at pH 6.5 and 8.5 via ultrafiltration. Trypsin was added at 1 mg of chymotrypsin/100 mg of CutC protein. Digestion was performed for 6 h at room temperature. The CutC molecular mass after cleavage was characterized by both SDS-PAGE and MALDI-TOF-MS.
CutD Purification and Reconstitution
Cell lysis was performed as described for CutC. After lysate centrifugation, the pellet was resuspended in 7 m urea with 5 mm DTT. CutD was purified by nickel affinity chromatography using a 1-ml HisTrap column as described for CutC, except all of the buffers were supplemented with 7 m urea. After purification, the CutD solution was sparged with argon and dialyzed overnight in anaerobic solution containing 150 mm KCl, 25 mm MOPS/KOH (pH 7.5), 5 mm DTT, 20 mm Na2S, and 25 mm (NH4)2Fe(SO4)2. After dialysis, the CutD solution was desalted and purified from precipitates on a 1-ml Zeba desalting column, equilibrated with 25 mm Tris-HCl (pH 7.5) and 50 mm NaCl. All steps after CutD purification with the HisTrap column were performed in an argon atmosphere in a disposable glove bag (Sigma-Aldrich).
In Vitro Activity Assay
The activity assay was performed in 25 mm Tris-HCl (pH 8.0), 50 mm NaCl, 3 mm SAM, 1 mm choline, 5 mm sodium dithionite, 5 mg/ml CutC (untreated or treated with chymotrypsin), and 4 mg/ml CutD. All stock solutions were sparged beforehand with argon for 10–15 min in a disposable glove bag. Samples with all components except SAM, CutC, or CutD were incubated as negative controls. Incubation was performed in an argon atmosphere at room temperature for 15 h. After incubation, the solution was mixed with 100% (50:50, v/v) formic acid and stored at −20 °C. The TMA quantity was analyzed by ultra performance liquid chromatography (UPLC)-MS/MS.
In Vivo Activity Assay
For the in vivo activity assay, 0.5 ml of E. coli BL21(DE3) night cultures harboring pRSF-CutC, pET-CutD, empty pET vector, and cotransfected pRSF-CutC and pET-CutD were inoculated in 15 ml of LB medium in Hungate tubes (Chemglass). The LB medium was sparged beforehand with argon for 30 min and supplemented with 0.4 m NaCl, 1 mm choline chloride, and 0.069 mg/ml ferric ammonium citrate as described previously (11). The cultures were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside after reaching A540 > 0.3. Incubation was performed for an additional 18 h at 37 °C with shaking. The cell culture was centrifuged, the supernatant was mixed with 100% (50:50, v/v) formic acid, and the TMA quantity was analyzed by UPLC-MS/MS.
CutC Crystallization and Data Collection
The protein was concentrated by ultrafiltration to 16 mg/ml in 20 mm Tris-HCl (pH 8.0). To obtain the substrate-bound form, choline chloride was added to a final concentration of 5 mm. CutC was crystallized using the sitting drop method, mixing 1 μl of protein solution with 1 μl of precipitant (20% (v/w) PEG 3350, 20–60 mm potassium/sodium tartrate, and 100 mm Bistris (pH 8.5)). CutC crystals without choline were obtained in slightly different conditions (20% (v/w) PEG 3350, 100–160 mm potassium/sodium tartrate, and 100 mm Bistris ((pH 6.5). Crystals appeared overnight. Crystals were soaked in cryoprotectant (30% glycerol in mother liquid) and frozen in liquid nitrogen. Data were collected at MAX-lab Synchrotron beamline I911-3 (Lund, Sweden). The best diffracting crystals were of the choline-bound form and diffracted up to 2.4 Å resolution. The best crystals without choline diffracted to 3.0 Å resolution.
MALDI-TOF-MS
Crystallization drop solution (1 μl) was mixed with 1 μl of 0.1% TFA and 1 μl of matrix solution containing 15 mg/ml 2,5-dihydroxyacetophenone in 20 mm ammonium citrate and 75% ethanol. Then, 1 μl of the obtained mixture was loaded on the target plate, dried, and analyzed using a Bruker Daltonics Autoflex mass spectrometer.
UPLC-MS/MS
Amount of TMA was measured by UPLC-MS/MS. A Micromass Quattro Micro tandem mass spectrometer in positive ionization electrospray mode with a Waters Acquity UPLC system was used to perform the analysis. Chromatographic separation was achieved on an Acquity UPLC BEH HILIC column (2.1 × 50 mm, 1.7 μm) in gradient mode with 10 mm ammonium acetate buffer (pH 4.0) as mobile phase A and acetonitrile as mobile phase B. The flow rate was 0.4 ml/min with a column temperature of 50 °C. The gradient program was 0 min at 8% A, 3.5 min at 8% A, 4.5 min at 60% A, 10 min at 60% A, 10.2 min at 8% A, and 12 min at 8% A. The ion source parameters were a capillary voltage of 2.8 kV, source temperature of 120 °C, and desolvation gas temperature of 350 °C at a flow rate of 700 liter/h. The cone voltage was 30 V, and the collision energy was 15 eV. Quantification was performed by integration of the multiple-reaction monitoring trace of TMA (60.1 ≫ 45.0 Da).
Structure Determination
Data were indexed with MOSFLM (24) and scaled with SCALA (25) from the CCP4 suite (26). Choline-bound CutC crystals exhibited an orthorhombic lattice and space group P212121 with 8 molecules in the asymmetric unit. CutC crystals without choline exhibited a monoclinic lattice and space group P21 with 4 molecules in the asymmetric unit. The choline-bound structure was determined by molecular replacement in MOLREP (27), using a CutC homology model built from GD (Protein Data Bank ID 1R9D) (16). The choline-free structure was determined in MOLREP using coordinates from the determined choline-bound structure. Models were further built manually with Coot (28) and refined with REFMAC5 (29). Data processing, refinement, and validation statistics are shown in Table 1.
TABLE 1.
Data processing and refinement statistics
Values in parentheses are for the highest resolution bin. PDB, Protein Data Bank; r.m.s.d., root mean square deviation.
| CutC |
||
|---|---|---|
| Choline-bound | Choline-free | |
| PDB ID | 5A0U | 5A0Z |
| Space group | P212121 | P21 |
| Wavelength (Å) | 1.000 | 0.984 |
| Resolution (Å) | 2.4 | 3.0 |
| Unit cell dimensions (Å) | a = 89.4, b = 221.9, c = 419.5 | a = 103.6, b = 154.6, c = 120.8 |
| Chains in asymmetric unit | 8 | 4 |
| Highest resolution bin (Å) | 2.40–2.46 | 3.00–3.08 |
| Rmerge | 0.173 (0.512) | 0.108 (0.378) |
| Total No. of observations | 923,068 | 117,615 |
| Total No. unique | 304,571 | 66,881 |
| I/σ | 4.7 (1.8) | 5.2 (1.9) |
| Completeness (%) | 93.57 (87.81) | 88.47 (74.82) |
| Multiplicity | 3.0 (2.7) | 1.8 (1.8) |
| R-factor | 0.19 (0.294) | 0.23 (0.312) |
| Rfree | 0.25 (0.341) | 0.28 (0.335) |
| Protein atoms | 50,144 | 23,203 |
| Ligand atoms | 56 | |
| Solvent molecules | 1976 | |
| Average Wilson B-factor | 15.8 | 26.6 |
| Atomic B-factor for protein | 28.4 | 37.6 |
| Atomic B-factor for ligand | 27.0 | |
| Atomic B-factor for solvent | 22.4 | |
| r.m.s.d. from ideal | ||
| Bond lengths (Å) | 0.012 | 0.014 |
| Bond angles | 1.565° | 1.721° |
| Ramachandran outliers (%) | 0.41 | 0.86 |
Results
Spontaneous and Induced Cleavage of the N-terminal Domain
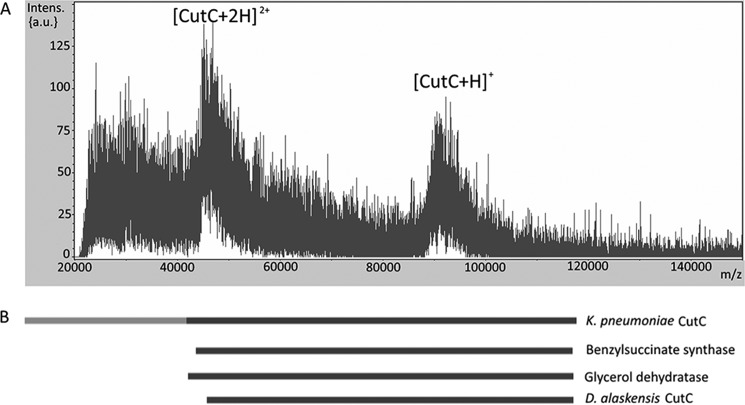
During full-length CutC crystallization trials, protein crystals appeared after what turned out to be a spontaneous partial degradation. Mass spectrometry analysis of the crystallization drops (Fig. 1A) proved that the molecular mass of the protein of interest was reduced from the expected 124 kDa to 90–92 kDa, corresponding to a loss of 305–325 N-terminal amino acids. The 124-kDa full-length CutC protein proved to be very unstable, with the N-terminal part starting to degrade almost immediately after purification by nickel affinity chromatography. To ensure the reproducibility and homogeneity of the protein for obtaining better crystals, in further experiments, full-length CutC was treated with chymotrypsin and purified by gel filtration on a Superdex 200 column. Chymotrypsin treatment resulted in a protein that had the same length as after spontaneous degradation. The exact biological role of the N-terminal domain, which contains ∼305–325 residues, remains unknown because the truncated protein contains the active site, has the same oligomerization state, and exhibits observable (although reduced) TMA-producing activity, as demonstrated below. Analogously, N-terminal truncations of up to 52 amino acids had no effect on the activity and oligomerization state of D. alaskensis CutC (12). The truncated C-terminal part of CutC also aligns well with other full-length GREs. Residues 334–1128 in the BLASTP alignment (Fig. 1B) show 93% coverage with full-length CutC from D. alaskensis (11, 12), 98% coverage with full-length GD (16), and 96% coverage with full-length BSS (17), which indicates that the N-terminal domain is disposable for the catalytic function of the enzyme.
FIGURE 1.
A, MALDI-TOF-MS analysis of CutC from a crystallization drop. Approximately 45,000–47,000 and 90,000–92,000 m/z large peaks can be distinguished. These peaks correspond to the double- and single-protonated CutC states, respectively. B, BLASTP alignment of K. pneumoniae CutC with other full-length GREs. The CutC N-terminal domain (residues 1–334), not visible in our crystal structures, is shown in light gray. Intens. (a.u.), intensity (absorbance units).
Enzymatic Activity
CutC/CutD from our study was tested for activity both in vivo and in vitro. The in vivo experiments consisted of anaerobic coexpression of CutC and CutD in E. coli BL21(DE3) cells in the presence of choline (Fig. 2A). The amount of TMA was higher when the CutC and CutD enzymes were expressed together rather than separately. Enzymatic activity was also tested in vitro with purified CutC and CutD proteins (Fig. 2B). Both untreated and chymotrypsin-treated CutC enzymes showed higher TMA-producing activities in the presence of SAM compared with the control without SAM, although the activity for chymotrypsin-treated protein was reduced. Dependence on SAM is a fundamental attribute of GRE catalysis because it is crucial for the generation of glycyl radical. The data also showed that both CutC and CutD are essential for catalysis, thus confirming that these two enzymes perform catalysis together.
FIGURE 2.
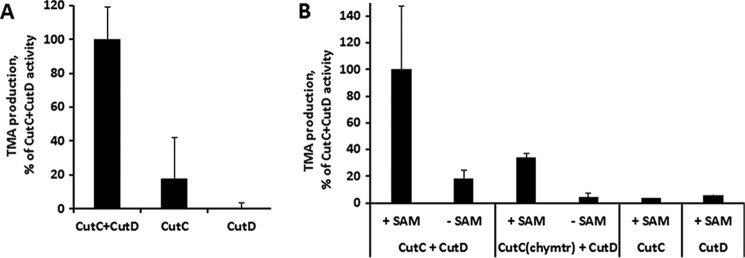
A, CutC enzymatic activity in vivo. Data are normalized to the empty vector and show elevated TMA production when cutC and cutD genes are coexpressed compared with separate cutC and cutD expression. B, CutC enzymatic activity in vitro. Data show TMA production in the presence of SAM for both full-length and chymotrypsin-treated (chymtr) CutC, but only when both SAM and CutD are present. Results are means ± S.D. of two independent experiments.
Quality of the Model, Overall Structure, and Oligomerization State
Electron density for the protein chain is visible starting from residue 334 in one of the eight monomers in the asymmetric unit, whereas an additional 3–4 residues are invisible in other molecules. As judged from mass spectrometry, for both the choline-bound and choline-free forms, an additional 10–30 N-terminal amino acids are present in the crystal in a disordered state and are not visible in the electron density map. For the choline-free form, there are several additional disordered regions that are clearly visible in the choline-bound form. These flexible regions are somewhat different among different molecules in the asymmetric unit, but there are several common regions around residues 937–946, 980–984, 1006–1017, and 1028–1040, for which no interpretable electron density for any chain could be observed.
The 10-β/α barrel structure, which is typical of other GREs, is also characteristic of CutC (Fig. 3). The monomer can be divided into three subdomains: N-terminal half-barrel (residues 334–771), C-terminal half-barrel (residues 771–1079), and glycine loop domain (residues 1079–1128). Gel filtration analysis (Fig. 4) suggested that CutC in solution both before and after treatment with chymotrypsin exists in oligomers. In the crystal structures of both the choline-bound and choline-free forms, a dimer with 2-fold symmetry can be distinguished (Fig. 3). Dimerization would be expected because similar dimers with 2-fold symmetry have been characterized for the majority of GREs (12, 14–17). For this dimer, AREAIMOL calculated a total contact surface of 14,525.5 Å2 (for the choline-bound form), thus confirming that it is likely the biological CutC dimer.
FIGURE 3.
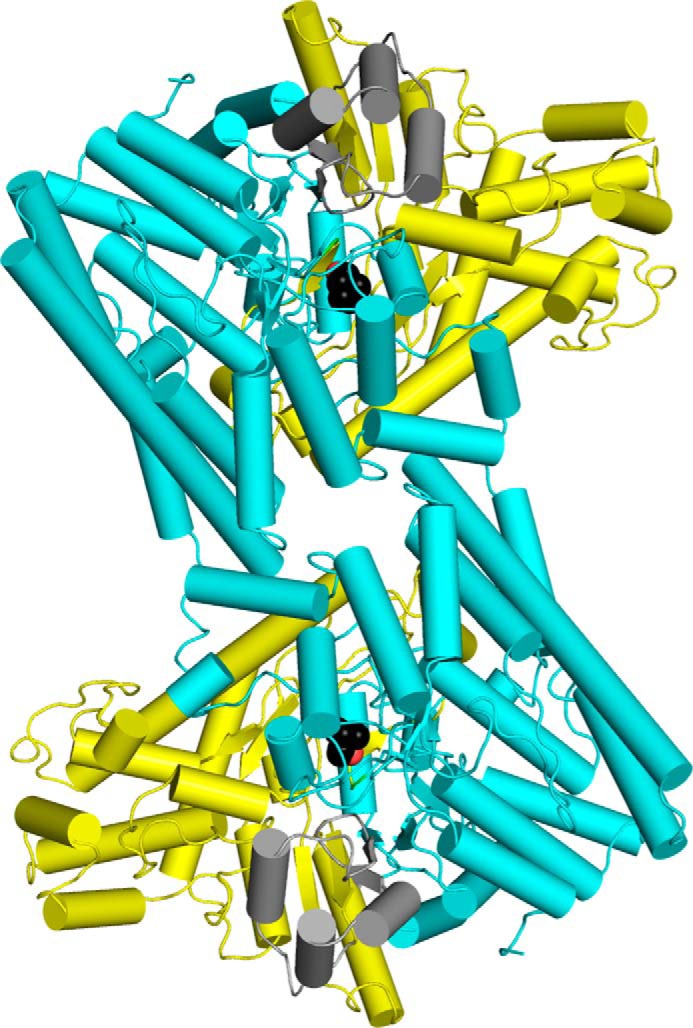
Structure of the CutC 2-fold dimer. Cyan, N-terminal half-barrel; yellow, C-terminal half-barrel; gray, glycine loop domain. Choline is displayed as a sphere model, with black carbon atoms and red oxygen atoms. This image was created using PyMOL.
FIGURE 4.
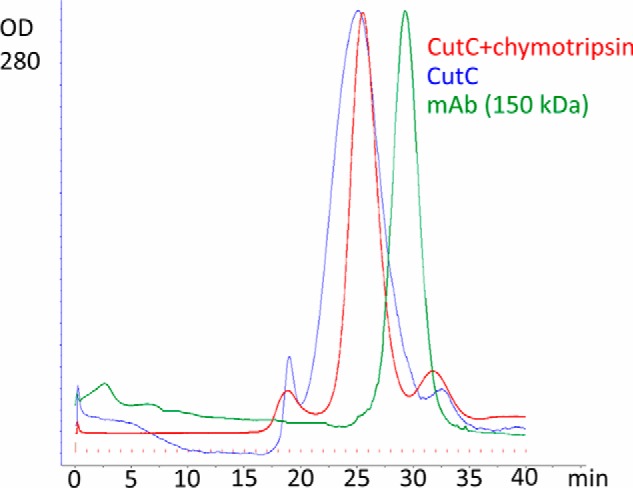
Gel filtration of untreated and chymotrypsin-treated CutC on a Superdex 200 column. Human monoclonal IgG antibody, larger than a CutC monomer but smaller than a CutC dimer, was used as a marker.
Active Site
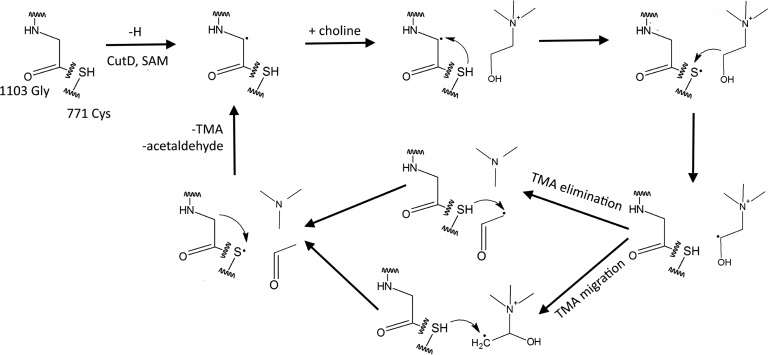
CutC Cys-771 and Gly-1103 align almost perfectly with the GD catalytic dyad Cys-433 and Gly-763, thus strongly suggesting that these 2 residues form glycyl and thiyl radicals that are essential for catalysis (Fig. 5). Theoretical and practical studies have suggested previously that, in CutC, the glycyl radical abstracts the hydrogen atom from cysteine to create a thiyl radical. This thiyl radical then further abstracts the hydrogen atom from the choline C1 atom, causing molecular rearrangement and TMA elimination either directly or through intermediate TMA migration to the generated radical site at C1 (Fig. 6) (12, 30). Cys-771 in the active site is positioned appropriately for thiyl radical hydrogen abstraction from the choline C1 atom, thus confirming the previous studies. Additionally, the choline C1 atom aligns with the glycerol carbon atom, from which the hydrogen abstraction is shown to occur (16). The choline OH group is located at the appropriate distance for hydrogen bond formation with the acid group of Glu-773. The corresponding Glu-435 of GD aligns nearly identically with Glu-773 of CutC, and the former analogously forms a hydrogen bond with the hydroxyl group of glycerol. The positively charged quaternary amine nitrogen of choline is located at the appropriate distance to interact electrostatically with the side chain of Asp-498 and to form a cation-π interaction with the nearby Phe-677.
FIGURE 5.
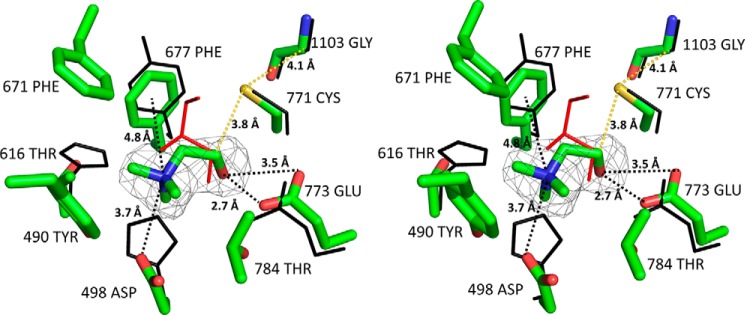
Stereoview alignment of CutC and GD (Protein Data Bank ID 1R9D) active sites. CutC and choline are shown as stick models, and GD (black) and glycerol (red) are shown as line models. An Fo − Fc map for choline is displayed at the 3σ level. Images were created with PyMOL.
FIGURE 6.
Current proposed reaction mechanism of CutC choline lyase. Dots indicate radical formation.
The active site pocket in the choline-bound form is completely closed and mostly fits the choline surface (Fig. 7A). Therefore, in this form, there is very little space left for designing substrate-like inhibitors with additional chemical groups. However, in the choline-free form, there is a possible tunnel to the outside that is formed by the movement of flexible regions, as discussed below (Fig. 7B). The C1 atom is the closest choline carbon atom to this tunnel, and it is thus the potential site for designing analog inhibitors. Such an inhibitor with additional atoms at C1 could potentially bind to the active site and prevent the CutC enzyme from transitioning to the active state.
FIGURE 7.
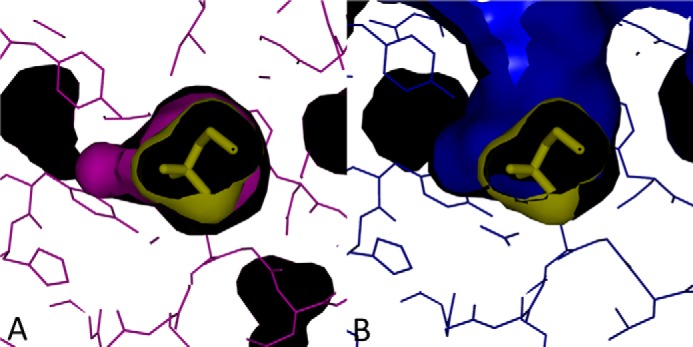
Surface models of active site pockets. A, choline-bound form. The choline-bound CutC form is displayed in purple, and choline is displayed in yellow (stick and surface models). B, choline-free form. The choline-free CutC form is displayed in purple, and choline is displayed in yellow (stick and surface models). Choline in the active site was modeled using data from the choline-bound form. Images were created with PyMOL.
Choline-bound and Choline-free Forms of CutC
The choline-bound and choline-free CutC forms have noticeable conformational differences (Fig. 8A). In the choline-free form, the CutC conformational change caused the N-terminal half-barrel and glycine loop domain to move away in opposite directions from a gap in which flexible regions, fixed in the choline-bound form, are located. In the choline-free form, the C-terminal half-barrel and glycine loop domain (α helices 20, 21, 23–26, 28, and 30–32 and β strands 7 and 8) are shifted up to 4 Å, but in the N-terminal half-barrel, they are shifted only up to 1.5 Å (Fig. 8A). A very similar conformational change has been observed for BSS, another GRE (17). Two forms of BSS, BSSαβγ and BSSαγ, have been crystallized, and they differ in a very similar manner to the choline-bound and choline-free forms of CutC, respectively (17). BSSαβγ consists of the main α subunit and two small β and γ subunits, whereas BSSαγ contains only the main α subunit and a small γ subunit. Similar to choline-free CutC, BSSαγ has a more noticeable conformational shift in the C-terminal half-barrel and glycine loop domain compared with the N-terminal half-barrel (Fig. 8B).
FIGURE 8.
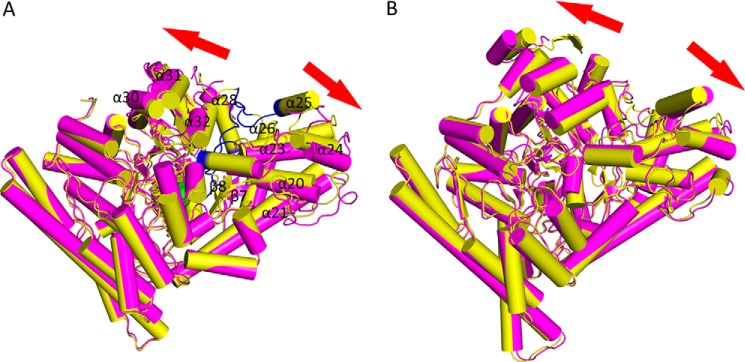
A, choline-bound (yellow) and native (purple) CutC forms. Regions present in the choline-bound form but absent in the native form electron density map are colored in blue. Choline is displayed as a sphere model, with green carbon atoms and red oxygen atoms. B, BSS α subunit, BSSαβγ (yellow), and BSSαγ (purple) (13). Images were created with PyMOL.
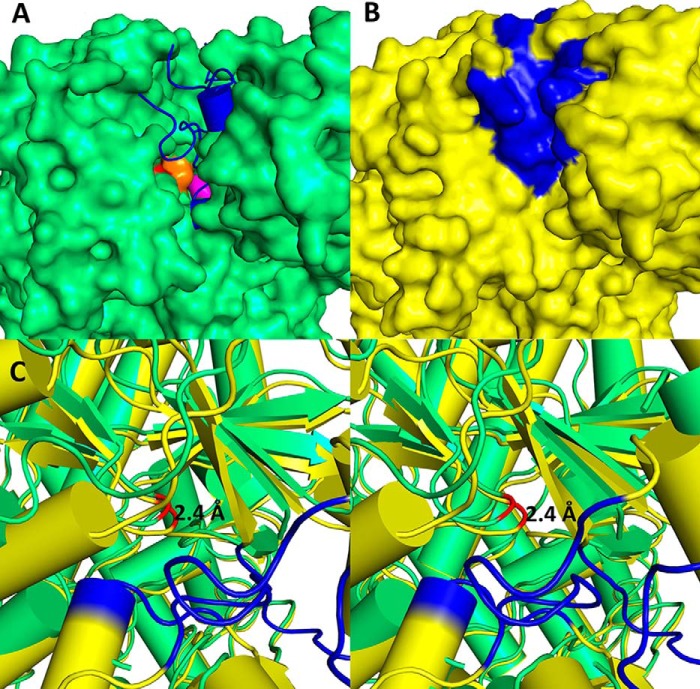
In the choline-free form, the flexible regions expose the active site, including Gly-1103 (Fig. 9A). In the choline-bound form, the active site is completely shielded from the outer environment via ordering of flexible regions (Fig. 9B), which suggests that conformational changes must be associated with increasing availability of the active site. The glycine loop is shifted 2.4 Å toward to the top of the protein (Fig. 9C). Similar movement was also observed for BSS and thought to be associated with making glycine accessible to the activating enzyme (17).
FIGURE 9.
A, surface model of the choline-free CutC form (green). The catalytic dyad Gly-1103 is shown as a red surface, and Cys-771 as an orange surface. Choline in the active site (purple surface) and disordered regions (blue) were modeled using data from the choline-bound form. B, surface model of the choline-bound CutC form (yellow). Regions disordered in the choline-free form are displayed in blue. C, stereoview alignment of glycine loops for the choline-bound (yellow) and choline-free (green) CutC forms. Gly-1103 in both forms is displayed in red. Regions disordered in the choline-free form are displayed in blue. Images were created with PyMOL.
Resistance to chymotrypsin treatment was observed for the BSSαβγ complex, but not for the BSSαγ complex. Analogously, CutC resistance to 6 h of chymotrypsin treatment was noticeable in the presence of choline, but in its absence (Fig. 10). The chymotrypsin resistance that was observable at pH 6.5 and 8.5 also excluded the possible influence of pH under different crystallization conditions. The CutC structure in the absence of choline appears to be open in both solution and the crystal, and the binding of choline to the active site appears to change the conformation to a stable, closed, more structured, and chymotrypsin-resistant form.
FIGURE 10.
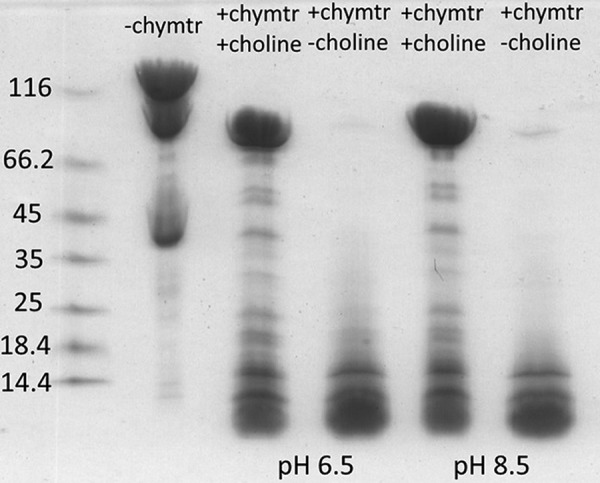
Chymotrypsin treatment of CutC for 6 h at room temperature and SDS-PAGE analysis. Marker sizes are shown on the left in kilodaltons. Partial degradation was noticeable for CutC even without the addition of chymotrypsin (chymtr).
Discussion
Recent studies indicate that the influence of the microbiota on human organism functions is very diverse. The association of the microbiota with diseases such as cancer (31), diabetes (32), and cardiovascular diseases (33) has been demonstrated. TMAO is a metabolite created in the host liver from microbiota-produced TMA, and it has been associated with several diseases (3–10). TMA production in the intestine can be suppressed by antibiotics (34, 35), but this effect is not complete, and long-term antibiotic treatment may lead to an imbalance in the microbiota (36). An alternative solution would be to develop specific enzymatic inhibitors that block TMA production but do not cause as much of an imbalance in the microbiota as antibiotics do. More information about the structures of TMA-producing enzymes from bacteria in the human microbiota therefore could be significant for finding ways to block TMA/TMAO formation, thus providing new microbiota-linked drug targets to treat cardiovascular diseases and trimethylaminuria. CutC choline lyase is very appropriate as a target of such studies, as it produces TMA from a common nutrient, choline, and can be found in the genomes of several microbiota representatives (11, 12, 19). Although there is too little free space in the choline-bound form of CutC to design substrate analog inhibitors, the choline-free form may contain a possible tunnel made by movement of disordered regions that could be used for this purpose.
The nature of the catalysis of CutC and other GREs is strictly anaerobic because oxygen reacts with glycyl radicals, inactivating the enzymes. However, the glycyl radical in GREs has proven to be very stable, lasting up to several days in vitro (37). Several protective strategies have been developed to prevent oxidative enzyme inactivation. One such strategy is using enzymes that act similarly to “spare parts”, reattaching the oxygen-cleaved C-terminal domains (38). The determined structures of certain GREs indicate another protective mechanism: shielding glycine from the outer environment by burying it in the protein core. However, it is unclear how the activating enzyme generates the glycyl radical and how the ligand gains access to the ligand-binding site. Co-crystallization studies with the activating enzyme of PFL and a PFL glycine loop-mimicking peptide have suggested that the active site glycine interacts with the activating enzyme directly (39). This means that a local movement of the glycine loop out of the protein core, a global structural change, or a combination of both must happen to achieve radical formation. PFL has been shown to exist in solution in both open and closed forms, and the equilibrium between these forms is thought to be modulated by the activating enzyme (40). Direct structural evidence for conformational changes, similar to the changes observed for K. pneumoniae CutC, has also been observed for BSS (17). For BSS, this conformational shift is regulated by an accessory β subunit. However, no accessory subunit or activating enzyme was present during the crystallization and chymotrypsin treatment of K. pneumoniae CutC, showing that, at least for CutC, no help from other proteins is required for conformational changes to occur. Chymotrypsin tolerance in the presence of choline also demonstrated that conformational changes occur in solution as well as in crystalline form. These results suggest that, in the open form, the active site is freely accessible to choline and that binding of choline is the only critical factor that transforms the structure from the open to closed form.
Interestingly, CutC is the only GRE characterized so far for which ligand binding to the ligand-binding site causes such an effect on the conformation in the crystal structure. Two other GREs, PFL and GD, have been crystallized in both ligand-bound and ligand-free states (14, 16), but no significant conformational changes in crystal structure between these two states have ever been observed. Additionally, both the ligand-bound and ligand-free GD states align better with the closed choline-bound CutC state. This raises the question of whether there is a correlation between the observed choline-induced CutC conformational changes and the accessibility of the active site glycine to the activating enzyme. It would be an inefficient strategy for a GRE to enter a stable and closed conformation independently of the presence or absence of a glycyl radical in the active site. An alternative explanation would be that the activating enzyme is able to induce CutC conformational changes partially or completely independently of the presence or absence of choline. Choline-driven conformational changes in this case could serve only as an additional regulatory mechanism, perhaps increasing affinity for the substrate.
Other results regarding the CutC active site, the mechanism of catalysis, and the oligomerization state are consistent with data from studies on other GREs. CutC has a typical 10-β/α barrel fold, and dimerization is observed. A catalytic dyad is formed by glycine and cysteine, which are located appropriately for hydrogen abstraction from the choline C1 atom, and the choline in the ligand-binding site is coordinated by glutamic acid, aspartic acid, and phenylalanine residues that were previously mapped with site-directed mutagenesis in D. alaskensis CutC and shown to be essential for catalysis (12, 30).
Author Contributions
G. K. designed and constructed vectors for protein expression, expressed and purified proteins, characterized enzymatic activity, crystallized protein, determined protein x-ray structure, and wrote the paper. J. K., S. G., M. M.-K., E. L., and M. D. characterized enzymatic activity. K. T. conceived and coordinated the study, determined protein x-ray structure, and wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank the MAX-lab staff for support.
This work was supported by State Research Program BIOMEDICINE and BioStruct-X Grant Proposal ID 7869. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5A0U and 5A0Z) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TMA
- trimethylamine
- TMAO
- trimethylamine oxide
- GRE
- glycyl radical enzyme
- SAM
- S-adenosylmethionine
- PFL
- pyruvate formate-lyase
- GD
- glycerol dehydratase
- BSS
- benzylsuccinate synthase
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- UPLC
- ultra performance liquid chromatography.
References
- 1. Hayward H. R., Stadtman T. C. (1960) Anaerobic degradation of choline. II. Preparation and properties of cell-free extracts of Vibrio cholinicus. J. Biol. Chem. 235, 538–543 [PubMed] [Google Scholar]
- 2. Sandhu S. S., Chase T., Jr. (1986) Aerobic degradation of choline by Proteus mirabilis: enzymatic requirements and pathway. Can. J. Microbiol. 32, 743–750 [DOI] [PubMed] [Google Scholar]
- 3. Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., Li L., Smith J. D., DiDonato J. A., Chen J., Li H., Wu G. D., Lewis J. D., Warrier M., Brown J. M., Krauss R. M., Tang W. H., Bushman F. D., Lusis A. J., Hazen S. L. (2013) Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Treacy E. P., Akerman B. R., Chow L. M., Youil R., Bibeau C., Lin J., Bruce A. G., Knight M., Danks D. M., Cashman J. R., Forrest S. M. (1998) Mutations of the flavin-containing monooxygenase gene (FMO3) cause trimethylaminuria, a defect in detoxication. Hum. Mol. Genet. 7, 839–845 [DOI] [PubMed] [Google Scholar]
- 5. Brown J. M., Hazen S. L. (2014) Metaorganismal nutrient metabolism as a basis of cardiovascular disease. Curr. Opin. Lipidol. 25, 48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y. M., Wu Y., Schauer P., Smith J. D., Allayee H., Tang W. H., DiDonato J. A., Lusis A. J., Hazen S. L. (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tang W. H., Wang Z., Levison B. S., Koeth R. A., Britt E. B., Fu X., Wu Y., Hazen S. L. (2013) Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trøseid M., Ueland T., Hov J. R., Svardal A., Gregersen I., Dahl C. P., Aakhus S., Gude E., Bjørndal B., Halvorsen B., Karlsen T. H., Aukrust P., Gullestad L., Berge R. K., Yndestad A. (2015) Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 277, 717–726 [DOI] [PubMed] [Google Scholar]
- 9. Tang W. H., Wang Z., Kennedy D. J., Wu Y., Buffa J. A., Agatisa-Boyle B., Li X. S., Levison B. S., Hazen S. L. (2015) Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao X., Liu X., Xu J., Xue C., Xue Y., Wang Y. (2014) Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 118, 476–481 [DOI] [PubMed] [Google Scholar]
- 11. Craciun S., Balskus E. P. (2012) Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 21307–21312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craciun S., Marks J. A., Balskus E. P. (2014) Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem. Biol. 9, 1408–1413 [DOI] [PubMed] [Google Scholar]
- 13. Shisler K. A., Broderick J. B. (2014) Glycyl radical activating enzymes: structure, mechanism, and substrate interactions. Arch. Biochem. Biophys. 546, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Becker A., Fritz-Wolf K., Kabsch W., Knappe J., Schultz S., Volker Wagner A. F. (1999) Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat. Struct. Biol. 6, 969–975 [DOI] [PubMed] [Google Scholar]
- 15. Logan D. T., Andersson J., Sjöberg B. M., Nordlund P. (1999) A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science 283, 1499–1504 [DOI] [PubMed] [Google Scholar]
- 16. O'Brien J. R., Raynaud C., Croux C., Girbal L., Soucaille P., Lanzilotta W. N. (2004) Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43, 4635–4645 [DOI] [PubMed] [Google Scholar]
- 17. Funk M. A., Judd E. T., Marsh E. N., Elliott S. J., Drennan C. L. (2014) Structures of benzylsuccinate synthase elucidate roles of accessory subunits in glycyl radical enzyme activation and activity. Proc. Natl. Acad. Sci. U.S.A. 111, 10161–10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagner A. F., Frey M., Neugebauer F. A., Schäfer W., Knappe J. (1992) The free radical in pyruvate formate-lyase is located on glycine-734. Proc. Natl. Acad. Sci. U.S.A. 89, 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romano K. A., Vivas E. I., Amador-Noguez D., Rey F. E. (2015) Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 6, e02481–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuka J., Liepinsh E., Makrecka-Kuka M., Liepins J., Cirule H., Gustina D., Loza E., Zharkova-Malkova O., Grinberga S., Pugovics O., Dambrova M. (2014) Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting l-carnitine microbial degradation. Life. Sci. 117, 84–92 [DOI] [PubMed] [Google Scholar]
- 22. Dambrova M., Liepinsh E., Kalvinsh I. (2002) Mildronate: cardioprotective action through carnitine lowering effect. Trends Cardiovasc. Med. 12, 275–279 [DOI] [PubMed] [Google Scholar]
- 23. Dambrova M., Skapare-Makarova E., Konrade I., Pugovics O., Grinberga S., Tirzite D., Petrovska R., Kalvins I., Liepins E. (2013) Meldonium decreases the diet-increased plasma levels of trimethylamine N-oxide, a metabolite associated with atherosclerosis. J. Clin. Pharmacol. 53, 1095–1098 [DOI] [PubMed] [Google Scholar]
- 24. Leslie A. G. W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 25. Evans P. R. (1997) SCALA. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 33 [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vagin A, Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Thibodeaux C. J., van der Donk W. A. (2012) Converging on a mechanism for choline degradation. Proc. Natl. Acad. Sci. U.S.A. 109, 21184–21185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zitvogel L., Galluzzi L., Viaud S., Vétizou M., Daillère R., Merad M., Kroemer G. (2015) Cancer and the gut microbiota: an unexpected link. Sci. Transl. Med. 7, 271ps1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He C., Shan Y., Song W. (2015) Targeting gut microbiota as a possible therapy for diabetes. Nutr. Res. 35, 361–367 [DOI] [PubMed] [Google Scholar]
- 33. Howitt M. R., Garrett W. S. (2012) A complex microworld in the gut: gut microbiota and cardiovascular disease connectivity. Nat. Med. 18, 1188–1189 [DOI] [PubMed] [Google Scholar]
- 34. Fraser-Andrews E. A., Manning N. J., Ashton G. H. S., Eldridge P., McGrath J., Menagé Hdu P. (2003) Fish odour syndrome with features of both primary and secondary trimethylaminuria. Clin. Exp. Dermatol. 28, 203–205 [DOI] [PubMed] [Google Scholar]
- 35. Treacy E., Johnson D., Pitt J. J., Danks D. M. (1995) Trimethylaminuria, fish odour syndrome: a new method of detection and response to treatment with metronidazole. J. Inherit. Metab. Dis. 18, 306–312 [DOI] [PubMed] [Google Scholar]
- 36. Jernberg C., Löfmark S., Edlund C., Jansson J. K. (2010) Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156, 3216–3223 [DOI] [PubMed] [Google Scholar]
- 37. Walsby C. J., Ortillo D., Yang J., Nnyepi M. R., Broderick W. E., Hoffman B. M., Broderick J. B. (2005) Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-Sam” protein superfamily. Inorg. Chem. 44, 727–741 [DOI] [PubMed] [Google Scholar]
- 38. Wagner A. F., Schultz S., Bomke J., Pils T., Lehmann W. D., Knappe J. (2001) YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem. Biophys. Res. Commun. 285, 456–462 [DOI] [PubMed] [Google Scholar]
- 39. Vey J. L., Yang J., Li M., Broderick W. E., Broderick J. B., Drennan C. L. (2008) Structural basis for glycyl radical formation by pyruvate formate lyase activating enzyme. Proc. Natl. Acad. Sci. U.S.A. 105, 16137–16141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng Y., Veneziano S. E., Gillispie G. D., Broderick J. B. (2010) Pyruvate formate-lyase, evidence for an open conformation favored in the presence of its activating enzyme. J. Biol. Chem. 285, 27224–27231 [DOI] [PMC free article] [PubMed] [Google Scholar]