Background: Because peroxidasin generates HOBr to form sulfilimine cross-links in collagen IV, any peroxidase producing HOBr may cross-link collagen IV.
Results: Among animal peroxidases, only peroxidasin efficiently cross-linked collagen IV requiring its catalytic and immunoglobulin domains.
Conclusion: Peroxidasin uniquely reacts HOBr with collagen IV to catalyze sulfilimine bond formation.
Significance: Insight into how peroxidasin cross-links collagen IV is critical to understand basement membrane function.
Keywords: basement membrane, extracellular matrix, myeloperoxidase, peroxidase, protein cross-linking, collagen IV, peroxidasin, sulfilimine bond
Abstract
The collagen IV sulfilimine cross-link and its catalyzing enzyme, peroxidasin, represent a dyad critical for tissue development, which is conserved throughout the animal kingdom. Peroxidasin forms novel sulfilimine bonds between opposing methionine and hydroxylysine residues to structurally reinforce the collagen IV scaffold, a function critical for basement membrane and tissue integrity. However, the molecular mechanism underlying cross-link formation remains unclear. In this work, we demonstrate that the catalytic domain of peroxidasin and its immunoglobulin (Ig) domains are required for efficient sulfilimine bond formation. Thus, these molecular features underlie the evolutionarily conserved function of peroxidasin in tissue development and integrity and distinguish peroxidasin from other peroxidases, such as myeloperoxidase (MPO) and eosinophil peroxidase (EPO).
Introduction
The collagen IV sulfilimine bond and peroxidasin represent a dyad critical for tissue development found in animal basement membranes (1). Collagen IV forms a mesh-like structure consisting of oligomerized triple helical protomers (2). The trimeric C-terminal non-collagenous (NC1) domains of two protomers associate head-to-head to form the NC1 hexamer, which is reinforced by sulfilimine bonds between opposing methionine and hydroxylysine residues (3). For example, in the predominant, vertebrate α121 collagen IV network, protomers, consisting of two α1 and one α2 chains, come together with α1 NC1 domains associating with α1 domains and reciprocally α2 NC1 domains engaging one another. Sulfilimine bonds may bridge the NC1 hexamer to form homo-dimeric (α1-α1 or α2-α2) subunits each with up to two cross-links. Thus, a total of zero to six sulfilimine cross-links may reinforce a collagen IV NC1 hexamer (3).
Peroxidasin and its formation of sulfilimine cross-links in collagen IV are critical for tissue development as loss of peroxidasin function in Drosophila and Caenorhabditis elegans leads to disordered, fragile basement membranes and tissues with early lethality (4, 5). Peroxidasin uses hydrogen peroxide (H2O2) and bromide (Br−) ions, to form HOBr as a reactive intermediate to form sulfilimine cross-links in collagen IV. Indeed, the role of Br− as a catalytic cofactor in this reaction represents the first known essential function for the trace element bromine (6).
Peroxidasin is a multidomain protein consisting of a catalytic peroxidase domain and non-catalytic leucine-rich repeat (LRR)3, Ig, and von Willebrand factor type C (vWFC) protein-protein interaction domains (7). Previous work in our group revealed that peroxidasin arises in Cnidaria alongside the collagen IV sulfilimine cross-link and is evolutionarily conserved throughout the animal kingdom (1). Furthermore, peroxidasin and collagen IV expression reflect the broad distribution of basement membranes in nearly all tissues (8). Conversely, thyroid peroxidase, lactoperoxidase, eosinophil peroxidase (EPO), and myeloperoxidase (MPO) are found only in vertebrates and exhibit tissue restricted expression patterns in these animals (9, 10). Thus, the ubiquity of peroxidasin within and between animal species suggests that functional redundancy with vertebrate heme peroxidases in normal physiology is improbable.
From a mechanistic perspective, a critical question arises as to whether vertebrate heme peroxidases capable of producing HOBr, such as EPO and MPO, can cross-link collagen IV in pathologic states, where they may associate with basement membrane (11, 12). For instance, MPO has been shown to interact with subendothelial basement membranes and both MPO and EPO can cross-link solubilized collagen IV in vitro raising the possibility of biochemical redundancy (4, 6, 13). In this work, we found that MPO and EPO poorly cross-link collagen IV, even when experimentally deposited into basement membrane. We therefore hypothesized that the LRR, Ig, and vWFC domains found in peroxidasin, but not in related animal heme peroxidases, uniquely allow peroxidasin to form sulfilimine bonds in collagen IV. Indeed, the catalytic and Ig domains are required for cross-linking activity, which distinguishes peroxidasin from other animal heme peroxidases.
Experimental Procedures
Cloning of Peroxidasin Deletion Constructs
Full-length peroxidasin open reading frame (ORF) cloned in the pCDNA-V5-His-TOPO vector (kindly provided by Dr. Miklos Geiszt, Semmelweis University, Budapest, Hungary) was used as the starting construct. The construct lacking the C-terminal vWFC domain (−vWFC) was created by PCR amplification of the ORF at the N terminus and a point between the catalytic and vWFC domains using flanking KpnI and NotI sites. The forward primer was 5′-GGGTGTCCGAGCGAATTCCGCTGCCTGTGC-3′ and the reverse primer was 5′-CATTGCGGCCGCAGGTCCTACAGTCTTCACAGCAGT-3′. Using PCR-based mutagenesis as previously described (14), EcoRI sites flanking the deletion of interest were inserted in either the full-length peroxidasin ORF or the −vWFC construct (Table 1). Deletion constructs were then created with EcoRI digestion, removal of the intervening fragment, and re-ligation to produce the construct. A 2-residue Gly-Ser insertion at the junction occurred as an expected byproduct. The entire ORF of all of the constructs were sequenced to ensure the absence of unwanted mutations.
TABLE 1.
Insertion points for EcoRI sites to create PXDN deletion constructs
Numbers indicate flanking residues for EcoRI site insertion within either full-length peroxidasin or the -vWFC construct.
| Deletion construct | Template | EcoRI insertion points (human PXDN residue) |
|---|---|---|
| −LRR | Full-length | 38–39 and 232–233 |
| −Ig | Full-length | 232–233 and 612–613 |
| Pxd-Ig | −vWFC | 38–39 and 232–233 |
| Pxd-Ig-7 aa | −vWFC | 38–39 and 239–240 |
| Pxd | −vWFC | 38–39 and 612–613 |
“Overlay” Assay for Collagen IV Sulfilimine Bond Formation
PFHR-9 cells were cultured in DMEM with high glucose + 5% fetal bovine serum at 5% CO2 for 5 days post-confluence in the presence of 50 μg/ml of ascorbic acid to promote collagen hydroxylation and 50 μm phloroglucinol to inhibit peroxidasin-mediated sulfilimine bond formation (4). The PFHR-9 cells were then removed using two detergent extractions at room temperature for 5–10 min with hypotonic buffer (10 mm Tris-Cl, pH 7.5, + 0.1 mm CaCl2 + 0.1% bovine serum albumin) + 1% Triton X-100. Cellular debris was further cleared with two washes with hypotonic buffer + 0.1% sodium deoxycholate. To remove endogenous peroxidasin, the matrix was extracted with 4 m guanidine hydrochloride + 50 mm Tris-Cl, pH 7.5, for 15 min at room temperature and then washed extensively with 1× PBS. These PFHR-9 matrix plates were either stored under sterile conditions at −80 °C or used immediately thereafter. HEK293T cells were plated onto PFHR-9 matrix and transfected with the cDNAs of interest using Lipofectamine LTX per the manufacturer's instructions (Life Technologies). The cells were then incubated for 48 h in the presence of 5 μm hematin and 5 mm sodium butyrate to maximize functional peroxidase. After incubation, the culture media was collected and extracellular matrix isolated for analysis.
To isolate extracellular matrix (ECM), the cells and underlying ECM were scraped into deoxycholate buffer (1% sodium deoxycholate, 10 mm Tris-Cl, pH 7.5, and 1 mm EDTA-Na) including protease inhibitors (0.5 mm phenylmethylsulfonyl fluoride, 10 μg/ml of leupeptin, and 1 μg/ml of pepstatin) and sonicated to eliminate viscosity. The resulting lysate was centrifuged at 20,000 × g for 15 min and the resulting supernatant was designated as the cell lysate and the insoluble pellet was further processed to isolate ECM. The pellet was resuspended in high salt buffer (1 m NaCl, 50 mm Tris-Cl, pH 7.5, and 0.5 mm PMSF) with sonication and extracted for 10–15 min at 4 °C. The insoluble material was again collected with centrifugation and rinsed briefly with low salt buffer (10 mm Tris-Cl, pH 7.5). After repeat centrifugation, the resulting pellet was operationally defined as ECM or basement membrane.
To delineate sulfilimine cross-link content of the collagen IV network, the ECM was sonicated in collagenase buffer (100 mm NaCl, 50 mm Tris-Cl, pH 7.5, 25 mm 6-aminocaproic acid, 5 mm benzamidine, 0.5 mm PMSF, and 50 μm phloroglucinol) and digested with collagenase (50 μg/ml; CLSPA grade, Worthington Biochemical Corp., Lakewood, NJ) overnight at 37 °C. The digested material was centrifuged at 20,000 × g for 30 min and the supernatant with solubilized NC1 hexamer was collected. This material underwent SDS-PAGE under non-reducing conditions followed by Coomassie Blue staining to visualize and quantify sulfilimine cross-linked dimeric and non-cross-linked monomeric subunits. The average number of cross-links per hexamer was determined using the density of the single (D1) and double cross-linked (D2) and non-cross-linked (M) subunit bands.
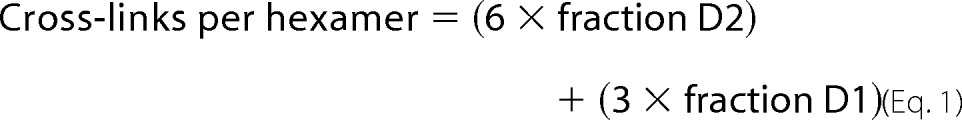 |
In parallel experiments, the isolated ECM underwent immunoblotting to determine the presence of MPO, EPO, and PXDN. In this case, the ECM was sonicated into SDS-PAGE sample buffer with 50 mm dithiothreitol (DTT) and heated at 80 °C for 15 min. The sample was then centrifuged (20,000 × g for 10 min) and the supernatant underwent immunoblot analysis (see below). In a subset of experiments (Fig. 3), the ECM first underwent collagenase digestion and then soluble and insoluble fractions were isolated by centrifugation (20,000 × g for 30 min). These fractions were then solubilized in SDS-PAGE sample buffer with DTT and underwent immunoblotting for MPO, EPO, PXDN, and collagen IV.
FIGURE 3.
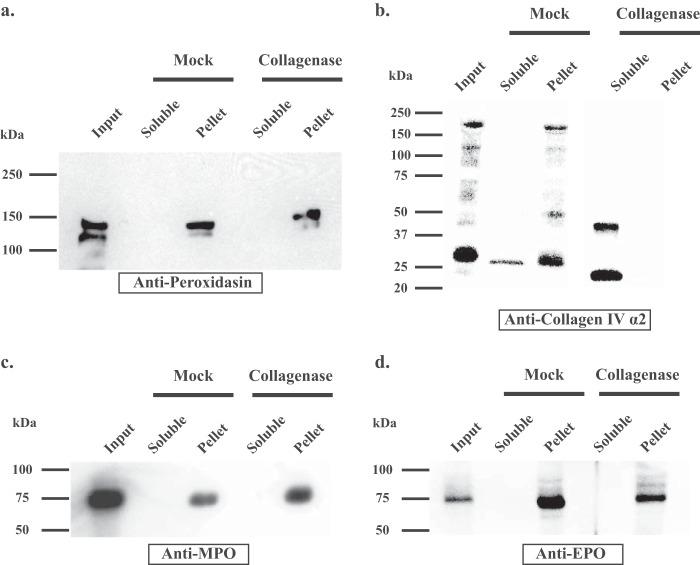
Peroxidasin, MPO, and EPO do not directly interact with collagen IV. a, PFHR-9 matrix was isolated after overlay with HEK293T cells transiently transfected with PXDN. The matrix (input) was collagenase or mock digested and the soluble and insoluble (pellet) fractions underwent SDS-PAGE under reducing conditions (with 50 mm DTT) and immunoblotted with anti-PXDN antibody. PXDN remained in the insoluble pellet despite collagenase digestion. b, in the same experiment as in a, the fractions were immunoblotted with anti-collagen IV α-2 NC1 domain antibody, which demonstrates that collagenase digestion solubilized the NC1 domain of collagen IV. After collagenase treatment, α-2 NC1 immunoreactivity is only present in the soluble fraction, whereas the mock digestion-insoluble pellet mimics input matrix with detection of partially solubilized full-length and degraded collagen IV. c, MPO and d, EPO also remain in the collagenase digest pellet as seen with PXDN in a.
Culture media was harvested to determine whether transfected peroxidases could be detected by immunoblotting and peroxidase activity. After addition of SDS-PAGE sample buffer with 50 mm DTT, media samples directly underwent immunoblotting. For measuring peroxidase activity, media was first precipitated with 40% ammonium sulfate. Precipitated protein was solubilized in cetyltrimethylammonium bromide buffer (150 mm NaCl, 50 mm Tris-Cl, pH 7.5, 0.2% cetyltrimethylammonium bromide, and protease inhibitors) at 1/10th the original media volume. 25 μl of the solubilized precipitate was combined with 250 μl of developing buffer (0.4 m sodium acetate, pH 5.4, 0.1% cetyltrimethylammonium bromide, 2 mm tetramethylbenzidine, and 50 μm H2O2) and A650 was measured as an indicator of peroxidase-mediated oxidation of tetramethylbenzidine.
Immunoblotting
Western blotting was conducted using standard techniques including 5% nonfat milk plus 0.05% Tween 20 for blocking nonspecific antibody binding. The following primary antibodies and dilutions were utilized. 1) Anti-human PXDN rabbit polyclonal antibody developed against peptide sequence of IREKLKRLYGSTLNI in our laboratory at 1:1000. 2) Anti-human/mouse MPO mouse monoclonal (R&D Systems, Minneapolis, MN) at 1:1000. 3) Anti-EPO rabbit polyclonal (Aviva Systems Biology, San Diego, CA) at 1:1000. 4) Anti-actin goat polyclonal (Santa Cruz Biotechnology, Dallas, TX) at 1:200. 5) Anti-collagen IV α2 chain (H22 rat monoclonal; Chondrex, Inc., Redmond, WA) at 1:1000. Bound primary antibody was detected using species-specific, horseradish peroxidase-conjugated secondary antibodies followed by enhanced chemiluminescence detection per the manufacturer's instructions (SuperSignalTM West Pico Substrate, Life Technologies).
Results
Among Animal Heme Peroxidases, Peroxidasin Uniquely Forms Sulfilimine Bonds in the Collagen IV Network
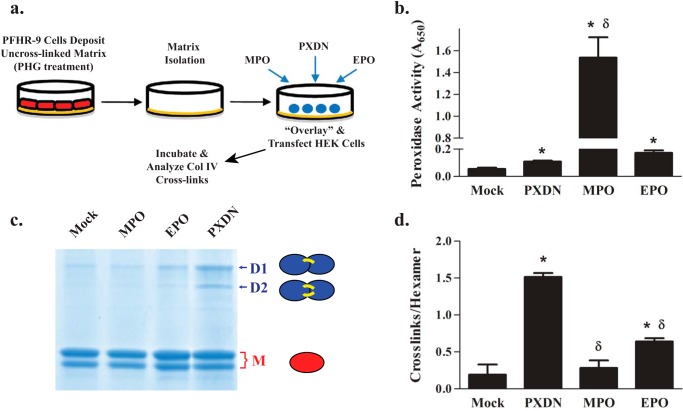
We had previously demonstrated that peroxidasin, but not MPO, cross-links the insoluble collagen IV network using an overlay method wherein transfected cells are plated on uncross-linked collagen IV (4) (Fig. 1a). After this initial work, we discovered the central role of Br− in peroxidasin function (6). Because MPO forms both HOCl and HOBr and peroxidasin generates HOBr, the inability of MPO to form sulfilimine cross-links may simply reflect inefficient HOBr production (4, 10). To explore this possibility, we tested the ability of EPO, known to predominantly form HOBr (4, 12), to cross-link collagen IV. Although EPO formed sulfilimine bonds marginally better than mock control and MPO, EPO was far inferior to peroxidasin suggesting that HOBr production alone is not the pivotal feature that distinguishes PXDN from MPO and EPO (Fig. 1, b–d).
FIGURE 1.
Peroxidasin uniquely forms sulfilimine cross-links in collagen IV. a, experimental design of overlay experiments. PFHR-9 cells were grown in the presence of 50 μm phloroglucinol (peroxidase inhibitor) to deposit an uncross-linked collagen IV network. PFHR-9 cells were removed by detergent extraction and the resultant matrix treated with 4 m guanidine-Cl to remove endogenous PXDN. HEK293T cells were plated on this matrix, transiently transfected with various constructs, and the underlying matrix isolated for analysis of collagen IV sulfilimine bond content 48 h thereafter. b, transient transfections yielded functional PXDN, MPO, and EPO as determined by tetramethylbenzidine oxidation and measurement of absorbance at 650 nm, the absorbance maximum of the oxidation product. c, PFHR-9 matrix was isolated after overlay, collagenase digested, and the resulting NC1 hexamers underwent SDS-PAGE to delineate sulfilimine cross-linked dimeric subunits (D1 and D2 represent single and double cross-linked forms, respectively) and uncross-linked monomeric subunits (M). Each of these band regions (D1, D2, and M) may appear as single bands (arrows) or doublets (brackets) due to unidentified variability in electrophoretic migration (4, 26, 27). d, the number of cross-links per collagen IV NC1 hexamer was determined for PXDN, MPO, and EPO using densitometric quantitation of the dimeric and monomeric subunits from SDS-PAGE of collagenase-digested PFHR-9 matrix as shown in c. Data were analyzed using analysis of variance followed by post hoc pairwise comparisons with Tukey's correction for multiple comparisons (*, p < 0.05 compared with mock; δ, p < 0.05 compared with PXDN, n = 5).
Peroxidases Universally Associate with Extracellular Matrix, but Not Collagen IV, in the Overlay Assay
Because halide oxidation preference (Br− versus Cl−) did not account for peroxidasin-mediated sulfilimine bond formation, we hypothesized that peroxidasin incorporates into extracellular matrix to produce HOBr near its collagen IV substrate, whereas MPO and EPO remain as soluble secreted proteins, whose hypohalous acid products are quenched by side reactions with other proteins before reaching their collagen IV target (15). To test this hypothesis, we used immunoblotting to determine whether peroxidasin, MPO, and EPO were present in the detergent and high-salt insoluble extracellular matrix fraction after overlay with transfected cells. Peroxidasin, MPO, and EPO all associated with extracellular matrix (Fig. 2, a–c). The absence of actin immunoreactivity ruled against the possibility of intracellular contamination as a cause for the association of peroxidasin, MPO, and EPO with extracellular matrix (Fig. 2d). Thus, MPO and EPO fail to cross-link collagen IV even when these peroxidases artificially associate with basement membrane in the overlay assay. These data suggest that extracellular matrix association and HOBr production per se are insufficient to cross-link collagen IV.
FIGURE 2.
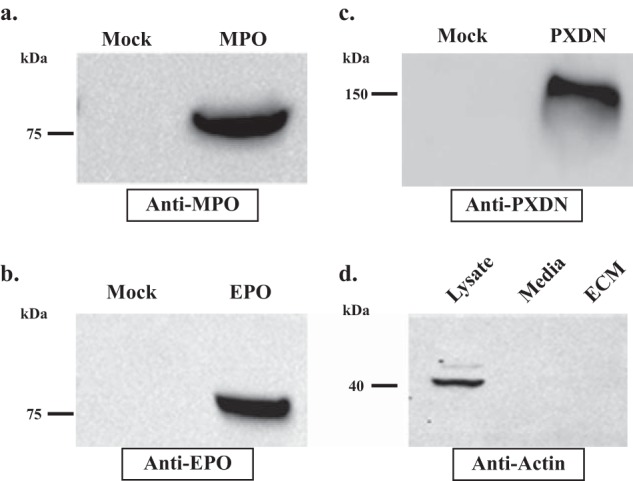
Peroxidasin, MPO, and EPO associate with extracellular matrix. a–c, immunoblotting of isolated PFHR-9 matrix after overlay with transiently transfected HEK293T cells demonstrates the presence of MPO, EPO, and PXDN in the detergent and high salt-insoluble ECM fraction. d, actin immunoblotting reveals the presence of actin only in the deoxycholate soluble cell lysate, but not in the culture media or detergent-high salt insoluble matrix fraction. Blots are representative of 4 independent experiments.
An alternative possibility is that peroxidasin uniquely binds to collagen IV, whereas MPO and EPO associate with other basement membrane components. But, solid phase and solution binding assays did not demonstrate an interaction between peroxidasin, MPO, or EPO with collagen IV using peroxidase concentrations up to 1 μm (data not shown). To verify this unexpected finding, we hypothesized that if peroxidasin is tightly bound to collagen IV in the insoluble extracellular matrix, then destruction of the collagen IV network with collagenase should solubilize peroxidasin. However, after collagenase treatment, peroxidasin remained insoluble (Fig. 3a), as did MPO and EPO (Fig. 3, c and d). As expected, collagenase treatment successfully solubilized the NC1 domain of the collagen IV network, the relevant site for peroxidasin action (Fig. 3b). Thus, we conclude that peroxidasin does not form a stable complex with the collagen IV network near the NC1 domain.
Peroxidase and Ig Domains of Peroxidasin Reconstitute Sulfilimine Bond Formation
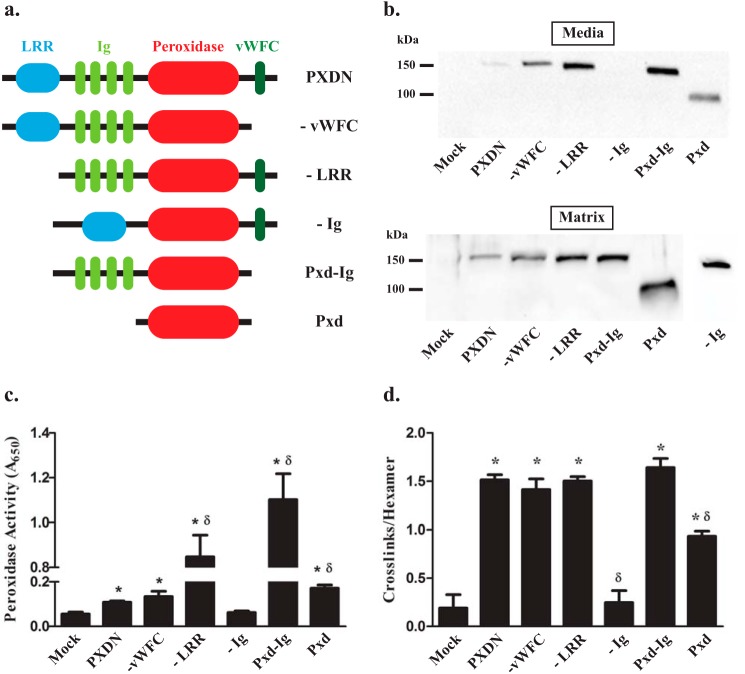
A distinctive feature of peroxidasin is the combination of protein-protein interaction domains, specifically LRR, Ig, and vWFC, along with a catalytic peroxidase domain (7). Thus, we hypothesized that one or more non-catalytic domains of peroxidasin would be required to efficiently form sulfilimine bonds. To test the requirement of the non-catalytic, protein-protein interaction domains of peroxidasin for sulfilimine bond formation, we created several domain deletion constructs (Fig. 4a) and examined their ability to cross-link basement membrane collagen IV in the overlay assay. Except for the construct lacking Ig domains (−Ig), all of the mutant peroxidasin deletion variants exhibited expression and colorimetric peroxidase activity in the culture media (Fig. 4, b and c). As suggested by the general association of peroxidases with extracellular matrix, all of the constructs, including −Ig, demonstrated basement membrane incorporation, because they were designed to include the peroxidase domain (Fig. 4c). The peroxidase domain of peroxidasin alone appeared to support collagen IV cross-linking although significantly less than full-length peroxidasin. When the Ig domains were added to the peroxidase domain, sulfilimine bond forming activity was completely restored to the level observed with full-length peroxidasin. Any construct that contained both peroxidase and Ig domains exhibited normal collagen IV cross-linking activity. Similarly, the construct lacking the Ig domain (−Ig) failed to form sulfilimine cross-links, although proper folding and function were uncertain, because the protein was not found in culture media to measure colorimetric peroxidase activity (Fig. 4d). Taken together, these data suggest that the Ig domain is required to recapitulate physiologic sulfilimine bond formation.
FIGURE 4.
The immunoglobulin and peroxidase domains of peroxidasin are required for sulfilimine bond formation. a, schematic depiction of the domain structure of peroxidasin and its deletion constructs. b, immunoblotting of media and matrix fractions under reducing conditions after transient transfection of overlaid HEK293T with peroxidasin constructs demonstrates adequate secretion and matrix incorporation of all constructs except the −Ig construct. Blots are representative of 5 independent experiments. c, all of the constructs, except −Ig, exhibit tetramethylbenzidine colorimetric peroxidase activity greater than mock transfection (*, p < 0.05; n = 7) with some constructs demonstrating peroxidase activity greater than wild type peroxidasin (δ, p < 0.05; n = 7). d, all of the constructs formed sulfilimine cross-links in collagen IV, quantified as cross-links per hexamer, better than mock control except for the −Ig construct (*, p < 0.05, n = 5). The catalytic domain only construct (Pxd) formed more cross-links than mock, but was significantly less than wild type Pxdn (δ, p < 0.05, n = 5). Data were analyzed using analysis of variance followed by post hoc pairwise comparisons with Tukey's correction for multiple comparisons.
The N-terminal Ig Domain of Peroxidasin Is Required for Sulfilimine Bond Formation
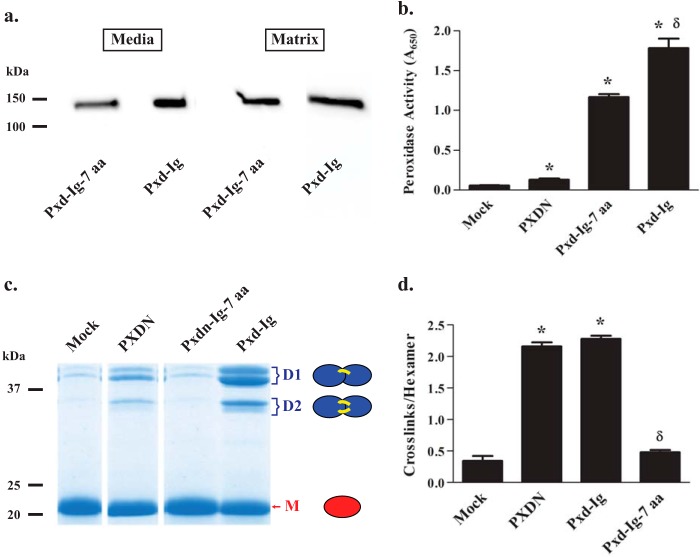
We hypothesized that peroxidasin may behave similarly to other proteins with multiple Ig domains where the N-terminal domain is most involved in interactions with other proteins (16–19). Thus, we focused our attention on the N-terminal Ig domain of peroxidasin and decided to create progressive deletions from the N-terminal end of the peroxidase-Ig construct (Pxd-Ig; Fig. 4a). The first construct lacking 7 N-terminal amino acids (VATITPE) of Pxd-Ig (Pxd-Ig-7 aa) was completely unable to form sulfilimine cross-links in collagen IV despite adequate peroxidase activity and basement membrane incorporation (Fig. 5). Thus, a relatively small N-terminal portion of the Ig domains is critical for sulfilimine bond formation.
FIGURE 5.
The N-terminal 7 residues of the peroxidasin immunoglobulin domains are necessary to cross-link collagen IV. a, immunoblotting, under reducing conditions, reveals the deletion construct lacking 7 N-terminal amino acids from the first immunoglobulin domain (Pxd-Ig-7 aa) is adequately expressed in cell culture media and incorporated into matrix. Blots are representative of 3 independent experiments. b, Pxd-Ig-7 aa demonstrates tetramethylbenzidine oxidation greater than full-length peroxidasin, but slightly less than the peroxidase-immunoglobulin construct (Pxd-Ig; ANOVA followed by post-hoc Tukey's pairwise comparison; *, p < 0.05 compared with PXDN; δ, p < 0.05 Pxd-Ig-7 aa versus Pxd-Ig, n = 3). c, SDS-PAGE of NC1 hexamers (non-reducing conditions) isolated after overlay with mock, PXDN, Pxd-Ig-7 aa, and Pxd-Ig reveal the formation of cross-linked dimeric subunits (D1 and D2 representing single and double cross-linked forms, respectively) primarily with PXDN and Pxd-Ig, whereas mock and Pxd-Ig-7 aa exhibit primarily monomeric, uncross-linked subunits (M). Each of these band regions (D1, D2, and M) appear as single bands (arrows) or doublets (brackets) due to unidentified variability in electrophoretic migration (4, 26, 27). d, the number of cross-links per collagen IV NC1 hexamer was determined for mock, PXDN, Pxd-Ig-7 aa, and Pxd-Ig-transfected cells using densitometric quantitation of the dimeric and monomeric subunits from SDS-PAGE of collagenase-digested PFHR-9 matrix as shown in c. Data were analyzed using analysis of variance followed by post hoc pairwise comparisons with Tukey's correction for multiple comparisons (*, p < 0.05 compared with mock; δ, p < 0.05 compared with PXDN, n = 3).
Discussion
Prior to this work, we assumed that the unique ability of PXDN to cross-link collagen IV may reflect selective HOBr production along with universal expression, paralleling collagen IV, within and among animals. Our assumption predicted that any cell secreting a HOBr producing peroxidase, such as EPO, would cross-link an underlying collagen IV network. However, EPO failed to efficiently cross-link collagen IV even when our experimental paradigm successfully created a non-physiologic association between EPO and basement membrane. These data led us to hypothesize that the non-catalytic domains of peroxidasin are required for cross-link formation. Using deletion constructs, we demonstrated that both the catalytic and N-terminal Ig domain of peroxidasin are essential to form sulfilimine cross-links, evolutionarily conserved post-translational modifications in collagen IV.
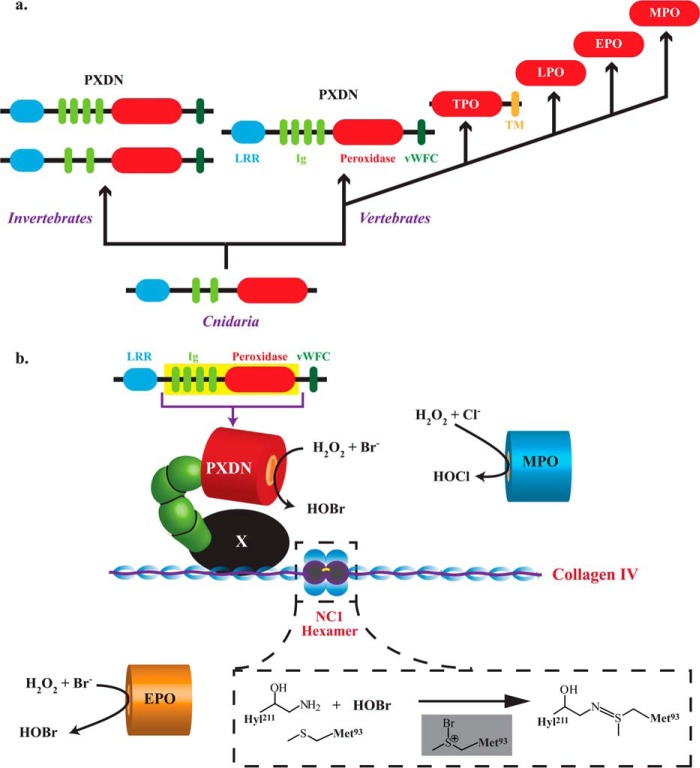
We have previously demonstrated that peroxidasin is conserved throughout the animal kingdom (1). Domain analysis of peroxidasin cDNA sequences from ancient animals, such as Cnidaria, revealed that the minimal structure of peroxidasin includes only 2 of 4 Ig domains consistent with this work, which found a requirement limited to a short, N-terminal portion of these domains in human peroxidasin (Fig. 6a). Taken together, evolutionary conservation, widespread expression in tissues, and the Ig domains of peroxidasin allow for it to form sulfilimine cross-links in collagen IV with no apparent redundancy with other animal heme peroxidases.
FIGURE 6.
Evolution of animal peroxidases and model for peroxidasin-mediated sulfilimine bond formation in collagen IV. a, PXDN is the ancestral animal heme peroxidase found in Cnidaria that extends into invertebrates and vertebrates with additional Ig and vWFC domains. In vertebrates, peroxidasin most likely gave rise to thyroid peroxidase (TPO) via gene duplication, deletion of non-catalytic domains, and fusion with a transmembrane (TM) domain. Lactoperoxidase (LPO), EPO, and MPO arose from successive gene duplications after loss of the TPO TM domain. b, the N-terminal Ig domain of peroxidasin is proposed to interact with an unknown intermediary matrix protein (in black denoted as X) to form a ternary complex with collagen IV such that HOBr generated by its catalytic domain is placed in a spatially productive position to form sulfilimine cross-links in the NC1 hexamer. Peroxidasin-derived HOBr oxidizes Met-93 to create a bromosulfonium intermediate leading to sulfilimine bond formation (inset). MPO and EPO associate with basement membrane but do not produce HOBr (MPO) or do so in a spatially inefficient manner (EPO).
The Ig domain superfamily is widely found in prokaryotes and eukaryotes. Bacterial Ig domains are often in enzymatically active proteins, whereas animal Ig domains are typically found as cell surface adhesion receptors primarily involved in immune and nervous system development and function (20, 21). Ig domains are categorized into 4 sets based on structural similarity to the analogous antibody domains: constant (C1 and C2 sets), variable (V set), and intermediate (I-set) (21). The Ig domains of peroxidasin are classified within the I-set, consistent with the hypothesis that the I-set is the primordial Ig domain of the animal kingdom (21, 22). Akin to bacterial Ig domain proteins, peroxidasin is the only known animal protein to combine an Ig domain with a catalytic domain, suggesting a functionally intermediate position for peroxidasin between bacterial and animal Ig domain superfamily proteins.
We initially considered a simple model wherein peroxidasin interacts with collagen IV via its N-terminal Ig domain to produce HOBr in close spatial proximity to its substrate, the sulfilimine cross-linked residues of the collagen IV NC1 hexamer. However, we found evidence against a stable association directly between peroxidasin and collagen IV although we cannot rule out a transient interaction. Thus, we hypothesize that peroxidasin associates with another basement membrane component to possibly form a ternary complex with collagen IV to facilitate HOBr delivery to the NC1 substrate. Presumably the N-terminal Ig domain modifies the spatial orientation of this complex to facilitate sulfilimine bond formation (Fig. 6b). The model implies that HOBr may diffuse a short distance before encountering its collagen IV substrate rather than being delivered directly without free diffusion. Indeed, recent work based on homology between peroxidasin and MPO suggests that the heme cavity of peroxidasin may not be large enough to accommodate collagen IV for direct HOBr delivery. The authors of this work hypothesize that a relatively short diffusion distance and inefficient bromide oxidation allows for sulfilimine bond formation with minimal collateral oxidative damage (23).
Notably, MPO has been previously shown to associate with endothelial basement membranes and interact with perlecan, collagen IV, and fibronectin (13, 24, 25). We failed to demonstrate a direct interaction between collagen IV and MPO, which may reflect technical differences (24). No prior work has examined an association between EPO and basement membrane, which we demonstrate herein.
The collagen IV sulfilimine cross-link and peroxidasin are critical for the integrity and tissue development of basement membranes. Based on this work, we envision a mechanism involving a ternary complex between peroxidasin, another basement membrane component, and collagen IV with peroxidasin generating diffusible HOBr to catalyze sulfilimine bond formation. In this model, the N-terminal Ig domain of peroxidasin is critical for the efficient delivery of HOBr to its collagen IV substrate.
Author Contributions
I. A. E.-T. conducted and analyzed the experiments. B. G. H. and G. B. conceived the work, designed the study, and wrote the manuscript. All authors approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK18381 (to B. G. H.), R01 DK 18381-38S1 (to I. A. E-T.), and K08 DK 097306 (to G. B.) and a Burroughs-Wellcome Fund Career Award for Medical Scientists (13030995) and developmental funds from the Vanderbilt University Division of Nephrology & Hypertension (to G. B.). The authors declare that they have no conflicts of interest with the contents of this article.
- LRR
- leucine-rich repeat
- vWFC
- von Willebrand factor C
- ECM
- extracellular matrix
- MPO
- myeloperoxidase
- EPO
- eosinophil peroxidase
- PXDN
- peroxidasin.
References
- 1. Fidler A. L., Vanacore R. M., Chetyrkin S. V., Pedchenko V. K., Bhave G., Yin V. P., Stothers C. L., Rose K. L., McDonald W. H., Clark T. A., Borza D. B., Steele R. E., Ivy M. T., Aspirnauts, Hudson J. K., Hudson B. G. (2014) A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc. Natl. Acad. Sci. U.S.A. 111, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khoshnoodi J., Pedchenko V., Hudson B. G. (2008) Mammalian collagen IV. Microsc. Res. Tech. 71, 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vanacore R., Ham A. J., Voehler M., Sanders C. R., Conrads T. P., Veenstra T. D., Sharpless K. B., Dawson P. E., Hudson B. G. (2009) A sulfilimine bond identified in collagen IV. Science 325, 1230–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhave G., Cummings C. F., Vanacore R. M., Kumagai-Cresse C., Ero-Tolliver I. A., Rafi M., Kang J. S., Pedchenko V., Fessler L. I., Fessler J. H., Hudson B. G. (2012) Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat. Chem. Biol. 8, 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gotenstein J. R., Swale R. E., Fukuda T., Wu Z., Giurumescu C. A., Goncharov A., Jin Y., Chisholm A. D. (2010) The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development 137, 3603–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCall A. S., Cummings C. F., Bhave G., Vanacore R., Page-McCaw A., Hudson B. G. (2014) Bromine is an essential trace element for assembly of collagen IV scaffolds in tissue development and architecture. Cell 157, 1380–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson R. E., Fessler L. I., Takagi Y., Blumberg B., Keene D. R., Olson P. F., Parker C. G., Fessler J. H. (1994) Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 13, 3438–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petérfi Z., Donkó A., Orient A., Sum A., Prókai A., Molnár B., Veréb Z., Rajnavolgyi E., Kovács K. J., Müller V., Szabó A. J., Geiszt M. (2009) Peroxidasin is secreted and incorporated into the extracellular matrix of myofibroblasts and fibrotic kidney. Am. J. Pathol. 175, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zamocky M., Jakopitsch C., Furtmüller P. G., Dunand C., Obinger C. (2008) The peroxidase-cyclooxygenase superfamily: Reconstructed evolution of critical enzymes of the innate immune system. Proteins 72, 589–605 [DOI] [PubMed] [Google Scholar]
- 10. Obinger C. (2006) Chemistry and biology of human peroxidases. Arch. Biochem. Biophys. 445, 197–198 [DOI] [PubMed] [Google Scholar]
- 11. Senthilmohan R., Kettle A. J. (2006) Bromination and chlorination reactions of myeloperoxidase at physiological concentrations of bromide and chloride. Arch. Biochem. Biophys. 445, 235–244 [DOI] [PubMed] [Google Scholar]
- 12. Weiss S. J., Test S. T., Eckmann C. M., Roos D., Regiani S. (1986) Brominating oxidants generated by human eosinophils. Science 234, 200–203 [DOI] [PubMed] [Google Scholar]
- 13. Baldus S., Eiserich J. P., Mani A., Castro L., Figueroa M., Chumley P., Ma W., Tousson A., White C. R., Bullard D. C., Brennan M. L., Lusis A. J., Moore K. P., Freeman B. A. (2001) Endothelial transcytosis of myeloperoxidase confers specificity to vascular ECM proteins as targets of tyrosine nitration. J. Clin. Invest. 108, 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhave G., Zhu W., Wang H., Brasier D. J., Oxford G. S., Gereau R. W., 4th. (2002) cAMP-dependent protein kinase regulates desensitization of the capsaicin receptor (VR1) by direct phosphorylation. Neuron 35, 721–731 [DOI] [PubMed] [Google Scholar]
- 15. Weiss S. J. (1989) Tissue destruction by neutrophils. N. Engl. J. Med. 320, 365–376 [DOI] [PubMed] [Google Scholar]
- 16. Brondijk T. H., de Ruiter T., Ballering J., Wienk H., Lebbink R. J., van Ingen H., Boelens R., Farndale R. W., Meyaard L., Huizinga E. G. (2010) Crystal structure and collagen-binding site of immune inhibitory receptor LAIR-1: unexpected implications for collagen binding by platelet receptor GPVI. Blood 115, 1364–1373 [DOI] [PubMed] [Google Scholar]
- 17. Horii K., Kahn M. L., Herr A. B. (2006) Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI. Blood 108, 936–942 [DOI] [PubMed] [Google Scholar]
- 18. Leitinger B. (2011) Transmembrane collagen receptors. Annu. Rev. Cell Dev. Biol. 27, 265–290 [DOI] [PubMed] [Google Scholar]
- 19. Kiselyov V. V., Berezin V., Maar T. E., Soroka V., Edvardsen K., Schousboe A., Bock E. (1997) The first immunoglobulin-like neural cell adhesion molecule (NCAM) domain is involved in double-reciprocal interaction with the second immunoglobulin-like NCAM domain and in heparin binding. J. Biol. Chem. 272, 10125–10134 [DOI] [PubMed] [Google Scholar]
- 20. Bork P., Holm L., Sander C. (1994) The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242, 309–320 [DOI] [PubMed] [Google Scholar]
- 21. Smith D. K., Xue H. (1997) Sequence profiles of immunoglobulin and immunoglobulin-like domains. J. Mol. Biol. 274, 530–545 [DOI] [PubMed] [Google Scholar]
- 22. Buljan M., Bateman A. (2009) The evolution of protein domain families. Biochem. Soc. Trans. 37, 751–755 [DOI] [PubMed] [Google Scholar]
- 23. Soudi M., Paumann-Page M., Delporte C., Pirker K. F., Bellei M., Edenhofer E., Stadlmayr G., Battistuzzi G., Boudjeltia K. Z., Furtmüller P. G., Van Antwerpen P., Obinger C. (2015) Multidomain human peroxidasin 1 is a highly glycosylated and stable homotrimeric high spin ferric peroxidase. J. Biol. Chem. 290, 10876–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kubala L., Kolářová H., Viteček J., Kremserová S., Klinke A., Lau D., Chapman A. L., Baldus S., Eiserich J. P. (2013) The potentiation of myeloperoxidase activity by the glycosaminoglycan-dependent binding of myeloperoxidase to proteins of the extracellular matrix. Biochim. Biophys. Acta 1830, 4524–4536 [DOI] [PubMed] [Google Scholar]
- 25. Rees M. D., Whitelock J. M., Malle E., Chuang C. Y., Iozzo R. V., Nilasaroya A., Davies M. J. (2010) Myeloperoxidase-derived oxidants selectively disrupt the protein core of the heparan sulfate proteoglycan perlecan. Matrix Biol. 29, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weber S., Dölz R., Timpl R., Fessler J. H., Engel J. (1988) Reductive cleavage and reformation of the interchain and intrachain disulfide bonds in the globular hexameric domain NC1 involved in network assembly of basement membrane collagen (type IV). Eur. J. Biochem. 175, 229–236 [DOI] [PubMed] [Google Scholar]
- 27. Weber S., Engel J., Wiedemann H., Glanville R. W., Timpl R. (1984) Subunit structure and assembly of the globular domain of basement-membrane collagen type IV. Eur. J. Biochem. 139, 401–410 [DOI] [PubMed] [Google Scholar]