Background: Hac1 protein, encoded by a cytoplasmically spliced mRNA, activates the unfolded protein response to maintain cellular protein homeostasis and alleviate endoplasmic reticulum stress.
Results: Under non-stress conditions, translation initiation on the HAC1 mRNA is repressed.
Conclusion: Base-pairing interaction between the 5′ leader and intron represses translation initiation on the HAC1 mRNA.
Significance: A unique mechanism of intron-mediated inhibition of ribosomal scanning.
Keywords: endoplasmic reticulum stress (ER stress), eukaryotic initiation factor 4A (eIF4A), eukaryotic translation initiation, ribosome, translation, unfolded protein response (UPR), HAC1
Abstract
The Hac1 transcription factor in yeast up-regulates a collection of genes that control protein homeostasis. Base-pairing interactions between sequences in the intron and the 5′-untranslated region (5′ UTR) of the HAC1 mRNA represses Hac1 protein production under basal conditions, whereas cytoplasmic splicing of the intron by the Ire1 kinase-endonuclease, activated under endoplasmic reticulum stress conditions, relieves the inhibition and enables Hac1 synthesis. Using a random mutational screen as well as site-directed mutagenesis, we identify point mutations within the 5′ UTR-intron interaction site that derepress translation of the unspliced HAC1 mRNA. We also show that insertion of an in-frame AUG start codon upstream of the interaction site releases the translational block, demonstrating that an elongating ribosome can disrupt the interaction. Moreover, overexpression of translation initiation factor eIF4A, a helicase, enhances production of Hac1 from an mRNA containing an upstream AUG start codon at the beginning of the base-paired region. These results suggest that the major block of translation occurs at the initiation stage. Supporting this interpretation, the point mutations that enhanced Hac1 production resulted in an increased percentage of the HAC1 mRNA associating with polysomes versus free ribosomal subunits. Thus, our results provide evidence that the 5′ UTR-intron interaction represses translation initiation on the unspliced HAC1 mRNA.
Introduction
In eukaryotic cells mRNA translation begins with the ordered assembly of the small (40S) ribosomal subunit, initiator methionyl-tRNA (Met-tRNAiMet), and initiation factors at the 5′-end of an mRNA (1, 2). The resulting preinitiation complex (PIC)3 then travels along the 5′ mRNA leader in search of a translation start codon in a process referred to as ribosomal scanning. Typically, the first AUG codon encountered by the scanning PIC is selected as the translation start site in part due to base-pairing interactions between the anticodon loop of the Met-tRNAiMet and the AUG codon. After selection of the start codon, initiation factors dissociate from the PIC, and the 60S ribosomal subunit joins yielding an 80S ribosomal complex (2). The 80S complex then enters the elongation phase of translation and decodes the mRNA to produce a polypeptide.
In addition to alterations in the activity of initiation factors, translation is also controlled by cis-acting mRNA elements (3). For example, binding of the translation factor eIF4E to the 5′-m7G (7-methylguanosine)-cap and the poly(A)-binding protein PABP to the 3′-poly(A) tail facilitates ribosome recruitment to an mRNA (4, 5). In addition, nucleotide sequence context (the Kozak consensus motif) flanking an AUG or non-canonical start codon impacts the efficiency with which a scanning ribosome selects a translation start site (3). Moreover, secondary structure in the 5′ UTR can interfere with PIC binding or scanning and thereby block translation (3, 6–8). Translation of mRNAs with more secondary structure shows a greater requirement for the RNA helicase eIF4A (1). Additionally, out-of-frame AUG start codons and upstream open reading frames (ORFs) in the 5′ leader can restrict access of the ribosome to the main ORF. A classic example is the yeast GCN4 mRNA in which short upstream ORFs potentiate translation of the main GCN4 ORF and confer heightened dependence on translation factor activity (9). In addition to these important roles of sequences in the 5′ UTR, sequence elements in the 3′ UTR can also modulate translation. Interaction of RNA-binding proteins such as the GAIT complex (10) and sex-lethal, Smaug, or CPEB (11, 12), with elements in the 3′ UTR of select mRNAs represses translation.
A unique strategy is observed in regulating translation of the yeast HAC1 mRNA involving the 5′ UTR and the intron (13). The intron (spanning nucleotides G661 to G913, with the adenine of AUG start codon assigned as 1) is not spliced in the nucleus by the spliceosome but instead is retained in the mRNA that is exported to the cytoplasm (14). Previous work established that base-pairing interactions between elements in the intron and the 5′ UTR repress translation of the unspliced HAC1 mRNA (13). Under conditions of endoplasmic reticulum (ER) stress, the endoribonuclease Ire1 is activated and cleaves both exon-intron boundaries of the HAC1 mRNA in the cytoplasm (15, 16). The two exons are then ligated by tRNA ligase (17), resulting in an altered ORF with a new codon starting at nucleotide G661 and a UAG stop codon at nucleotide 963 (see Fig. 1A). Finally, the 3′ UTR has been reported to positively regulate translation of the spliced HAC1 mRNA (18). The Hac1 protein produced from the spliced mRNA is a transcription factor and primary effector of the unfolded protein response (UPR) (19, 20) that activates expression of a set of genes involved in maintaining protein homeostasis and alleviating ER stress (16, 21–24). As the Hac1 proteins produced from both the spliced and unspliced mRNA are functional transcription factors (15), a key element in the UPR is repression of translation of the unspliced Hac1 mRNA.
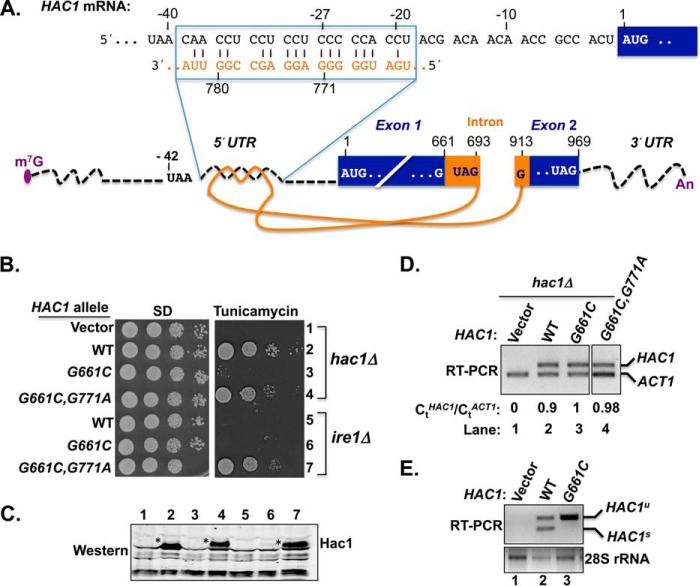
FIGURE 1.
Intron point mutation derepressed translation of unspliced HAC1 mRNA. A, schematic diagram of HAC1 mRNA. The m7G (7-methylguanosine) cap, 5′ and 3′ UTRs (black dotted lines), exons (dark blue boxes), intron (solid orange line), and poly(A) tail (An) are shown. The intron spans nucleotides G661 to G913 and interacts with the 5′ UTR. The nucleotide sequence of the 5′ UTR-intron interaction is shown at the top. The adenine of the HAC1 AUG start codon is assigned as position +1 with positive and negative values for the downstream and upstream nucleotides, respectively. Two in-frame HAC1 UAG stop codons are shown; one located in the intron (nucleotide positions +691 to +693) and the second located in exon 2 (nucleotide positions +967 to +969). B, yeast growth assay. Transformants of hac1Δ or ire1Δ yeast strains carrying an empty vector or expressing the indicated HAC1 allele were grown in SD medium to saturation, and 5 μl of serial dilutions (of A600 = 1.0, 0.1, 0.01, and 0.001) were spotted on SD medium or SD medium containing 0.4 μg/ml tunicamycin and incubated 3 d at 30 °C. C, immunoblot analysis of Hac1 protein. Strains described in panel B were grown in SD medium and treated with 5 mm DTT to induce ER stress, and WCEs were prepared and subjected to SDS-PAGE followed by Western blot analysis using a polyclonal antibody raised against recombinant Hac1 protein. Lanes are numbered according to row numbers in panel B. D, analysis of HAC1 mRNA levels. Total RNA was extracted from the hac1Δ strains expressing the indicated HAC1 alleles and grown under non-stress condition. The RNA was used as a template for RT and Q-PCR analyses of HAC1 and ACT1 mRNAs as described under “Experimental Procedures.” The ratio of threshold values (Ct) for HAC1 and ACT1 mRNA are shown. E, RT-PCR analysis of HAC1 mRNA splicing. A hac1Δ strain carrying an empty vector or expressing WT HAC1 or the HAC1-G661C mutant was grown in SD medium and treated with 5 mm DTT, and then total RNA was extracted and used as a template for RT-PCR analysis of 28S rRNA or of unspliced (HAC1u) and spliced (HAC1s) HAC1 transcripts as previously described (25).
Despite much progress in understanding the regulation of HAC1 mRNA translation, the molecular mechanism is not yet clear. To gain mechanistic insights into HAC1 mRNA translation, we conducted a genetic screen to identify intragenic mutations that enable a splicing-deficient and translationally inert HAC1 mRNA (i.e. HAC1-G661C) to produce a Hac1 protein capable of stimulating the ER stress response. From this screen we identified a single base mutation (i.e. G771A) in the intron that is predicted to disrupt the base-pairing interaction with the 5′ UTR (13), suggesting that disruption of the 5′ UTR-intron interaction leads to translation of the HAC1-G661C,G771A mRNA without splicing. Consistent with the notion that the 5′ UTR-intron interaction interferes with ribosomal scanning and translation initiation on the HAC1 mRNA, introduction of an upstream in-frame AUG codon before the 5′ UTR-intron base-pairing region relieved the translational block on the HAC1 mRNA. Taken together, in contrast to previously proposed models, our data reveal a unique mechanism of intron-mediated inhibition of ribosomal scanning.
Experimental Procedures
Yeast Strains and Plasmids
The ire1Δ (MATa his3-Δ1 leu2-Δ0 met5-Δ0 ura3-Δ0 ire1::KanMX) and hac1Δ (MATa his3-Δ1 leu2-Δ0 met5-Δ0 ura3-Δ0 hac1::KanMX) yeast strains were obtained from the yeast genome deletion collection. Strain J751 (MATa his3-Δ1 leu2-Δ0 met5-Δ0 ura3-Δ0 ire1:NAT) was derived by replacing KanMX4 in the ire1Δ strain with the nourseothricin-resistance (NAT) marker gene. The hac1Δ ire1Δ (MATa his3-Δ1 leu2-Δ0 met5-Δ0 ura3-Δ0 ire1:NAT hac1::KanMX) double deletion strain J772 was constructed by replacing HAC1 with KanMX4 in strain J751. Strain J1167 (MATa his3-Δ1 leu2-Δ0 met5-Δ0 ura3-Δ0 ire1:NAT HAC1-G767C,G768C) was constructed by integration of plasmid pC4070 (linearized with AscI) into strain J751. After selection of segregants on 5-fluoroorotic acid, the replacement of the WT HAC1 allele with the HAC1-G767C,G768C intron mutant was confirmed by PCR. For the UPR assay, yeast growth was tested on SD medium containing the ER stress inducer tunicamycin (0.4 μg/ml). Mutations were created by site-directed mutagenesis or by fusion PCR. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pRS316 | Low copy URA3 vector | (46) |
| pC3641 | Destroy SacI site in pRS316 | This study |
| pRS306 | URA3 integrating vector | (46) |
| D63 | HAC1 in pRS316 (insert at Kpn1/BamH1 sites) | (25) |
| D470 | HAC1-G661C in pRS316 | This study |
| D803 | HAC1-G661C,G771A in pRS316 | This study |
| pC4058 | HA-HAC1 in pC3641 | This study |
| D1278 | HAC1-C−27G in D63 | This study |
| D1279 | HA-HAC1-G771C in D63 | This study |
| pC4059 | HA-HAC1-C−23G in pC4058 | This study |
| pC4060 | HA-HAC1-C−23G,C−24G in pC4058 | This study |
| pC4062 | HA-HAC1-G767C in pC4058 | This study |
| pC4063 | HA-HAC1-G767C,G768C in pC4058 | This study |
| D1280 | HAC1-C−27G,G771C in D63 | This study |
| pC4070 | HA-HAC1-G767C,G768C in pRS306 | This study |
| D1116 | HAC1-A1G,A7G in pRS316 (HAC1-A1G) | This study |
| D1118 | HAC1-AUG−42,A1G,A7G in pRS316 (AUG−42,A1G) | This study |
| D1117 | HAC1-AUG−33, A1G,A7G in pRS316 (AUG−33,A1G) | This study |
| D1119 | HAC1-AUG−60, A1G,A7G in pRS316 (AUG−60,A1G) | This study |
| D860 | IRE1 in pRS316 | This study |
| B3353 | TIF2 (eIF4A) in YEplac181 (High copy LEU2) | (47) |
| FJZ051 | TIF3 (eIF4B) in YEplac181 | (36) |
| B3355 | CDC33 (eIF4E) in YEplac181 | (47) |
| B3995 | TIF1-D170E (eIF4A-D170E) in YEplac181 | (34) |
Reverse Transcription (RT) and Quantitative (Q) PCR Analyses
RNA isolation and RT-PCR analyses were performed essentially as described previously (25). Yeast cells were grown in synthetic complete medium at 30 °C till the A600 reached ∼0.5–06. When desired, DTT (5 mm) was added to the medium to induce ER stress, and cells were grown for an additional 4 h. Total RNA was isolated using an RNeasy kit (Qiagen), and first strand cDNA was synthesized using Superscript-III reverse transcriptase (Invitrogen) and random primers. The synthetic cDNA was subjected to RT-PCR and Q-PCR analyses. For RT-PCR, the synthetic cDNA was amplified by PCR using primers specific for HAC1 (forward primer 5′-CGCAATCGAACTTGGCTATCCCTACC-3′, corresponding to nucleotides +35 to +60 of the HAC1 open reading frame (ORF), and reverse primer 5′-GGGTAGACTGTTTCCCGC-3′, corresponding to nucleotides +604 to +621) or ACT1 (forward primer 5′-CTGAAAGAGAAATTGTCCGTG-3′, corresponding to nucleotides +919 to +939 of the ACT1 ORF (numbering includes the intron), and reverse primer 5′-CTTGTGGTGAACGATAGATGG 3′, corresponding to nucleotides +1408 to +1428). For analysis of HAC1 mRNA splicing, the same forward primer was used along with a reverse primer (5′-CCCACCAACAGCGATAATAACGAG-3′) that corresponded to nucleotides +1002 to +1025. PCR products were then resolved by agarose gel electrophoresis. For Q-PCR, the synthetic cDNA was amplified on a real time PCR machine (Bio-Rad CFX 3.1) using primers specific for HAC1 (forward primer 5′-CGCAATCGAACTTGGCTATCCC-3′ and reverse primer 5′-CTCTCGAGATACTGCAGATG-3′) or ACT1 (forward primer 5′-CTGAAAGAGAAATTGTCCGTG-3′ and reverse primer5′-GTGATGACTTGACCATCTGG-3′). Experiments were repeated twice, and each sample was run in triplicate. Data were analyzed using CFX 3.1 software, and the threshold (Ct) value was calculated. The Ct value represents the average value of triplicate samples, and these average values were used to calculate the CtHAC1/CtACT1 ratio.
Western Blot Analysis
Yeast cells were grown in synthetic complete medium to A600 = 0.6. When desired, 5 mm DTT was then added to the medium to induce ER stress, and cells were incubated an additional 4 h before harvesting. Cells were harvested by centrifugation, mixed with 2 volumes of 20% (w/v) trichloroacetic acid, and then broken by agitation with glass beads. Proteins were extracted with SDS Loading Buffer (2% SDS, 2 mm EDTA, 50 mm Tris-HC1 (pH 6.8), 10% glycerol, 0.01% bromphenol blue), and after neutralization with 1 m Tris base, samples were boiled for 5 min and then subjected to SDS-PAGE and immunoblot analysis using antibodies raised against recombinant Hac1 protein or yeast eIF2α or using anti-HA antibodies (Cell Signaling Technology or Roche Applied Science).
Polysome and Northern Analysis
Cells were grown to mid-log phase, treated with cycloheximide (100 μg/ml), incubated for 5 min, and harvested. Cell pellets were resuspended in 5 ml of Polysome Buffer (20 mm Tris-HCl (pH 8.0), 140 mm KCl, 5 mm MgCl2, 100 μg/ml cycloheximide), pelleted, and then resuspended in 1 ml of Polysome Buffer containing 1% Triton X-100 (Sigma), 1 mm DTT, EDTA-free Protease Inhibitor tablet (Roche Applied Science) and 4 μl of RNaseOUT (Invitrogen). Cells were broken by vigorous mixing with glass beads at 4 °C using a vortex mixer. WCEs were clarified by centrifugation, and the equivalent of A600 = 25 units was layered on top of 7–47% (w/v) sucrose gradients made in Polysome Buffer containing 1 mm DTT and EDTA-free Protease Inhibitor tablets. Samples were sedimented at 39,000 rpm for 150 min at 4 °C in an SW41 rotor (Beckman Coulter). Gradients were fractionated while scanning at A254.
To isolate RNA, glycogen was added to the sucrose gradient fractions; the fractions were then mixed with ethanol and stored at −20 °C overnight. RNA pellets were collected by sedimentation, resuspended, and then extracted twice with phenol:chloroform. The RNA was then precipitated in ethanol, washed with 70% ethanol, and resuspended in 30 μl of Loading Buffer (21 μl of H2O, 3 μl of formaldehyde, 3 μl of 10× MOPS buffer, 3 μl of 10× RNA loading dye). Equal volumes (25 μl) were used for Northern analysis.
To visualize HAC1 by Northern analysis, RNA samples were separated on a 1.2% agarose, 5% formaldehyde gel in MOPS buffer, transferred to a NyTran membrane (Whatman) overnight in 20× SSC buffer, and then UV cross-linked to the membrane in a Stratalinker (Stratagene). After prehybridization in 12 ml UltraHYB buffer (Ambion) for 2 h at 42 °C, the membrane was incubated with randomly labeled [32P]dCTP probes for HAC1 and ADH1 overnight at 42 °C while rotating. The HAC1 (primers oCB1 (5′-GTCTACGGCAGGTAGCGTCGT-3′) and oCB25 (5′-AATTCGCAATCGAACTTGGCTATCCC-3′)) and ADH1 (primers oCB105 (5′-ATGTCTATCCCAGAAAC-3′) and oCB106 (5′-ACCCAAGACTCTGTAACC-3′)) probes were generated by PCR followed by random-labeling with [32P]dCTP probes (Lofstrand). Blots were washed at 42 °C for 2 × 10 min in 2× SSC, 0.1% SDS buffer followed by 2 × 15 min in 0.1× SSC, 0.1% SDS buffer. After the final wash, the RNA was imaged and quantitated using phosphorimaging.
Results
Genetic Screen Identifies a Single Mutation in the HAC1 Intron That Bypasses the Requirement for mRNA Splicing to Derepress HAC1 Production
Under conditions of ER stress, the endonuclease Ire1 cleaves the phosphodiester bonds after nucleotides G661 and G913 (Fig. 1A) to remove the intron from the HAC1 mRNA (16, 26). After ligation of the exons, translation of the spliced mRNA yields a Hac1 protein of 238 amino acids (13), also known as the Hac1i isoform (“i” for induced; produced from the ER stress-induced, spliced mRNA). In the absence of splicing, the first in-frame stop codon (UAG) after G661 is located at nucleotide positions 691–693 (see Fig. 1A). This alternate 693-nucleotide ORF in the unspliced mRNA can be translated into a functional Hac1 protein of 230 amino acids (27), known as the Hac1u isoform (“u” for uninduced; produced under non-stress conditions). However, translation of the unspliced mRNA and, thus, production of Hac1u is repressed by the intron (Fig. 1A, orange), which interacts with the 5′ UTR and somehow blocks translation of the HAC1 ORF (13, 28). To gain mechanistic insights into the intron-mediated translational repression, we screened for intragenic suppressor mutations in a splicing-deficient and translationally inert HAC1-G661C allele (see below), expecting that the second-site mutations would allow ribosomes to bypass the 5′ UTR-intron interaction and produce a Hac1 protein from the unspliced mRNA.
Whereas a hac1Δ yeast strain carrying an empty vector grew as well on synthetic dextrose (SD) medium as the same strain expressing HAC1 from a low copy number plasmid (Fig. 1B, rows 1 and 2), growth on SD medium containing the ER stress inducer tunicamycin required HAC1 expression (Fig. 1B, rows 1 and 2). These results are consistent with the previous reports showing that Hac1 function is essential for the ER stress response (21, 22). Mutation of the first Ire1-cleavage site (i.e. G661C) impaired yeast cell growth on tunicamycin medium (Fig. 1B, row 3), suggesting that the G661C mutation reduced the HAC1 mRNA level, mRNA splicing, and/or translation.
To determine whether the G661C mutation caused a reduction in HAC1 mRNA levels and/or mRNA splicing, total RNA was extracted from these cells and then used as a template to amplify the HAC1 mRNA as well as an endogenous ACT1 mRNA (housekeeping control) by RT-PCR as described under “Experimental Procedures.” As shown in Fig. 1D, similar amounts of ACT1 mRNA were detected in each RNA sample (lanes 1–3). As expected, no HAC1 mRNA was detected in the hac1Δ strain transformed with an empty vector (Fig. 1D, lane 1), whereas similar amounts of HAC1 mRNA were detected in the strains expressing WT HAC1 (lane 2) or the HAC1-G661C mutant (lane 3). Thus, the tunicamycin-sensitive (TmS) phenotype of the strain expressing the HAC1-G661C mutant is likely not due to reduced HAC1 mRNA levels.
To determine whether the G661C mutation impaired mRNA splicing, RT-PCR was performed using primers that readily distinguish the unspliced (HAC1u) and spliced (HAC1s) transcripts. Again, as expected, no HAC1 mRNA was detected in the RNA sample prepared from the hac1Δ strain carrying an empty vector (Fig. 1E, upper panel, lane 1). Consistent with the Ire1 endonuclease functioning at basal levels in unstressedcells, both HAC1u and HAC1s transcripts were amplified from cells expressing WT HAC1 (Fig. 1E, upper panel, lane 2). In cells expressing the HAC1-G661C allele, the vast majority of mRNA was unspliced (HAC1u; Fig. 1E, lane 3), consistent with the idea that the G661C mutation in the first cleavage site impaired HAC1 mRNA splicing and consequently reduced the ER stress response. The defective splicing of the HAC1-G661C mRNA is supported by previous studies showing that mutation of this guanine residue at the cleavage site impairs Ire1 cleavage of the HAC1 mRNA both in vivo and in vitro (16, 29).
To confirm that the G661C mutation eliminated Hac1 protein expression, WCEs were prepared from the cells exposed to DTT to induce ER stress (“Experimental Procedures”) and then subjected to immunoblot analysis using an antibody against the Hac1 protein. As expected, the Hac1 protein was detected in the extract obtained from cells expressing the WT HAC1 allele (Fig. 1C, lane 2). In contrast, no Hac1 protein was detected in cells expressing the HAC1-G661C mutant (lane 3). These results are consistent with the model (13, 15) that translation of the unspliced HAC1 mRNA is blocked.
To screen for intragenic suppressors of the splicing- and translation-defective HAC1-G661C allele, a plasmid bearing the HAC1-G661C variant was amplified in Escherichia coli XL-1 red cells (Stratagene) that are deficient in three DNA repair pathways. The mutagenized pool of plasmids was purified and used to transform a hac1Δ yeast strain. Transformants were screened for the ability to grow on medium containing tunicamycin. The HAC1 plasmid was isolated from the tunicamycin-resistant (TmR) colonies, re-tested in a hac1Δ strain to confirm that the TmR phenotype was associated with the plasmid, and then sequenced to detect the mutation. A single second-site suppressor mutation, G771A, was found to restore the ability of the HAC1-G661C variant to activate the ER stress response (Fig. 1B, row 4).
The base G771 is located in the region of the intron that is predicted to form base-pairing interactions with the 5′ UTR of the HAC1 mRNA (13). In the alignment shown in Fig. 1A, the G771 base in the intron is base-paired with C−27 in the 5′ UTR. Given the location of this suppressor mutation, we hypothesized that the G771A mutation disrupted the 5′ UTR-intron interaction leading to derepression of HAC1 mRNA translation in the absence of splicing. To test the possibility that the G771A mutation enabled translation of unspliced HAC1 mRNA, plasmids encoding WT HAC1, the splice-site mutant HAC1-G661C, or the suppressor mutant HAC1-G661C,G771A were introduced into a yeast strain lacking the Ire1 endonuclease. The transformants were then tested for the ability to activate the UPR as detected by growth on medium containing tunicamycin. As shown in the Fig. 1B (rows 5 and 6), the ire1Δ strain expressing WT HAC1 or HAC1-G661C failed to grow on the medium containing tunicamycin, consistent with the notion that Ire1-mediated splicing of HAC1 mRNA is essential for the ER stress response (13, 15, 21). In contrast, the HAC1-G661C,G771A mutant conferred a TmR phenotype when expressed in the ire1Δ strain (Fig. 1B, row 7), suggesting that the G771A mutation derepressed HAC1 mRNA translation in the absence of splicing. Consistent with this interpretation, immunoblot analysis revealed substantial Hac1 protein levels without an increase in the level of the HAC1 mRNA (Fig. 1D, lane 4) when the HAC1-G661C,G771A mutant was expressed in either hac1Δ (Fig. 1C, lane 4) or ire1Δ (Fig. 1C, lane 7) cells. These data demonstrate that a single base mutation in the intron is sufficient to eliminate translational control of the HAC1 mRNA.
Translational Control of the HAC1 mRNA Is Sensitive to Alteration of a Single Base Pair Interaction between the Intron and 5′ UTR
To gain further insights into HAC1 translational control and the importance of base-pairing contacts between the intron and the 5′ UTR of the HAC1 mRNA, mutations designed to disrupt single or double base-pairing interactions were introduced into the intron or the 5′ UTR of a human influenza hemagglutinin (HA) epitope-tagged version of HAC1. In this construct, the HA tag was inserted between residues Ser-10 and Asn-11 of the HAC1 ORF. Importantly, the HA-tagged HAC1 allele functioned like untagged WT HAC1 and complemented the TmS phenotype of a hac1Δ strain (Fig. 2A, row 2) in an IRE1-dependent manner (Fig. 2B, row 2). As shown in Fig. 2B, the single C−23G (row 3) and the double C−23G,C−24G (row 4) mutations, located in the 5′ UTR of the HA-HAC1 mRNA and designed to disrupt the base-pairing interaction with the intron (Fig. 1A), suppressed the TmS phenotype of an ire1Δ hac1Δ double-mutant strain. As no HAC1 mRNA splicing takes place in the ire1Δ hac1Δ strain, these results indicate that, like the G771A mutation in the intron (Fig. 1B, row 7), point mutations in the HAC1 5′ UTR can derepress Hac1 production. Mutations in other intron residues involved in the base-pairing interactions also led to prominent TmR phenotypes. As shown in Fig. 2B, the G767C (row 5) and G767C,G768C (row 6) mutations in HA-HAC1 conferred strong TmR phenotypes in the ire1Δ hac1Δ strain, suggesting that mutations in either the 5′ UTR or the intron allowed ribosomes to produce the Hac1 protein from the unspliced mRNA.
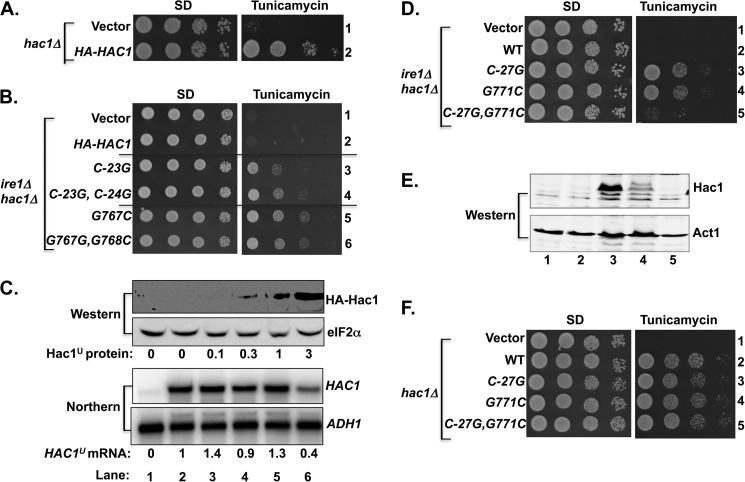
FIGURE 2.
Base-pairing interaction between intron and 5′ UTR repressed HAC1 mRNA translation. A and B, yeast growth assays. Transformants of ire1Δ hac1Δ yeast strain carrying an empty vector or expressing the indicated HA-tagged HAC1 allele were grown in SD medium to saturation, and 5 μl of serial dilutions (of A600 = 1.0, 0.1, 0.01, and 0.001) were spotted on SD medium or SD medium containing 0.4 μg/ml tunicamycin and incubated for 3 days at 30 °C. C, analysis of HAC1 mRNA and protein expression in transformants of the ire1Δ hac1Δ strain. Strains described in panel A were grown in SD medium (unstressed conditions), and WCEs were prepared and subjected to SDS-PAGE followed by Western analysis using polyclonal antibodies against the HA epitope and translation initiation factor eIF2α (upper two panels), and total RNA was extracted and subjected to Northern analysis using probes specific for the HAC1 and ADH1 mRNAs (lower two panels). Lanes are numbered according to row numbers in panel A. The levels of Hac1u protein were determined by quantitative densitometry and normalized to the levels of eIF2α protein. HAC1u mRNA levels were normalized to ADH1 mRNA levels, with the WT ratio (lane 2) set to 1.0. D and F, yeast growth assays. Transformants of the ire1Δ hac1Δ strain carrying an empty vector or expressing the indicated HAC1 allele were grown as described in panels A and B. E, analysis of Hac1 protein expression. WCEs were prepared and subjected to SDS-PAGE followed by Western analysis using polyclonal anti-Hac1 and anti Act1 antibodies. Lanes are numbered according to the row numbers in panel D.
Immunoblot and Northern analyses were used to assess the impact of the 5′ UTR and intron mutations on Hac1 protein and mRNA levels. As the cells lack Ire1 protein and no splicing of the HAC1 mRNA will take place, extracts were prepared from cells grown in the absence of ER stress. Whereas no HA-Hac1 was detected in the extracts from cells expressing WT HA-HAC1 (Fig. 2C, upper panel, lane 2), very low or low levels of HA-Hac1 protein were observed in the extracts prepared from cells expressing the C−23G or C−23G,C−24G mutant alleles, respectively, of HA-HAC1 (Fig. 2C, upper panel, lanes 3 and 4). Thus, low level expression of HA-Hac1 protein from the HA-HAC1-C−23G allele was apparently sufficient to confer a TmR phenotype. Consistent with the more pronounced TmR phenotype, the HA-Hac1 protein was readily detected in extracts prepared from the ire1Δ hac1Δ strain expressing the G767C (Fig. 2C, lane 5) and G767C,G768C (lane 6) mutant form of HA-HAC1. Northern analyses revealed that the levels of HA-HAC1 mRNA were similar in cells expressing the WT or various mutant alleles (Fig. 2C, lower panel, lanes 3–6 versus 2), suggesting that differences in protein expression were not due to altered mRNA levels. These results provide further support for the hypothesis that mutations designed to disrupt the 5′ UTR-intron interaction derepress Hac1 production by enabling translation of the unspliced HAC1 mRNA.
The random mutational screen (Fig. 1B) revealed that the G771A mutation in the HAC1 intron derepressed Hac1 synthesis. In the unspliced HAC1 mRNA, G771 is predicted to form a base-pairing interaction with C−27 (Fig. 1A). To test the importance of this predicted base pair, three additional HAC1 mutants were generated. The single mutants HAC1-C−27G and HAC1-G771C were made to destroy the pairing and the double mutant HAC1-C−27G,G771C was constructed to restore the pairing in the opposite orientation. As shown in Fig. 2D, both HAC1-C−27G (row 3) and HAC1-G771C mutants (row 4) conferred a TmR phenotype in the ire1Δ hac1Δ strain. In contrast, the HAC1-C−27G,G771C double mutant (Fig. 2D, row 5) functioned like WT HAC1 (row 2) and conferred a TmS phenotype in the ire1Δ hac1Δ strain. As expected, the Hac1 protein was detected only in the extracts obtained from cells expressing the HAC1-C−27G mutant (Fig. 2E, Western, lane 3) and the HAC1-G771C mutant (lane 4). The mutual suppression of the HAC1-C−27G and HAC1-G771C mutants in the HAC1-C−27G,G771C double mutant is consistent with the proposed base-pairing interaction between these residues and further demonstrates that repression of HAC1 expression by the 5′ UTR-intron interaction is sensitive to loss of a single base pair interaction.
When expressed in a hac1Δ strain (IRE1 intact in the chromosome), the HAC1-C−27G, HAC1-G771C, and HAC1-C−27G,G771C mutants, like WT HAC1, conferred a TmR phenotype (Fig. 2F, rows 2–5). Based on the results of these mutational analyses, we conclude that 5′ UTR-intron interaction and, in particular, the C−27/G771 base pair plays an important role in repressing HAC1 mRNA translation.
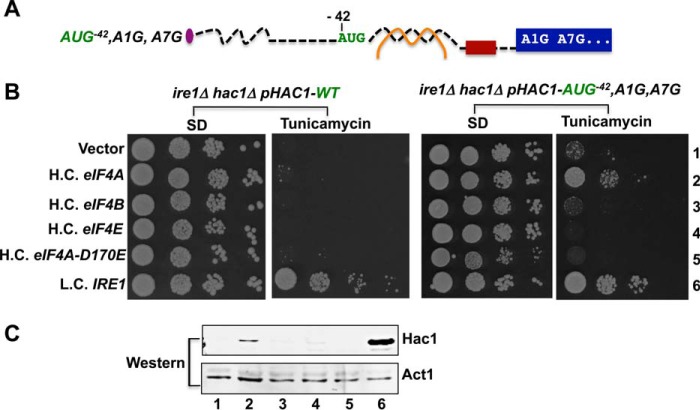
Introduction of an In-frame AUG Start Codon Upstream of the Secondary Structure in the 5′ UTR Derepresses HAC1 Synthesis
The secondary structure in the 5′ UTR could impair Hac1 synthesis by blocking ribosomal scanning to the start codon or, as has been proposed (13), by impairing translation elongation. To differentiate between these models, AUG start codons were inserted at three sites in the 5′ UTR of the HAC1 gene (Fig. 3A). All three of the introduced AUG codons were in-frame with the HAC1 ORF and thus would be predicted to encode Hac1 proteins with N-terminal extensions. To exclusively monitor translation initiating at the inserted AUG codon, the adenine of the authentic HAC1 start codon was mutated to guanine, generating the HAC1-A1G allele, and the adenine of the AUG codon encoding Met3 was likewise altered to guanine to generate the HAC1-A7G mutation (Fig. 3A). As shown in Fig. 3B, the HAC1-A1G,A7G mutant allele failed to complement the TmS phenotype of a hac1Δ strain (row 3). As the A1G,A7G mutation did not reduce HAC1 mRNA levels (Fig. 3D, lane 3), this result is consistent with the notion that, even following splicing, scanning ribosomes fail to initiate at the HAC1 start site (a GUG codon in the A1G mutant), and no Hac1 protein was produced (Fig. 3C, lane 2).
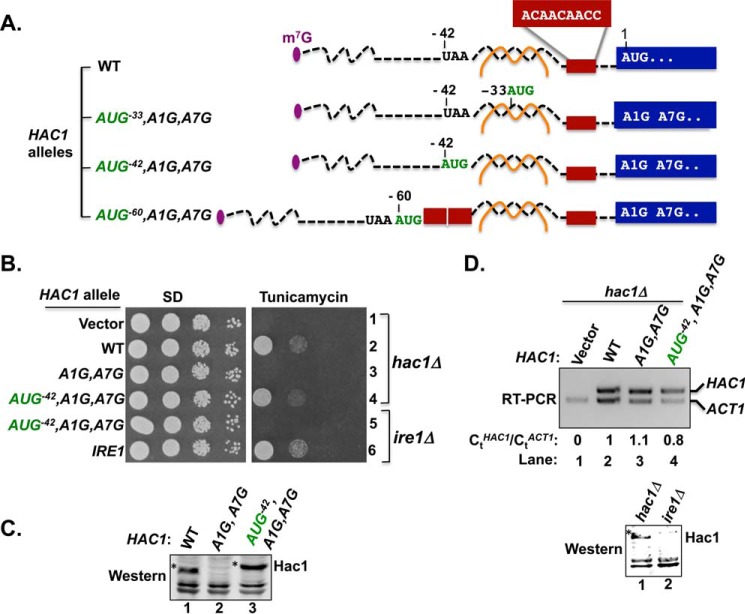
FIGURE 3.
Insertion of an AUG codon immediately upstream of the 5′ UTR-intron interaction in the HAC1 mRNA failed to derepress translation. A, schematic diagrams showing the 5′ UTRs of HAC1, HAC1-AUG−33,A1G, HAC1-AUG−42,A1G, and HAC1-AUG−60,A1G alleles. Color schemes are the same as in Fig. 1A. The brown box represents the nine-nucleotide sequence 5′-−15ACAACAACC−7-3′. The A1G mutation converts the HAC1 start codon to GUG and also includes a second mutation A7G that alters the Met3 codon. The nucleotides −33CCU−31 or −42UAA−40 were replaced by an AUG codon in the HAC1-AUG−30,A1G or HAC1-AUG−42 allele, respectively. In the HAC1-AUG−60 allele, a 21-nucleotide sequence consisting of an AUG codon plus two repeats of the 9 nucleotides represented by the red box was inserted upstream of position −39. B, yeast growth assay. Transformants of hac1Δ or ire1Δ yeast strains carrying an empty vector or expressing the indicated HAC1 or IRE1 allele were grown in SD medium to saturation, and 5 μl of serial dilutions (of A600 = 1.0, 0.1, 0.01, and 0.001) were spotted on SD medium or SD medium containing 0.4 μg/ml tunicamycin and incubated for 3 days at 30 °C. C, immunoblot analysis of Hac1 protein. Strains described in panel B (rows 2, 3, and 4) were grown in SD medium and treated with 5 mm DTT to induce ER stress, and WCEs were prepared and subjected to SDS-PAGE followed by Western analysis using a polyclonal antibody raised against recombinant Hac1 protein. D, RT-PCR and Western analysis of HAC1 expression. Upper panel, total RNA was extracted from the hac1Δ strains expressing the indicated HAC1 alleles after growth under non-stress conditions. The RNA was used as a template for RT-PCR and Q-PCR analysis of HAC1 and ACT1 mRNAs as described under “Experimental Procedures.” The ratio of threshold values (Ct) from Q-PCR analysis of HAC1 and ACT1 mRNAs are shown. Lower panel, hac1Δ and ire1Δ strains expressing HAC1-AUG−42,A1G were grown in SD medium and treated with 5 mm DTT to induce ER stress, and then WCEs were prepared and subjected to SDS-PAGE followed by Western analysis using polyclonal anti-Hac1 antibodies.
We first characterized the effect of replacing nucleotides −42UAA−40, immediately upstream of the 5′ UTR-intron interaction (see Figs. 1A and 3A), with an AUG codon to generate the HAC1-AUG−42,A1G,A7G allele. As shown in Fig. 3B, the HAC1-AUG−42,A1G,A7G allele conferred a TmR phenotype when expressed in a hac1Δ strain (row 4), suggesting that a functional Hac1 protein with an extra 14 amino acids at the N terminus was expressed from the AUG−42 codon. To directly examine Hac1 production, Western analyses were performed on WCEs prepared from ER-stressed cells using an antibody prepared against recombinant Hac1 protein. Consistent with the introduction of 14 extra N-terminal residues, the Hac1 protein in the HAC1-AUG−42,A1G,A7G cells was of higher molecular weight than the WT Hac1 protein (Fig. 3C, lanes 1 and 3). It is important to note that these experiments were performed using a hac1Δ strain in which the chromosomal IRE1 gene was intact. Thus, it is likely that the HAC1-AUG−42,A1G,A7G mRNA was spliced in these cells, and we conclude that a 14-residue N-terminal extension does not impair Hac1 production or the ability of Hac1 to promote yeast cell growth on medium containing tunicamycin. However, the intent of these experiments was to determine whether the introduced AUG−42 start codon could promote translation of the unspliced HAC1 mRNA. Therefore, we expressed the HAC1-AUG−42,A1G,A7G allele in an ire1Δ strain where the mRNA will not be spliced. As shown in Fig. 3B, the HAC1-AUG−42,A1G,A7G allele failed to complement the TmS phenotype of the ire1Δ strain (Fig. 3B, row 5), and Western analyses showed that the Hac1 protein was not produced (Fig. 3D, lower panel, lane 2). These results indicate that the unspliced HAC1-AUG−42,A1G,A7G mRNA is translationally repressed, and we propose that the adjacent secondary structure from the 5′ UTR-intron interaction may preclude scanning ribosomes from accessing the AUG−42 start codon or may impede formation of a productive 80S ribosome on the AUG−42 codon.
To further test the ability of inserted AUG start codons to overcome translational repression on the unspliced HAC1 mRNA, two additional mutants were constructed. First, the nucleotides −33CCU−31 in the HAC1-A1G,A7G allele were substituted by an AUG triplet to generate the HAC1-AUG−33,A1G,A7G allele (Figs. 1A and 3A). As this mutation is predicted to disrupt two base pair interactions between the 5′ UTR and the intron (Fig. 1A) and as we previously saw that disruption of a single base pair interaction was sufficient to derepress HAC1 mRNA translation (Figs. 1 and 2), the AUG−33 mutation was expected to likewise derepress HAC1 expression. Accordingly, as shown in Fig. 4A, row 3, and in contrast to WT HAC1 (row 1), the HAC1-AUG−33,A1G,A7G allele conferred a TmR phenotype in the ire1Δ hac1Δ strain, and Hac1 protein was detected in extracts from cells expressing the mutant but not the WT protein (Fig. 4B, lane 3 versus 1). These experiments with the HAC1-AUG−33,A1G,A7G mutant provide further support for the hypothesis that disruption of the secondary structure formed by the base-pairing interaction between the 5′ UTR and intron is sufficient to derepress translation of the unspliced HAC1 mRNA.
FIGURE 4.
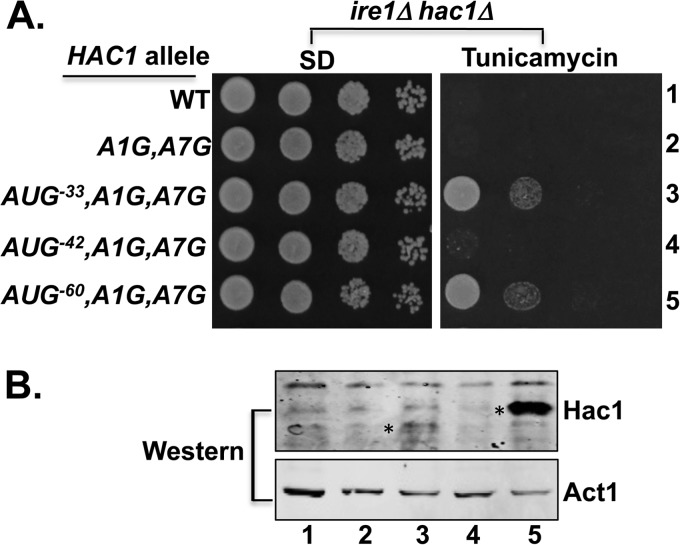
Insertion of an AUG codon at position −60 derepressed translation of unspliced HAC1 mRNA. A, yeast growth assays. Transformants of an ire1Δ hac1Δ strain carrying an empty vector or expressing the indicated HAC1 allele were grown in SD medium to saturation, and 5 μl of serial dilutions (of A600 = 1.0, 0.1, 0.01, and 0.001) were spotted on SD medium or SD medium containing 0.4 μg/ml tunicamycin and incubated for 3 days at 30 °C. B, immunoblot analysis of Hac1 protein. Strains from panel A were grown under non-stressed conditions in SD medium, and WCEs were prepared and subjected to SDS-PAGE followed by Western analysis using polyclonal antibodies against recombinant Hac1 or Act1 protein. Lanes are numbered according to row numbers in panel A, and an asterisk is positioned on the left side of the Hac1 protein in lanes 3 and 5.
Because the AUG−42 insertion may have failed to derepress HAC1 mRNA translation due to the close proximity of AUG−42 to the 5′ UTR-intron secondary structure, the third mutant was constructed by inserting the AUG codon further upstream from the secondary structure. To avoid inserting the AUG codon too close to the 5′ end of the mRNA, where it may be susceptible to being skipped by scanning ribosomes (30, 31), the HAC1-AUG−60,A1G,A7G allele was generated by inserting an AUG codon followed by 18 additional nucleotides at position −39 (Figs. 1A and 3A). The inserted nucleotides consist of two 9-nucleotide repeats of the sequence normally found between positions −15 and −7 (5′-−15ACAACAACC−7-3′) of the HAC1 mRNA. As this sequence is not part of the 5′ UTR-intron interaction and is rich in adenine and cytosine residues, it is predicted to be unstructured and thus serve as a simple spacer between the inserted AUG start codon and the base-paired element in the HAC1 5′ UTR. In this new HAC1-AUG−60,A1G,A7G allele, the inserted AUG−60 start codon is located ∼26 nucleotides from the 5′ cap (32) and ∼22 nucleotides upstream of the start of the base-paired elements.
Interestingly, when introduced into the ire1Δ hac1Δ strain, the HAC1-AUG−60,A1G,A7G allele conferred a TmR phenotype (Fig. 4A, row 5). Consistent with this growth phenotype, Western analysis of WCEs from the strain grown under non-ER stress conditions revealed a prominent Hac1 protein signal (Fig. 4B, lane 5). Importantly, the Hac1 protein in the HAC1-AUG−60,A1G,A7G strain migrated more slowly in SDS-PAGE than the Hac1 protein produced in the HAC1-AUG−33,A1G,A7G strain (Fig. 4B, lane 3), consistent with the longer N-terminal extension for the protein initiating at AUG−60. As the HAC1-AUG−60,A1G,A7G mRNA is unspliced in the ire1Δ hac1Δ strain, and thus the 5′ UTR-intron interaction is intact, the ability of AUG−60 codon insertion to derepress Hac1 production leads to two important conclusions. First, in contrast to the previously proposed model of a translation elongation block on the unspliced HAC1 mRNA (13), elongating ribosomes are able to translate through the 5′ UTR-intron interaction. Second, the data are consistent with a model in which the 5′ UTR-intron interaction impedes translation initiation by blocking ribosomes scanning to the native AUG start codon.
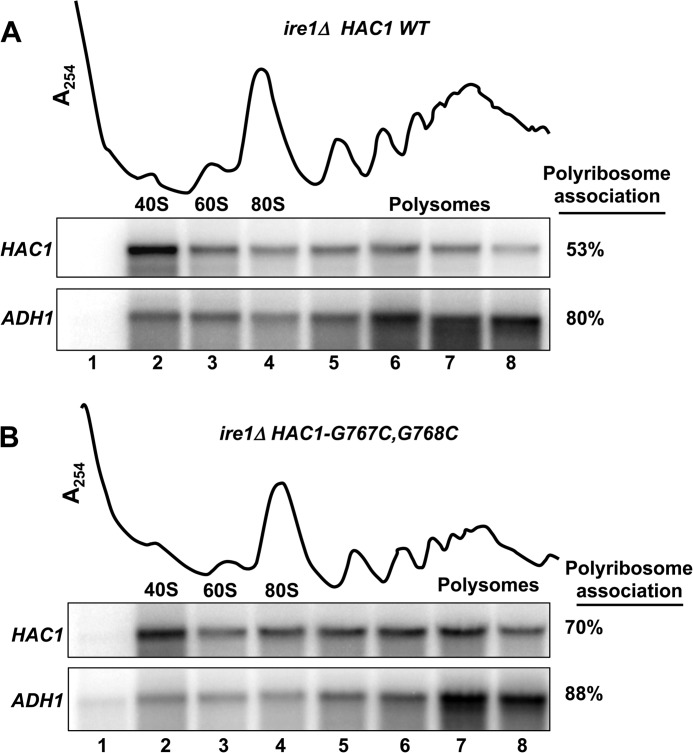
Disruption of the 5′ UTR-Intron Interaction Results in Greater Association of HAC1 mRNA with Polysomes
The genetic suppressor and AUG insertion mutant studies support the model that ribosomal scanning on the WT unspliced HAC1 mRNA is paused at the 5′ UTR-intron interaction site. Accordingly, the point mutations that weaken the 5′ UTR-intron interaction allow scanning ribosomes to traverse the 5′ UTR and initiate translation at the AUG start codon of HAC1. If this model is correct, few ribosomes should associate with the unspliced WT HAC1 mRNA, whereas the HAC1 mRNA containing the point mutations that disrupt the 5′ UTR-intron interaction should be readily translated and thus show a greater association with polysomes. To test this possibility, the WT chromosomal HAC1 allele (translationally repressed) in the ire1Δ strain J751 was replaced with the HAC1-G767C,G768C mutant (translationally active; see Fig. 2, B and C, lane 6) to create strain J1167. Next, WCEs from strains J751 and J1167 were subjected to velocity sedimentation in 7–47% sucrose gradients, and then the gradients were fractionated while being scanned at 254 nm (see “Experimental Procedures”) to visualize the free 40S and 60S ribosomal subunits, 80S monosomes, and polyribosomes (Fig. 5). To monitor the sedimentation of mRNAs in the gradients, RNA was isolated from the gradient fractions and analyzed by Northern blot using probes to detect the HAC1 mRNA and the ADH1 mRNA. As shown in Fig. 5A, the WT HAC1 mRNA in unstressed ire1Δ cells showed strong association with 40S subunits (lane 2) and only ∼53% of the HAC1 mRNA associated with polysomes. In contrast, the ADH1 mRNA was primarily (∼80%) associated with the polysome fractions. Disruption of the 5′ UTR-intron base-pairing interaction in the HAC1-G767C,G768C mutant resulted in more (∼70%) of the HAC1 mRNA associating with polysomes (Fig. 5B, lanes 5–8). These data provide independent support for the idea that the 5′ UTR-intron interaction blocks ribosomal scanning, resulting in a substantial fraction of the HAC1 mRNA associating with a single 40S subunit. Accordingly, disruption of the secondary structure releases the initiation block and enables more ribosomes to translate the HAC1 mRNA, resulting in more of the mRNA associating with polyribosomes.
FIGURE 5.
Disruption of the 5′ UTR-intron interaction increased the relative association of HAC1 mRNA with polysomes. WCEs from strains J751 (HAC1 ire1Δ, panel A) and J1167 (HAC1-G767C,G768C ire1Δ, panel B) were resolved by velocity sedimentation in 7–47% sucrose gradients. Gradients were fractionated while scanning at A254 (upper tracings), and the positions of the 40S and 60S subunits, 80S ribosomes, and polysomes are indicated. Total RNA was extracted from each fraction and subjected to Northern analysis using probes for HAC1 and ADH1 as indicated. The amounts of HAC1 mRNA in the gradient fractions were quantified, and the percentage of HAC1 mRNA associating with polysomes (fractions 4–8) was calculated.
Overexpression of eIF4A Restores the UPR and Derepresses Hac1 Production in Yeast Expressing the HAC1-AUG−42,A1G,A7G Variant
Whereas insertion of an AUG codon at position −60 far upstream of the secondary structure in the HAC1 mRNA led to constitutive derepression of Hac1 synthesis (Fig. 4), the insertion of an AUG codon at position −42 immediately before the secondary structure failed to derepress Hac1 synthesis in the absence of splicing (Fig. 3). As our data indicate that the secondary structure formed by the base-pairing interactions between the intron and 5′ UTR in the HAC1 mRNA interferes with translation initiation, we reasoned that scanning ribosomes lack sufficient secondary structure unwinding ability to access the AUG−42 start codon. However, because the AUG−42 codon is located just upstream of the secondary structure, the scanning ribosome may only need to partially melt the secondary structure to access the AUG−42 codon. Accordingly, we reasoned that overexpression of the translation factors that function in mRNA binding andscanning might derepress Hac1 production on the HAC1-AUG−42,A1G,A7G mRNA. Consistent with this hypothesis, overexpression of eIF4A, an RNA helicase that promotes melting of RNA secondary structures (1) conferred a tunicamycin-resistant phenotype in an ire1Δ hac1Δ strain expressing the HAC1-AUG−42,A1G,A7G allele (Fig. 6B, right panels, row 2). This derepression of HAC1 synthesis was dependent on eIF4A helicase activity as overexpression of the eIF4A-D170E mutant that lacks helicase activity (33, 34) failed to support growth on tunicamycin medium (Fig. 6B, right panels, row 5). Moreover, overexpression of eIF4E, the mRNA cap-binding protein (1), or eIF4B, a factor that promotes eIF4A activity in mRNA remodeling and scanning (1, 35, 36) (35, 36), failed to promote growth of the strain expressing the HAC1-AUG−42,A1G,A7G allele on tunicamycin medium (Fig. 6B, right panels, rows 2, 3, and 4). Consistent with these growth phenotypes, Western blot analyses demonstrated that overexpression of eIF4A, but not eIF4E, eIF4B, or non-functional eIF4A-D170E, promoted synthesis of Hac1 in from the HAC1-AUG−42,A1G,A7G mRNA (Fig. 6C). These data support the model that increasing the cellular amount of eIF4A enables the scanning ribosome to melt secondary structure on the HAC1-AUG−42,A1G,A7G mRNA and gain access to the AUG−42 start codon. In contrast to the ability of eIF4A overexpression to derepress translation of the HAC1-AUG−42,A1G,A7G mRNA, overexpression of eIF4A failed to confer a tunicamycin-resistant phenotype in cells expressing the wild-type HAC1 mRNA (Fig. 6B, left panel, row 2). Thus, we propose that overexpression of eIF4A can promote the modest level of secondary structure melting required for the scanning ribosome to access AUG−42 but is not sufficient for the ribosome to traverse through the entire base-paired region and access the wild-type HAC1 start codon.
FIGURE 6.
Overexpression of translation initiation factor eIF4A derepressed HAC1-AUG−42,A1G,A7G mRNA translation. A, schematic diagram showing the 5′ UTR of the HAC1-AUG−42,A1G,A7G allele. Color schemes are the same as in Fig. 1A. B, yeast growth assays. An ire1Δ hac1Δ strain was transformed with a low copy number URA3 plasmid expressing HAC1 (left panels) or the HAC1-AUG−42,A1G,A7G allele (right panels). The resulting strains were then transformed with high copy number (H.C.) LEU2 plasmids expressing eIF4A, eIF4B, eF4E, or eIF4A-D170E or with a low copy-number (L.C.) LEU2 plasmid encoding IRE1. C, immunoblot analysis of Hac1 protein. Strains from the right panels in B were grown under stressed conditions in SD medium, and WCEs were prepared and subjected to SDS-PAGE followed by Western analysis using a polyclonal antibody raised against recombinant Hac1 protein. Lanes are numbered according to row numbers in panel B.
Discussion
The Hac1 transcription factor is the primary regulator of the UPR (19, 20). Under conditions of ER stress, HAC1 expression is increased. The Hac1 protein then promotes the expression of a variety of proteins including ER chaperones that help alleviate the stress conditions. As production of Hac1 and stimulation of the UPR in the absence of ER stress would be wasteful, it is not surprising that cells have adopted a mechanism to limit Hac1 synthesis in unstressed cells. Intriguingly, in yeast the mechanism to limit HAC1 expression under non-stressed conditions is linked to the unique cytoplasmic splicing of the HAC1 mRNA (13, 15, 21). The primary HAC1 transcript containing an intron that disrupts the main ORF is exported to the cytoplasm with the intron intact (14). Translation of the unspliced HAC1 mRNA will terminate at a stop codon in the intron, whereas translation of the spliced HAC1 mRNA will generate a protein with an altered C terminus that terminates in the second exon. Previous studies (15, 27, 37) and the results in this report demonstrate that both forms of the Hac1 protein are functional and enable cells to grow under ER stress conditions. However, production of Hac1 protein from the unspliced mRNA is inhibited by base-pairing interactions between sequences in the intron and in the 5′ UTR of the HAC1 mRNA. Within a span of 19 nucleotides, 16 form base-pairing interactions between the 5′ UTR and intron including a stretch of 11 consecutive base pairs. Splicing of the HAC1 mRNA removes the intron and thus eliminates the base-pairing interactions and enables HAC1 mRNA translation. As previously demonstrated by Rüegsegger et al. (13), replacement of the 5′ UTR base-pairing element with the sequence from the intron or replacement of the intron element with the sequence from the 5′ UTR enabled constitutive translation of the HAC1 mRNA. Moreover, when the 5′ UTR and intron elements were swapped simultaneously to allow the base-pairing interaction to occur, but with opposite orientation compared with the original context, HAC1 mRNA translational repression was restored. Although these studies clearly demonstrated that the intron played an important role in limiting HAC1 mRNA translation, the molecular mechanism of the repression was not clarified.
To gain insights into the mechanism of the translational repression of the unspliced HAC1 mRNA, we employed two mutational strategies. First, the cleavage site between exon 1 and the intron was mutated to eliminate splicing of the HAC1 mRNA, and then a second-site suppressor mutation was identified that restored Hac1 synthesis. This suppressor mutation disrupted a single base pair in the 5′ UTR-intron interaction. Likewise, using a traditional site-directed mutagenesis approach, we showed that altering a single base pair interaction between the 5′ UTR and intron was sufficient to derepress HAC1 mRNA translation in cells lacking the Ire1 endonuclease and was thus unable to splice the HAC1 mRNA. Although Rüegsegger et al. (13) showed that eliminating all 16 base-pairing interactions between the 5′ UTR and intron eliminated the translational repression, our studies reveal that loss of a single base pair interaction within or at the end of the 11 consecutive base pairs is sufficient to allow translation of the unspliced HAC1 mRNA. Thus, it appears that the HAC1 mRNA structure is poised on the threshold of strength required for translational repression, perhaps indicating that further strengthening of this interaction with additional base pair interactions might be deleterious. Likewise, it seems possible that conditions or factors that promote melting of mRNA secondary structures might be able to derepress translation of the unspliced HAC1 mRNA.
Secondary structural elements in the 5′ UTR of mRNAs have previously been shown to inhibit translation (6, 7, 38). As these structured mRNAs failed to associate with ribosomes, it was concluded that the 5′ UTR structures blocked 40S ribosome loading and/or ribosomal scanning to the start codon on the mRNA. Accordingly, the 5′ UTR-intron interaction would be predicted to impair translation initiation on the HAC1 mRNA. At odds with this hypothesis, Rüegsegger et al. (13) reported that unspliced HAC1 mRNA co-sedimented with polysomes on sucrose gradients, leading to a proposal that the 5′ UTR-intron interaction impedes translation elongation. We sought to directly test whether the 5′ UTR-intron interaction could block an elongating ribosome. Whereas inserting an AUG start codon immediately upstream of the interaction site failed to derepress translation of the unspliced HAC1 mRNA (AUG−42 in Figs. 3 and 4), insertion of an AUG codon further upstream (AUG−60) enabled Hac1 synthesis in the absence of splicing (Fig. 4). In this latter construct, the inserted AUG−60 codon was positioned ∼26 residues from the cap and ∼22 residues upstream of the secondary structure. These gaps between the cap, AUG codon, and secondary structure provide sufficient room for a 43S PIC to bind at the cap, scan to AUG−60 start codon, and assemble a translationally competent 80S ribosome. Notably, toe-printing assays have shown that the leading edge of the ribosome is typically around 16 nucleotides downstream of the P site (39, 40), so the 22-nucleotide gap between AUG−60 and the 5′ UTR secondary structure provides the space needed for an 80S ribosome. The ability of ribosomes initiating at AUG−60 to elongate through the 5′ UTR-intron secondary structure is consistent with previous reports demonstrating that elongating ribosomes can melt secondary structures (7, 41). Moreover, these results demonstrate that the 5′ UTR-intron interaction does not block translation elongation, and we therefore conclude that translation of the unspliced HAC1 mRNA is blocked at translation initiation.
Polysome profiles provide an additional means to characterize translation defects. When translation initiation is impaired, fewer ribosomes associate with an mRNA, and the mRNA sediments as a free messenger ribonucleoprotein particle (mRNP) or perhaps associated with 40S subunits and smaller polysomes. On the other hand, if translation elongation is impaired, the mRNA will readily bind to ribosomes and sediment with large polysomes in sucrose gradient analyses. Rüegsegger et al. (13) found that the WT unspliced HAC1 mRNA co-sedimented with free 40S subunits as well as with polysomes, and mutation of the 5′ UTR-intron interaction resulted in loss of the association with 40S subunits and a shift of the majority of the mRNA to the polysome fractions. Likewise, Mori et al. (42) reported that the unspliced HAC1 mRNA in unstressed cells co-sedimented with ribosomal subunits, whereas the spliced HAC1 mRNA in tunicamycin-treated cells sedimented with large polysomes. In our studies we found that the association of the HAC1 mRNA with polysomes was influenced by the amount of HAC1 mRNA in the cell. Overexpression of HAC1 mRNA resulted in a relative shift in the sedimentation of the mRNA such that a greater fraction co-sedimented with polysomes.4 As this result did not correlate with increased synthesis of Hac1 protein, we reasoned that the overexpressed mRNA might not be translated but, rather, associated with different complexes that co-sedimented with polysomes. To avoid this possible complication, we generated isogenic strains expressing WT HAC1 or a translationally derepressed HAC1 intron mutant. Whereas the unspliced HAC1 mRNA was roughly equally distributed between a free state, possibly a messenger ribonucleoprotein or associated with 40S particles, and a form that co-sedimented with polysomes (Fig. 5A), disruption of the 5′ UTR-intron interaction resulted in a relative shift of the HAC1 mRNA to co-sediment with polysomes (Fig. 5B). The limited association of unspliced HAC1 mRNA with polysomes is consistent with the results of ribosomal profiling studies in which the HAC1 mRNA in unstressed cells was not found to be associated with many ribosomes (43, 44). Taken together, the results of the polysome profile analyses and site-directed mutagenesis studies are consistent with the notion that the 5′ UTR-intron interaction represses translation of the unspliced HAC1 mRNA by blocking translation initiation and that splicing relieves this block to promote HAC1 mRNA translation.
As 5′ UTR secondary structure can block scanning of the 43S PIC after binding to the mRNA (7, 45), we reasoned that overexpression of translation factors that promote scanning might stimulate HAC1 mRNA translation. Accordingly, we found that overexpression of the RNA helicase eIF4A could derepress HAC1-AUG−42,A1G,A7G mRNA translation in the absence of splicing (Fig. 6). As overexpression of eIF4A failed to confer a tunicamycin-resistant phenotype in cells expressing wild-type HAC1 mRNA (Fig. 6), we propose that overexpression of eIF4A modestly increases the ability of the scanning ribosome to melt secondary structures. This boost in melting power is not sufficient to enable a ribosome to scan through the entire base-paired region formed between the HAC1 intron and 5′ UTR but may help melt the beginning of the base-paired region to enable the scanning ribosome to access a start codon at position −42. The finding that overexpression of a translation initiation factor can promote Hac1 synthesis provides further support for the notion that the base-pairing between the intron and 5′ UTR of the HAC1 mRNA interferes with translation initiation. Moreover, these studies suggest that HAC1 translational control can be used as a tool to gain insights into translation initiation. Much like GCN4 translational control has provided novel insights into the factors that promote Met-tRNAiMet binding to the 40S ribosome (9), HAC1 translational control may provide new insights into the factors that assist ribosomes in melting and scanning through secondary structures in the 5′ UTR of an mRNA.
Author Contributions
L. S., C. B., M. A. M., and M. D. performed and analyzed the experiments. M. D. and T. D. wrote the paper.
Acknowledgment
We thank Dr. Colin G. Scanes (UW-Milwaukee) for reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants 1R15GM101575-01 (to M. D.) and by the Intramural Research Program of the NIH (NICHD; to T. E. D.). This work was also supported by Research Growth Initiative (UW-Milwaukee Graduate School; to M. D.). The authors declare that they have no conflicts of interest with the contents of this article.
C. Bolinger, unpublished data.
- PIC
- preinitiation complex
- ER
- endoplasmic reticulum
- WCE
- whole cell extract
- NAT
- nourseothricin-resistance marker genemarker gene
- Q-PCR
- quantitative PCR
- SD
- synthetic dextrose
- TmS
- tunicamycin-sensitive
- TmR
- tunicamycin-resistant
- UPR
- unfolded protein response.
References
- 1. Hinnebusch A. G. (2014) The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 [DOI] [PubMed] [Google Scholar]
- 2. Hinnebusch A. G., Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kozak M. (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37 [DOI] [PubMed] [Google Scholar]
- 4. Tarun S. Z. Jr., Sachs A. B. (1995) A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9, 2997–3007 [DOI] [PubMed] [Google Scholar]
- 5. Tarun S. Z. Jr., Sachs A. B. (1997) Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol. Cell. Biol. 17, 6876–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kozak M. (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 83, 2850–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kozak M. (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol. Cell. Biol. 9, 5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867–19870 [PubMed] [Google Scholar]
- 9. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 10. Mukhopadhyay R., Jia J., Arif A., Ray P. S., Fox P. L. (2009) The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gebauer F., Preiss T., Hentze M. W. (2012) From cis-regulatory elements to complex RNPs and back. Cold Spring Harb. Perspect. Biol. 4, a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mendez R., Richter J. D. (2001) Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529 [DOI] [PubMed] [Google Scholar]
- 13. Rüegsegger U., Leber J. H., Walter P. (2001) Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107, 103–114 [DOI] [PubMed] [Google Scholar]
- 14. Chapman R. E., Walter P. (1997) Translational attenuation mediated by an mRNA intron. Curr. Biol. 7, 850–859 [DOI] [PubMed] [Google Scholar]
- 15. Kawahara T., Yanagi H., Yura T., Mori K. (1997) Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8, 1845–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sidrauski C., Walter P. (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 17. Sidrauski C., Cox J. S., Walter P. (1996) tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405–413 [DOI] [PubMed] [Google Scholar]
- 18. Anshu A., Mannan M. A., Chakraborty A., Chakrabarti S., Dey M. (2015) A novel role for protein kinase Kin2 in regulating HAC1 mRNA translocation, splicing and translation. Mol. Cell. Biol. 35, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 20. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 21. Cox J. S., Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 22. Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. (1996) Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1, 803–817 [DOI] [PubMed] [Google Scholar]
- 23. Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 24. Patil C. K., Li H., Walter P. (2004) Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLos Biol. 2, E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mannan M. A., Shadrick W. R., Biener G., Shin B. S., Anshu A., Raicu V., Frick D. N., Dey M. (2013) An ire1-phk1 chimera reveals a dispensable role of autokinase activity in endoplasmic reticulum stress response. J. Mol. Biol. 425, 2083–2099 [DOI] [PubMed] [Google Scholar]
- 26. Cox J. S., Shamu C. E., Walter P. (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 27. Mori K., Ogawa N., Kawahara T., Yanagi H., Yura T. (2000) mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 97, 4660–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hooks K. B., Griffiths-Jones S. (2011) Conserved RNA structures in the non-canonical Hac1/Xbp1 intron. RNA Biol. 8, 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gonzalez T. N., Sidrauski C., Dörfler S., Walter P. (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 18, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kozak M. (1991) A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1, 111–115 [PMC free article] [PubMed] [Google Scholar]
- 31. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pause A., Sonenberg N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11, 2643–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmid S. R., Linder P. (1991) Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 11, 3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Özeş A. R., Feoktistova K., Avanzino B. C., Fraser C. S. (2011) Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 412, 674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou F., Walker S. E., Mitchell S. F., Lorsch J. R., Hinnebusch A. G. (2014) Identification and characterization of functionally critical, conserved motifs in the internal repeats and N-terminal domain of yeast translation initiation factor 4B (yeIF4B). J. Biol. Chem. 289, 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox J. S., Chapman R. E., Walter P. (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pelletier J., Sonenberg N. (1985) Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526 [DOI] [PubMed] [Google Scholar]
- 39. Pestova T. V., Borukhov S. I., Hellen C. U. (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394, 854–859 [DOI] [PubMed] [Google Scholar]
- 40. Anthony D. D., Merrick W. C. (1992) Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 267, 1554–1562 [PubMed] [Google Scholar]
- 41. Takyar S., Hickerson R. P., Noller H. F. (2005) mRNA helicase activity of the ribosome. Cell 120, 49–58 [DOI] [PubMed] [Google Scholar]
- 42. Mori T., Ogasawara C., Inada T., Englert M., Beier H., Takezawa M., Endo T., Yoshihisa T. (2010) Dual functions of yeast tRNA ligase in the unfolded protein response: unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol. Biol. Cell 21, 3722–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guydosh N. R., Green R. (2014) Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156, 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pestova T. V., Kolupaeva V. G. (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de la Cruz J., Iost I., Kressler D., Linder P. (1997) The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]