Background: Yin Yang 1 is a transcription factor capable of activating or repressing its target genes, depending on context and cofactors.
Results: YY1 is tyrosine-phosphorylated by Src family kinases, leading to down-regulation of its activity.
Conclusion: Tyrosine phosphorylation of YY1 affects its ability to regulate gene expression.
Significance: This is a novel post-translational modification of YY1 regulating its function.
Keywords: gene regulation, phosphotyrosine, protein phosphorylation, retrovirus, Src, transcription factor
Abstract
Yin Yang 1 (YY1) is a member of the GLI-Krüppel class of DNA and RNA binding transcription factors that can either activate or repress gene expression during cell growth, differentiation, and embryogenesis. Although much is known about YY1 interacting proteins and the target promoters regulated by YY1, much less is known about YY1 regulation through post-translational modifications. In this study we show that YY1 is tyrosine-phosphorylated in multiple cell types. Using a combination of pharmacological inhibition, kinase overexpression, and kinase knock-out studies, we demonstrate that YY1 is a target of multiple Src family kinases in vitro and in vivo. Moreover, we have identified multiple sites of YY1 phosphorylation and analyzed the effect of phosphorylation on the activity of YY1-responsive retroviral and cellular promoters. Phosphorylation of tyrosine 383 interferes with DNA and RNA binding, leading to the down-regulation of YY1 activity. Finally, we provide the first evidence that YY1 is a downstream target of epidermal growth factor receptor signaling in vivo. Taken together, the identification of YY1 as a target of Src family kinases provide key insights into the inhibitory role of tyrosine kinases in modulating YY1 activity.
Introduction
Yin Yang 1 (YY1)2 is a ubiquitously expressed DNA/RNA binding transcription factor that takes its name from its dual activity on the adeno-associated virus (AAV) P5 promoter, where the presence of adenoviral protein E1A converts YY1 from a transcriptional repressor to an activator (1). In mice, targeted deletion of YY1 results in peri-implantation lethality, highlighting its essential role in embryogenesis (2). YY1 binds to DNA through the recognition of a specific consensus sequence, leading to the activation or repression of many cellular genes including c-Myc, c-Fos, β-actin, IFN-γ, CREB, Sp1, and others (3, 4). Moreover, YY1 has been shown to physically interact with numerous proteins required for cell growth, apoptosis, and cancer progression, including p53, Mdm2, Ezh2, and Rb (3, 4). In Xenopus oocytes, YY1 has been demonstrated to associate with structurally divergent RNA species (5), raising the possibility that YY1 can bind both DNA and RNA. Highlighting this dual binding property, YY1 was shown to tether Xist, a noncoding RNA (lncRNA), to DNA regions on the X chromosome, leading to X chromosome inactivation in maternal cells (6). Therefore, it appears that the zinc finger domain of YY1 can simultaneously bind different nucleic acid motifs in Xist RNA and DNA (6).
Apart from its function on cellular promoters, YY1 plays a critical role in the transcriptional regulation of retroviral promoters. For example, YY1 binds to and represses the HIV-1 promoter, thereby maintaining the virus in a latent state (7, 8). In the case of Moloney murine leukemia virus (MLV), YY1 directly binds to the negative control region (NCR) in the viral long terminal repeat (LTR) (9), leading to transcriptional silencing of MLV in mouse embryonic cells (10). Consistent with its dual role in transcriptional activation, YY1 potentiates expression from the human T-lymphotropic virus 1 (HTLV-1) promoter, likely through association with the HTLV-1 RNA.3
The sequence-specific DNA binding activity of YY1 is mediated by four C2H2-type zinc finger motifs (amino acids 298–414) located in the C terminus (11). Besides their role in DNA binding, portions of the zinc finger motifs (amino acid 333–397) together with a region rich in alanine and glycine (amino acid 157–201) contribute to transcriptional repression (12, 13). The N-terminal region contains several unusual features also required for transcriptional activation. This includes two stretches of acidic residues (amino acids 16–29 and amino acids 43–53) followed by a glycine-rich region (amino acids 54–69), 11 consecutive histidine residues (amino acids 70–80), and sequences rich in proline and glutamine (amino acids 80–100) (12, 14–16).
Despite the wealth of knowledge garnered about YY1 interacting proteins and target promoters regulated by YY1, much less is known about YY1 regulation. Based on previous studies, YY1 appears to be a target of several posttranslational modifications, some of which contribute to changes in YY1 activity. Sumoylation of YY1 by PIASy on lysine 288 negatively affects the transcriptional activity of YY1 (17), whereas O-linked glycosylation of YY1 disrupts its interaction with Rb, leading to enhanced DNA binding (18). Moreover, YY1 also undergoes a complex acetylation/deacetylation cycle that changes its affinity for DNA and capacity to interact with histone deacetylases (19). In prostate cancer cells, nitric oxide (NO) has been shown to facilitate S-nitrosylation of YY1 on cysteine residues, resulting in decreased DNA binding and up-regulation of Fas expression (20). Serine/threonine phosphorylation of YY1 occurs during the cell cycle. Specifically, threonine 39 is a target of Polo-like kinase 1 (Plk1), but the functional importance of this modification remains to be determined (21). Phosphorylation of serine 180 and 184 by Aurora B kinase has been shown to have effects on DNA binding and YY1's ability to be acetylated by histone acetyltransferase p300 (22). Finally, phosphorylation of threonine 348 and 378 in the DNA binding domain of YY1 by an unknown kinase leads to decreased DNA binding (23).
In this study we provide evidence that YY1 is a target of tyrosine phosphorylation in various cell types by members of the Src family kinases. Using mutagenesis and biochemical analysis, we were able to identify sites of YY1 phosphorylation and characterize the role of each site in regulating YY1 activity. We show that phosphorylation of YY1 in its DNA binding domain interferes with its ability to bind DNA or RNA, thereby down-regulating its ability to repress or activate gene expression from known retroviral and cellular promoters. Lastly, we demonstrate a role for epidermal growth factor receptor (EGFR) signaling in mediating YY1 phosphorylation. Taken together, our findings provide insights into the control of YY1-mediated transcription by cellular tyrosine kinases.
Experimental Procedures
Cell Lines, Transfections, and Antibodies
Cell lines including human embryonic kidney (HEK)-293 (CRL-1573), HeLa (CCL-2), Jurkat (TIB-152), Rat2 (CRL-1764), F9 embryonic carcinoma (CRL-1720), L-929 (CCL-1), and TE671 (CRL-8805) cells were purchased from American Type Culture Collection. Ba/F3 cells were a gift from Dr. Alan Tall (Columbia University). All cells except Jurkat and Ba/F3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mm glutamine, 1000 units/ml penicillin, and 100 mg/ml streptomycin. Jurkat cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 2 mm l-glutamine, and antibiotics. Ba/F3 cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS and 1.6 ng/ml recombinant IL-3. All cells were maintained in a 37 °C incubator with 5% CO2. Transient transfection studies in HEK-293T cells were carried out using polyethyleneimine, and Jurkat cell transfections were carried out using Lipofectamine LTX (Life Technologies) according to the manufacturer's instructions. Mouse monoclonal antibodies used in this study include: anti-FLAG M2 (Sigma), anti-YY1 (clone H-10, Santa Cruz Biotechnology), anti-HA (clone 16B12, Covance), anti-GAPDH (clone 6C5, EMD Millipore), and anti-phosphotyrosine (clone PY99, Santa Cruz Biotechnology). Rabbit polyclonal antibodies used in this study include: anti-YY1 (C-20, Santa Cruz Biotechnology), anti-Lamin B1 (H-90, Santa Cruz Biotechnology), and anti-phospho-ERK1/2 (#9101, Cell Signaling Technology).
Plasmids, Cloning, and Mutagenesis
HA-tagged wild-type YY1 was a generous gift from Dr. Yang Shi (Harvard Medical School). To generate FLAG-tagged YY1 or various tyrosine kinase constructs, respective cDNAs were PCR-amplified and cloned into BamHI/XhoI sites of pCDNA3.1 3×-FLAG vector (Invitrogen). pEGFP-N1-human Lyn was a gift from Dr. Anna Huttenlocher (Addgene plasmid #35958) (24). Generation of YY1 phosphorylation mutants was accomplished using GeneArt® site-directed mutagenesis kit (Invitrogen) according to the manufacturer's instructions. All mutations were verified by DNA sequencing. To generate HTLV-1 LTR luciferase reporter constructs, full-length HTLV-1 LTR was PCR-amplified and cloned into HindIII sites of pGL3-Basic vector (Promega) via Sequence and Ligation Independent Cloning (SLIC) as described (25). shRNA containing pLKO.1 lentiviral vectors were purchased from Thermo Scientific (Pittsburg, PA). The sequence of the YY1 shRNA hairpin is AACTTCTTATTACAACCGTCG (mature antisense).
Drug Treatments, Immunoprecipitation, and Immunoblotting
For studies involving pervanadate treatment, cells were treated with 0.2 mm Na3VO4 for 30 min at 37 °C. For experiments involving Src family kinase or BCR-Abl inhibition, cells were pretreated with 10 μm SU6656 (EMD Millipore) or 10 μm imatinib (Sigma) for 1 h, respectively, before pervanadate addition. For EGF stimulation experiments, HeLa cells were serum-starved for 16 h before stimulation with 100 ng/ml EGF (Invitrogen) for the indicated time points. For immunoprecipitation, cells were washed twice with ice-cold 1× Tris-buffered saline (TBS) and lysed for 30 min on ice in high salt IPH buffer (50 mm Tris-HCl, pH 8, 400 mm NaCl, 0.5 mm EDTA, 5 mm MgCl2, 1 mm CaCl2, 1 mm Na3VO4, 0.5% Nonidet P-40) supplemented with protease and phosphatase inhibitors (Roche Applied Science). After centrifugation at 16,100 × g for 20 min, 5 μg of specific antibodies were added to the clarified lysate and allowed to mix end-over-end at 4 °C overnight. Next day, 25 μl of protein A or G Dynabeads (Invitrogen) were added for 2 h. Captured antibody-antigen complexes were first washed 3 times in high salt IPH buffer followed by 2 washes in low salt IPH buffer containing 150 mm NaCl. Bound proteins were resolved on 10% SDS-PAGE gel, transferred to nitrocellulose, blocked for 1 h at room temperature, and incubated overnight with the respective primary antibodies. The next day blots were washed with 1× TBS, incubated with HRP conjugated secondary antibodies (Amersham Biosciences), and developed using ECL (Amersham Biosciences).
Nuclear/Cytoplasmic Fractionation
Jurkat cells were treated with 0.2 mm Na3VO4 for 30 min at 37 °C. Cells were then subjected to nuclear/cytoplasmic fractionation using the EpiSeeker Nuclear Extraction kit (Abcam) according to the manufacturer's instructions.
Recombinant YY1 Purification
A plasmid expressing His6-YY1 was generated by inserting human YY1 cDNA (Dr. Yang Shi, Harvard Medical School) into the pQE-80L vector. Purification of recombinant YY1 from BL21-DE3 cells was carried out as described (1).
In Vitro Kinase Assay
In vitro phosphorylation of YY1 using purified Lyn was performed using 50 ng of bacterially expressed, purified YY1 together with 10 ng of recombinant Lyn (SignalChem) in 25 μl of kinase assay buffer (SignalChem) for 30 min at 30 °C. The reaction was terminated by the addition of Laemmli buffer, and products were subjected to SDS-PAGE and immunoblotting.
RNA Isolation and Real-time Quantitative RT-PCR
Total RNA was prepared with the RNeasy Mini kit (Qiagen), treated with DNase I (Ambion), and subjected to cDNA synthesis using SuperScript® III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative RT-PCR was performed using SYBR Green Master Mix (Roche Applied Science) using the following PCR conditions: 10 min at 95 °C followed by 40 cycles of 30s at 95 °C and 1 min at 60 °C. For all reactions relative expression was quantified using the threshold cycle (2−ΔΔCT) method (26) normalized to β-actin gene. The following primer sets were used: β-actin (forward, CATGTACGTTGCTATCCAGGC; reverse, CTCCTTAATGTCACGCACGAT), c-Fos (forward, CCGGGGATAGCCTCTCTTACT; reverse, CCAGGTCCGTGCAGAAGTC).
Electrophoretic Mobility Shift Assay (EMSA)
5′-Biotin-labeled double-stranded DNA probes were incubated with 500 ng of purified YY1 protein in EMSA binding buffer (10 mm Tris, pH 7.5, 50 mm KCl, 1 mm DTT, 2.5% glycerol, 5 mm MgCl2, 50 ng/μl Poly (dI·dC), 0.05% Nonidet P-40) at room temperature for 30 min. DNA-protein complexes were resolved on 6% DNA retardation gels (Novex) and developed using the LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer's instructions.
In Vitro RNA-Protein Pulldown Assay
The HTLV-1 R region or control RNA was transcribed in vitro from linearized plasmid DNA using T7 polymerase in the presence of Biotin-16-UTP (Roche Applied Science). Synthesized transcripts were treated with DNase for 1 h at 37 °C, purified with RNeasy kit (Qiagen), and renatured by heating and slow cooling. 2 μg of renatured RNA immobilized on Dynabeads MyOne Streptavidin (Invitrogen) were incubated with 1 μg of His6-YY1 for 2 h at room temperature in high salt phosphate buffered saline (PBS) containing 500 mm NaCl, 2 mm MgCl2, 0.2 mm ZnCl2, 15 mm β-mercaptoethanol, 100 units/ml RNase inhibitor, 0.1 mg/ml yeast tRNA (Ambion), 0.05% BSA, and 0.2% Nonidet P-40. Beads were then washed four times using the same binding buffer and twice with regular PBS and subjected to SDS-PAGE and immunoblotting with anti-His6 antibodies.
Generation of Stable YY1 shRNA Knockdown Cell Lines
RNAi knockdown in F9 EC cells was performed using shRNA containing pLKO.1 lentiviral vectors purchased from Thermo Scientific (Pittsburg, PA). Briefly, vesicular stomatitis virus glycoprotein-pseudotyped shRNA lentiviral particles were generated by cotransfection of HEK-293T cells using polyethyleneimine with pLKO.1-shRNA, p8.91 (encoding HIV gag-pol), and pMD.G (encoding VSV-G) plasmid DNAs using a 2:1:1 ratio of plasmids. Viral preparations were harvested 48 h post-transfection, filtered through a 0.45-μm filter (Pall Acrodisc), and used to infect F9 cells for 6 h. Infected F9 cells were then allowed to recover in fresh media for 36 h, after which puromycin (2 μg/ml) was added to select for stable knockdown clones. For YY1 rescue experiments, puromycin-resistant cells were reinfected with lentiviral particles harboring shRNA-resistant YY1-rescuing constructs (derivatives of pLVX-IRES-Neo, Clontech) and subjected to selection with G418 (0.5 mg/ml) for 2 weeks. Surviving cells were harvested, and YY1 levels were quantified by immunoblotting.
Retroviral Transduction Assays
Generation of Moloney murine leukemia virus-based vector genomes encoding Zeocin resistance and retroviral transduction assays were performed as described previously (27).
Dual Luciferase Assays
HEK-293A cells were seeded at 150,000 cells per well in a 24-well plate the day before transfection. The next day cells in each well were transfected with 400 ng of empty vector or various YY1 expression constructs, 100 ng of HTLV-1 LTR firefly luciferase construct, and 10 ng of HSV-TK Renilla luciferase control plasmid using Lipofectamine LTX (Life Sciences) at a 1:4 ratio. Twenty-four hours later cells were lysed in 100 μl of 1× Reporter Lysis Buffer (Promega) followed by quantification of luciferase activity using POLARstar Omega multimode plate reader (BMG Labtech) according to the manufacturer's instructions. Relative luciferase expression was calculated by first dividing the firefly luciferase signal by the renilla luciferase signal in a given cell, and the resulting ratio was then normalized to the ratio obtained from wild-type YY1-transfected cells (set to 1).
Results
YY1 Is Tyrosine-phosphorylated in Multiple Cell Types
To address whether YY1 is a target of tyrosine phosphorylation in vivo, Jurkat cells were treated with pervanadate to inhibit endogenous tyrosine phosphatases, and YY1 phosphorylation was assayed by immunoprecipitation and Western blotting using an anti-phosphotyrosine antibody. Tyrosine phosphorylation of YY1 was at very low levels in untreated cells but became readily detectable upon pervanadate treatment (Fig. 1A, left panel), whereas total YY1 levels remained unaffected (right panel). We next examined YY1 phosphorylation in a panel of cell lines. Consistently, similar increases in YY1 phosphorylation were observed upon pervanadate treatment (Fig. 1B). YY1 is apparently a target of tyrosine phosphorylation across multiple cell types.
FIGURE 1.
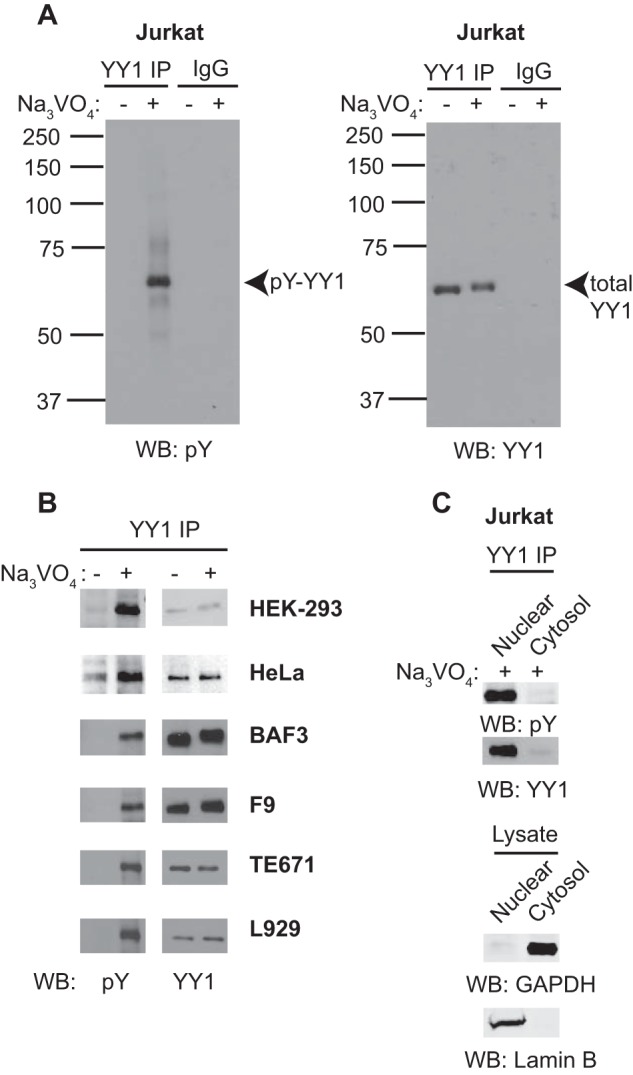
YY1 is tyrosine-phosphorylated in multiple cell types. A, cell lysates prepared from Jurkat cells treated with or without 200 μm pervanadate (Na3VO4) for 30 min to preserve tyrosine phosphorylation on proteins were subjected to immunoprecipitation (IP) with an anti-YY1 antibody or nonspecific IgG. Bound proteins were subjected to 10% SDS-PAGE and Western blotting (WB) with anti-phosphotyrosine (pY) or anti-YY1 antibodies. Data shown are representative of five independent experiments. B, cell lysates prepared from various cell lines treated with or without 200 μm pervanadate (Na3VO4) for 30 min were subjected to immunoprecipitation and Western blotting as indicated. Data shown are representative of two independent experiments. C, nuclear or cytoplasmic extract prepared from Jurkat cells treated with 200 μm pervanadate (Na3VO4) for 30 min were subjected to immunoprecipitation and Western blotting as indicated.
To address the subcellular localization of phosphorylated YY1, pervanadate-treated Jurkat cells were subjected to nuclear/cytoplasmic fractionation followed by YY1 immunoprecipitation and phosphotyrosine immunoblotting. The majority of total and phosphorylated YY1 appears in the nuclear fraction (Fig. 1C, top panels). The purity of our nuclear and cytoplasmic extracts was confirmed using anti-Lamin B and GAPDH antibodies, respectively.
YY1 Phosphorylation Is Mediated by Src Family Kinases
Src family kinases form the largest subfamily of nonreceptor protein tyrosine kinases, with nine members found in most mammals. Certain family members such as c-Src, Fyn, and Yes are widely expressed in various cell types, whereas Hck, Lyn, and Lck are confined to cells of hematopoietic lineages (28). Based on our observation that YY1 phosphorylation can be induced in many cell types (Fig. 1B), we hypothesized that Src family kinases may play a role in this process. To test this we examined the effect of Src family kinase and c-Abl inhibition on pervanadate-induced YY1 phosphorylation. Jurkat cells were pretreated with SU6656, a small-molecule inhibitor of the Src family kinases, or imatinib, an inhibitor of c-Abl, before pervanadate addition, followed by YY1 immunoprecipitation and phosphotyrosine immunoblotting. In contrast to vehicle-treated control cells, SU6656 treatment completely abolished pervanadate-induced YY1 phosphorylation (Fig. 2A), whereas imatinib resulted in a partial reduction. These observations suggest that YY1 may be a substrate of Src family kinases and/or c-Abl.
FIGURE 2.
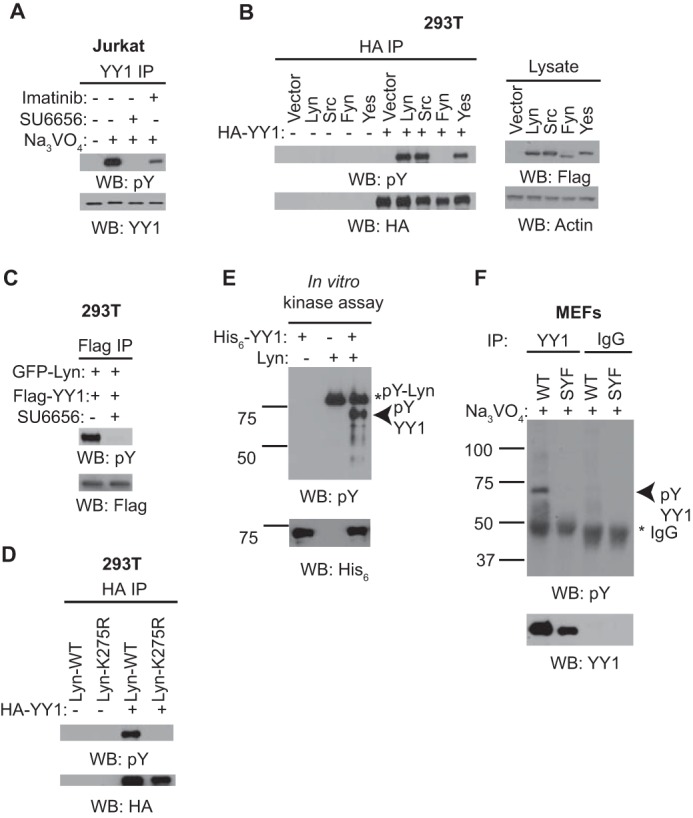
YY1 phosphorylation is mediated by Src family kinases. A, cell lysates prepared from Jurkat cells pretreated with 10 μm SU6656 or 10 μm imatinib for 1 h before pervanadate (Na3VO4) addition were subjected to immunoprecipitation (IP) and Western blotting (WB) as indicated. Data shown are representative of three independent experiments. B, cell lysates prepared from HEK-293T cells transfected with FLAG-tagged tyrosine kinase constructs together with or without HA-tagged YY1 for 48 h were subjected to immunoprecipitation with anti-HA antibody and Western blotting as indicated. The lysate lanes represent 2% of the material used for each immunoprecipitation reaction. Data shown are representative of three independent experiments. C, HEK-293T cells transfected with GFP-tagged Lyn together with FLAG-tagged YY1 for 48 h were subjected to 10 μm SU6656 treatment before cell lysis followed by immunoprecipitation with anti-FLAG antibodies and Western blotting as indicated. D, similar experiment as described in Fig. 2B, in which a kinase inactive Lyn (K275R) was included. Data shown are representative of three independent experiments. Data shown are representative of three independent experiments. E, in vitro kinase assay using 50 ng of purified His6-YY1 together with 10 ng of GST-Lyn kinase were subjected to 10% SDS-PAGE and Western blotting as indicated. Data shown are representative of four independent experiments. F, a similar experiment as described in Fig. 1A using wild-type (WT) or Src/Yes/Fyn (SYF) knock-out mouse embryonic fibroblasts (MEFs). Data shown are representative of two independent experiments.
Given that SU6656 targets multiple members of the Src family kinase, notably Src, Yes, Lyn, and Fyn (29), we reasoned that overexpression of each individual kinase may induce YY1 phosphorylation detectable even in the absence of pervanadate. To test this, plasmids expressing FLAG-tagged Src, Yes, Lyn, and Fyn were co-transfected with a construct expressing HA-tagged YY1 into HEK-293T cells, and the levels of phosphorylated YY1 were scored by immunoprecipitation and phosphotyrosine immunoblotting. We detected robust phosphorylation of YY1 upon overexpression of Src, Lyn, and Yes, but not Fyn, indicating the involvement of some, but not all Src family kinases (Fig. 2B). In addition, using Lyn as a representative member of the Src family kinases, we observed loss of YY1 phosphorylation upon the treatment of wild-type Lyn-overexpressing cells with SU6656 (Fig. 2C) or in cells overexpressing a kinase-dead Lyn mutant (K275R) (Fig. 2D), suggesting the requirement for intact kinase activity. To determine if Lyn can directly phosphorylate YY1 in vitro, purified GST-tagged Lyn was incubated with bacterially expressed His6-YY1 in the presence of ATP, and the extent of YY1 phosphorylation was monitored by phosphotyrosine immunoblotting. Purified Lyn readily phosphorylated recombinant YY1 in vitro (Fig. 2E).
Finally, to further confirm the involvement of Src family kinases in YY1 phosphorylation, we examined the extent of pervanadate-induced YY1 phosphorylation in wild-type versus mouse fibroblasts deficient in Src, Yes, and Fyn (30) (Fig. 2F). These cells, being fibroblasts, also lack Lyn kinase (30). Consistently, although YY1 phosphorylation was detectable in wild-type mouse fibroblasts, pervanadate treatment failed to induce YY1 phosphorylation in Src, Yes, and Fyn knockout cells. Collectively, these results suggest that multiple Src family kinases may be responsible for YY1 phosphorylation in vivo. The contribution of individual family members to YY1 phosphorylation would presumably depend on the Src family kinase signature and expression levels within each cell type.
Characterizing the Sites of YY1 Phosphorylation
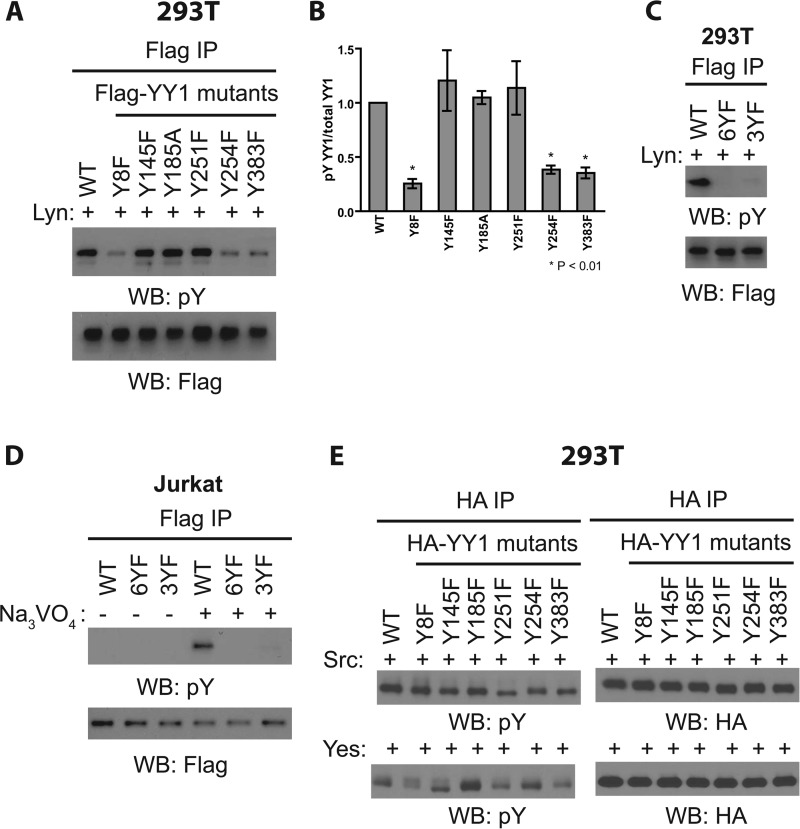
YY1 protein contains a total of six tyrosine residues. To investigate the sites of Src family kinase-induced YY1 phosphorylation in vivo, we introduced mutations altering each of the six tyrosine to nonphosphorylatable phenylalanine residues in the protein (Tyr to Phe) and asked whether single mutations affected the phosphorylation of YY1 in Src family kinase-overexpressing cells. In the case of Lyn overexpression, single mutations at either tyrosine 8, 254, or 383 severely reduced Lyn-mediated YY1 phosphorylation, suggesting that these three sites may be targets of Lyn in vivo (Fig. 3, A and B). Mutations of Tyr-145, Tyr-185, and Tyr-251 had no effect on overall phosphorylation levels (Fig. 3, A and B). The phosphorylation of the three critical sites may occur cooperatively, as the single mutations at each site greatly reduced the overall YY1 phosphorylation (Fig. 3, A and B). Lyn-induced phosphorylation of YY1 was not observed in the Y8F/Y254F/Y383F triple mutant, as was the case for a mutant in which all six tyrosines were mutated (6YF) (Fig. 3C). Similarly, the triple YF mutant expressed in Jurkat cells, where Lyn is expressed at high levels, failed to undergo robust phosphorylation upon pervanadate treatment (Fig. 3D).
FIGURE 3.
Characterizing the sites of YY1 phosphorylation. A, cell lysates prepared from HEK-293T cells transfected with GFP-Lyn together with WT or nonphosphorylatable FLAG-YY1 mutants for 48 h were subjected to immunoprecipitation (IP) with anti-FLAG antibody and Western blotting (WB) as indicated. Data shown are representative of three independent experiments. B, densitometry measurements of the levels of tyrosine-phosphorylated YY1 (pY) normalized to total YY1 (pY YY1/total YY1, y axis) as shown in Fig. A. Results shown are the means ± S.E. from three independent experiments. Student's t test was used for statistical analysis. * denotes p < 0.01 with respect to wild-type YY1. C, similar experiment as A using Y8F, Y254F, Y383F (3YF) triple nonphosphorylatable YY1 mutant and YY1 mutant in which all six tyrosines were mutated to phenylalanine (6YF). Data shown are representative of three independent experiments. D, Jurkat cells were transfected with wild-type or nonphosphorylatable FLAG-YY1 mutants for 48 h before pervanadate treatment followed by cell lysis, immunoprecipitation with anti-FLAG antibody, and Western blotting as indicated. Data shown are representative of three independent experiments. E, cell lysates prepared from HEK-293T cells transfected with FLAG-Src or FLAG-Yes together with WT or nonphosphorylatable HA-YY1 mutants for 48 h were subjected to immunoprecipitation with anti-HA antibody and Western blotting as indicated. Data shown are representative of two independent experiments.
The profile of the response of the YY1 phosphorylation pattern to mutation was different in cells overexpressing various members of the Src family kinases. For example, Y251F, Y254F, and Y383F mutations resulted in only a modest reduction in YY1 phosphorylation in Src-overexpressing cells (Fig. 3E, upper panels). In contrast, the overall phosphorylation of a different set of mutants, Y8F, Y251F, and Y383F, was reduced upon Yes overexpression (Fig. 3E, lower panels). In all cases the total levels of YY1 protein were unaffected by the mutations (Fig. 3, A and E). Collectively, these results suggest that different Src family kinases have different requirements for individual residues for the overall YY1 phosphorylation.
Functional Analysis of YY1 Phosphorylation Mutants Using Retroviral and Cellular Readouts
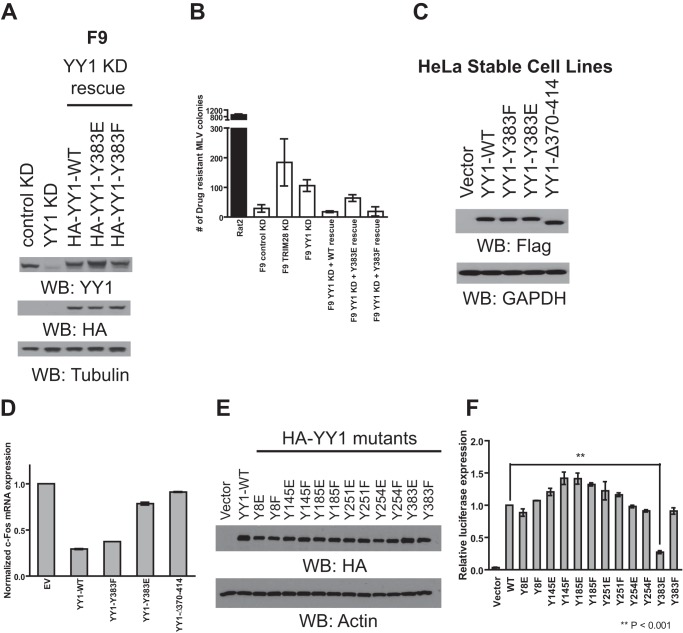
Previously it was shown that phosphorylation of threonine 348 and 378 in the DNA binding domain of YY1 resulted in decreased DNA binding (23). Given that tyrosine 383 also resides in the DNA binding domain and is a target of Lyn, Src, and Yes induced phosphorylation in vivo (Fig. 3, A and E), we first tested the effect of phosphomimetic (Y383E) and nonphosphorylatable (Y383F) YY1 mutant overexpression on the regulation of YY1-responsive retroviral promoters. YY1 is known to function as a transcriptional repressor of the Moloney MLV promoter in F9 mouse embryonic carcinoma cells (10). Using this cell line as a readout of YY1 activity, we infected F9 YY1 shRNA knockdown cells (Fig. 4A) with MLV reporter viruses containing a Zeocin resistance gene under the control of the MLV promoter. Infected cells were selected with Zeocin for 2 weeks, after which the number of surviving colonies was determined. Compared with control knockdown cells, YY1 knockdown caused a 3-fold reduction in MLV repression, as indicated by the increased number of drug-resistant colonies (Fig. 4B). As a positive control, a 5-fold reduction in MLV repression was observed in TRIM28 knockdown cells as reported previously (31). Importantly, YY1-mediated MLV repression was efficiently restored upon the introduction of shRNA resistant wild-type and Y383F mutant YY1 but not Y383E YY1 (Fig. 4B). This suggests that phosphorylation of tyrosine 383 and the phosphomimetic Y383E mutation interfere with YY1 activity.
FIGURE 4.
Functional analysis of YY1 phosphorylation mutants using retroviral and cellular readouts. A, lysate prepared from F9 cells stably transduced with either control shRNA or YY1-specific shRNA were subjected to Western blotting (WB) as indicated (left two). To rescue YY1 expression, YY1-shRNA knockdown cells were stably transduced again with either HA wild type, Y383E, or Y383F YY1, and lysates from these cells were subjected to Western blotting as indicated (third to fifth lanes). B, Rat2 and various F9 knockdown (KD) cell lines were infected with VSV-G-pseudotyped MLV particles carrying the Zeocin resistance gene under the control of the MLV promoter. Reporter gene expression in each cell line was assessed by colony counting after Zeocin selection. The graph shows the number of Zeocin-resistant colonies after drug selection. Results shown are the means ± S.D. from two independent experiments performed in triplicate. C, lysate prepared from HeLa cells stably transduced with retroviral expression vectors containing wild-type or mutant YY1 were subjected to Western blotting as indicated. D, total RNA prepared from HeLa cells outlined in Fig. 4C was subjected to quantitative RT-PCR using the indicated primers. Levels of c-fos transcripts were first normalized using the 2−ΔΔCT method to the value obtained for the β-actin gene; the obtained values were then normalized to the values obtained from HeLa cells stably transduced with vector control (set to 1). Results shown are means ± S.D. from two independent experiments performed in duplicate. E, cell lysates prepared from HEK-293A cells transfected with HA-tagged wild type or mutant YY1 were subjected to Western blotting as indicated. F, HEK-293A cells were transfected with a combination of three plasmids, empty vector or various YY1 mutants, HTLV-1 LTR firefly luciferase, and HSV-TK renilla luciferase control plasmid. Twenty-four hours later the activity of the HTLV-1 promoter was determined by dual luciferase assay. Relative luciferase expression was calculated by first dividing the firefly luciferase signal by the renilla luciferase signal in a given cell, and the resulting ratio was then normalized to the ratio obtained from wild-type YY1-transfected cells (set to 1). Results shown are the means ± S.E. from four independent experiments performed in duplicates. Student's t test was used for statistical analysis. ** denotes p < 0.001.
We next analyzed the effect of Tyr-383 mutant overexpression on previously documented YY1-responsive cellular promoters. HeLa cells stably expressing FLAG-tagged wild-type, Y383E, or Y383F YY1 were generated (Fig. 4C) followed by RNA isolation and RT-PCR analysis of a YY1-responsive mRNA c-fos (32, 33) (Fig. 4D). In parallel, HeLa cells expressing empty vector or YY1Δ370–414, a mutant defective in DNA binding (16), were used as controls. Compared with empty vector control, both wild-type and Y383F YY1 overexpression resulted in a 4–5-fold decrease in c-fos mRNA levels, consistent with previous reports (32, 33) (Fig. 4D). Little to no transcriptional repression was seen with the YY1Δ370–414 or Y383E mutant (Fig. 4D), further suggesting that phosphorylation of tyrosine 383 may interfere with YY1's ability to regulate certain cellular promoters.
Given YY1's dual role in transcriptional activation, we also analyzed the ability of the Y383E mutant to transactivate expression from the HTLV-1 promoter in HEK-293A cells.3 These cells were co-transfected with HTLV-1 firefly luciferase construct together with empty vector, wild type, or Y383E or Y383F YY1 mutants as well as a thymidine kinase (TK) promoter-driven renilla luciferase control plasmid for normalization of transfection efficiency. Transfected cells were harvested twenty-four hours later, lysed, and subjected to dual luciferase assays. Overexpression of wild-type or Y383F YY1 caused a potent 29-fold activation of the HTLV-1 promoter compared with empty vector control (Fig. 4F). Overexpression of the Y383E mutant showed impaired transactivation (seven-fold compared with vector control) (Fig. 4F), again consistent with the notion that phosphorylation of this residue may impair YY1 activity. Surprisingly, phosphomimetic and nonphosphorylatable mutations at the other five tyrosines in YY1 had no negative effect on the transactivation of the HTLV-1 promoter (Fig. 4F), suggesting that phosphorylation at these sites does not affect YY1 activation function. Taken together, our findings suggest that phosphorylation of tyrosine 383 may interfere with YY1's ability to act as a transcriptional regulator.
Phosphomimetic Y383E Mutation or Phosphorylation of YY1 at Tyrosine 383 Impairs Its Ability to Bind DNA and RNA
To test the possibility that phosphorylation of tyrosine 383 alters its ability to bind DNA or RNA, we expressed recombinant His6-tagged wild type, Y383E, and Y383F YY1 in bacteria and purified them using nickel chromatography (Fig. 5A). Next, we used EMSAs to assess the ability of YY1 mutants to bind consensus DNA sequences. Two different double-stranded DNA oligonucleotides were used as probes in our EMSAs, a MLV-NCR probe encompassing a portion of the MLV promoter (10), and an AAV-P5 probe containing a portion of the adeno-associated virus promoter (1) (Fig. 5B). Wild-type and Y383F YY1 were equally capable of binding to the MLV-NCR DNA, whereas no binding was observed for the Y383E mutant (Fig. 5B). This lack of binding is consistent with our observation that wild-type and Y383F, but not Y383E, can rescue YY1-mediated MLV silencing in F9 cells (Fig. 4B). Moreover, a similar pattern of binding was observed for the AAV-P5 probe (Fig. 5B).
FIGURE 5.
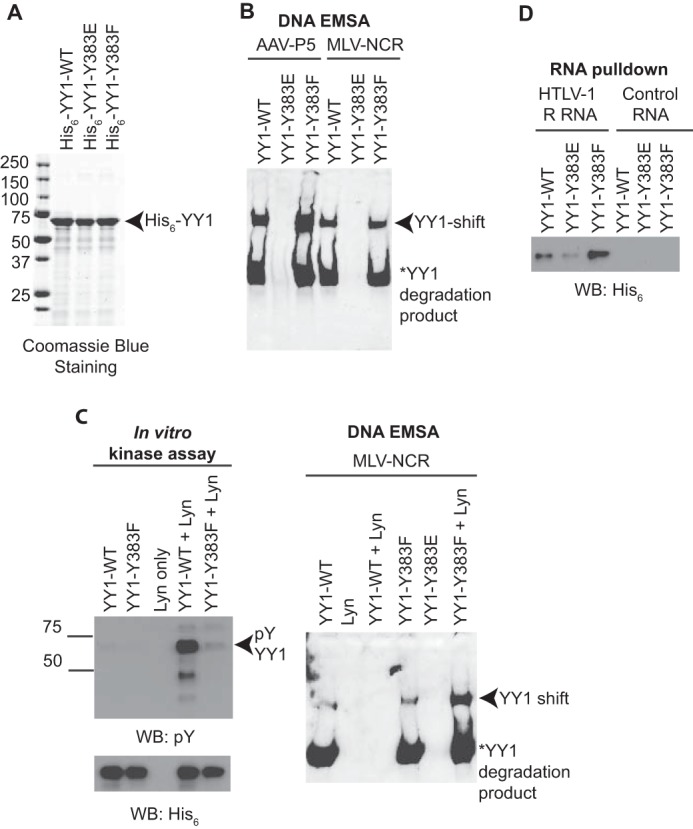
Phosphomimetic Y383E mutation or phosphorylation of YY1 at tyrosine 383 impairs its ability to bind DNA and RNA. A, SDS-PAGE and Coomassie Blue staining of affinity purified His6-tagged recombinant YY1 produced in Escherichia coli. B, EMSA were performed using biotin-labeled MLV-NCR or AAV-P5 oligonucleotides and bacterially purified His6-tagged YY1. Formation of the YY1-DNA complex is indicated by the arrow. Data shown are representative of three independent experiments. C, in vitro phosphorylated YY1 (left panel) was used for EMSA using biotin-labeled MLV-NCR probe (right panel). Formation of the YY1-DNA complex is indicated by the arrow. Data shown are representative of two independent experiments. WB, Western blot. D, bacterially purified His6-tagged YY1 was subjected to an in vitro RNA binding assay using biotin-labeled HTLV-1 R RNA or control RNA. Formation of the YY1-RNA complex was monitored by Western blotting with anti-His6 antibody.
Mutating tyrosine 383 to a glutamic acid could alter the structure of YY1 and cause the protein to misfold, resulting in defective DNA binding, differently than Tyr-383 phosphorylation. To address this possibility, we repeated the EMSA experiment using in vitro phosphorylated wild-type or Y383F YY1 (Fig. 5C, left panel). Interestingly, although purified Lyn induced robust phosphorylation of wild-type YY1, phosphorylation of the Y383F mutant was drastically reduced (Fig. 5C, left panel). This observation is consistent with our in vivo phosphorylation experiments (Fig. 3, A and B) and further suggests that phosphorylation of YY1 by Lyn at different tyrosines may occur in a cooperative manner such that elimination of one site greatly reduces the probability that other sites will become phosphorylated. Importantly, MLV-NCR binding was abolished upon the phosphorylation of wild-type but not Y383F YY1 (Fig. 5C, right panel), further demonstrating that the addition of a natural phosphate at tyrosine 383 impairs DNA binding.
We next asked whether reduced nucleic acid binding could also account for the reduced transactivation of the HTLV-1 promoter seen with the Y383E mutant (Fig. 4F). As shown in Fig. 4F, compared with wild-type YY1, overexpression of the Y383E mutant showed a 4-fold reduction in transactivation of the HTLV-1 promoter. Given that transactivation of the HTLV-1 promoter may require YY1 binding to the HTLV-1 RNA,3 we postulated that the decreased transactivation seen with the Y383E mutant may be a result of reduced RNA binding. To test this, we carried out in vitro RNA binding assays. Biotin-labeled HTLV-1 or control RNA immobilized on streptavidin beads was incubated with recombinant His6-tagged wild type, Y383E, or Y383F YY1. RNA-protein complexes were resolved by SDS-PAGE, and the extent of RNA binding was quantified by immunoblotting with an anti-His6 antibody. Analogous to the case for DNA binding (Fig. 5, B and C), RNA binding was greatly reduced in the Y383E mutant in comparison with wild-type or Y383F YY1 (Fig. 5D). Together, our data suggest that phosphorylation of tyrosine 383 interferes with YY1's ability to make contacts with both DNA and RNA, thus providing a mechanistic basis for the reduction in YY1 activity.
YY1 Is Downstream of EGFR Signaling in HeLa Cells
Src and Src family member kinases are important in tyrosine kinase receptor signaling (34). For example, activation of EGFR leads to c-Src activation (35), which in turn phosphorylates EGFR and propagates signaling downstream of the receptor (36, 37). Based on our proposed role for Src family kinases in phosphorylating YY1, we hypothesized that YY1 phosphorylation may be downstream of physiologic signaling pathways such as EGFR. To test this, we treated serum-starved HeLa cells with recombinant EGF for various times and monitored YY1 phosphorylation by immunoprecipitation and phosphotyrosine immunoblotting. To test for the efficiency of EGF stimulation, cell lysates were probed with antibodies specific for phospho-ERK1/2, a known downstream target of EGFR. EGF stimulation resulted in robust phosphorylation of ERK1/2 (Fig. 6, lower panels). Moreover, increased YY1 phosphorylation was observed at all the time points tested (Fig. 6, upper panels), suggesting that YY1 is a novel downstream target of EGFR signaling.
FIGURE 6.
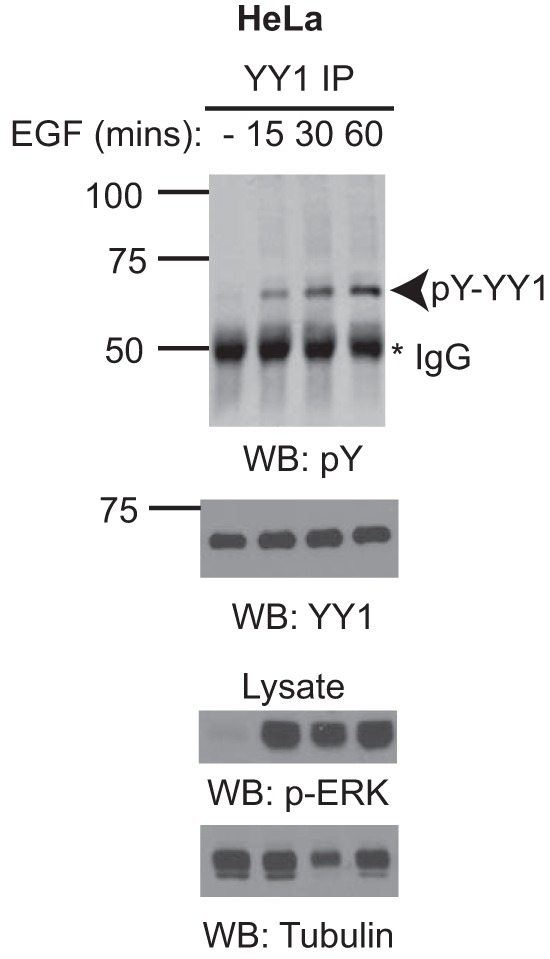
YY1 is downstream of EGFR signaling in HeLa cells. Serum-starved HeLa cells were stimulated with 100 ng/ml recombinant EGF for the indicated time points. Cell lysates were prepared and subjected to immunoprecipitation (IP) and Western blotting (WB) as indicated. The lysate lanes represent 2% of the material used for each immunoprecipitation reaction. Data shown are representative of two independent experiments.
Discussion
In this study we demonstrate that YY1 is a target of tyrosine phosphorylation in various cell types in vivo. Using a combination of kinase inhibitors, kinase overexpression, and kinase knock-out studies, we arrived at the conclusion that YY1 phosphorylation is likely mediated by multiple Src family kinases (Fig. 2). The nine Src family kinases exhibit different tissue expression patterns. For example, c-Src, Fyn, and Yes are ubiquitously expressed (30), whereas Lyn shows preferential expression in hematopoietic cells (38). Given the conservation of YY1 phosphorylation across different cell types (Fig. 1), we postulate that depending on the Src kinase signature within a cell, different cell types may utilize different members of the kinase family for phosphorylating YY1. Thus, this mechanism of YY1 regulation may function in a wide range of tissues and settings.
In Lyn-overexpressing cells, overall YY1 phosphorylation was greatly reduced by single mutations at either tyrosine 8, 254, or 383, suggesting that these three sites are preferentially phosphorylated by Lyn and that phosphorylation of these sites may occur in a cooperative manner, as mutations at each site greatly reduced the likelihood that the other sites become phosphorylated in vivo (Fig. 3, A and B). Phosphorylation of bacterially purified Y383F YY1 using recombinant Lyn was also significantly reduced in vitro (Fig. 5C). However, in both Src- and Yes-overexpressing cells, YY1 was preferentially phosphorylated on different residues (Fig. 3E), suggesting that the sites of YY1 phosphorylation in a given cell type may depend on the expression pattern of Src family kinases within that cell. Importantly, it appears that tyrosine 383 is a target of all three kinases (Fig. 3, A and E).
To examine the functional importance of YY1 phosphorylation sites uncovered by our mutagenesis analysis, we tested the effect of introducing phosphomimetic and nonphosphorylatable mutations on the ability of YY1 to regulate gene expression driven by both retroviral and cellular promoters. Our observation that overexpression of Y383E, but not Y383F, affected the ability of YY1 to either transactivate the HTLV-1 promoter or repress the MLV or endogenous c-fos promoter (Fig. 4, B, D, and F) highlighted the functional importance of this tyrosine residue.
YY1 contains four C2H2 zinc finger domains, each of which is composed of conserved cysteines, histidines, and hydrophobic residues folding to form a ββα structure surrounding a central zinc ion (39). Tyr-383 is located at the very start of the fourth zinc finger, before the first cysteine of the CCHH cluster chelating the zinc, and is not the highly conserved bulky hydrophobic residue in the center of the zinc finger. A closer inspection of the co-crystal structure of YY1 bound to the AAV P5 promoter initiator element (39) (Fig. 7) reveals that tyrosine 383 together with valine 384 and critical cysteine 385 participate in the formation of the first β-sheet of the fourth zinc finger. Hence, one would predict that the addition of a phosphate at this position may perturb the first β-sheet, thus interfering with zinc and/or nucleic acid binding. We cannot rule out the possibility that phosphorylation-induced changes in protein folding may also affect YY1's affinity for other transcriptional cofactors such as CTCF (40) and Smads (41).
FIGURE 7.
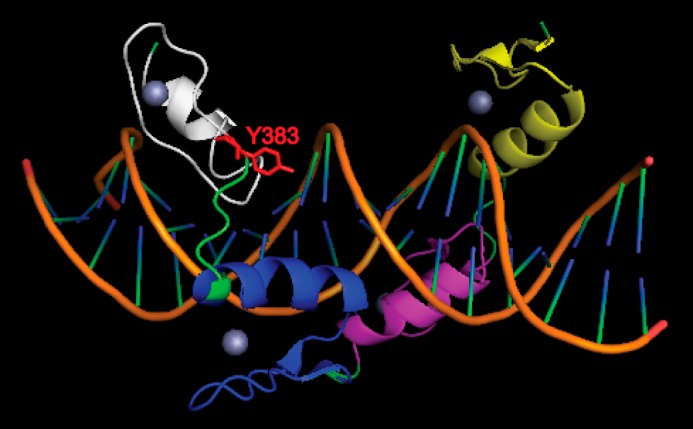
Structure of YY1 DNA binding domain in complex with AAV P5 promoter initiator element. The DNA is shown in orange. The zinc fingers of YY1 are shown as yellow, purple, blue, and white from the N to C termini. Zinc ions are depicted as spheres. The critical tyrosine 383 residue with its side chain is shown in red. Note that structure shown is based on previous report (39).
The importance of Tyr-383 is supported by biochemical assays showing reduced DNA (Fig. 5, B and C) and RNA (Fig. 5D) binding by the Y383E mutant. Apart from tyrosine 383, it is surprising that phosphomimetic mutations at the other five tyrosines failed to affect YY1 function in the context of HTLV-1 transactivation. For example, tyrosine 185 lies in close proximity to serine 180 and serine 184, sites previously shown to be a target of Aurora B kinase, leading to changes in DNA binding and YY1 acetylation (22). Despite this proximity, the Y185E and Y185F mutants maintained full transactivation activity on the HTLV-1 promoter (Fig. 4F). Therefore, the simplest explanation is that phosphorylation of the other five tyrosines is not important for YY1's activation function.
Inactivation of gene-specific transcription factors often affects control of cell division and mitosis. One mechanism of transcription factor inactivation during the cell cycle involves serine/threonine phosphorylation of the consensus TGEKP zinc finger linker sequence in C2H2 transcription factors such as Sp1, Ikaros (42), and YY1 (23, 43), leading to reduced DNA binding. YY1 activity may also be similarly controlled during the cell cycle by phosphorylation of tyrosine 383. To our knowledge, regulation of a C2H2 transcription factor through tyrosine phosphorylation of its zinc finger has not been documented. In YY1, tyrosine 383 makes up part of the consensus YXC motif that is also found at the start of many other C2H2 transcription factors (44), thereby raising that possibility that other C2H2 family members may be regulated in a similar manner.
YY1 is a stable, constitutive, and widely expressed protein across many cell types (16). This makes regulation of YY1 through post-translational modifications an attractive means of controlling its activity. The levels of tyrosine-phosphorylated YY1 is low under basal conditions but becomes readily detectable upon the inhibition of endogenous tyrosine phosphatases (Fig. 1) or the activation of cytosolic tyrosine kinases via EGFR signaling (Fig. 6). Given that protein phosphorylation is dictated by the balance between kinase and phosphatase activities, our data suggest that the majority of YY1 is unphosphorylated and available for DNA binding under steady-state conditions, leading to persistent activation or repression of YY1-responsive genes. However, the presence of extracellular stimuli such as EGF allows for dynamic changes in the DNA binding capacity of YY1 through tyrosine phosphorylation, thereby permitting temporal fine-tuning of YY1-responsive promoters.
Under steady-state conditions, YY1 is predominantly a nuclear protein with a minor cytoplasmic presence (16). Cellular fractionation experiments using pervanadate-treated Jurkat cells showed high levels of tyrosine-phosphorylated YY1 in the nucleus and trace amounts in the cytoplasm (Fig. 1C). This is consistent with emerging evidence showing that beside cytoplasmic localization, a fraction of the Lyn (45), Src, Fyn, and Yes kinases (46) is present in the nucleus. Nevertheless, the interplay between YY1 phosphorylation and cellular localization is a topic of future studies.
In summary, our work has revealed a novel means of regulating YY1 function through tyrosine phosphorylation. Further studies will be necessary to address the specific tyrosine phosphatase(s) responsible for YY1 dephosphorylation as well as the localization and dynamics of tyrosine-phosphorylated YY1 in vivo. These studies will provide us with a better understanding of the complex interplay between tyrosine kinases and phosphatases in regulating YY1 function.
Author Contributions
G. Z. W. and S. P. G. designed the study and wrote the paper. G. Z. W. performed the experiments. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgment
We thank Dr. Yang Shi (Harvard Medical School) for generosity with reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA30488 (NCI). The authors declare that they have no conflicts of interest with the contents of this article.
G. Z. Wang and S. P. Goff, unpublished observations.
- YY1
- Yin Yang 1
- AAV
- adeno-associated virus
- NCR
- negative control region
- LTR
- long terminal repeat
- HTLV-1
- human T-lymphotropic virus 1
- EGFR
- epidermal growth factor receptor
- TK
- thymidine kinase.
References
- 1. Shi Y., Seto E., Chang L. S., Shenk T. (1991) Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377–388 [DOI] [PubMed] [Google Scholar]
- 2. Donohoe M. E., Zhang X., McGinnis L., Biggers J., Li E., Shi Y. (1999) Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19, 7237–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon S., Akopyan G., Garban H., Bonavida B. (2006) Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25, 1125–1142 [DOI] [PubMed] [Google Scholar]
- 4. Shi Y., Lee J. S., Galvin K. M. (1997) Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332, F49–F66 [DOI] [PubMed] [Google Scholar]
- 5. Belak Z. R., Ficzycz A., Ovsenek N. (2008) Biochemical characterization of Yin Yang 1-RNA complexes. Biochem. Cell Biol. 86, 31–36 [DOI] [PubMed] [Google Scholar]
- 6. Jeon Y., Lee J. T. (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernhard W., Barreto K., Raithatha S., Sadowski I. (2013) An upstream YY1 binding site on the HIV-1 LTR contributes to latent infection. PloS ONE 8, e77052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Margolis D. M., Somasundaran M., Green M. R. (1994) Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J. Virol. 68, 905–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flanagan J. R., Becker K. G., Ennist D. L., Gleason S. L., Driggers P. H., Levi B. Z., Appella E., Ozato K. (1992) Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol. Cell. Biol. 12, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlesinger S., Lee A. H., Wang G. Z., Green L., Goff S. P. (2013) Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 4, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hariharan N., Kelley D. E., Perry R. P. (1991) Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc. Natl. Acad. Sci. U.S.A. 88, 9799–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bushmeyer S., Park K., Atchison M. L. (1995) Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem. 270, 30213–30220 [DOI] [PubMed] [Google Scholar]
- 13. Galvin K. M., Shi Y. (1997) Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17, 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee T. C., Zhang Y., Schwartz R. J. (1994) Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9, 1047–1052 [PubMed] [Google Scholar]
- 15. Lee J. S., See R. H., Galvin K. M., Wang J., Shi Y. (1995) Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 23, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Austen M., Lüscher B., Lüscher-Firzlaff J. M. (1997) Characterization of the transcriptional regulator YY1: the bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272, 1709–1717 [DOI] [PubMed] [Google Scholar]
- 17. Deng Z., Wan M., Sui G. (2007) PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol. Cell. Biol. 27, 3780–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiromura M., Choi C. H., Sabourin N. A., Jones H., Bachvarov D., Usheva A. (2003) YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation). J. Biol. Chem. 278, 14046–14052 [DOI] [PubMed] [Google Scholar]
- 19. Yao Y. L., Yang W. M., Seto E. (2001) Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21, 5979–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hongo F., Garban H., Huerta-Yepez S., Vega M., Jazirehi A. R., Mizutani Y., Miki T., Bonavida B. (2005) Inhibition of the transcription factor Yin Yang 1 activity by S-nitrosation. Biochem. Biophys. Res. Commun. 336, 692–701 [DOI] [PubMed] [Google Scholar]
- 21. Rizkallah R., Alexander K. E., Kassardjian A., Lüscher B., Hurt M. M. (2011) The transcription factor YY1 is a substrate for Polo-like kinase 1 at the G2/M transition of the cell cycle. PloS ONE 6, e15928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kassardjian A., Rizkallah R., Riman S., Renfro S. H., Alexander K. E., Hurt M. M. (2012) The transcription factor YY1 is a novel substrate for Aurora B kinase at G2/M transition of the cell cycle. PloS ONE 7, e50645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rizkallah R., Hurt M. M. (2009) Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol. Biol. Cell 20, 4766–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoo S. K., Starnes T. W., Deng Q., Huttenlocher A. (2011) Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature 480, 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li M. Z., Elledge S. J. (2012) SLIC: a method for sequence- and ligation-independent cloning. Methods Mol. Biol. 852, 51–59 [DOI] [PubMed] [Google Scholar]
- 26. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Wang G. Z., Wolf D., Goff S. P. (2014) EBP1, a novel host factor involved in primer binding site-dependent restriction of moloney murine leukemia virus in embryonic cells. J. Virol. 88, 1825–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas S. M., Brugge J. S. (1997) Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 29. Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. (2000) SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 20, 9018–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stein P. L., Vogel H., Soriano P. (1994) Combined deficiencies of Src, Fyn, and Yes tyrosine kinases in mutant mice. Genes Dev. 8, 1999–2007 [DOI] [PubMed] [Google Scholar]
- 31. Wolf D., Goff S. P. (2007) TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 [DOI] [PubMed] [Google Scholar]
- 32. Gualberto A., LePage D., Pons G., Mader S. L., Park K., Atchison M. L., Walsh K. (1992) Functional antagonism between YY1 and the serum response factor. Mol. Cell. Biol. 12, 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Natesan S., Gilman M. (1995) YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol. Cell. Biol. 15, 5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bromann P. A., Korkaya H., Courtneidge S. A. (2004) The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 23, 7957–7968 [DOI] [PubMed] [Google Scholar]
- 35. Weernink P. A., Rijksen G. (1995) Activation and translocation of c-Src to the cytoskeleton by both platelet-derived growth factor and epidermal growth factor. J. Biol. Chem. 270, 2264–2267 [DOI] [PubMed] [Google Scholar]
- 36. Stover D. R., Becker M., Liebetanz J., Lydon N. B. (1995) Src phosphorylation of the epidermal growth factor receptor at novel sites mediates receptor interaction with Src and P85 α. J. Biol. Chem. 270, 15591–15597 [DOI] [PubMed] [Google Scholar]
- 37. Biscardi J. S., Maa M. C., Tice D. A., Cox M. E., Leu T. H., Parsons S. J. (1999) c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr-845 and Tyr-1101 is associated with modulation of receptor function. J. Biol. Chem. 274, 8335–8343 [DOI] [PubMed] [Google Scholar]
- 38. Hibbs M. L., Tarlinton D. M., Armes J., Grail D., Hodgson G., Maglitto R., Stacker S. A., Dunn A. R. (1995) Multiple defects in the immune system of Lyn-deficient mice, culminating in autoimmune disease. Cell 83, 301–311 [DOI] [PubMed] [Google Scholar]
- 39. Houbaviy H. B., Usheva A., Shenk T., Burley S. K. (1996) Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. U.S.A. 93, 13577–13582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Donohoe M. E., Zhang L. F., Xu N., Shi Y., Lee J. T. (2007) Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell 25, 43–56 [DOI] [PubMed] [Google Scholar]
- 41. Kurisaki K., Kurisaki A., Valcourt U., Terentiev A. A., Pardali K., Ten Dijke P., Heldin C. H., Ericsson J., Moustakas A. (2003) Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 23, 4494–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dovat S., Ronni T., Russell D., Ferrini R., Cobb B. S., Smale S. T. (2002) A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 16, 2985–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rizkallah R., Alexander K. E., Hurt M. M. (2011) Global mitotic phosphorylation of C2H2 zinc finger protein linker peptides. Cell Cycle 10, 3327–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brayer K. J., Segal D. J. (2008) Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50, 111–131 [DOI] [PubMed] [Google Scholar]
- 45. Radha V., Nambirajan S., Swarup G. (1996) Association of Lyn tyrosine kinase with the nuclear matrix and cell-cycle-dependent changes in matrix-associated tyrosine kinase activity. Eur. J. Biochem. 236, 352–359 [DOI] [PubMed] [Google Scholar]
- 46. Takahashi A., Obata Y., Fukumoto Y., Nakayama Y., Kasahara K., Kuga T., Higashiyama Y., Saito T., Yokoyama K. K., Yamaguchi N. (2009) Nuclear localization of Src-family tyrosine kinases is required for growth factor-induced euchromatinization. Exp. Cell Res. 315, 1117–1141 [DOI] [PubMed] [Google Scholar]