Background: Mitochondrial DNA (mtDNA) mutations accumulate with age, but how this impacts cardiac progenitor cell (CPC) function is unknown.
Results: mtDNA mutations disrupt mitochondrial function and reduce differentiation potential of CPCs.
Conclusion: Preserving mtDNA integrity is critical for CPC homeostasis and regenerative potential.
Significance: Increased understanding of pathways regulating progenitor cell function is critical for development of effective cell therapies.
Keywords: aging, glycolysis, heart failure, mitochondria, mitochondrial DNA (mtDNA), mitophagy, stem cells, cardiac progenitor cells
Abstract
Transfer of cardiac progenitor cells (CPCs) improves cardiac function in heart failure patients. However, CPC function is reduced with age, limiting their regenerative potential. Aging is associated with numerous changes in cells including accumulation of mitochondrial DNA (mtDNA) mutations, but it is unknown how this impacts CPC function. Here, we demonstrate that acquisition of mtDNA mutations disrupts mitochondrial function, enhances mitophagy, and reduces the replicative and regenerative capacities of the CPCs. We show that activation of differentiation in CPCs is associated with expansion of the mitochondrial network and increased mitochondrial oxidative phosphorylation. Interestingly, mutant CPCs are deficient in mitochondrial respiration and rely on glycolysis for energy. In response to differentiation, these cells fail to activate mitochondrial respiration. This inability to meet the increased energy demand leads to activation of cell death. These findings demonstrate the consequences of accumulating mtDNA mutations and the importance of mtDNA integrity in CPC homeostasis and regenerative potential.
Introduction
The heart contains a pool of c-kit-positive cardiac progenitor cells (CPCs)3 that has the potential to repair the myocardium after injury by secreting paracrine factors as well as producing new cardiac myocytes and vascular structures (1). However, the functional role of endogenous CPCs in the myocardium and how they contribute to repair are still unclear and controversial. Beltrami et al. (2) initially reported that injection of CPCs into the infarct region leads to generation of new vessels and cardiac myocytes by the CPCs. More recently, it was reported that endogenous CPCs are necessary for both cardiac homeostasis and regeneration after injury (3). This study found that cardiac damage induced by isoproterenol treatment leads to CPC activation and commitment to the myocardial cell lineages, including cardiac myocytes. In contrast, a recent study found that differentiation of endogenous CPCs into cardiac myocytes occurs at a very low rate even after injury (4), whereas another study reported that new cardiac myocytes are generated only from pre-existing myocytes and that endogenous CPCs do not play a critical role in myocardial homeostasis and repair (5). Although the functional role of endogenous CPCs is controversial, it is clear that infusion of CPCs into the injured myocardium leads to repair and improved function. For instance, in animal models of myocardial infarction or doxorubicin-induced cardiomyopathy, injection of CPCs reduces injury and improves left ventricular function (6–8). More importantly, utilization of autologous CPCs in patients with ischemic cardiomyopathy in the SCIPIO clinical trial is showing promising results (9, 10). Studies have found that CPC function is reduced with age, which reduces their regenerative capacity (11–13). Because stem cell therapies for cardiovascular disease primarily target geriatric patients, the autologous CPCs might have reduced regenerative capacity once they are transplanted back into the patient's heart. The exact mechanisms underlying the age-related changes in CPC function are still unclear. Therefore, a deeper understanding of the biological processes that regulate CPC function and survival is needed so that more effective therapeutic strategies to repair the heart can be developed.
Mitochondria regulate several key processes including metabolism, heme synthesis, and cell death. Mitochondria are also responsible for producing energy via oxidative phosphorylation (OXPHOS) for cellular development, differentiation, and growth (14–16). Mitochondria contain their own DNA that is replicated independently of the nuclear DNA. The mitochondrial DNA (mtDNA) only encodes 37 genes, and 13 of them are subunits of the respiratory complexes or the ATP synthase involved in OXPHOS. Unfortunately, mtDNA is more susceptible to genetic mutations than nuclear DNA because it is constantly exposed to reactive oxygen species generated by the respiratory chain in the mitochondrial inner membrane. Studies have reported that mtDNA mutations and deletions accumulate with age in various tissues in humans and rodents, which can lead to impaired mitochondrial function (17–21). Mutations in mtDNA in the heart have also been demonstrated after treatment with cardiotoxic therapies such as doxorubicin (22) or nucleoside reverse transcriptase inhibitors (23) and after myocardial infarction (24). The contribution of mtDNA mutations to aging has been confirmed by studies in mice expressing a proofreading-deficient mitochondrial DNA polymerase γ (POLG). These mice accumulate mtDNA mutations in cells at a faster rate than wild-type mice, which leads to an accelerated aging phenotype and reduced lifespan (25, 26). Moreover, accumulation of mtDNA mutations in the heart is associated with increased oxidative damage and apoptosis, which results in the development of cardiomyopathy at 13–14 months of age (27).
Recent studies have also implicated mitochondria as critical regulators of stem cell function. Embryonic, neuronal, and mesenchymal stem cells have been reported to contain few immature mitochondria that are clustered in the perinuclear region and to rely on glycolysis for energy production (28–30). However, differentiation of stem cells requires metabolic reprogramming to meet the increased energy demand that occurs concomitantly with a shift from cytosolic anaerobic glycolysis to mitochondrial respiration (29, 31, 32). To date, no attention has been given to the role of mitochondria in CPCs and how accumulation of mtDNA mutations impacts these cells. In this study, we demonstrate that functional mitochondria are critical for CPC function and survival. Accumulation of mtDNA mutations in CPCs leads to disruption of mitochondrial function, reduced proliferation, and increased susceptibility to stress. The mutant CPCs are also unable to transition from glycolysis to oxidative phosphorylation in response to differentiation, which instead leads to activation of cell death. Thus, our studies highlight the critical role of mitochondria for CPC function and the consequences of accumulating mtDNA mutations.
Experimental Procedures
CPC Isolation and Culture
All experimental procedures were performed in accordance with institutional guidelines and approved by the Institutional Animal Care and Use Committee of the University of California San Diego. For experiments in Fig. 1, c-kit-positive CPCs were isolated from 2-month-old male wild-type FVB mice. For all other experiments CPCs were isolated from male mice homozygous for the PolgD257A mutation on a C57Bl/6 background (25), and corresponding control CPCs were isolated from wild-type, sex-matched litter mates. All c-kit-positive adult CPCs were isolated and cultured as previously described (33). To activate differentiation, CPCs were incubated in α-minimal essential medium containing 10 nm dexamethasone for up to 7 days.
FIGURE 1.
Activation of the differentiation program is associated with induction of mitochondrial biogenesis and expansion of the mitochondrial network in CPCs. A, mitochondrial content increased in CPCs after 7 days of incubation in DM. The CPCs with increased mitochondrial content were also positive for the cardiac myocyte lineage marker GATA-4. Scale bar = 20 μm. B, quantitation of GATA-4 positive CPCs at day 0 and day 7 (n = 4). C, real-time qPCR analysis of GATA-4 (n = 6), GATA-6 (n = 3), and PECAM1 (n = 4) gene expression at day 0 and day 7. D, phosphorylation of adenosine monophosphate-activated protein kinase (AMPK) in CPCs after incubation in DM (n = 3). E, real-time qPCR analysis of the mitochondrial biogenesis regulator PGC-1α (n = 4) gene expression at day 0 and day 7. F, real-time qPCR of mtDNA content at day 0 and day 7 (n = 8). Data are normalized to genomic DNA. G, representative Western blot of mitochondrial OXPHOS proteins and quantitation of bands (n = 6). *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus day 0.
Primary Myoblast Isolation and Culture
Primary myoblasts were isolated from hind limb skeletal muscle of 3–4-month-old WT and POLG mice litter mates using a protocol adapted from rat skeletal muscle (34). Muscles were digested in 0.20% type IA collagenase and 0.25% trypsin in DMEM (Gibco), and cells then cultured in growth medium (20% FBS and 1% penicillin-streptomycin in DMEM) on a bed of Matrigel™ (on coverslip, 1 mg/ml, Basement Membrane Matrix; BD Biosciences). Myoblast differentiation was induced 5 days later with 2% horse serum and 1% penicillin-streptomycin in DMEM for 2–4 days.
Echocardiography
Echocardiography was performed as previously described (35) using a Vevo770 In Vivo Micro-Imaging System with an RMV707B 15–45 MHz imaging transducer (VisualSonics Inc.), and parameters were quantified using the VisualSonics software.
Quantitative Polymerase Chain Reaction
Genomic DNA was extracted using GenElute Mammalian Genomic DNA Miniprep Kit (Sigma) and PCR-amplified with TaqMan Universal Master Mix II. 18S rRNA was used as a control for nuclear DNA, and d-loop was used for mtDNA quantitation using primer sequences previously described (36). For gene expression assays, RNA was extracted using the RNeasy Mini kit (Qiagen). cDNA was synthesized using the QuantiTect Reverse Transcription kit (Qiagen) following the manufacturer's protocols. Taqman primers for PGC-1α, PGC-1β, and GATA-4 were from Integrated DNA Technologies. TaqMan primers for 18S, GATA-6, PECAM1, COX1 (Complex IV subunit 1), Cox4i1 (Complex IV subunit 4), and the TaqMan Universal Master Mix II were from Applied Biosystems. qPCR was carried out on a CFX96 real-time PCR detection system (Bio-Rad). Relative amounts of mRNA were normalized to 18S, and -fold change in gene expression was calculated using the 2(−ΔΔCt) method.
Western Blot
Samples were prepared in lysis buffer containing 50 mm Tris-HCl, 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100, and protease inhibitor mixture (Roche Applied Science) and run on Invitrogen NuPAGE Bis-Tris gels. The membranes were probed with the following antibodies: MitoProfile Total OXPHOS Rodent WB antibody mixture (MitoSciences), PGC-1 (Abcam), COXIV subunit 4 (Invitrogen), adenosine monophosphate-activated protein kinase (AMPK), p-AMPK, hexokinase I, hexokinase II, PKM1/2, GAPDH, LC3A/B (Cell Signaling Technology), DLP1 (BD Biosciences), MFN1, MyoD (Santa Cruz Biotechnology), MFN2, Tubulin (Sigma), and actin (GeneTex). Membranes were imaged using a ChemiDoc XRS+ System (Bio-Rad).
Mitochondrial Respiration Measurements
Intact mitochondria were isolated from adult mouse hearts, and oxygen consumption measurements were taken at 30 °C using an Oxygraph apparatus (Hansatech Instruments). Complex I- and Complex II-dependent oxygen consumption were measured as previously described (35). Complex IV-dependent respiration was measured by the addition of 1 μm antimycin A and 0.4 mm TMPD plus 1 mm ascorbate. Oligomycin (5 μg/ml) was added to inhibit ATP synthase followed by carbonyl cyanide p-trifluoromethoxyphenylhydrazone (1 μm) to obtain maximal respiration rate. Mitochondrial oxygen consumption of monolayers of CPCs was measured using the Seahorse XF96 analyzer (Seahorse Bioscience), adapted from a previously described protocol (36). CPCs were plated at 2 × 104 or 1 × 104 cells per well 24 h before measurement of O2 consumption rates. Assays were run in unbuffered, serum-free DMEM containing 10 mm glucose, 3 mm glutamine, and 1 mm pyruvate. CPCs were allowed to equilibrate on the plate before the addition of 2 μm oligomycin to measure ATP-linked respiration. Three successive additions of 500 nm carbonyl cyanide p-trifluoromethoxyphenylhydrazone were added to measure maximal respiration. Data were analyzed using Wave for Desktop software (Seahorse Bioscience).
Immunofluorescence Microscopy
Cells were fixed, permeabilized, and blocked as previously described (35). CPCs were stained with antibodies against Tom20 (Santa Cruz Biotechnology), COXIV (Invitrogen), GATA-4 (Santa Cruz Biotechnology), or LAMP2 (Sigma). Myoblasts were stained with antibodies against MyoD and NCAM (Santa Cruz Biotechnology). Cells were incubated in Alexa Fluor 488 or 594 secondary antibodies (Life Technologies) followed by Hoechst 33342 (10 μg/ml, Life Technologies) to stain nuclei. Cells were imaged using a Carl Zeiss Axio Observer Z1, and Z-stacks were acquired using a high resolution AxioCam MRm digital camera, a 63× oil immersion objective, and Zeiss AxioVision 4.8 software (Carl Zeiss). A minimum of 100 cells per group were counted for each condition.
Brightfield Microscopy
Myoblasts were imaged at pre- and post-differentiation using a Carl Zeiss Axio Observer Z1 at 10× magnification. Myotube diameter was quantified using ImageJ software.
Transmission Electron Microscopy
Adult mouse hearts and CPCs were fixed, sectioned, and mounted as previously described (35). Sections were examined on a TECNAI G2 Spirit BioTWIN Transmission Electron Microscope equipped with an Eagle 4k HS digital camera (FEI).
Proliferation Assay
CPCs were seeded in a 12-well tissue culture plate, and cell number was determined by counting using a hemocytometer 24 and 72 h later. CPCs were also seeded in a 96-well black wall clear bottom plate (Corning), and cell number was measured with the CyQUANT NF Cell Proliferation Assay kit (Life Technologies). Fluorescence measurements were made using a SpectraMax M3 microplate reader and SoftMax Pro 5.4 software (Molecular Devices).
Cell Death Assay
CPCs were treated with H2O2 (Sigma), doxorubicin hydrochloride (Sigma), sunitinib malate (BioVision), or 2-deoxyglucose (Sigma). Cells were stained with Hoechst 33342 (10 μg/ml, Life Technologies) and YO-PRO-1 iodide (100 nm, Life Technologies) for 15 min and immediately imaged using a Carl Zeiss Axio Observer Z1 at 10× magnification, AxioCam MRm digital camera, and Zeiss AxioVision 4.8 software (Carl Zeiss).
Glycolysis Assay
CPCs were seeded in a 12-well tissue culture plate and incubated in growth medium for 4 days. l-Lactate concentration in the growth medium was measured using a Glycolysis Cell-based Assay kit (Cayman Chemical) according to the manufacturer's protocol. Colorimetric measurements were made using a SpectraMax M3 microplate reader and SoftMax Pro 5.4 software (Molecular Devices).
ATP Assay
ATP levels were measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) according to the manufacturer's protocol. CPCs (40,000 cells/100 μl) were added to a 96-well black wall clear bottom plate (Corning). Luminescent measurements were made using a SpectraMax M3 microplate reader and SoftMax Pro 5.4 software (Molecular Devices).
Assessment of Mitochondrial Membrane Potential
CPCs were incubated with tetramethylrhodamine methyl ester (25 nm, Invitrogen) and Hoechst 33342 (10 μg/ml, Life Technologies) for 15 min at 37 °C. Live cells were examined using a Carl Zeiss Axio Observer Z1 at 40× magnification. Fluorescence intensity was quantified using ImageJ software.
Assessment of Autophagy and Mitophagy
To assess baseline autophagic flux, CPCs were treated with DMSO or 50 nm bafilomycin A1 (EMD Millipore) for 2 h before homogenization and Western blot analysis. To visualize lysosomes, CPCs were fixed and stained with anti-LAMP2 (Sigma). To assess mitochondrial autophagy, CPCs were infected with an adenovirus encoding GFP-LC3 as previously described (37). Cells were treated with DMSO or 50 nm bafilomycin A1 for 3 h, fixed, and stained with anti-Tom20. Cells were imaged at 63× magnification, and Z-stacks were acquired using a Carl Zeiss Axio Observer Z1. LAMP2 staining was quantified using ImageJ software. Mitophagy was assessed by analyzing colocalization between GFP-LC3-positive autophagosomes and Tom20-labeled mitochondria.
Statistical Analyses
All values are expressed as the means ± S.E. Student's t test was used to compare two sets of data. p values < 0.05 were considered statistically significant.
Results
Activation of the Differentiation Program in CPCs Leads to Induction of Mitochondrial Biogenesis
Differentiation is a process that requires energy, which is primarily supplied by mitochondria via oxidative phosphorylation (16). When examining mitochondria in wild-type CPCs from FVBN mice, we found that undifferentiated CPCs contained few mitochondria that were localized primarily in the perinuclear region (Fig. 1A). Incubation of CPCs in differentiation medium (DM) for 7 days led to increased cell size and expansion of the mitochondrial network (Fig. 1A). Next, we confirmed that incubation in DM led to up-regulation of cardiac lineage markers in CPCs, consistent with previous studies (2, 33, 38, 39). We observed an increase in cells positive for GATA-4, a transcription factor that regulates myocardial differentiation, growth, and survival, after incubation in DM. We found that few cells expressed GATA-4 at Day 0, but there was a significant increase in CPCs stained positively for GATA-4 after 7 days in DM, which correlated with an expansion of their mitochondria (Fig. 1, A and B). Real time qPCR confirmed a significant increase in GATA-4 at day 7 (Fig. 1C). There were also significant increases in mRNA encoding the smooth muscle marker GATA-6 and the endothelial marker PECAM1 (Fig. 1C). These results indicate that lineage commitment in CPCs is accompanied by expansion of mitochondria.
Next, we assessed whether the increased energy demand associated with activation of differentiation led to activation of mitochondrial biogenesis. First we confirmed that adenosine monophosphate activated protein kinase (AMPK), a major regulator of mitochondrial biogenesis in response to energy depletion, was activated during differentiation (Fig. 1D). We also found that PGC-1α, a member of the PPARγ co-activator (PGC) family of transcriptional co-activators, was significantly induced in CPCs after incubation in DM (Fig. 1E). The activation of adenosine monophosphate-activated protein kinase and induction of PGC-1α correlated with an increase in mtDNA content (Fig. 1F) and increased levels of mitochondrial OXPHOS proteins (Fig. 1G) in the CPCs in DM. These studies suggest that initiation of differentiation in CPCs leads to activation of mitochondrial biogenesis and enhanced mitochondrial oxidative phosphorylation to adapt to increased energy demand.
mtDNA Mutations Impair CPC Proliferation and Increase Susceptibility to Cell Death
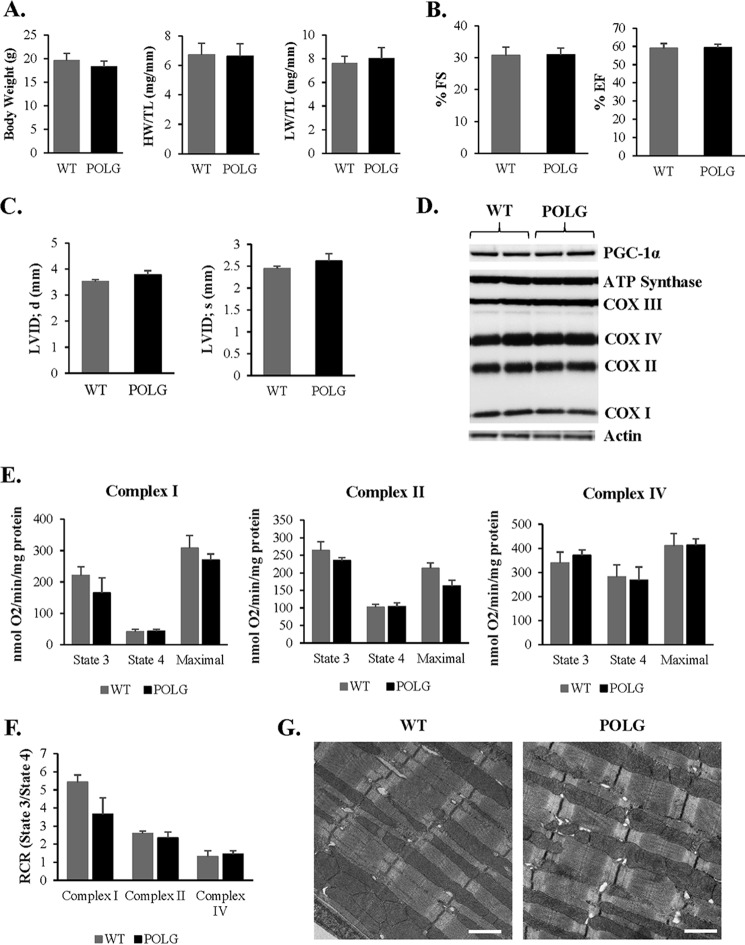
To further explore the role of mitochondria in CPC function and survival, we isolated CPCs from 2-month-old mice with a proofreading-defective POLG and their wild-type (WT) litter mates. The POLG mice undergo premature aging and develop cardiomyopathy with a reduced lifespan due to accumulation of mtDNA mutations (25). However, at 2 months of age, we observed no significant differences in body, heart, or lung weights and cardiac function in POLG mice when compared with age-matched WT litter mates (Fig. 2, A–C). We also found no differences in cardiac mitochondrial content or function at this age between WT and POLG mice (Fig. 2, D–F). Analysis of mitochondrial ultrastructure by transmission electron microscopy confirmed similar mitochondrial content and morphology in WT and POLG hearts at this age (Fig. 2G).
FIGURE 2.
No differences in cardiac and mitochondrial function in WT and POLG mice at 2 months. A, average body weight, heart weight/tibia length (HW/TL), and lung weight/tibia length (LW/TL) ratios were measured in age-matched WT and POLG litter mates (n = 10). Echocardiography measurements at 2 months of age show no significant differences in % fractional shortening (% FS) (B) and % ejection fraction (% EF), left ventricular internal dimension at diastole (LVID; d), and left ventricular internal dimension at systole (LVID; s) (n = 7) (C). D, representative Western blot of mitochondrial biogenesis and OXPHOS proteins in the heart at 2 months. E, mitochondria were isolated from the hearts of 2-month-old mice. Substrates and inhibitors specific to each respiratory complex were added, and oxygen consumption was measured using an oxygen electrode (n = 3). F, respiratory control ratio (RCR) was calculated by dividing state 3/state 4 oxygen consumption. G, transmission electron micrographs show normal mitochondrial ultrastructure in the hearts of WT and POLG mice. Scale bar = 1 μm.
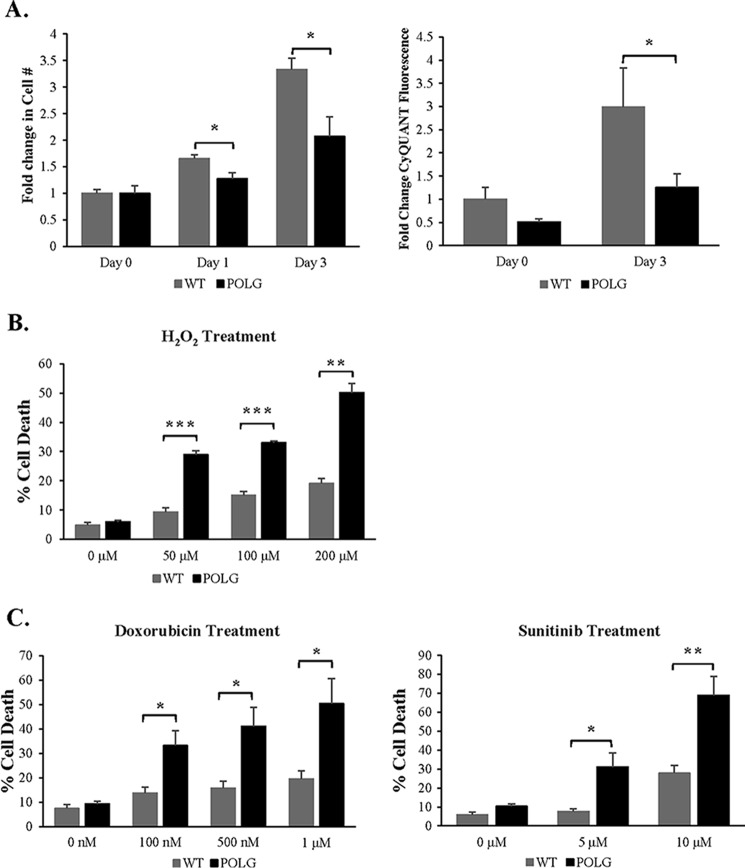
We then investigated whether CPCs from these hearts were affected by the mutated POLG. Proliferation is important for self-renewal and maintenance of the CPC population in the heart, and we found that the POLG CPCs had significantly reduced proliferation rates compared with WT CPCs (Fig. 3A). In addition, the POLG CPCs were more sensitive to cell death after treatment with H2O2 (Fig. 3B). Treatment with doxorubicin, an anthracycline topoisomerase inhibitor, or sunitinib, a tyrosine kinase inhibitor, also resulted in increased cell death in POLG CPCs (Fig. 3C). Thus, these data suggest that accumulation of mtDNA mutations in the POLG CPCs reduces their proliferative capacity and increases their susceptibility to stress.
FIGURE 3.
Mutant CPCs exhibit reduced proliferation and increased sensitivity to stress. A, WT and POLG CPCs were incubated in growth medium for up to 3 days, and proliferation was determined by assessing changes in cell number (n = 4) or by CyQUANT fluorescence (n = 9). WT and POLG CPCs were treated with H2O2 (B) and doxorubicin or sunitinib (C) for 24 h. Cell death was assessed using YO-PRO-1 staining (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
mtDNA Mutations Contribute to Abnormal Mitochondrial Morphology and Reduced Differentiation Potential
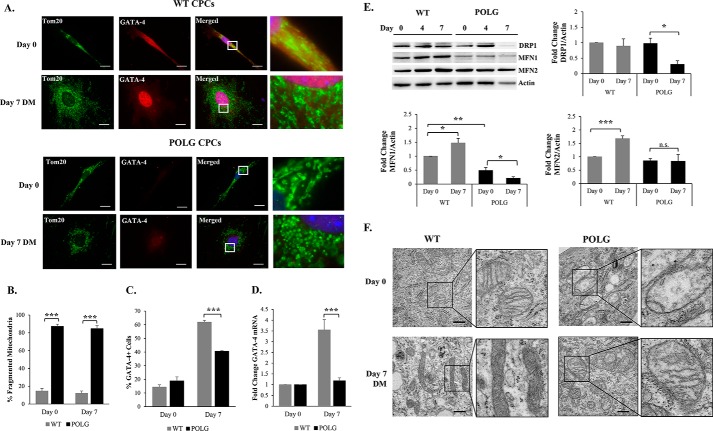
Alterations in mitochondrial morphology allow cells to adapt to changes in the cellular environment, including differentiation (40). When examining the effect of accumulating mtDNA mutations on mitochondrial morphology, we discovered that the mutant POLG CPCs contained fragmented mitochondria under baseline conditions (Fig. 4, A and B). Moreover, although WT CPCs increased in cell size and expanded their mitochondrial network after incubation in DM, POLG CPCs maintained their fragmented mitochondria, which clustered in the perinuclear region and failed to expand in response to the differentiation stimulus (Fig. 4A). The abnormal mitochondrial morphology was also associated with reduced activation of the differentiation program in the POLG CPCs. We observed significantly fewer GATA-4-positive POLG CPCs compared with WT after incubation in DM (Fig. 4C). The POLG CPCs also had significantly reduced levels of GATA-4 transcripts compared with WT CPCs after incubation in DM (Fig. 4D). Thus, these data demonstrate that accumulation of mtDNA mutations leads to abnormal mitochondrial morphology and impaired activation of the differentiation program in CPCs.
FIGURE 4.
Mutant CPCs have abnormal mitochondrial morphology and reduced activation of the differentiation program. A, the mitochondrial network in POLG CPCs has a fragmented appearance at both day 0 and day 7. Scale bar = 20 μm. Quantitation of fragmented mitochondria (n = 3) (B) and GATA-4-positive CPCs (n = 3) (C). D, real-time qPCR analysis of GATA-4 gene expression at day 0 and day 7 in WT and POLG CPCs (n = 4). E, representative Western blots and band quantitation of DRP1 (n = 4), MFN1 (n = 5), and MFN2 (n = 4) in WT and POLG CPCs. F, transmission electron micrographs of WT and POLG CPCs under baseline conditions and after incubation in DM. CPCs contain immature mitochondria. Mitochondrial electron density increased in WT CPCs after 7 days in DM. POLG CPCs had abnormal mitochondrial structure and reduced cristae. Scale bar = 500 nm. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
The fragmented mitochondrial morphology in POLG CPCs prompted us to investigate whether proteins involved in mitochondrial dynamics were altered in the POLG CPCs. Surprisingly, we saw a reduction in the mitochondrial fission protein DRP1 in POLG CPCs after 7 days in DM (Fig. 4E). Interestingly, mitochondrial fusion proteins MFN1 and MFN2 were increased in WT CPCs during differentiation but not in POLG CPCs (Fig. 4E). Thus, there is a shift in the balance between fission and fusion proteins toward fusion in WT CPCs, but not in POLG CPCs, upon activation of differentiation. Furthermore, we analyzed mitochondrial ultrastructure in CPCs by transmission electron microscopy and saw drastic differences between WT and POLG CPCs. Mitochondria in undifferentiated WT CPCs had underdeveloped cristae that became more electron-dense upon activation of the differentiation program (Fig. 4F). In contrast, mitochondria in POLG CPCs were swollen with abnormal cristae structure under baseline conditions and failed to increase cristae density after differentiation. Taken together, these data suggest that accumulation of mtDNA mutations leads to impaired mitochondrial dynamics and differentiation program in the CPCs.
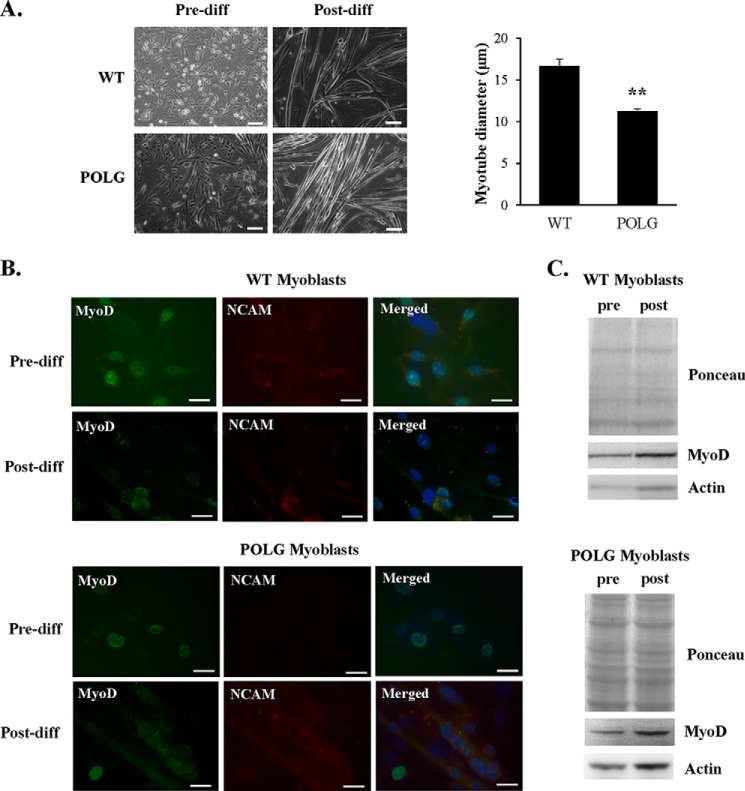
Next, we investigated whether other progenitor cells were similarly affected by accumulation of mtDNA mutations. Satellite cells that reside on the basal lamina of the myofiber are important for skeletal muscle maintenance and facilitate repair by giving rise to myoblasts that fuse to form myofibers. Studies have reported that although the number of satellite cells declines with age, these cells still retain their myogenic potential (41, 42). We found that myoblasts isolated from WT and POLG litter mates formed myotubes in culture upon differentiation. However, myotube diameter in the POLG mice was significantly smaller than WT (Fig. 5A). Both WT and POLG myoblasts expressed the muscle phenotypic marker MyoD pre- and post-differentiation and the adhesion marker NCAM after differentiation (Fig. 5, B and C). These data suggest that myoblasts from POLG mice are less affected by the presence of a mutant POLG. These myoblasts have sufficient capacity to adapt to metabolic changes during differentiation in order to successfully fuse and form myotubes. However, the smaller myofiber size suggests that defective POLG may affect growth signaling in differentiating myoblasts.
FIGURE 5.
POLG myoblasts generate smaller skeletal muscle fibers. A, WT and POLG myoblasts have similar morphology pre-differentiation (Pre-diff). After 4 days of differentiation, POLG myotubes had significantly smaller diameter than WT (n = 4). Scale bar = 100 μm. B, both WT and POLG myoblasts express phenotypic muscle marker MyoD pre- and post-differentiation and NCAM post-differentiation. Scale bar = 20 μm. C, Western blot of MyoD protein levels pre- and post-differentiation. **, p < 0.01 versus WT.
POLG CPCs Have Impaired Mitochondrial Biogenesis and Energetics
The abnormal mitochondrial morphology and distribution in POLG CPCs are indicators of defective mitochondrial function. Because the primary function of mitochondria is to supply the cell with energy via oxidative phosphorylation, we compared OXPHOS protein levels between WT and POLG CPCs. As previously observed in Fig. 1, incubation of WT CPCs in DM resulted in increased expression of OXPHOS proteins. In contrast, POLG CPCs had reduced levels of select OXPHOS proteins, most notably in Complex III and IV subunits (Fig. 6A). In fact, we observed no increase in nuclear -and mitochondria-encoded subunits of Complex IV in the POLG CPCs after incubation in DM for 7 days (Fig. 6B). The baseline protein levels were already significantly lower in POLG CPCs compared with WT at day 0. Interestingly, we found that Complex IV Subunit 1 (Cox1) and Subunit 4 (Cox4i1) transcripts were similar in WT and POLG CPCs at day 0 (Fig. 6C), suggesting that the differences in protein levels between WT and POLG CPCs may be attributed to reduced protein stability in the POLG CPCs. We also found that, in contrast to WT CPCs, the POLG CPCs failed to induce mitochondrial biogenesis factors PGC-1α or PGC-1β to the same extent as WT CPCs during differentiation (Fig. 6D). This suggests that accumulation of mtDNA mutations also impairs the mitochondrial biogenesis response.
FIGURE 6.
Mutant CPCs have reduced levels of both nuclear and mitochondrial encoded OXPHOS proteins and reduced activation of mitochondrial biogenesis. A, representative Western blots revealed reduced levels of selective OXPHOS subunits in POLG CPCs. B, quantitation of mitochondria-encoded complex IV Subunit 1 (n = 5) and nuclear-encoded subunit 4 protein expression (n = 5). C, real-time qPCR analysis of Cox1 (complex IV Subunit 1) and Cox4i1 (complex IV subunit 4) gene expression at baseline (n = 3). D, real-time qPCR analysis of PGC-1α (n = 3) and PGC-1β (n = 3) gene expression at day 0 and day 7 in WT and POLG CPCs. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
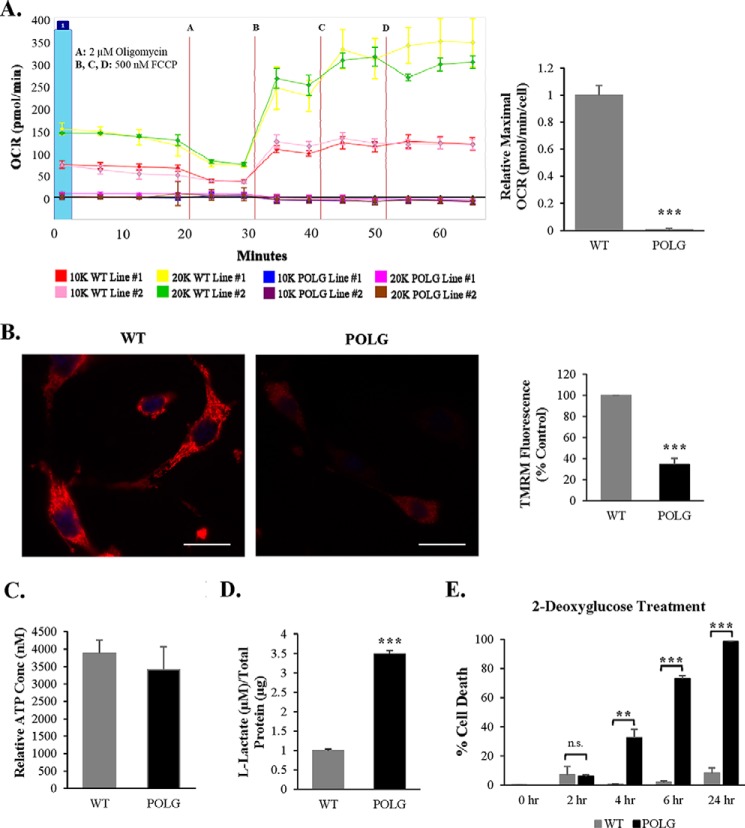
Our data indicated that POLG CPCs have abnormal mitochondrial structure and contain reduced levels of critical proteins involved in OXPHOS. Therefore, we assessed the level of mitochondrial respiration in WT and POLG CPCs by measuring oxygen consumption using the Seahorse XF Analyzer. In WT CPCs, increased oxygen consumption rate corresponded with increased cell number in response to the mitochondrial uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone. Surprisingly, POLG CPCs had no detectable oxygen consumption regardless of cell number (Fig. 7A), indicating that these cells have shut down mitochondrial respiration. The OXPHOS complexes are also responsible for generating the mitochondrial membrane potential, which is used by the F0F1-ATPase for ATP synthesis (43). We confirmed that the POLG CPCs had significantly reduced mitochondrial membrane potential as assessed by reduced tetramethylrhodamine methyl ester (TMRM) staining (Fig. 7B). Tetramethylrhodamine methyl ester uptake by mitochondria is dependent on the membrane potential. However, cellular ATP levels were similar in WT and POLG CPCs (Fig. 7C), suggesting that POLG CPCs utilize alternative mechanisms to generate energy. Therefore, we examined whether the POLG CPCs relied on glycolysis to support their energy demands by assessing extracellular lactate, a glycolytic byproduct. We found that the POLG CPCs had significantly increased extracellular l-lactate compared with WT CPCs (Fig. 7D), and the addition of 2-deoxyglucose to the growth medium to inhibit glycolysis resulted in rapid death of the POLG CPCs (Fig. 7E). This suggests that POLG CPCs depend solely on glycolysis to meet their energy demands.
FIGURE 7.
Mutant CPCs have impaired mitochondrial respiration and reduced membrane potential. A, mitochondrial respiration was measured in WT and POLG CPCs using a Seahorse XF analyzer. Maximal oxygen consumption rate (OCR) was normalized to cell number (n = 4). Cell seeding density: 10K = 10,000; 20K = 20,000. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone. B, representative fluorescent images and quantitation of tetramethylrhodamine methyl ester (TMRM) fluorescence in WT and POLG CPCs (n = 3). Scale bar = 25 μm. Units are arbitrary. C, analysis of cellular ATP content in WT and POLG CPCs (n = 3). D, l-lactate present in growth medium was measured in WT and POLG CPCs (n = 3). E, 2-deoxyglucose (10 nm) was added to the growth medium to inhibit glycolysis in WT and POLG CPCs. Cell death was assessed using YO-PRO-1 staining (n = 3). **, p < 0.01; ***, p < 0.001 versus WT; n.s., not significant.
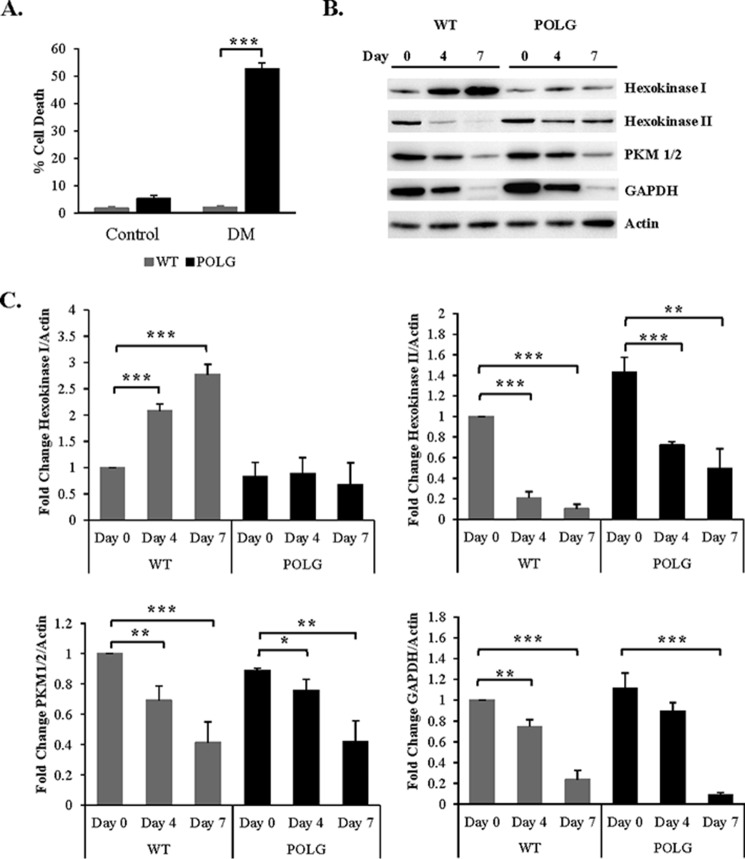
Studies have found that differentiation is associated with a metabolic transition from glycolysis to oxidative phosphorylation (29, 32). Because our data indicate that the POLG CPCs do not utilize oxidative phosphorylation for ATP production and differentiation is associated with an increase in energy demand, we examined the effect of differentiation on cell survival in these cells. We observed that a significant number of POLG CPCs underwent cell death in DM (Fig. 8A). To gain further insight into the metabolic switch that occurs during differentiation, we analyzed levels of glycolytic enzymes in WT and POLG CPCs. We found that mitochondria-associated hexokinase I increased in WT during differentiation but remained low in POLG CPCs. Hexokinase II, PKM1/2, and GAPDH decreased with time upon incubation in DM in both WT and POLG CPCs (Fig. 8, B and C). Taken together, these results suggest that both WT and POLG CPCs down-regulate cytosolic glycolysis during differentiation. However, because POLG CPCs are unable to concurrently activate oxidative phosphorylation, the resulting energy deficiency combined with increased energy demand leads to activation of cell death.
FIGURE 8.
Activation of differentiation leads to down-regulation of cytosolic glycolytic enzymes in both WT and POLG CPCs. A, WT and POLG CPCs were incubated in differentiation medium, and cell death was assessed using YO-PRO-1 staining 4 days later (n = 3). B, representative Western blots of glycolytic enzymes after incubation in DM. C, quantitation of hexokinase I, hexokinase II, PKM1/2, and GAPDH at different time points after differentiation (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
POLG CPCs Have Increased Mitophagy
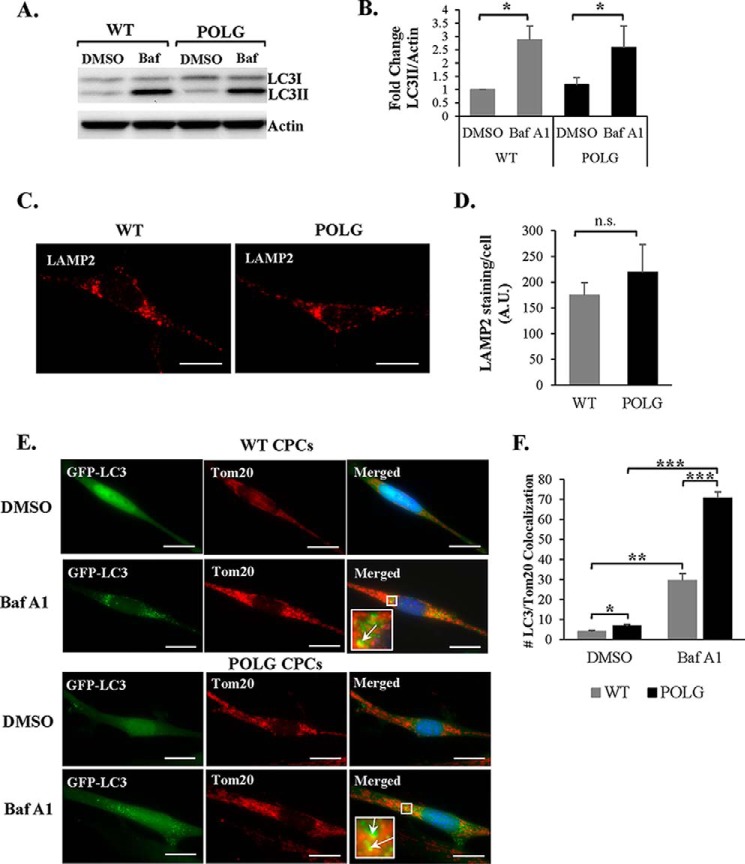
Loss of mitochondrial membrane potential is a known trigger for mitochondrial autophagy or mitophagy. Mitophagy is a process by which defective mitochondria are sequestered in autophagosomes and subsequently degraded in lysosomes (44). Because mitochondria in the POLG CPCs are impaired, we examined whether these cells had increased levels of mitophagy. First, we assessed whether autophagic flux was altered in POLG CPCs. To assess flux, cells were treated with vehicle or bafilomycin A1 to inhibit the fusion between autophagosomes and lysosomes. We found that both WT and POLG CPCs accumulated similar levels of the autophagy marker LC3II in the presence of bafilomycin A1 (Fig. 9, A and B). This suggests that autophagic flux is not different and that a similar number of autophagosomes are formed at steady state in both WT and POLG CPCs. We also assessed whether there were differences in lysosomal activity by LAMP2 staining, but we found no significant difference in lysosomal content between WT and POLG CPCs (Fig. 9, C and D). These data suggest that baseline autophagic flux is not altered in POLG CPCs.
FIGURE 9.
Mutant CPCs have normal autophagic flux but increased mitophagy. A, representative Western blots show similar levels of LC3II protein in both WT and POLG CPCs after treatment with 50 nm bafilomycin A1 (Baf) for 2 h. B, quantitation of LC3II protein levels (n = 3). Representative fluorescent images (C) and quantitation of LAMP2 staining in WT and POLG CPCs (n = 3) (D) are shown. Scale bar = 20 μm. A.U., arbitrary units. Representative fluorescent images (E) and quantitation of GFP-LC3 and Tom20 colocalization per cell in WT and POLG CPCs (n = 3) (F) are shown. Cells were treated with DMSO or 50 nm bafilomycin A1 for 3 h before fixation. Scale bar = 20 μm. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
Next, we assessed the level of mitophagy in WT and POLG CPCs by monitoring Tom20-labeled mitochondria sequestered in GFP-LC3-positive autophagosomes. Cells were treated with bafilomycin A1 to prevent degradation of autophagosomes containing mitochondria, as observed by increased formation of GFP-LC3 puncta in both WT and POLG CPCs (Fig. 9E). In addition, we found that POLG CPCs exhibited a significantly higher number of GFP-LC3-positive autophagosomes that co-localized with Tom20-labeled mitochondria compared with WT (Fig. 9F). This suggests that the rate of mitophagy is higher in POLG CPCs under baseline conditions. Interestingly, we also noted that the GFP-LC3-positive puncta were in general smaller in POLG CPCs than WT CPCs, but why this occurs is currently unknown.
Discussion
This study provides important new insights into the critical role of mitochondria in CPC survival and execution of the differentiation program. We demonstrate that acquiring mtDNA mutations causes a disruption in mitochondrial function, which leads to a decline in the replicative and regenerative capacities of the CPCs. Our data show that the mutant POLG CPCs rely solely on glycolysis for energy production and are unable to transition from glycolysis to mitochondrial respiration. Activation of the differentiation program leads to deactivation of glycolysis in both WT and POLG CPCs. However, because the mutant CPCs are unable to meet the increased energy demand, they undergo cell death instead. Thus, our findings demonstrate for the first time the importance of mtDNA integrity in CPC homeostasis and regenerative potential as well as the consequences of accumulating mtDNA mutations.
Aging is associated with accumulation of mtDNA damage in various tissues including the heart, and studies in mice have confirmed that mtDNA mutations are a contributor rather than a cause of the aging process. Mice with a mutant POLG exhibit multiple premature aging characteristics, including kyphosis, osteoporosis, hair loss, and anemia (25, 26). These phenotypes arise gradually along with mtDNA mutations, confirming that the accumulation of mtDNA mutations directly contributes to the phenotype. Moreover, the mice develop these phenotypes at an age where heart function is still unaffected (25), suggesting that post-mitotic tissues are less susceptible to mtDNA mutations than tissues with more rapid cellular turnover. Our study suggests that the susceptibility to mtDNA mutations also differs between cells within the same tissue. Adult cardiomyocytes are more protected against accumulation of mtDNA mutations than CPCs, and this protection might be due to their senescent phenotype compared with the proliferating CPCs.
In addition, the finding that cardiac structure and function are not affected by the presence of dysfunctional CPCs in 2-month-old POLG mice indicates that a functional pool of CPCs is not required to maintain cardiac homeostasis in young mice. This is consistent with the findings in mice with mutations in the gene-encoding c-kit (45). The Kit mutant mice have normal cardiac function at 4 months of age, but at 12 months of age they display cardiac hypertrophy and significantly impaired LV function compared with WT mice, suggesting that functional CPCs are more important for cardiac homeostasis in the aging myocardium. Interestingly, this study also observed that the Kit mutant mice had reduced myocardial vascularization. Because some studies suggest that CPCs generate very few myocytes (4, 5), it is possible that the c-kit+ CPCs are important for maintaining the vasculature in the aging myocardium rather than forming new myocytes.
Aging is coupled with an increase in senescence and apoptosis of CPCs, which gradually reduces the pool of functional CPCs within the myocardium (46). Decreased CPC number and senescence of the remaining CPCs is associated with the development of cardiac dysfunction and failure with age (47). Thus, our study suggests that accumulation of mtDNA mutations in CPCs with age contributes to their functional decline, and it is possible that the presence of dysfunctional POLG CPCs contributes to the more rapid aging cardiac phenotype in these mice compared with WT mice. After a myocardial infarction, endogenous CPCs migrate to the border zone where they participate in the repair process (33, 48). Future studies need to determine whether POLG mice are more susceptible to stress due to the pool of non-functional CPCs.
Stem and progenitor cell populations also exhibit different susceptibility to mtDNA mutations. For instance, hematopoietic stem cells, and their downstream progeny in POLG mutator mice have similar levels of mtDNA mutations. However, the mtDNA mutations have little effect on the hematopoietic stem cells, whereas the progenitors have impaired differentiation (49). It is speculated that this difference in susceptibility is due to the relative quiescent metabolism and reduced proliferative property of the hematopoietic stem cells. In addition, mtDNA mutations led to reduced self-renewal capacity of embryonic neuronal stem cells. These cells had a slight reduction in respiratory chain complexes I and IV but no differences in complexes II and III when compared with WT stem cells (50). In contrast, we found a significant reduction in several mitochondrial respiratory complexes in adult POLG CPCs, which may be due to differences between embryonic versus adult cells. Although CPCs exist in niches in the heart where they are relatively quiescent, they still undergo proliferation to maintain the pool. Also, by isolating and expanding the CPCs in culture, we increase the frequency of mtDNA mutations and accelerate their aging phenotype.
Our data suggest that mutations in mtDNA-encoded subunits in the POLG CPCs affect assembly of the respiratory complexes. We noted a significant reduction in both nuclear and mitochondria-encoded Complex IV subunits in the POLG CPCs. It is well established that mutations in one subunit can affect the stability of the entire respiratory complex. Genetic alterations leading to destabilization of one complex can also lead to secondary loss of the other complexes (51–53). Therefore, mutations in only one mtDNA-encoded subunit can have severe consequences for mitochondrial function and cell survival. We also found that mtDNA mutations affect mitochondrial dynamics. It has been reported that mitochondrial fusion is required for differentiation of embryonic stem cells into myocytes, and disruption of mitochondrial fusion proteins MFN1 and MFN2 in mouse embryonic stem cells impairs their differentiation into cardiac myocytes (54). Our data also show that there is an increase in MFN1 and MFN2 proteins, whereas there is a corresponding decrease in DRP1 upon differentiation of WT CPCs, suggesting that the balance is shifted toward fusion in these cells. In contrast, the balance between fission and fusion is disrupted in POLG CPCs, and the mitochondria have adopted a fragmented morphology. Thus, accumulation of mtDNA mutations also impairs mitochondrial dynamics in the CPCs.
Differentiation of cells is associated with a large increase in energy demand (55), and our data demonstrate that there is an increase in proteins involved in OXPHOS and a corresponding decrease in enzymes involved in cytosolic glycolysis in CPCs. The most surprising finding in our study is that the POLG CPCs rely solely on glycolysis for energy production instead of mitochondrial respiration. We observed no significant difference in cellular ATP levels between WT and mutant POLG CPCs, suggesting that the undifferentiated POLG CPCs do not rely on mitochondria for ATP production and can compensate for the defect in mitochondrial respiration via glycolysis. Another surprising finding is that the mutant POLG CPCs are unable to switch to mitochondrial respiration in response to differentiation but still turn off the glycolytic pathway. It is likely that the failure to activate mitochondrial respiration leads to activation of the cell death observed upon differentiation of the POLG CPCs. This is in contrast to iPS cells generated from POLG mice, which become hyperactive in glycolytic activity during differentiation to compensate for the defects in OXPHOS (56). Overall, this suggests that acquiring mtDNA mutations have differential effects in the undifferentiated versus the differentiated CPC. CPCs do not require a lot of energy and can rely on glycolysis for self-renewal and proliferation. However, defective mtDNA will lead to a failure in making the metabolic switch to mitochondrial respiration during differentiation, and cell death will be activated instead.
Loss of mitochondrial membrane potential is a known activator of mitophagy (57, 58). Our data show that POLG CPCs exhibit increased mitophagy at baseline compared with WT. This is not surprising considering the degree of mitochondrial dysfunction in the mutant CPCs. Although mitophagy appears to be increased in the mutant CPCs, it is not sufficient to replace the dysfunctional mitochondria in these cells. It is known that mitochondrial degradation is coupled to mitochondrial biogenesis (59), but the underlying mechanism of this cross-talk is still unclear and under intense investigation. Our data suggest that the mitochondrial biogenesis program is impaired in these cells. Thus, it is possible that the reduced mitochondrial biogenesis also affects the rate of mitochondrial turnover. An imbalance will lead to inefficient turnover of aberrant mitochondria. However, the relationship between mitochondrial degradation and synthesis in the POLG CPCs needs to be further investigated.
The transplantation of autologous CPCs and many other stem cells to treat heart failure show promise in clinical trials (9, 10, 60–62). However, the fitness of the donor cell is a crucial parameter in cell transplantation therapies regardless of the cell type used. Reduced function of the transplanted cells will lead to poor retention and impaired regenerative capacity. Data from our study demonstrate that as mtDNA mutations accumulate with age in CPCs, their therapeutic potential is reduced. Also, as the aging CPCs accumulate mtDNA mutations, they will be less likely to survive transplantation into the hostile environment of a diseased heart. However, the mechanistic basis for the age-related decline in CPC function is very complex and involves multiple changes at the molecular and cellular levels. Although our studies suggest that accumulation of mtDNA damage is an important contributor to the functional decline of CPCs, additional factors will also contribute to the aging phenotype. For instance, reduced levels of the nucleolar protein nucleostemin contribute to senescence in CPCs (12), and defects in EphA2 signaling in aged CPCs impair their migration (13). Increased knowledge of the molecular basis for CPC function can be used to select for the most optimal autologous CPCs to be transplanted back into a patient to ensure maximal myocardial regeneration and repair. Overall, additional identification and characterization of specific pathways that contribute to dysfunction of CPCs, as well as other stem cell populations, will have great impact on the field of regenerative medicine.
Author Contributions
A. B. G. and A. M. O. designed the study, analyzed the experiments, and wrote the paper. A. M. O., E. R. G., D. A. K., I. L. B., S.-B. O., and A. N. M. designed, performed. and analyzed the experiments. T. A. P. generated the POLG mice. M. A. S. provided reagents and contributed to experimental design and analysis. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL087023, R01HL101217, and P01HL085577 (to A. B. G.), Institutional Training Grant T32GM007752 (NIGMS; University of California San Diego Graduate Training Program in Cellular and Molecular Pharmacology) and National Research Service Award Predoctoral Fellowship F31HL123309 (to A. M. O.), and R01HL067245, R37HL091102, R01HL105759, R01HL113656, R01HL113647, and R01HL122525 (to M. A. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- CPC
- cardiac progenitor cell
- POLG
- mitochondrial DNA polymerase gamma
- OXPHOS
- oxidative phosphorylation
- DM
- differentiation medium
- Tom20
- translocase of outer membrane
- COXIV
- mitochondrial complex IV (cytochrome c oxidase)
- PGC-1
- peroxisome proliferator-activated receptor γ coactivator 1
- DRP1
- dynamin-related protein 1
- MFN1/2
- mitofusin-1/2
- PKM1/2
- pyruvate kinase muscle isozymes 1/2
- LAMP2
- lysosome-associated membrane protein-2
- GFP-LC3
- green fluorescent protein-microtubule-associated protein 1 light chain 3
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qPCR
- quantitative PCR.
References
- 1. Leri A., Kajstura J., Anversa P. (2011) Role of cardiac stem cells in cardiac pathophysiology: a paradigm shift in human myocardial biology. Circ. Res. 109, 941–961 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2. Beltrami A. P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S., Kasahara H., Rota M., Musso E., Urbanek K., Leri A., Kajstura J., Nadal-Ginard B., Anversa P. (2003) Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114, 763–776 [DOI] [PubMed] [Google Scholar]
- 3. Ellison G. M., Vicinanza C., Smith A. J., Aquila I., Leone A., Waring C. D., Henning B. J., Stirparo G. G., Papait R., Scarfò M., Agosti V., Viglietto G., Condorelli G., Indolfi C., Ottolenghi S., Torella D., Nadal-Ginard B. (2013) Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 154, 827–842 [DOI] [PubMed] [Google Scholar]
- 4. van Berlo J. H., Kanisicak O., Maillet M., Vagnozzi R. J., Karch J., Lin S. C., Middleton R. C., Marbán E., Molkentin J. D. (2014) c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509, 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Senyo S. E., Steinhauser M. L., Pizzimenti C. L., Yang V. K., Cai L., Wang M., Wu T. D., Guerquin-Kern J. L., Lechene C. P., Lee R. T. (2013) Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Q., Guo Y., Ou Q., Chen N., Wu W. J., Yuan F., O'Brien E., Wang T., Luo L., Hunt G. N., Zhu X., Bolli R. (2011) Intracoronary administration of cardiac stem cells in mice: a new, improved technique for cell therapy in murine models. Basic Res. Cardiol. 106, 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fischer K. M., Cottage C. T., Wu W., Din S., Gude N. A., Avitabile D., Quijada P., Collins B. L., Fransioli J., Sussman M. A. (2009) Enhancement of myocardial regeneration through genetic engineering of cardiac progenitor cells expressing Pim-1 kinase. Circulation 120, 2077–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Angelis A., Piegari E., Cappetta D., Marino L., Filippelli A., Berrino L., Ferreira-Martins J., Zheng H., Hosoda T., Rota M., Urbanek K., Kajstura J., Leri A., Rossi F., Anversa P. (2010) Anthracycline cardiomyopathy is mediated by depletion of the cardiac stem cell pool and is rescued by restoration of progenitor cell function. Circulation 121, 276–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolli R., Chugh A. R., D'Amario D., Loughran J. H., Stoddard M. F., Ikram S., Beache G. M., Wagner S. G., Leri A., Hosoda T., Sanada F., Elmore J. B., Goichberg P., Cappetta D., Solankhi N. K., Fahsah I., Rokosh D. G., Slaughter M. S., Kajstura J., Anversa P. (2011) Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet 378, 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Chugh A. R., Beache G. M., Loughran J. H., Mewton N., Elmore J. B., Kajstura J., Pappas P., Tatooles A., Stoddard M. F., Lima J. A., Slaughter M. S., Anversa P., Bolli R. (2012) Administration of cardiac stem cells in patients with ischemic cardiomyopathy: the SCIPIO trial: surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation 126, S54–S64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mohsin S., Khan M., Nguyen J., Alkatib M., Siddiqi S., Hariharan N., Wallach K., Monsanto M., Gude N., Dembitsky W., Sussman M. A. (2013) Rejuvenation of human cardiac progenitor cells with Pim-1 kinase. Circ. Res. 113, 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hariharan N., Quijada P., Mohsin S., Joyo A., Samse K., Monsanto M., De La Torre A., Avitabile D., Ormachea L., McGregor M. J., Tsai E. J., Sussman M. A. (2015) Nucleostemin rejuvenates cardiac progenitor cells and antagonizes myocardial aging. J. Am. Coll. Cardiol. 65, 133–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goichberg P., Kannappan R., Cimini M., Bai Y., Sanada F., Sorrentino A., Signore S., Kajstura J., Rota M., Anversa P., Leri A. (2013) Age-associated defects in EphA2 signaling impair the migration of human cardiac progenitor cells. Circulation 128, 2211–2223 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Nunnari J., Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atamna H. (2004) Heme, iron, and the mitochondrial decay of ageing. Ageing Res. Rev. 3, 303–318 [DOI] [PubMed] [Google Scholar]
- 16. Xu X., Duan S., Yi F., Ocampo A., Liu G. H., Izpisua Belmonte J. C. (2013) Mitochondrial regulation in pluripotent stem cells. Cell Metab. 18, 325–332 [DOI] [PubMed] [Google Scholar]
- 17. Dai D. F., Santana L. F., Vermulst M., Tomazela D. M., Emond M. J., MacCoss M. J., Gollahon K., Martin G. M., Loeb L. A., Ladiges W. C., Rabinovitch P. S. (2009) Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation 119, 2789–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corral-Debrinski M., Horton T., Lott M. T., Shoffner J. M., Beal M. F., Wallace D. C. (1992) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 2, 324–329 [DOI] [PubMed] [Google Scholar]
- 19. Wanagat J., Wolff M. R., Aiken J. M. (2002) Age-associated changes in function, structure and mitochondrial genetic and enzymatic abnormalities in the Fischer 344 x Brown Norway F(1) hybrid rat heart. J. Mol. Cell Cardiol. 34, 17–28 [DOI] [PubMed] [Google Scholar]
- 20. Khaidakov M., Heflich R. H., Manjanatha M. G., Myers M. B., Aidoo A. (2003) Accumulation of point mutations in mitochondrial DNA of aging mice. Mutat. Res. 526, 1–7 [DOI] [PubMed] [Google Scholar]
- 21. Bratic A., Larsson N. G. (2013) The role of mitochondria in aging. J. Clin. Invest. 123, 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lebrecht D., Walker U. A. (2007) Role of mtDNA lesions in anthracycline cardiotoxicity. Cardiovasc. Toxicol. 7, 108–113 [DOI] [PubMed] [Google Scholar]
- 23. Frerichs F. C., Dingemans K. P., Brinkman K. (2002) Cardiomyopathy with mitochondrial damage associated with nucleoside reverse-transcriptase inhibitors. N. Engl. J. Med. 347, 1895–1896 [DOI] [PubMed] [Google Scholar]
- 24. Ide T., Tsutsui H., Hayashidani S., Kang D., Suematsu N., Nakamura K., Utsumi H., Hamasaki N., Takeshita A. (2001) Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ. Res. 88, 529–535 [DOI] [PubMed] [Google Scholar]
- 25. Kujoth G. C., Hiona A., Pugh T. D., Someya S., Panzer K., Wohlgemuth S. E., Hofer T., Seo A. Y., Sullivan R., Jobling W. A., Morrow J. D., Van Remmen H., Sedivy J. M., Yamasoba T., Tanokura M., Weindruch R., Leeuwenburgh C., Prolla T. A. (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484 [DOI] [PubMed] [Google Scholar]
- 26. Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., Törnell J., Jacobs H. T., Larsson N. G. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 [DOI] [PubMed] [Google Scholar]
- 27. Dai D. F., Chen T., Wanagat J., Laflamme M., Marcinek D. J., Emond M. J., Ngo C. P., Prolla T. A., Rabinovitch P. S. (2010) Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell 9, 536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W., Osenbroch P., Skinnes R., Esbensen Y., Bjørås M., Eide L. (2010) Mitochondrial DNA integrity is essential for mitochondrial maturation during differentiation of neural stem cells. Stem Cells 28, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 29. Chung S., Dzeja P. P., Faustino R. S., Perez-Terzic C., Behfar A., Terzic A. (2007) Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells Nat. Clin. Pract. Cardiovasc. Med. 4, S60–S67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen C. T., Shih Y. R., Kuo T. K., Lee O. K., Wei Y. H. (2008) Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26, 960–968 [DOI] [PubMed] [Google Scholar]
- 31. Spitkovsky D., Sasse P., Kolossov E., Böttinger C., Fleischmann B. K., Hescheler J., Wiesner R. J. (2004) Activity of complex III of the mitochondrial electron transport chain is essential for early heart muscle cell differentiation. FASEB J. 18, 1300–1302 [DOI] [PubMed] [Google Scholar]
- 32. Chung S., Arrell D. K., Faustino R. S., Terzic A., Dzeja P. P. (2010) Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J. Mol. Cell Cardiol. 48, 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fransioli J., Bailey B., Gude N. A., Cottage C. T., Muraski J. A., Emmanuel G., Wu W., Alvarez R., Rubio M., Ottolenghi S., Schaefer E., Sussman M. A. (2008) Evolution of the c-kit-positive cell response to pathological challenge in the myocardium. Stem Cells 26, 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luchessi A. D., Cambiaghi T. D., Hirabara S. M., Lambertucci R. H., Silveira L. R., Baptista I. L., Moriscot A. S., Costa-Neto C. M., Curi R. (2009) Involvement of eukaryotic translation initiation factor 5A (eIF5A) in skeletal muscle stem cell differentiation. J. Cell. Physiol. 218, 480–489 [DOI] [PubMed] [Google Scholar]
- 35. Kubli D. A., Zhang X., Lee Y., Hanna R. A., Quinsay M. N., Nguyen C. K., Jimenez R., Petrosyan S., Murphy A. N., Gustafsson A. B. (2013) Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J. Biol. Chem. 288, 915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rikka S., Quinsay M. N., Thomas R. L., Kubli D. A., Zhang X., Murphy A. N., Gustafsson Å. B. (2011) Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanna R. A., Quinsay M. N., Orogo A. M., Giang K., Rikka S., Gustafsson Å. B. (2012) Microtubule-associated protein 1 light chain 3 (LC3) Interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094–19104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linke A., Müller P., Nurzynska D., Casarsa C., Torella D., Nascimbene A., Castaldo C., Cascapera S., Böhm M., Quaini F., Urbanek K., Leri A., Hintze T. H., Kajstura J., Anversa P. (2005) Stem cells in the dog heart are self-renewing, clonogenic, and multipotent and regenerate infarcted myocardium, improving cardiac function. Proc. Natl. Acad. Sci. U.S.A. 102, 8966–8971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bearzi C., Rota M., Hosoda T., Tillmanns J., Nascimbene A., De Angelis A., Yasuzawa-Amano S., Trofimova I., Siggins R. W., Lecapitaine N., Cascapera S., Beltrami A. P., D'Alessandro D. A., Zias E., Quaini F., Urbanek K., Michler R. E., Bolli R., Kajstura J., Leri A., Anversa P. (2007) Human cardiac stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 14068–14073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kasahara A., Scorrano L. (2014) Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 24, 761–770 [DOI] [PubMed] [Google Scholar]
- 41. Shefer G., Van de Mark D. P., Richardson J. B., Yablonka-Reuveni Z. (2006) Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev. Biol. 294, 50–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shavlakadze T., McGeachie J., Grounds M. D. (2010) Delayed but excellent myogenic stem cell response of regenerating geriatric skeletal muscles in mice. Biogerontology 11, 363–376 [DOI] [PubMed] [Google Scholar]
- 43. Hatefi Y. (1985) The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54, 1015–1069 [DOI] [PubMed] [Google Scholar]
- 44. Kubli D. A., Gustafsson Å. B. (2012) Mitochondria and mitophagy: the yin and yang of cell death control. Circ. Res. 111, 1208–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ye L., Zhang E. Y., Xiong Q., Astle C. M., Zhang P., Li Q., From A. H., Harrison D. E., Zhang J. J. (2012) Aging kit mutant mice develop cardiomyopathy. PloS ONE 7, e33407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cesselli D., Beltrami A. P., D'Aurizio F., Marcon P., Bergamin N., Toffoletto B., Pandolfi M., Puppato E., Marino L., Signore S., Livi U., Verardo R., Piazza S., Marchionni L., Fiorini C., Schneider C., Hosoda T., Rota M., Kajstura J., Anversa P., Beltrami C. A., Leri A. (2011) Effects of age and heart failure on human cardiac stem cell function. Am. J. Pathol. 179, 349–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Torella D., Rota M., Nurzynska D., Musso E., Monsen A., Shiraishi I., Zias E., Walsh K., Rosenzweig A., Sussman M. A., Urbanek K., Nadal-Ginard B., Kajstura J., Anversa P., Leri A. (2004) Cardiac stem cell and myocyte aging, heart failure, and insulin-like growth factor-1 overexpression. Circ. Res. 94, 514–524 [DOI] [PubMed] [Google Scholar]
- 48. Huang C., Zhang X., Ramil J. M., Rikka S., Kim L., Lee Y., Gude N. A., Thistlethwaite P. A., Sussman M. A., Gottlieb R. A., Gustafsson A. B. (2010) Juvenile exposure to anthracyclines impairs cardiac progenitor cell function and vascularization resulting in greater susceptibility to stress-induced myocardial injury in adult mice. Circulation 121, 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Norddahl G. L., Pronk C. J., Wahlestedt M., Sten G., Nygren J. M., Ugale A., Sigvardsson M., Bryder D. (2011) Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell 8, 499–510 [DOI] [PubMed] [Google Scholar]
- 50. Ahlqvist K. J., Hämäläinen R. H., Yatsuga S., Uutela M., Terzioglu M., Götz A., Forsström S., Salven P., Angers-Loustau A., Kopra O. H., Tyynismaa H., Larsson N. G., Wartiovaara K., Prolla T., Trifunovic A., Suomalainen A. (2012) Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 15, 100–109 [DOI] [PubMed] [Google Scholar]
- 51. Schägger H., de Coo R., Bauer M. F., Hofmann S., Godinot C., Brandt U. (2004) Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279, 36349–36353 [DOI] [PubMed] [Google Scholar]
- 52. Acín-Pérez R., Bayona-Bafaluy M. P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C. T., Enríquez J. A. (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T. (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 361, 462–469 [DOI] [PubMed] [Google Scholar]
- 54. Kasahara A., Cipolat S., Chen Y., Dorn G. W. 2nd, Scorrano L. (2013) Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 342, 734–737 [DOI] [PubMed] [Google Scholar]
- 55. Gaspar J. A., Doss M. X., Hengstler J. G., Cadenas C., Hescheler J., Sachinidis A. (2014) Unique metabolic features of stem cells, cardiomyocytes, and their progenitors. Circ. Res. 114, 1346–1360 [DOI] [PubMed] [Google Scholar]
- 56. Wahlestedt M., Ameur A., Moraghebi R., Norddahl G. L., Sten G., Woods N. B., Bryder D. (2014) Somatic cells with a heavy mitochondrial DNA mutational load render induced pluripotent stem cells with distinct differentiation defects. Stem Cells 32, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 57. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Andres A. M., Stotland A., Queliconi B. B., Gottlieb R. A. (2015) A time to reap, a time to sow: mitophagy and biogenesis in cardiac pathophysiology. J. Mol. Cell. Cardiol. 78, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hare J. M., Fishman J. E., Gerstenblith G., DiFede Velazquez D. L., Zambrano J. P., Suncion V. Y., Tracy M., Ghersin E., Johnston P. V., Brinker J. A., Breton E., Davis-Sproul J., Schulman I. H., Byrnes J., Mendizabal A. M., Lowery M. H., Rouy D., Altman P., Wong Po Foo C., Ruiz P., Amador A., Da Silva J., McNiece I. K., Heldman A. W., George R., Lardo A. (2012) Comparison of allogeneic vs autologous bone marrow-derived mesenchymal stem cells delivered by transendocardial injection in patients with ischemic cardiomyopathy: the POSEIDON randomized trial. JAMA 308, 2369–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Heldman A. W., DiFede D. L., Fishman J. E., Zambrano J. P., Trachtenberg B. H., Karantalis V., Mushtaq M., Williams A. R., Suncion V. Y., McNiece I. K., Ghersin E., Soto V., Lopera G., Miki R., Willens H., Hendel R., Mitrani R., Pattany P., Feigenbaum G., Oskouei B., Byrnes J., Lowery M. H., Sierra J., Pujol M. V., Delgado C., Gonzalez P. J., Rodriguez J. E., Bagno L. L., Rouy D., Altman P., Foo C. W., da Silva J., Anderson E., Schwarz R., Mendizabal A., Hare J. M. (2014) Transendocardial mesenchymal stem cells and mononuclear bone marrow cells for ischemic cardiomyopathy: the TAC-HFT randomized trial. JAMA 311, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Karantalis V., DiFede D. L., Gerstenblith G., Pham S., Symes J., Zambrano J. P., Fishman J., Pattany P., McNiece I., Conte J., Schulman S., Wu K., Shah A., Breton E., Davis-Sproul J., Schwarz R., Feigenbaum G., Mushtaq M., Suncion V. Y., Lardo A. C., Borrello I., Mendizabal A., Karas T. Z., Byrnes J., Lowery M., Heldman A. W., Hare J. M. (2014) Autologous mesenchymal stem cells produce concordant improvements in regional function, tissue perfusion, and fibrotic burden when administered to patients undergoing coronary artery bypass grafting: The Prospective Randomized Study of Mesenchymal Stem Cell Therapy in Patients Undergoing Cardiac Surgery (PROMETHEUS) trial. Circ. Res. 114, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]