Background: The interleukin-4-inducing principle from Schistosoma mansoni eggs (IPSE/α-1) triggers basophils to release interleukin-4 and interleukin-13 in an IgE-dependent but antigen-independent way.
Results: Structural analysis identified IPSE/α-1 as a new member of the βγ-crystallin superfamily with a unique IgE-binding loop.
Conclusion: IPSE/α-1 activates basophils via IgE-binding crystallin folds.
Significance: Schistosomes use unique mechanisms to manipulate the host's immune response.
Keywords: crystal structure, crystallin, interleukin, nuclear magnetic resonance (NMR), Schistosoma mansoni, basophil
Abstract
The IL-4-inducing principle from Schistosoma mansoni eggs (IPSE/α-1), the major secretory product of eggs from the parasitic worm S. mansoni, efficiently triggers basophils to release the immunomodulatory key cytokine interleukin-4. Activation by IPSE/α-1 requires the presence of IgE on the basophils, but the detailed molecular mechanism underlying activation is unknown. NMR and crystallographic analysis of IPSEΔNLS, a monomeric IPSE/α-1 mutant, revealed that IPSE/α-1 is a new member of the βγ-crystallin superfamily. We demonstrate that this molecule is a general immunoglobulin-binding factor with highest affinity for IgE. NMR binding studies of IPSEΔNLS with the 180-kDa molecule IgE identified a large positively charged binding surface that includes a flexible loop, which is unique to the IPSE/α-1 crystallin fold. Mutational analysis of amino acids in the binding interface showed that residues contributing to IgE binding are important for IgE-dependent activation of basophils. As IPSE/α-1 is unable to cross-link IgE, we propose that this molecule, by taking advantage of its unique IgE-binding crystallin fold, activates basophils by a novel, cross-linking-independent mechanism.
Introduction
Schistosomes are parasitic worms causing a chronic disease with more than 200,000 deaths per year alone in sub-Saharan Africa (World Health Organization, Schistosomiasis Facts, 2012). During acute infection, schistosome eggs induce an inflammatory T helper type 2 (Th2) response characterized by high immunoglobulin (Ig) E titers and eosinophilia (1). Subsequently, the Th2 response is down-regulated by anti-inflammatory mechanisms to restrict host tissue damage (1). In this context, the cytokine interleukin (IL)-4 plays an important role, first as key inducer cytokine for a Th2 response (2) and second as a key factor for limiting extensive inflammation. The absence of IL-4 signaling in Schistosoma mansoni-infected mice, which are unable to produce IL-4 (IL-4−/− mice) or to react to IL-4 (IL-4 receptor α-chain−/− mice), is associated with massive intestinal inflammation and rapid death (3, 4). Recent studies suggest that basophils are involved in anti-helminth immunity (5, 6). They have been shown to migrate to parasite-affected tissues, where they become fully activated and represent the major source of IL-4. However, the mechanisms by which the parasites activate basophils remained elusive.
Previously, we identified a homodimeric 35-kDa glycoprotein secreted from live S. mansoni eggs, which induces the release of IL-4 from human and murine basophils (7, 8). This protein was called IPSE (IL-4-inducing principle from S. mansoni eggs) (7) but was subsequently renamed IPSE/α-1 (9) because it was found to be identical with the major excretory/secretory schistosome egg antigen α-1 described earlier (10). IPSE/α-1 has a unique sequence; apart from a predicted putative greek key motif, there is no similarity to any other known protein, except to translated cDNA sequences from Schistosoma japonicum and translated genomic sequences from Schistosoma haematobium (11). IPSE/α-1 triggers the release of IL-4 from basophils of nonsensitized healthy human donors, and intravenous injection of IPSE/α-1 into mice leads to basophil-IL-4 release in the liver, which, apart from the gut, is the major site of egg deposition during S. mansoni infection (8). Basophil activation by IPSE/α-1 was found to depend on the presence of IgE on the surface of the basophils irrespective of IgE's antigen specificity (7, 12). IgE-dependent, antigen-independent basophil activation is well known from multivalent lectins or B cell superantigens, which cross-link IgE via binding to its carbohydrate side chains or directly to its immunoglobulin backbone, respectively (13, 14). However, molecular details of the mechanism of action of IPSE/α-1 are not yet known.
Here, we report NMR and crystallographic structures of IPSEΔNLS, a monomeric deletion mutant of IPSE/α-1. IPSEΔNLS lacks a stretch of 10 amino acids at the C terminus comprising a putative nuclear localization sequence (NLS)7 (15) and Cys-132, which mediates a disulfide bridge for dimer formation of the native molecule (16). The structure consists of two symmetrically arranged greek key motifs and exhibits high structural similarity with members of the βγ-crystallin superfamily. Biochemical studies show that IPSE/α-1 is a general immunoglobulin-binding factor binding to all isotypes but with highest affinity to IgE. NMR chemical shift analysis combined with functional assays suggest that IPSE/α-1 interacts with IgE via a large surface area that includes a loop comprising positively charged and aromatic amino acids, which are critical for IgE binding. In contrast to lectins and B cell superantigens, our data obtained with soluble or immobilized molecules suggest that IPSE/α-1 is not able to cross-link IgE. Thus, we propose that IPSE/α-1 activates basophils by a novel IgE-dependent, antigen-independent mechanism that does not involve cross-linking of IgE.
Experimental Procedures
Sample Preparation
Unlabeled IPSE/α-1 protein has been prepared as described elsewhere (17). For preparation of uniformly 13C,15N- and 15N-labeled samples, bacteria were grown in M9 medium supplemented with 13C-labeled glucose and/or 15NH4Cl, respectively. For preparation of 2H/15N/13C-labeled IPSEΔNLS, the same protocol was employed but using [U-2H13C]glucose and growing bacteria in 100% D2O. Point mutations were introduced in pProExHTb-IPSEΔNLS and pProExHTb IPSE using the QuikChange site-directed mutagenesis protocol (Invitrogen). The corresponding proteins contain two additional residues at the N terminus from a tobacco etch virus protease cleavage site, which was introduced by the expression vector.
NMR spectroscopy was carried out on a Bruker Avance III 750 MHz spectrometer equipped with a TXI probehead, a Bruker Avance III 600 MHz spectrometer equipped with a cryogenic TCI probehead, or on an Avance I 900 MHz spectrometer equipped with a cryogenic TXI probehead. Unless stated otherwise, all experiments were carried out on a 1 mm uniformly 13C/15N-labeled IPSEΔNLS at 298 K. Spectra were processed with NMRPipe (18) and analyzed with Sparky3. For backbone and side chain assignment CBCA(CO)NH, CBCANH, and (H)CCH-TOCSY (19), spectra were recorded (20). Distance information was obtained from 15N- and 13C-edited NOESY spectra with a mixing time of 70 ms. 15N R1 and R2 relaxation rates and {1H}-15N heteronuclear NOE data were measured at 750 MHz proton Larmor frequency and 298 K as described (21). Relaxation delays of 0.0216, 0.054, 0.27, 0.81, 1.08, 1.4, and 1.62 s were used to measure R1 and 0.0144, 0.0288, 0.0576, 0.0864, 0.115, 0.173, and 0.173 were used to extract R2 rates. Errors were estimated from duplicate measurements of two delays and Monte Carlo simulation. Uncertainty of the ratio of R2/R1 relaxation rates was determined by error propagation. Residue-specific tumbling correlation times, τc, were calculated as described elsewhere (21).
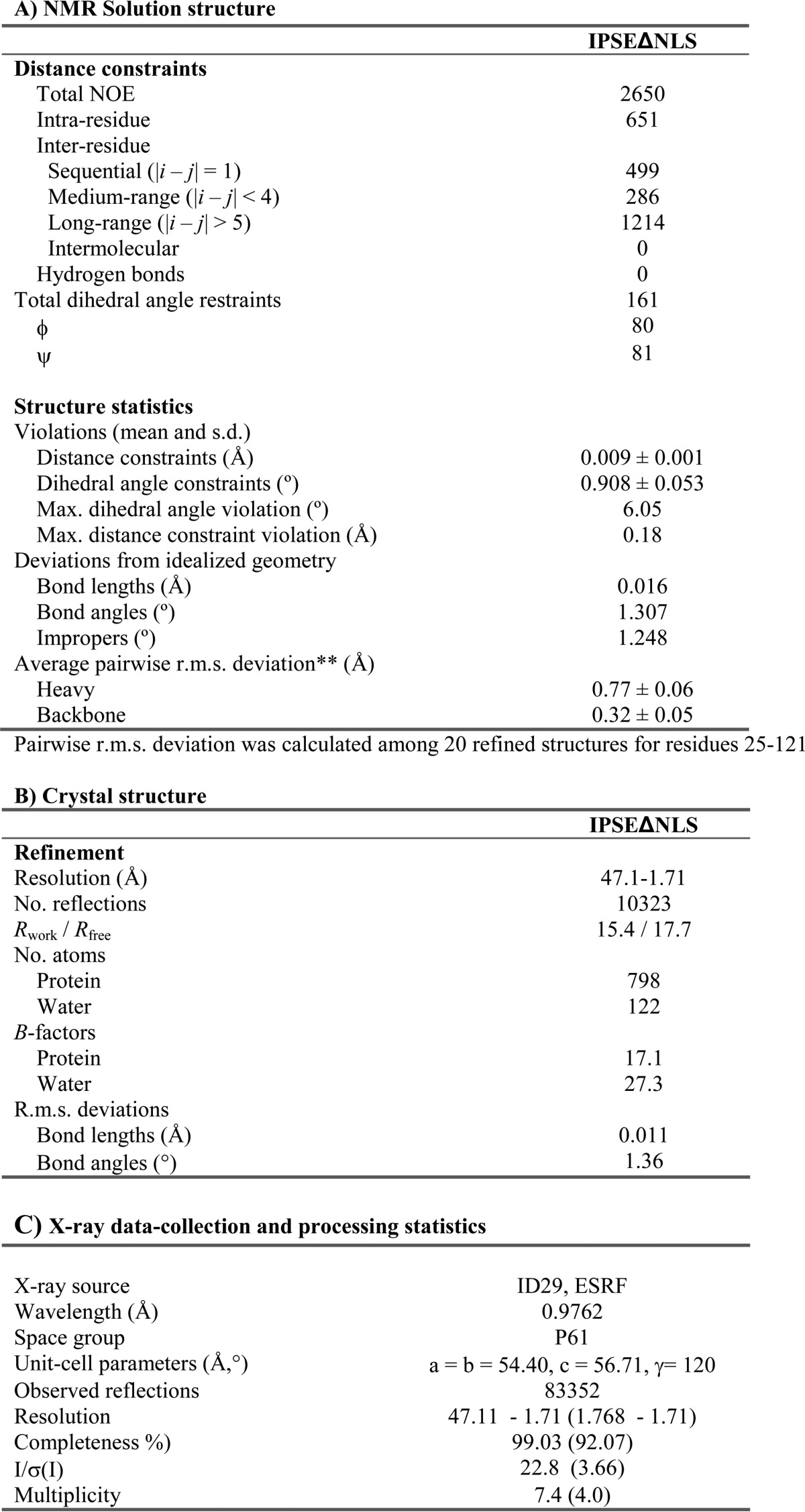
Structure calculation was performed by automated NOE cross-peak assignment using the software CYANA 3.0 (22). Automatically assigned NOEs and completeness of the NOE cross-peaks were inspected manually. Distance restraints from the CYANA calculation and TALOS+ (23)-derived torsion angles were used in a water refinement calculation (24) applying the RECOORD protocol (25). Ramachandran plot statistics after refinement were as follows: 91.1% of residues in most favored regions, 8.9% in additional allowed regions, 0% in generously allowed regions, and 0% in disallowed regions. Quality of the NMR-derived structure ensemble was validated using the iCING web server as well as PROCHECK (26) and WHATCHECK (27). Molecular images were generated using PyMOL (Delano Scientific).
NMR Titrations
Uniformly 2H/15N/13C-labeled IPSEΔNLS was stepwise added to 100 μm IgE (Dianova) at ratios of 0.5:1 and 1:1 (IPSEΔNLS/IgE), and chemical shifts were recorded using CRINEPT-HMQC spectra (28). A CRINEPT spectrum of 100 μm 2H/15N/13C-labeled IPSEΔNLS was recorded as a reference.
X-ray Structure Determination
IPSEΔNLS was solved by molecular replacement using Phaser as implemented in CCP4 6.1.131 with the modified NMR structure as a search model. Coordinates of IPSEΔNLS were refined using REFMAC 5.6.0119 alternating with rounds of manual model building using the program Coot. Initial crystallization trials were performed in 96-well sitting drop vapor diffusion plates at 20 °C at a protein concentration of 14 mg ml−1 using commercially available screens. Two conditions yielded small crystals, which were optimized through a combination of OptiSalt screening (Qiagen) and customized grid screens resulting in well diffracting crystals, which appeared after at least 1 week. Crystals were flash-frozen at 100 K in precipitant solution containing 15% glycerol. Diffraction data were collected at a wavelength of 0.9762 Å using an ADSC Q315R detector at ID29 at the ESRF. The data were indexed and integrated using DENZO and scaled with SCALEPACK (29). The crystals belong to the space group P61 with cell axes of a = b = 54.4 Å, c = 56.1 Å and diffracted to a resolution of 1.71 Å with one molecule in the asymmetric unit. The structure was determined by molecular replacement with PHASER (30) using the IPSE NMR structure with all the loops trimmed as a search model. The structure was refined using Refmac (31) with isotropic B-factors and manual rebuilding of protein was done with COOT (32). For more details see Table 1. The final model has no outliers in the Ramachandran plot, and 98.92% of all residues are in the favored region. A final check of the stereochemical quality of the final model was carried out using the program MolProbity (33).
TABLE 1.
Refinement statistics for IPSEΔNLS
Computational Modeling of a IPSEΔNLS·IgE Fab complex structure was performed using the ClusPro web server (34). The lowest energy structure of the IPSEΔNLS NMR structure ensemble and an IgE Fc structure from the protein database (PDB accession code 2WQR) were used for the rigid body docking. Unstructured terminal residues were removed from the IPSEΔNLS structure. Attractive restraints were introduced for residues that were significantly broadened or experienced chemical shift perturbations in the NMR titration. Repulsive restraints were used for unaffected residues.
Size Exclusion Chromatography
IPSE/α-1 dimer was mixed with IgE or IgG in a 1:1 molar ratio, preincubated for 30 min at room temperature, and applied to a Superdex 200 10/300 GL column (GE Healthcare). Chromatography was performed in PBS, pH 7.4, at 0.4 ml/min at the ÄKTA-Purifier 10 (GE Healthcare). Fractions of 0.5 ml were collected and analyzed by SDS-PAGE and silver staining.
Surface Plasmon Resonance
Experiments were performed on a BIAcore 3000 (GE Healthcare) and repeated at least twice. IgE or IgG was immobilized on a CM5 chip by standard amine coupling chemistry (for IPSE/α-1 dimer: IgE, 969 RU; IgG, 4435 RU; for IPSEΔNLS monomer: IgE, 5058 RU; IgG, 8428 RU). One flow cell was activated and deactivated without ligand for control purposes. Unspecific binding to the control flow cell was subtracted from the binding to the ligand-loaded flow cells. Running buffer was PBS, pH 7.5. Regeneration was performed with 0.075% SDS in PBS, pH 7.5. Measurements were performed under the following conditions: immobilized IgE, 100–700 nm IPSE/α-1 dimer with a flow rate of 10 μl/min for 10 min at 25 °C or 62.5–8000 nm IPSEΔNLS monomer with a flow rate of 10 μl/min for 10 min at 25 °C; immobilized IgG, 150–700 nm IPSE/α-1 dimer with a flow rate of 5 μl/min for 20 min at 25 °C or 125–16,000 nm IPSEΔNLS monomer with a flow rate of 10 μl/min for 10 min at 25 °C.
SDS-PAGE and Western blotting were performed according to standard protocols as described previously (7). Materials were obtained from the following suppliers: human IgG (Octagam); human IgE and human IgG Fab and Fc (Acris); human IgA (Dianova); human IgM (Medac). Immunoglobulins and immunoglobulin fragments were detected by alkaline phosphatase (AP)-labeled antibodies (diluted between 1:10,000 and 1:40,000) as follows: goat anti-human IgE, goat anti-human IgG (Fc), goat anti-human IgA, goat anti-human IgM, and goat anti-human Ig (κ light chain) (Dianova). Binding of IPSE/α-1 was detected by a monoclonal anti-IPSE/α-1 antibody 74–4B-7 followed by AP-goat anti-mouse IgG (Dianova) or using biotinylated IPSE/α-1 and subsequent incubation with AP-labeled streptavidin (SAV-AP) (Dianova). Dot blotting was performed using the SCR 96 D Minifold II-Dot Blotter (Schleicher & Schuell).
Sandwich dot blot was performed by dotting different amounts of human IgE (Acris) (4–0.5 pmol) onto nitrocellulose membrane. Following blocking of free binding sites, the membrane was incubated overnight either with IPSE/α-1 (125 pmol/ml), anti-IgE (30 pmol/ml; positive control), or with buffer (negative control). Following a washing step, membranes were incubated with biotinylated IgE (500 ng/ml) for 2 h and, subsequently, with alkaline phosphatase-labeled streptavidin diluted 1:10,000 for 1 h.
For biotinylation, 6 mg of IPSE/α-1 in PBS, pH 8.5, was mixed with 1 ml of a freshly prepared solution of N-hydroxysuccinimide-biotin (Sigma) in DMSO (1 mg/ml) and incubated for 4 h at room temperature followed by dialysis against PBS, pH 7.4. Biotinylated IPSE/α-1 was stored at −80 °C.
Peptide Scan
Overlapping 15-mer peptides were synthesized on cellulose membrane supports via the SPOT technique according to Roeckendorf et al. (35). After side chain deprotection of the peptides, the membrane was incubated for 10 min with 15 ml of 100% (v/v) ethanol and subsequently washed three times, for 10 min each time, with 15 ml of phosphate-buffered saline, pH 7.0 (PBS: 2.7 mm KCl, 1.5 mm KH2PO4, 136 mm NaCl, 8.1 mm Na2HPO4). For fake stripping, the membrane was incubated twice in an ultrasonic device, for 5 min each time, in 15 ml of N,N′-dimethylformamide, then for 5 min in 100 ml of double-distilled water, then twice, for 7 min each time, in 15 ml of denaturing buffer (8 m urea, 1% (w/v) SDS, 0.5% (v/v) β-mercaptoethanol·HOAc, pH 7.0), and finally three times, for 5 min each time, in PBS. The membrane was then blocked for 4 h with 25 ml of 1% (w/v) casein (Hammarsten grade, BDH) in PBST (PBS containing 0.05% (v/v) Tween 20). After a wash for 10 min with 20 ml of PBS, the membrane was incubated overnight in tight-fitting trays at 4 °C with 14.5 ml (2 ml/cm2 membrane) of 6.7–8.3 nmol/ml IgE (Acris) that had been labeled according the manufacturer's instructions with DY781-N-hydroxysuccinimidyl ester (IgE-DY781; Dyomics) in blocking buffer. Subsequently, the membrane was washed five times, for 10 min each time, with 25 ml of PBST and then for 10 min with 25 ml of PBS. Fluorescence was quantitated and normalized to the mean fluorescence (=100%) of the parental peptide IPSE62–76 (Fig. 7D, inset, spots D1 and D2) (Odyssey Infrared Imager, Odyssey software (version 1.2); Li-COR Biosciences).
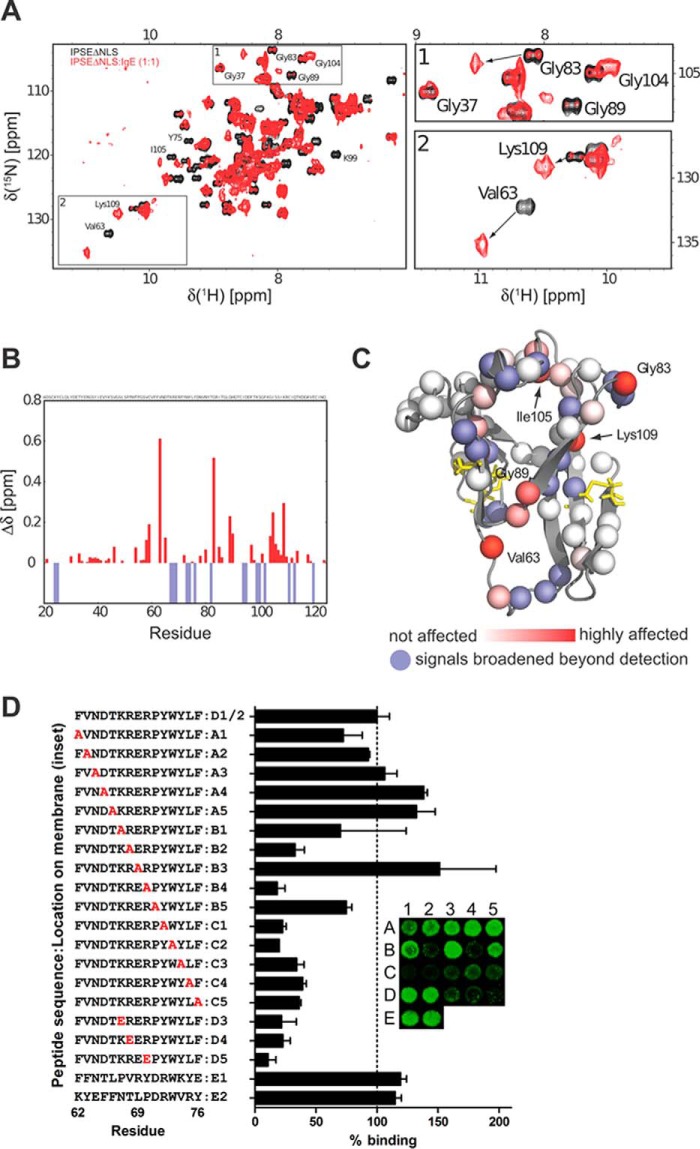
FIGURE 7.
Mapping the binding surface of IPSEΔNLS with IgE. A, 1H,15N NMR correlation spectra of IPSEΔNLS (black) and the IPSEΔNLS·IgE complex (molecular mass ∼200 kDa, red). The presence of NMR signals corresponding to free IPSEΔNLS at a molar ratio of ∼1:1 (IPSEΔNLS/IgE) suggests that IgE has only one IPSEΔNLS-binding site. (Note, the NMR signals of even a very small fraction of free IPSEΔNLS (10 kDa) are much stronger than those of the 200-kDa complex.) B, NMR chemical shift perturbation (CSP) versus residue number. Blue negative bars indicate amide signals that are broadened beyond detection. C, mapping of the IgE binding interface onto the structure of IPSEΔNLS. Nitrogen atoms of the backbone amide groups are represented as spheres and colored red according to CSP or blue if line broadening is observed in the NMR titration experiment. D, peptide scan. Binding of labeled IgE was to immobilized 15-mer IPSE-derived peptides. Binding (means ± S.E.) of IgE-DY781 to 15-mer peptides is expressed in relation to the IPSE-derived peptide 62–76 (inset, spots D1 and D2; mean fluorescence = 100%). Residues exchanged to Ala or Glu in the respective peptides are highlighted in red. Inset, representative example of a peptide library read-out.
Deglycosylation of Human IgG Fc Fragment by PNGase F Treatment
IgG Fc (Acris) was dialyzed against PBS, pH 7.5, containing 0.05 mm EDTA. 330 pmol of IgG Fc were incubated with 5000 milliunits of PNGase F (Roche Applied Science) in 100 mm sodium phosphate buffer containing 25 mm EDTA, pH 8.0, for 17 h at 37 °C. For control purposes, IgG Fc was incubated with buffer alone. SDS-PAGE and Western blotting were performed as described above. Carbohydrates were detected by AP-labeled Aleuria aurantia agglutinin (0.1 μg/ml; Vector Laboratories).
Double Immunodiffusion Assay (Ouchterlony)
The immunodiffusion assay was performed according to standard procedures. 1% agarose (HEEO ultra quality, Roth) in barbitone buffer, pH 8.2 (0.04 m 5,5-diethylbarbituric acid sodium salt and 0.017 m 5,5-diethylbarbituric acid, both Merck, conserved with 0.015% (w/v) merthiolate, Sigma), was pipetted onto carrier film (agarose gel support film; FMC). Using a gel punching device (CAMAG), holes were punched into the gel and each was filled with 8 μl of solution of binding partners (positions and concentrations see legend of Fig. 10D). After 48 h of incubation, the gels were pressed between two glass plates and several layers of pre-wetted filter paper with weights of 1 and 5 kg, respectively, for 10 min each. Gels were washed with 0.9% NaCl and distilled water, and dried at 40 °C before staining with Coomassie.
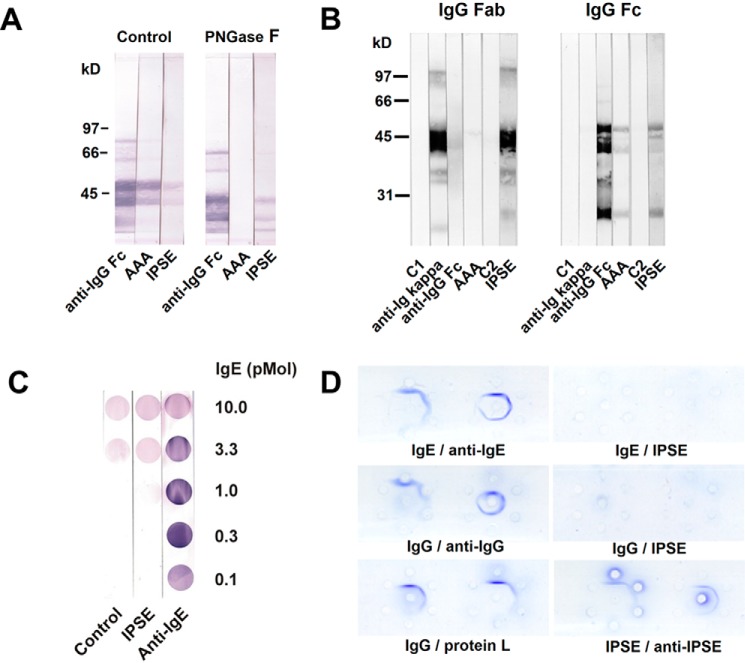
FIGURE 10.
Mode of interaction of IPSE/α-1 with immunoglobulins. A, binding of IPSE/α-1 to deglycosylated IgG Fc. Blotted mock-treated IgG Fc (Control) and IgG Fc deglycosylated with PNGase F were incubated with α-human IgG Fc or A. aurantia agglutinin or labeled IPSE/α-1. B, IPSE/α-1 binds to Fab and Fc fragments. Blotted Fab and Fc fragments of human IgG were detected by α-human IgG (Fc), by α-κ light chain (anti-Ig kappa), by A. aurantia agglutinin or by labeled IPSE/α-1. C1 = buffer control and C2 = SAV-AP. C, sandwich blots. Human IgE dotted at different amounts (10 to 0.1 pmol) was incubated with buffer (negative control) or α-IgE (positive control) or IPSE/α-1, followed by biotinylated IgE. α-IgE gives a clear signal with a maximum at 0.3 pmol/dot, whereas IPSE/α-1 does not exceed background level. D, double immunodiffusion assay (Ouchterlony). Binding partners were applied at various ratios. One partner was pipetted into the center well, the other one clockwise beginning at 12 o'clock in order of dilution in the surrounding wells: 1:1, 1:2, 1:4, 1.8, 1:16, and 1:32. The following starting concentrations were used: 0.5 mg/ml IgE and 1 mg/ml anti-IgE; 0.5 mg/ml IgG and 0.5 mg/ml anti-IgG; 0.5 mg/ml IgG and 0.5 mg/ml protein L (a B cell superantigen from P. magnus); 0.25 mg/ml IgE and 0.05 mg/ml IPSE/α-1 (corresponds to a molar ratio of ∼1:1); 0.25 mg/ml IgG and 0.05 mg/ml IPSE/α-1; and 0.05 mg/ml IPSE/α-1 and rabbit anti-IPSE antiserum (1:2). Precipitation arcs indicate complex formation.
Induction of IL-4 Release from Human Basophils
Basophils were purified from peripheral blood of healthy human donors to a mean purity of 99% (36). Purified basophils were cultured and stimulated as described previously (7). Incubation was performed in 100 μl of culture medium (Iscove's modified Dulbecco's medium supplemented with 100 units/ml penicillin G, 50 μg/ml transferrin, 5 μg/ml insulin, 100 μg/ml streptomycin, and 10% fetal calf serum (PAA)) at a concentration of 106 basophils/ml. Cells were stimulated in the presence of IL-3 (2.5 ng/ml) as a basophil survival factor with various stimuli at the concentrations indicated, and supernatants were collected after 18 h and stored at −20 °C until further investigation. Interleukin-4 release was measured in the culture supernatants by using a two-site sandwich ELISA (Eli-Pair, Diaclone) according to the manufacturer's protocol.
Accession Codes
The atomic coordinates for the NMR ensembles and the crystal structures of IPSEΔNLS have been deposited in the Protein Data Bank under accession numbers 4AKA and 4EL6, respectively. The chemical shift assignments have been deposited in the Biological Magnetic Resonance Data Bank under accession number 17405.
Results
Solution and Crystal Structure of IPSE/α-1
IPSE/α-1 has recently been identified to be the major excretory/secretory component of S. mansoni eggs (37). However, apart from a translated cDNA sequence from S. japonicum (accession number FN317557) and recently sequenced genes from S. hematobium predicted to encode excretory/secretory proteins (11), a BLAST search in the sequence database did not reveal any similar sequences. On the primary structure level, a (Y/F)XXXX(Y/F)XG motif followed by a serine ∼30 residues downstream suggested the presence of a greek key motif. Premature full-length IPSE/α-1 consists of 134 amino acids including an N-terminal signal peptide (residues 1–20), which is cleaved upon release of the mature IPSE/α-1 (residues 21–134). In the following, the residue numbering refers to the position in premature full-length IPSE/α-1. For this study, we cloned and expressed mature IPSE/α-1 (residues 21–134) and a monomeric deletion mutant of IPSE/α-1, IPSEΔNLS (residues 21–124), which lacks 10 amino acids at the C terminus, including a positively charged NLS (residues 125–131) (15) and Cys-132 being responsible for dimer formation (Fig. 1A). Expression in Escherichia coli of both wild-type IPSE/α-1 dimers and IPSEΔNLS monomers resulted in formation of inclusion bodies, which were solubilized by 8 m urea and refolded with physiological buffers containing the glutathione redox system GSH-GSSG to achieve dimerization. Two N-glycosylation sites present on the natural IPSE/α-1 were found not to be involved in basophil activation. E. coli-expressed nonglycosylated IPSE/α-1 exhibited the same functional activity as the glycosylated natural protein. Although neither the dimer nor the monomer expressed in E. coli were glycosylated, the wild-type IPSE/α-1 dimers aggregated after refolding at concentrations above 0.2 mg/ml. In contrast, the IPSEΔNLS monomers remained highly soluble at concentrations of >10 mg/ml and were thus suitable for structural studies (Fig. 1B).
FIGURE 1.
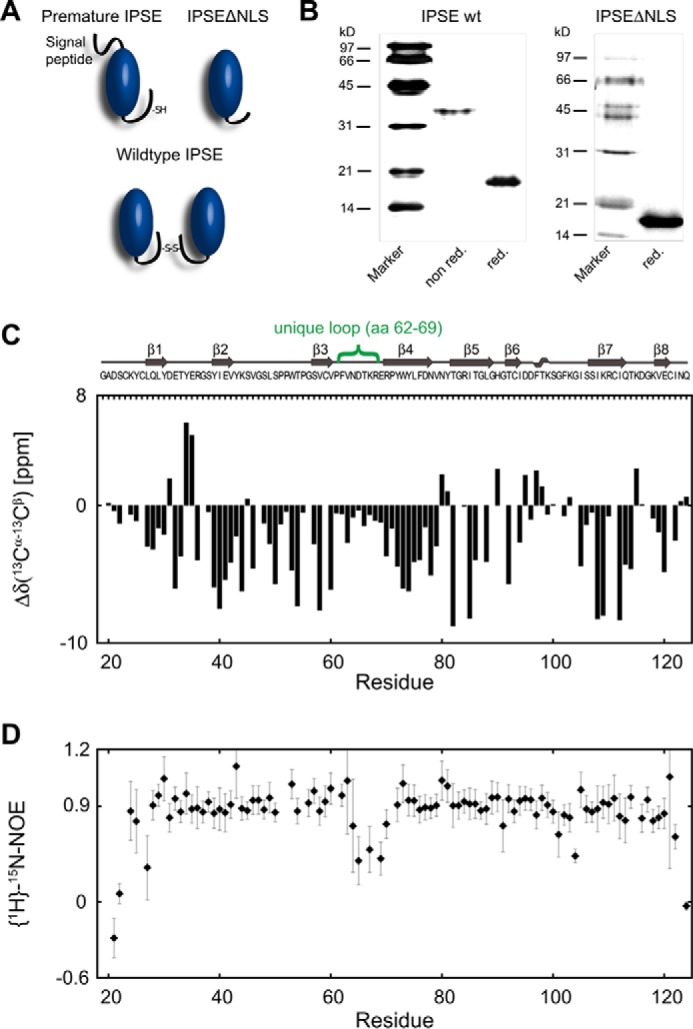
Biochemical and NMR analysis of proteins studied. A, schematic depiction of different mutants of IPSE/α-1. The monomeric IPSEΔNLS was used for structural analysis. B, comparison of recombinantly expressed and purified IPSE WT and IPSEΔNLS by SDS-PAGE and silver staining under nonreducing (non-red) and reducing (red) conditions. C, primary sequence, secondary structure, NMR 13C secondary chemical shifts; D, {1H}-15N heteronuclear NOE of IPSEΔNLS.
To elucidate the molecular basis for the function of IPSE/α-1, we performed structural analysis of IPSEΔNLS using NMR spectroscopy and x-ray crystallography. NMR secondary chemical shift analysis indicated an all-β-strand secondary structure of IPSEΔNLS (Fig. 1C). 15N NMR relaxation data showed that the polypeptide chain is rigid except for one loop (residues 62–69) that exhibits increased conformational flexibility (Fig. 1D). The tumbling correlation time τc ≈7 ns estimated from 15N R2/R1 relaxation rates indicates that IPSEΔNLS (12 kDa) is a monomer in solution. This is in line with earlier studies demonstrating that the C-terminal cysteine (Cys-132) absent in IPSEΔNLS is required for covalent dimerization of wild-type IPSE/α-1 (16). The 13Cβ secondary chemical shifts of six other cysteines in IPSEΔNLS reflect an oxidized state of the attached sulfur atoms, consistent with the presence of three intramolecular disulfide bridges.
The solution structure of IPSEΔNLS (Fig. 2A, left) was determined based on a large number of NOE-derived distance restraints and dihedral angle restraints derived from TALOS+ (Table 1) (23). NOE patterns and chemical shifts indicated the presence of disulfide bonds between cysteines 23 and 26, 59 and 93, as well as 111 and 121, which served as additional restraints in the structural calculation.
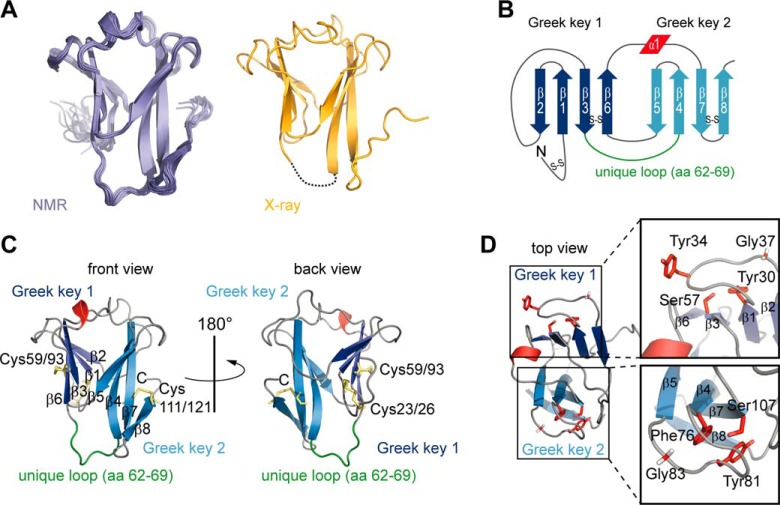
FIGURE 2.
Structure of IPSEΔNLS. A, NMR (left) and crystal (right) structure of IPSEΔNLS. B, secondary structure topology of IPSEΔNLS. C, ribbon representation of the NMR-derived structure of IPSEΔNLS. Cysteine residues involved in disulfide bridges and the unique loop of the IPSE crystallin fold are indicated. D, zoomed view highlighting amino acids of the greek key signature sequence (red sticks).
Crystals of IPSEΔNLS, which diffracted to a resolution of 1.71 Å, were obtained as described earlier (17). The crystallographic structure (Fig. 2A, right, and Table 1) exhibited unambiguous electron density for the three disulfide bridges indicated above, whereas residues 64–68 within the flexible loop (see above) were not visible in the crystal structure due to missing electron density. The crystal and the NMR structure of IPSEΔNLS were in excellent agreement and superimposed with a backbone coordinate root mean square deviation (r.m.s.d.) of 0.5 Å. The structure of IPSEΔNLS consists of a sandwich of two β-sheets comprising eight anti-parallel β-strands (β1 to β8), which form two greek key motifs arranged in a pseudo 2-fold symmetry (Fig. 2B). The loops between the first two β-strands of each motif (β1/β2 and β4/β5, respectively) represent a characteristic β-hairpin motif. The two β-sheets are formed by three strands of one greek key motif each and a fourth β-strand contributed from the other greek key, as is typically observed in crystallin folds (Fig. 2D).
IPSE/α-1 Adopts a Crystallin Fold
The βγ-crystallin superfamily includes structural proteins from the vertebrate eye lens and a few distant relatives found in lower eukaryotes and prokaryotes. Most of the eye lens proteins contain four greek key motifs arranged in two domains. The structures of the C-terminal domain of human eye lens γ-B-crystallin (PDB code 2JDF), which is the best characterized member of the βγ-crystallin superfamily (Fig. 3A, right) and IPSEΔNLS monomer (Fig. 3A, left), are superimposed with a backbone coordinate r.m.s.d. of 1.7 Å. Despite this strong structural similarity at the primary sequence level, the conservation of IPSE and the C- or N-terminal domain of human eye lens γ-B-crystallin was less than 15% identity (Fig. 3C). Structural similarity despite low sequence identity has also been reported for several other members of the βγ-crystallin superfamily in lower organisms (38). Another example, Spherulin 3A from Physarum polycephalum, which contains only one domain with two greek key motifs, dimerizes via a disulfide bridge near the N terminus to restore the two-domain crystallin fold. IPSE/α-1 was found to dimerize in a similar fashion, although the respective cysteines are located at the C terminus rather than at the N terminus (Fig. 3B). Irrespective of the overall low sequence similarity, the highly conserved signature sequence (Y/F)XXXX(Y/F)XG represents a hallmark of the crystallin fold. The aromatic residues in this motif interact with a conserved serine on the third β-strand of each greek key motif (corresponding to β3 and to β7 of greek key motifs 1 and 2, respectively). Interestingly, in greek key motif 1 of IPSE/α-1, the spacing between the two tyrosine residues in the signature YDETYERG is shortened by one residue compared with other crystallin sequences (Fig. 3C). As a result, the tyrosine rings cannot stack with each other so that Tyr-34 is solvent-exposed. However, the formation of the hairpin loop is not significantly affected (Fig. 2D).
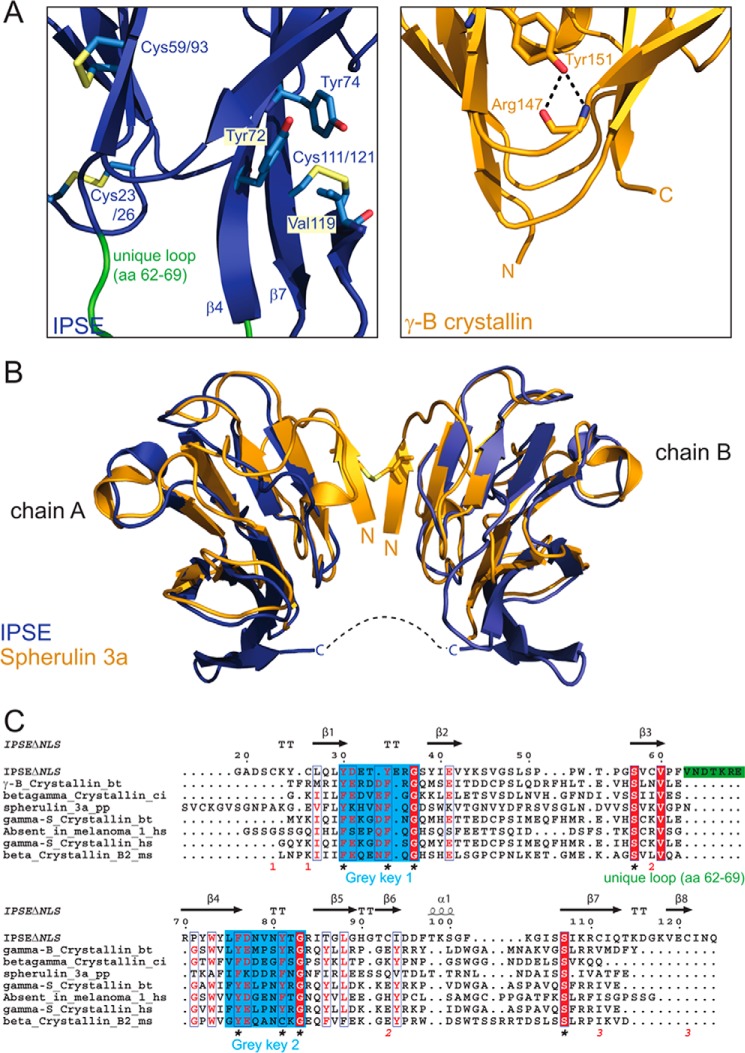
FIGURE 3.
IPSEΔNLS adopts a βγ-crystallin fold. A, zoomed view of IPSEΔNLS (blue, right) and human eye lens γ-B-crystallin (dark yellow, left). Shown are the residues of the tyrosine corner of γ-B-crystallin (Tyr-151 and Arg-147) and the orthogonal stacked Tyr-72 and -74 as well as the disulfide bridges of IPSEΔNLS (sticks) and the flexible loop (amino acids (aa) 62–69) (green). B, structure superposition of two IPSEΔNLS monomers (blue) with dimeric Spherulin 3A (orange). Structures align with a backbone r.m.s.d. of 1.8 Å. C, structure-based multiple sequence alignment of crystallin superfamily members. Above the sequences: secondary structure elements of IPSEΔNLS (except of disulfide bridges). Below the sequences: disulfide bridges are shown as red numbers; the flexible loop (amino acids 62–69) unique to IPSE/α-1 is depicted as a green bar; the two (F/Y)XXXX(F/Y)G signature sequences and the conserved serine residues characteristic of the crystallin fold are indicated by asterisks.
Although the fold of the human lens γ-B-crystallin is maintained by strong hydrophobic interactions in the protein core, the IPSE fold is stabilized by three disulfide bonds (23/26, 59/93, and 111/121) (Fig. 2, B and C), consistent with its high stability over a wide pH range and against 2 m urea. These disulfide bonds may compensate for the lack of a tyrosine corner (39), which is present in most γ-crystallins (see γ-B-crystallin, Tyr-151 in Fig. 3A, right). A typical feature of a tyrosine corner is a hydrogen bond between the hydroxyl group of the tyrosine side chain (Tyr-151 in Fig. 3A, right) and the peptide backbone two to five amino acids upstream stabilizing the formation of a β-hairpin. In contrast, the IPSE/α-1 fold and stability appear to depend on disulfide bridge formation, consistent with the observation that IPSEΔNLS rapidly precipitated under reducing conditions. Distinct from other proteins of the crystallin family, the β7 and the β4 strands of greek key motif 2 in IPSE/α-1 are elongated (Fig. 3C). Furthermore, orthogonal stacking of Tyr-74 and Tyr-72 and additional hydrophobic contacts lead to a curvature of this β-sheet (Fig. 3A, left). The long flexible loop between the two greek key motifs (residues 62–69; Fig. 3, A and C, colored in green) turned out to be a unique feature of IPSE/α-1 compared with other crystallins.
IPSE/α-1 Is an Immunoglobulin-binding Factor
Earlier, we demonstrated that the presence of IgE on the surface of basophils is essential for functional activity of IPSE/α-1. This prompted us to investigate the interaction of IPSE/α-1 with IgE, other immunoglobulin isotypes, and further serum components. All experiments described in the following were performed with the IPSE/α-1 dimer, which is the basophil-activating form. Western blots from whole human serum samples demonstrated that IPSE/α-1 binds to the immunoglobulin bands and to a band identified by the N terminus sequencing as transferrin, but not to other serum components (Fig. 4A). More detailed analysis confirmed that it binds to all human serum immunoglobulin isotypes (IgE, IgG, IgA, and IgM) in a nonantigen-specific manner (Fig. 4B). Thus, IPSE/α-1 is a general immunoglobulin-binding factor. Titration of IgE and IgG on dot blots suggested that IPSE/α-1 binds with higher affinity to IgE than to IgG (Fig. 4C). This finding was corroborated by size exclusion chromatography of preformed IPSE/α-1·IgE and IPSE/α-1·IgG complexes in solution. IPSE/α-1·IgG, in contrast to IPSE·α-1-IgE, completely dissociated upon the mechanical shear forces during size exclusion chromatography. This demonstrated that IPSE·α-1 binds with considerably lower affinity to IgG than to IgE (Fig. 5).
FIGURE 4.
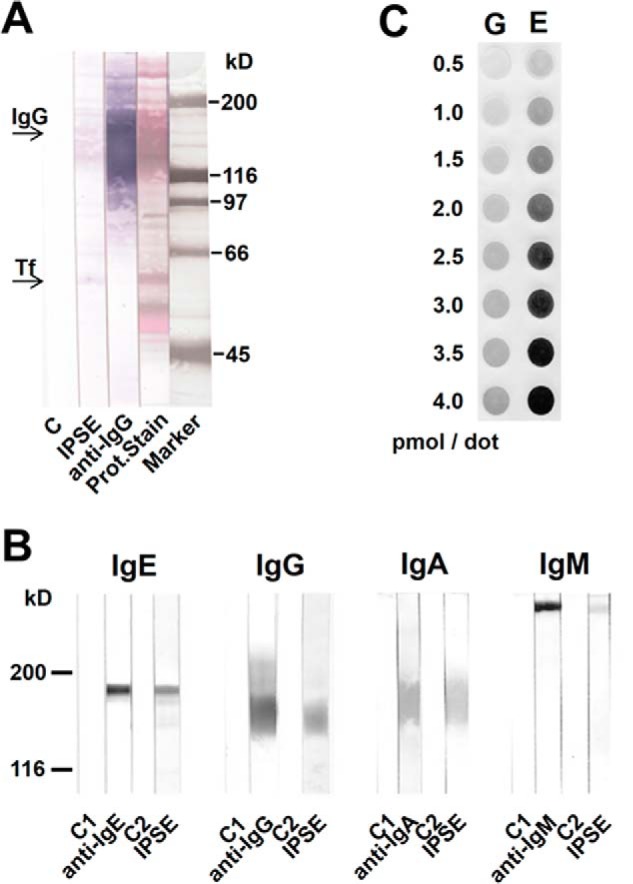
IPSE/α-1 is an immunoglobulin-binding protein with highest affinity to IgE. A, IPSE/α-1 binds to immunoglobulins and transferrin (Tf) in human serum. Western blots of human serum were incubated with IPSE/α-1 followed by labeled streptavidin (SAV-AP) or labeled α-human IgG. Proteins were stained with AuroDye and india ink. B, IPSE/α-1 binds to various immunoglobulin isotypes. Strips with blotted human polyclonal IgE, IgG, IgA, and IgM were incubated with the respective anti-isotype antibodies or IPSE/α-1 followed by SAV-AP. Controls: secondary antibody (C1), SAV-AP (C2). C, dotted IgG (G) and IgE (E) were incubated with labeled IPSE/α-1 followed by SAV-AP.
FIGURE 5.
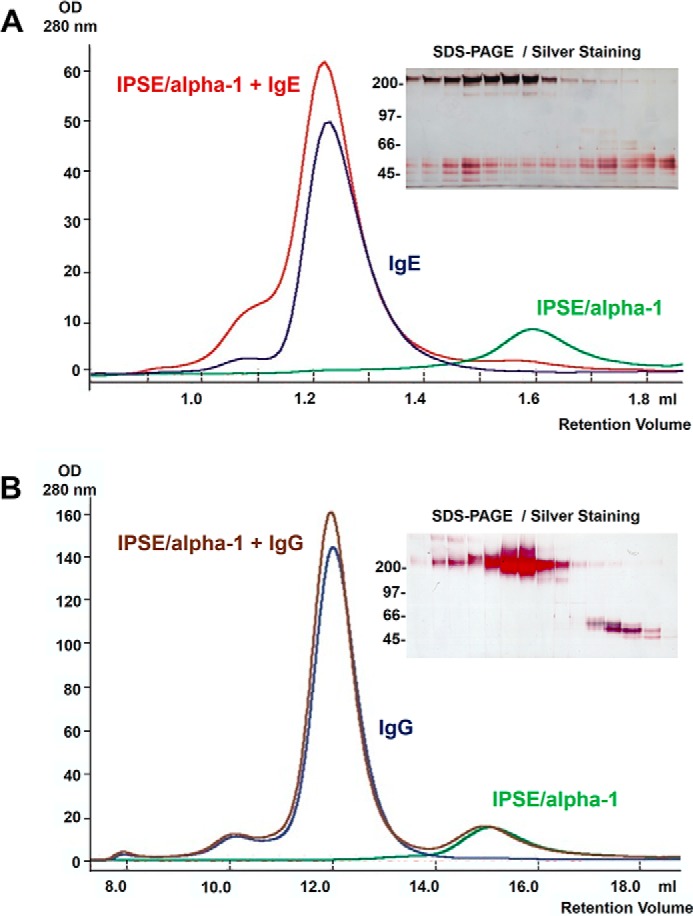
Size exclusion chromatography. Complexes of IPSE/α-1 and IgE (A) and complexes of IPSE/α-1 and IgG (ratio 1:1), respectively (B), were applied to a Superdex 75 column (red curve). For control, the same amount IPSE/α-1 alone (green curve) and IgE or IgG alone (blue curve) were run over the column. Fractions of the IPSE/α-1·IgE or IPSE/α-1·IgG complexes were analyzed by SDS-PAGE and silver staining. High molecular weight fractions contain IPSE/α-1 and IgE but not IgG, suggesting that IPSE/α-1 binds with higher affinity to IgE than to IgG.
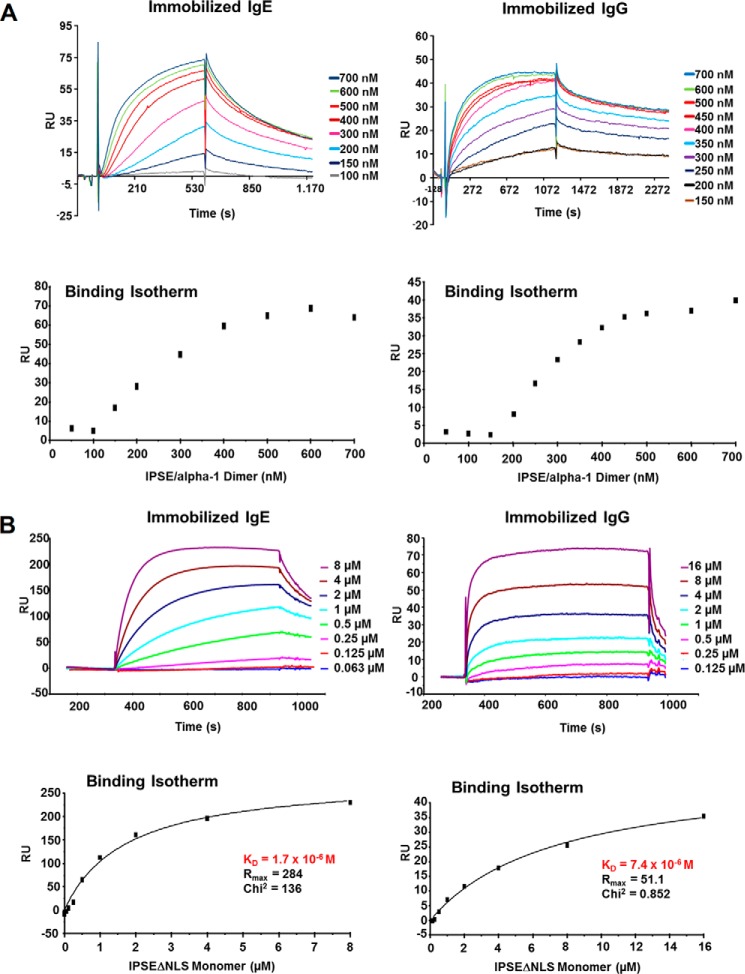
To quantify the binding affinity of IPSE/α-1 to IgE and IgG, we performed surface plasmon resonance (SPR) assays with IPSE/α-1 dimer and IPSEΔNLS monomer. SPR assay with the IPSE/α-1 dimer (Fig. 6A) showed that IPSE/α-1 clearly bind to immobilized immunoglobulins on the CM-5 sensor chip. However, the binding isotherms obtained upon analysis of the data showed a sigmoidal curve indicating a complex binding scenario of dimeric IPSE/α-1 to the dimeric immunoglobulins. As a consequence, simple curve fitting using the Langmuir (1:1) calculation was not applicable to determine quantitative affinity data. Thus, we performed additional SPR experiments with the monomeric mutant IPSEΔNLS (Fig. 6B). Fitting of the binding isotherms using a 1:1 binding model revealed a KD value of 1.7 μm for the binding of IPSEΔNLS to IgE and 7.4 μm for the binding to IgG, which corresponds to a 4-fold higher binding affinity to IgE compared with IgG. This corroborates the earlier observations that IPSE/α-1 binds with higher affinity to IgE than to IgG, although one should keep in mind that the overall binding affinity of the dimer to IgE might differ from that of the monomeric mutant.
FIGURE 6.
Surface plasmon resonance assays. Binding of IPSE/α-1 dimer (A) and IPSEΔNLS monomer (B) to IgE and IgG immobilized on a CM5 chip was analyzed by real time measurement. Curves were obtained after subtraction of the unspecific binding to the reference cell. Bound IPSE (RU) was estimated at the end of the association phase. Binding isotherms of the IPSE/α-1 dimer (A) showed a sigmoidal curve characteristic for multivalent positive cooperative binding. Thus, Langmuir (1:1) calculation of affinity constants was not applicable. For the IPSEΔNLS monomer (B) dissociation constants of 1.7 and 7.4 μm for IgE and IgG, respectively, were measured.
Analysis of the Interaction Sites of IPSE/α-1 with IgE
To map the binding interface of IPSE/α-1 with the IgE antibody, we compared NMR chemical shifts of IPSEΔNLS free and IPSEΔNLS bound to IgE at molar ratios of 0.5:1 and 1:1 (IPSEΔNLS/IgE). Given the high molecular mass of the complex (≈200 kDa), 2H labeling of IPSEΔNLS was required for sufficient sensitivity. Uniformly 2H/15N/13C-labeled IPSEΔNLS was titrated with unlabeled human IgE to monitor chemical shift perturbations (CSP) using 1H,15N correlation experiments. Upon binding of IPSEΔNLS to the 180-kDa IgE molecule, IPSEΔNLS signals experience extensive line broadening, which likely results from the low molecular tumbling rate and thus strongly increased NMR line widths in the high molecular weight complex. We therefore used relaxation-optimized NMR experiments (28) to detect IPSEΔNLS bound to IgE (Fig. 7A). A number of amide proton signals close to the interaction site remain broadened beyond detection, although for several other signals significant chemical shift differences are observed compared with free IPSEΔNLS. The binding kinetics of the IPSE/IgE interaction is slow at the NMR chemical shift time scale consistent with a low micromolar to nanomolar dissociation constant. The observed CSPs identify a large binding surface on one side of IPSEΔNLS that includes the flexible loop (residues 62–69) connecting the two greek key motifs (Fig. 7, A–C). The opposite side of the structure (β1 and β2 with the hairpin loop in greek key motif 1, and β7 and β8 in greek key motif 2) is largely unaffected by IPSE binding to IgE. The amide group of Val-63, which is in close proximity to the solvent-exposed aromatic side chain of Phe-62 in the flexible loop, experiences the strongest chemical shift perturbation. Another patch of strongly affected residues, e.g. Gly-83, is in close spatial proximity to the highly conserved Tyr-81. The NMR data thus indicated that the binding interface involves these aromatic side chains. Most of the interface is located in overall positively charged regions of the protein, especially the flexible loop (residues 62–69) and β-strand β4. This implies an essential role of electrostatic and charged interactions.
Peptide scan analysis of the sequence requirements in the peptide comprising residues 62–76 revealed that exchanges of positively charged amino acids to either alanine or glutamate lead to a drastic decrease in IgE binding (B1, B2, B4 and D3, D4, D5, respectively) (Fig. 7D). This highlights the importance of the positive charges for the IPSE/IgE interaction. In addition, exchange of the aromatic residues Tyr-72 and -74, Trp-73, Phe-76, and Leu-75 by alanines (C1, C2, C3, C4, and C5) strongly decreases the binding affinity to IgE, demonstrating the importance of additional hydrophobic interactions. Interestingly, scrambling of the amino acid sequence did not change the binding intensity significantly, suggesting that the overall charge and hydrophobicity might be even more important than the sequence itself. Taken together, the peptide scan and NMR titration demonstrated the importance of positively charged and aromatic amino acid side chains for enabling effective binding of IPSE/α-1 to IgE.
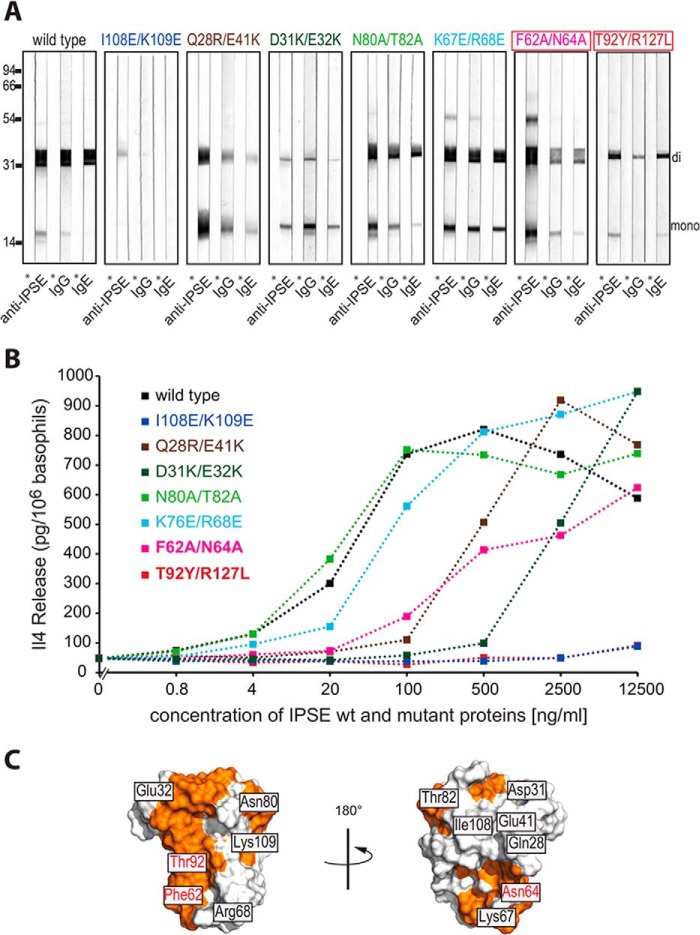
The residues relevant for the interaction of IPSE/α-1 with IgE as determined by NMR and peptide scan experiments were further investigated by mutational analysis (Fig. 8). Mutations were introduced both into IPSEΔNLS (monomeric due to lack of the inter-chain disulfide bond) and into wild-type IPSE/α-1 (dimeric). The monomeric IPSEΔNLS mutants were analyzed by NMR for structural integrity (Fig. 9, A and B); the dimeric IPSE/α-1 mutants were analyzed for immunoglobulin binding (Western blotting; Fig. 8A) and for basophil IL-4-releasing capacity (Fig. 8B).
FIGURE 8.
Mutational analysis of immunoglobulin binding and functional activity of IPSE/α-1. A, binding of monoclonal anti-IPSE antibody, human IgG, or human IgE to IPSE/α-1 wild-type and mutants, respectively, assessed by Western blots. Mutants in red boxes were analyzed by SPR for binding affinity with IgE. B, IL-4 release from human basophils induced by different concentrations of IPSE/α-1 wild-type and mutants, respectively. Shown is one representative result of three independent experiments. C, surface representation of IPSEΔNLS. Binding interface was derived from the NMR titration and the peptide scan (orange).
FIGURE 9.
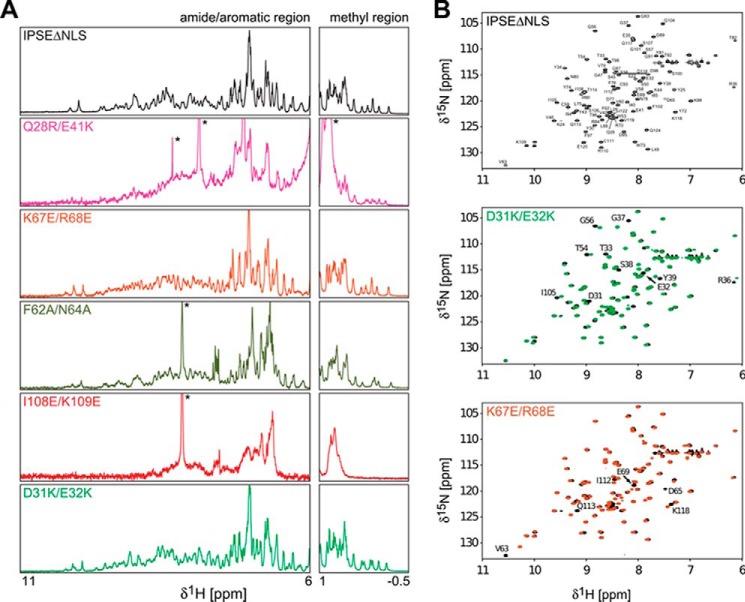
One-dimensional 1H NMR (A) and 1H,15N HSQC spectra (B) spectra of IPSEΔNLS and IPSEΔNLS mutants. A, spectral regions of amide and aromatic and methyl protons for the mutants Q28R/E41K, K67E/R68E, F62A/N64A, I108E/K109E, and D31K/E32K are shown. The good dispersion of amide proton signals and the presence of methyl proton signals below 0 ppm observed for all variants except I108E/K109E are indicative of the presence of tertiary folds. Signals arising from low molecular weight contaminations are marked with an asterisk. Spectra were recorded at 600 MHz 1H Larmor frequency. B, only signals of amides in close spatial proximity to the mutations are shifted in the spectra of D31K/E32K and K67E/R68E mutants. Spectra were recorded at 600 MHz 1H Larmor frequency.
The mutants I108E/K109E, Q28R/E41K, and D31K/E32K showed reduced immunoglobulin binding and loss of functional activity due to compromised in vitro folding. NMR analysis revealed that the monomeric IPSEΔNLS mutant I108E/K109E is unfolded in solution (Fig. 9A). In contrast, the monomeric mutants Q28R/E41K and D31K/E32K appeared folded in the NMR analysis. However, Western blot analysis of the corresponding dimeric mutant preparations revealed a shift in the dimer/monomer ratio toward the monomer (Fig. 8A). Thus, the reduced functional activity of these two preparations is potentially due to a reduced proportion of the functionally active dimer.
All other IPSE/α-1 mutants were structurally intact as assessed by NMR (monomeric IPSEΔNLS mutants) and by monitoring the intensity of dimeric IPSE/α-1 bands on Western blots. The mutants N80A/T82A and K67E/R68E displayed immunoglobulin binding and IL-4-inducing capacities similar to the wild-type IPSE/α-1. This is consistent with the fact that the respective residues are located outside (Asn-80 and Thr-82) or at the edge (Lys-67 and Arg-68) of the binding interface as assessed by NMR (Fig. 9C). The double mutant F62A/N64A exhibited reduced binding to IgG and IgE compared with the wild type and markedly reduced basophil-activating capacity. The pronounced effect of the amino acid exchange at positions 62 and 64 corroborates the critical role of the flexible loop (amino acids 62–69) as the core of the immunoglobulin interaction site. The mutant T92Y/R127L is identical with a factor from S. mansoni eggs termed SmCKBP (40). SmCKBP has been reported to bind to, and neutralize, chemokines but to be unable to activate basophils. In accordance with this previous report, recombinant T92Y/R127L displayed markedly reduced immunoglobulin binding together with a complete lack of IL-4-releasing capacity. These results were consistent with the position of Thr-92 within the binding surface as mapped by the NMR titrations (Figs. 8 and 9C). Taken together, the mutational analysis showed a good correlation between the ability of IPSE/α-1 to bind immunoglobulins and to activate basophils. Our results suggest that IgE binding involves a binding surface that includes the unique flexible loop of the IPSE/α-1 crystallin fold, although functional activity requires the presence of dimeric IPSE/α-1.
IPSE/α-1 Does Not Cross-link IgE
The finding that only dimeric but not monomeric IPSE/α-1 is able to activate basophils suggests that the functional activity of this molecule involves cross-linking of receptor-bound IgE, the classical mechanism seen with allergens, lectins, and B cell superantigens. Because an antigen-specific binding mode (such as exhibited by an allergen) could be excluded, we asked whether IPSE/α-1 interacts with the carbohydrate side chains (like a lectin) or with the Fab or Fc regions of immunoglobulins (like a B cell superantigen). We therefore performed Western blot experiments with Fab and Fc fragments of human IgG. IgG was used as a paradigmatic immunoglobulin, as this isotype is available in large quantities and is bound by IPSE/α-1.
A lectin-like binding of IPSE/α-1 to immunoglobulin glycan side chains could be excluded by comparing its binding to native (i.e. glycosylated) and to PNGase F-deglycosylated Fc fragments of IgG (Fab fragments of IgG are not glycosylated). Deglycosylation of IgG Fc was confirmed by Western blot analysis, as deglycosylated IgG Fc was found to band at a lower molecular weight than native IgG Fc (Fig. 10A). Moreover, the lectin A. aurantia agglutinin that binds to fucose residues on native IgG no longer detected deglycosylated IgG Fc. In contrast, IPSE/α-1 recognized both glycosylated and deglycosylated IgG Fc. This clearly rules out a lectin-like interaction between IPSE/α-1 and IgG, a finding that quite likely applies also to the other immunoglobulin isotypes (due to the highly conserved immunoglobulin domains).
To investigate whether IPSE/α-1 binds to the Fab or the Fc part of IgG, whole IgG and Fab and Fc fragments were analyzed for binding of IPSE/α-1 by Western blotting. IPSE/α-1 was found to bind to both Fab and Fc fragments (Fig. 10B).
Typically, IgE-dependent activation of basophils occurs via cross-linking of IgE on their surface as described for allergens, anti-IgE antibodies, lectins, or B cell superantigens. Thus, we expected the same mechanism for IPSE/α-1. However, methods to demonstrate cross-linking, such as sandwich dot blots and double immunodiffusion assay, showed that IPSE/α-1 was not able to cross-link IgE. By using sandwich dot blots, we investigated whether IPSE/α-1 cross-linked unlabeled membrane-bound IgE with labeled soluble IgE. Although the blot with anti-IgE (positive control) showed a clear positive signal, the blot with IPSE/α-1 signal did not exceed background (buffer control) levels indicating that IPSE/α-1 was not able to cross-link IgE (Fig. 10C). A background signal observed with ≥3.3 pmol pf IgE/dot likely resulted from the tendency of IgE to form aggregates at high concentrations. Analogous results were obtained with the double immunodiffusion assay. Binding partners were applied in different concentration ratios to wells punched in the agarose on microscope slides. After diffusion into the gel for 48 h, immunoprecipitation due to cross-linking of the binding partners was visualized by Coomassie staining. As expected, all ligand combinations used as positive controls (IgE/anti-IgE, IgG/anti-IgG, and IgG/protein L (a B cell superantigen from Peptostreptococcus magnus) and IPSE/α-1/anti-IPSE antiserum) yielded clearly visible precipitin lines, but neither IPSE/IgE nor IPSE/IgG formed complexes large enough to precipitate in the gel, suggesting that IPSE/α-1 did not cross-link IgE or IgG in solution (Fig. 10D). Of note, the small IPSE/α-1·IgE complexes formed at equivalence as assessed by size exclusion chromatography indicated a binding stoichiometry of one IPSE/α-1 dimer to one IgE molecule and thus confirmed that cross-linking did not take place (Fig. 5).
Discussion
Accumulating evidence suggests an important role of basophils for the immune response against helminth infection. However, the molecular mechanism of parasite-induced basophil activation is unknown for the majority of worm infections. In the case of schistosomiasis, IPSE/α-1 represents the active principle responsible for activation of basophils (7). To better understand the molecular mechanisms by which IPSE/α-1 induces basophil IL-4 release from nonsensitized healthy individuals, we determined the structure of this molecule and investigated its interaction with IgE.
The solution and crystal structures of the monomeric IPSEΔNLS classified IPSE/α-1 as a novel member of the βγ-crystallin superfamily. βγ-Crystallins, such as the eye lens γ-B-crystallins, are characterized by extremely high stability toward harsh conditions such as extreme pH (pH 1), high temperature (70 °C), and chaotropic agents (6 m urea) (38). For IPSE/α-1, such a stable structure might be important, because following secretion from schistosome eggs it has to withstand the aggressive environment of the inflammatory egg granuloma characterized by low pH and high levels of oxidative and other denaturing agents.
The structural and mutational analysis of the complex of IPSEΔNLS and IgE revealed a positively charged interaction interface, which includes the IPSE/α-1-unique loop. Computational modeling suggested that the negatively charged region on the Cϵ2 domain of IgE provides a suitable binding interface for the positively charged IPSE/α-1 (Fig. 11, A and B).
FIGURE 11.
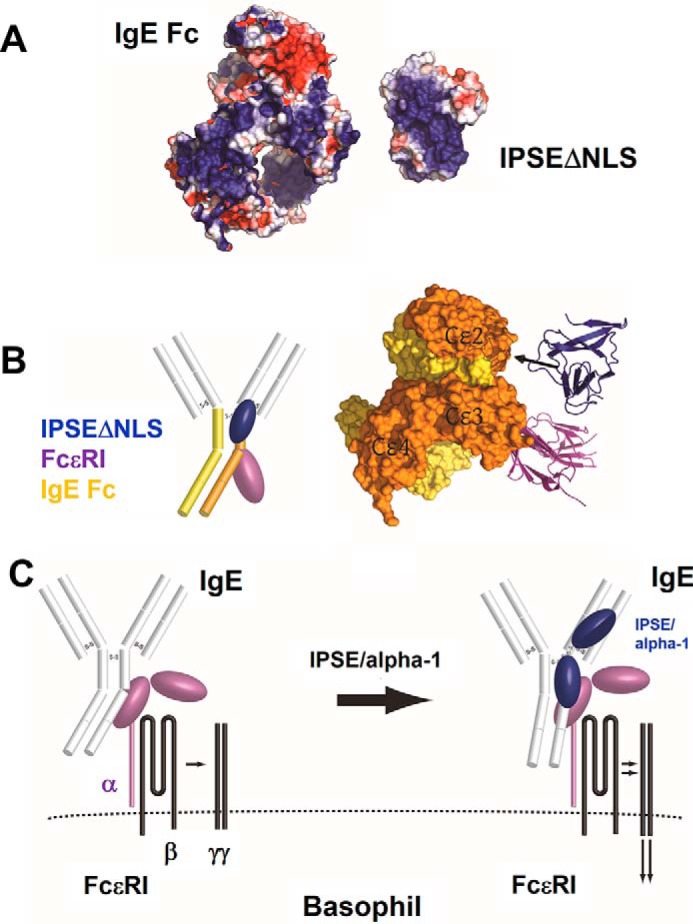
Model of the interaction of IPSE/α-1 with IgE. A putative novel mechanism of basophil activation is shown. A, surface representation of IgE Fc (PDB accession code 2Y7Q, left) and IPSEΔNLS (right) colored according to electrostatic surface potential at ±2 kB T/e for positive (blue) or negative (red) charge potential using the program APBS (48). B, proposed interaction between IPSEΔNLS and IgE (represented by the crystal structure of IgE Fc/FcϵRI, PDB accession code 2WQR). The positively charged binding interface of IPSEΔNLS mapped by NMR and mutational analysis presumably interacts with a negatively charged surface on the Cϵ2 domain of IgE, while the FcϵRI binds to the neighboring Cϵ3 domain of the IgE molecule. C, schematic model of IgE receptor activation on the surface of basophils. IPSE/α-1 may induce a structural rearrangement in the IgE molecule. This triggers the activation of the receptor signaling cascade.
We therefore developed a model for the mechanism of basophil activation by IPSE/α-1 (Fig. 11C), based on the computational modeling and our experimental data, specifically the inability of IPSE/α-1 to cross-link IgE, a putative 1:1 stoichiometry of IPSE/α-1·IgE complexes, and the binding of IPSE/α-1 to both Fab and Fc fragments: The first monomer of the IPSE/α-1 dimer binds the negatively charged surface of the Cϵ2 domain. Binding of the first IPSE/α-1 monomer to the IgE-Fc fragment facilitates binding of the second IPSE/α-1 monomer to the IgE-Fab. We hypothesize that this might trigger a conformational change of the IgE molecule in close proximity to the Fcϵ receptor and subsequent receptor activation without cross-linking. To validate this mechanism, further experiments are warranted to locate the binding site(s) of dimeric IPSE/α-1 on the IgE molecule by co-crystallization.
For the past 30 years, the receptor-mediated response to foreign antigens has been known to involve the high affinity IgE receptor on mast cells and basophils. Cross-linking of two or more IgE·receptor complexes on the cell surface stimulates degranulation of the cells and the release of mediators and cytokines (41). In contrast, IPSE/α-1 appeared to activate basophils by a novel mechanism that involves binding but not cross-linking receptor-bound IgE leaving the IgE “monomeric.” A role of IgE monomers in initiating cell signaling upon binding to FcϵRI has been controversially discussed (42, 43). High concentrations of monomeric IgE were reported to partially activate the FcϵRI, thereby triggering cellular events such as degranulation and cytokine production (42). Moreover, different monoclonal IgEs seem to differ in their ability to activate basophils, but the structural differences that account for the functional divergence are not known (43). In contrast, Schweitzer-Stenner and Pecht consider nonphysiological experimental conditions to cause the effects of monomeric IgE (44). Recently, however, the cross-linking model for cell activation has been challenged also for the T and B cell receptors (45, 46). It was shown that inactive T and B cell receptors are present in an oligomeric form, whereas binding of their respective ligands leads to dissociation into monomeric receptors and subsequent clustering of the monomeric receptors (dissociation model) (45, 47). Further studies are necessary to determine the mechanism of action of IPSE/α-1. Because the solution methods used in this study do not necessarily reproduce the cell-surface environment in which IPSE/α-1 acts, further studies should reveal the structural rearrangement of FcϵRI-bound IgE on the cell surface upon binding of IPSE/α-1.
In conclusion, the parasite-derived protein IPSE/α-1 is the first member of the βγ-crystallin superfamily discovered to have an immunomodulatory function. Additionally, this is the first report on the interaction of a crystallin fold with IgE. In contrast to other known IgE-dependent basophil activators, IPSE/α-1 does not cross-link IgE; instead it appears to activate basophils via a novel mechanism. Our results regarding the structure and the molecular function of the basophil IL-4-inducing factor IPSE/α-1 help to better understand the mechanism by which parasites modulate the host's immune system and may provide a starting point for pharmacological intervention in the future.
Author Contributions
N. H. M., H. H., M. S., and G. S. designed and coordinated this study and wrote the paper. N. H. M., K. T., T. M., and M. S, designed, performed, and analyzed NMR experiments, H. M. and J. M.-D. solved the crystal structure, and Si. B. performed and analyzed the experiments shown in Figs. 4, A and B, and 10, A, B, and D. D. B. provided technical assistance and performed experiments shown in Fig. 8. A. M. performed experiments shown in Fig. 5 and G. S. the experiments shown in Figs. 4C and 10C. S. B. and A. F. designed and performed the peptide scan (Fig. 7D). T. K., T. W., and G. S. designed, performed, and analyzed the SPR assays (Fig. 6). All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to Bernd Simon (EMBL Heidelberg, Germany), Gülden Demiraslan and Ulrike Schütz (Munich, Germany), Sven Mueller-Loennies and Niels Röckendorf (RCB, Borstel, Germany), and Prof. Brian J. Sutton and Prof. James M. McDonnell (King's College, London, UK) for support and discussions. We thank the Bavarian NMR Centre for NMR time, and the ESRF for access to measurements at beamline ID29. Editorial support was provided by David Young, Young Medical Communications and Consulting Ltd.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants EXC114 and SFB 594 (to M. S) and the Bavarian Ministry of Sciences, Research, and the Arts in the framework of the Bavarian Molecular Biosystems Research Network (to T. M.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4AKA and 4EL6) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Submitted to the BMRB Data Bank under accession number 17405.
- NLS
- nuclear localization sequence
- r.m.s.d.
- root mean square deviation
- PNGase F
- peptide:N-glycosidase F
- AP
- alkaline phos
- SPR
- surface plasmon resonance
- CSP
- chemical shift perturbation.
References
- 1. Schramm G., Haas H. (2010) Th2 immune response against Schistosoma mansoni infection. Microbes Infect. 12, 881–888 [DOI] [PubMed] [Google Scholar]
- 2. Seder R. A., Paul W. E., Davis M. M., Fazekas de St Groth B. (1992) The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 176, 1091–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brunet L. R., Finkelman F. D., Cheever A. W., Kopf M. A., Pearce E. J. (1997) IL-4 protects against TNF-α 32 -mediated cachexia and death during acute schistosomiasis. J. Immunol. 159, 777–785 [PubMed] [Google Scholar]
- 4. Herbert D. R., Orekov T., Perkins C., Rothenberg M. E., Finkelman F. D. (2008) IL-4Rα expression by bone marrow-derived cells is necessary and sufficient for host protection against acute schistosomiasis. J. Immunol. 180, 4948–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sullivan B. M., Liang H. E., Bando J. K., Wu D., Cheng L. E., McKerrow J. K., Allen C. D., Locksley R. M. (2011) Genetic analysis of basophil function in vivo. Nat. Immunol. 12, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Panhuys N., Prout M., Forbes E., Min B., Paul W. E., Le Gros G. (2011) Basophils are the major producers of IL-4 during primary helminth infection. J. Immunol. 186, 2719–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schramm G., Falcone F. H., Gronow A., Haisch K., Mamat U., Doenhoff M. J., Oliveira G., Galle J., Dahinden C. A., Haas H. (2003) Molecular characterization of an interleukin-4-inducing factor from Schistosoma mansoni eggs. J. Biol. Chem. 278, 18384–18392 [DOI] [PubMed] [Google Scholar]
- 8. Schramm G., Mohrs K., Wodrich M., Doenhoff M. J., Pearce E. J., Haas H., Mohrs M. (2007) Cutting edge: IPSE/α-1, a glycoprotein from Schistosoma mansoni eggs, induces IgE-dependent, antigen-independent IL-4 production by murine basophils in vivo. J. Immunol. 178, 6023–6027 [DOI] [PubMed] [Google Scholar]
- 9. Schramm G., Gronow A., Knobloch J., Wippersteg V., Grevelding C. G., Galle J., Fuller H., Stanley R. G., Chiodini P. L., Haas H., Doenhoff M. J. (2006) IPSE/α-1: a major immunogenic component secreted from Schistosoma mansoni eggs. Mol. Biochem. Parasitol. 147, 9–19 [DOI] [PubMed] [Google Scholar]
- 10. Dunne D. W., Jones F. M., Doenhoff M. J. (1991) The purification, characterization, serological activity and hepatotoxic properties of two cationic glycoproteins (α1 and ω1) from Schistosoma mansoni eggs. Parasitology 103, 225–236 [DOI] [PubMed] [Google Scholar]
- 11. Young N. D., Jex A. R., Li B., Liu S., Yang L., Xiong Z., Li Y., Cantacessi C., Hall R. S., Xu X., Chen F., Wu X., Zerlotini A., Oliveira G., Hofmann A., et al. (2012) Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 44, 221–225 [DOI] [PubMed] [Google Scholar]
- 12. Falcone F. H., Dahinden C. A., Gibbs B. F., Noll T., Amon U., Hebestreit H., Abrahamsen O., Klaucke J., Schlaak M., Haas H. (1996) Human basophils release interleukin-4 after stimulation with Schistosoma mansoni egg antigen. Eur. J. Immunol. 26, 1147–1155 [DOI] [PubMed] [Google Scholar]
- 13. Watanabe T., Inoue T., Ochi H., Terashima M., Asano Y., Nakatani T. (1999) Lipid A directly inhibits IL-4 production by murine Th2 cells but does not inhibit IFN-γ production by Th1 cells. Eur. J. Immunol. 29, 413–418 [DOI] [PubMed] [Google Scholar]
- 14. Patella V., Florio G., Petraroli A., Marone G. (2000) HIV-1 gp120 induces IL-4 and IL-13 release from human FcϵRI+ cells through interaction with the VH3 region of IgE. J. Immunol. 164, 589–595 [DOI] [PubMed] [Google Scholar]
- 15. Kaur I., Schramm G., Everts B., Scholzen T., Kindle K. B., Beetz C., Montiel-Duarte C., Blindow S., Jones A. T., Haas H., Stolnik S., Heery D. M., Falcone F. H. (2011) Interleukin-4-inducing principle from Schistosoma mansoni eggs contains a functional C-terminal nuclear localization signal necessary for nuclear translocation in mammalian cells but not for its uptake. Infect. Immun. 79, 1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wuhrer M., Balog C. I., Catalina M. I., Jones F. M., Schramm G., Haas H., Doenhoff M. J., Dunne D. W., Deelder A. M., Hokke C. H. (2006) IPSE/α-1, a major secretory glycoprotein antigen from schistosome eggs, expresses the Lewis X motif on core-difucosylated N-glycans. FEBS J. 273, 2276–2292 [DOI] [PubMed] [Google Scholar]
- 17. Mayerhofer H., Schramm G., Hatzopoulos G. N., Mueller-Dieckmann C., Haas H., Mueller-Dieckmann J. (2009) Cloning, expression, purification, crystallization and preliminary x-ray crystallographic analysis of interleukin-4-inducing principle from Schistosoma mansoni eggs (IPSE/α-1). Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 594–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 19. Sattler M., Schleucher J., Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nuclear Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 20. Meyer N. H., Schramm G., Sattler M. (2011) (1)H, (13)C and (15)N chemical shift assignments of IPSEΔNLS. Biomol. NMR Assign. 5, 225–227 [DOI] [PubMed] [Google Scholar]
- 21. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 22. Güntert P. (2009) Automated structure determination from NMR spectra. Eur. Biophys. J. 38, 129–143 [DOI] [PubMed] [Google Scholar]
- 23. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Refinement of protein structures in explicit solvent. Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 25. Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Güntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. (2005) RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 26. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 27. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature. 381, 272. [DOI] [PubMed] [Google Scholar]
- 28. Riek R., Wider G., Pervushin K., Wüthrich K. (1999) Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc. Natl. Acad. Sci. U.S.A. 96, 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Röckendorf N., Bade S., Hirst T. R., Gorris H. H., Frey A. (2007) Synthesis of a fluorescent ganglioside GM1 derivative and screening of a synthetic peptide library for GM1 binding sequence motifs. Bioconjug. Chem. 18, 573–578 [DOI] [PubMed] [Google Scholar]
- 36. Haisch K., Gibbs B. F., Körber H., Ernst M., Grage-Griebenow E., Schlaak M., Haas H. (1999) Purification of morphologically and functionally intact human basophils to near homogeneity. J. Immunol. Methods 226, 129–137 [DOI] [PubMed] [Google Scholar]
- 37. Mathieson W., Wilson R. A. (2010) A comparative proteomic study of the undeveloped and developed Schistosoma mansoni egg and its contents: The miracidium, hatch fluid and secretions. Int. J. Parasitol. 40, 617–628 [DOI] [PubMed] [Google Scholar]
- 38. Jaenicke R., Slingsby C. (2001) Lens crystallins and their microbial homologs: structure, stability, and function. Crit. Rev. Biochem. Mol. Biol. 36, 435–499 [DOI] [PubMed] [Google Scholar]
- 39. Hemmingsen J. M., Gernert K. M., Richardson J. S., Richardson D. C. (1994) The tyrosine corner: a feature of most greek key β-barrel proteins. Protein Sci. 3, 1927–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith P., Fallon R. E., Mangan N. E., Walsh C. M., Saraiva M., Sayers J. R., McKenzie A. N., Alcami A., Fallon P. G. (2005) Schistosoma mansoni secretes a chemokine binding protein with antiinflammatory activity. J. Exp. Med. 202, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holowka D., Sil D., Torigoe C., Baird B. (2007) Insights into immunoglobulin E receptor signaling from structurally defined ligands. Immunol. Rev. 217, 269–279 [DOI] [PubMed] [Google Scholar]
- 42. Kitaura J., Xiao W., Maeda-Yamamoto M., Kawakami Y., Lowell C. A., Kawakami T. (2004) Early divergence of Fcϵ receptor I signals for receptor up-regulation and internalization from degranulation, cytokine production, and survival. J. Immunol. 173, 4317–4323 [DOI] [PubMed] [Google Scholar]
- 43. Kitaura J., Song J., Tsai M., Asai K., Maeda-Yamamoto M., Mocsai A., Kawakami Y., Liu F. T., Lowell C. A., Barisas B. G., Galli S. J., Kawakami T. (2003) Evidence that IgE molecules mediate a spectrum of effects on mast cell survival and activation via aggregation of the FcϵRI. Proc. Natl. Acad. Sci. U.S.A. 100, 12911–12916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schweitzer-Stenner R., Pecht I. (2005) Death of a dogma or enforcing the artificial: monomeric IgE binding may initiate mast cell response by inducing its receptor aggregation. J. Immunol. 174, 4461–4464 [DOI] [PubMed] [Google Scholar]
- 45. Yang J., Reth M. (2010) Oligomeric organization of the B-cell antigen receptor on resting cells. Nature 467, 465–469 [DOI] [PubMed] [Google Scholar]
- 46. Lillemeier B. F., Mörtelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T., Davis M. M. (2010) TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang J., Reth M. (2010) The dissociation activation model of B cell antigen receptor triggering. FEBS Lett. 584, 4872–4877 [DOI] [PubMed] [Google Scholar]
- 48. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]