Background: Innate immune signaling requires multiple mechanisms to suppress signaling in the absence of stimulation.
Results: TNF receptor associated factor 6 (TRAF6) activity is regulated by reversible arginine methylation.
Conclusion: Arginine methylation of TRAF6 inhibits signaling in the absence of Toll-like receptor ligands.
Significance: Reversible TRAF6 methylation is a novel mechanism that controls innate immune responses.
Keywords: endotoxin, hepatocyte, innate immunity, macrophage, TNF receptor associated factor (TRAF), toll-like receptor (TLR), Jumonji domain-containing protein 6, arginine methylation, post-translational modification, protein arginine methyltransferase
Abstract
Arginine methylation is a common post-translational modification, but its role in regulating protein function is poorly understood. This study demonstrates that, TNF receptor-associated factor 6 (TRAF6), an E3 ubiquitin ligase involved in innate immune signaling, is regulated by reversible arginine methylation in a range of primary and cultured cells. Under basal conditions, TRAF6 is methylated by the methyltransferase PRMT1, and this inhibits its ubiquitin ligase activity, reducing activation of toll-like receptor signaling. In response to toll-like receptor ligands, TRAF6 is demethylated by the Jumonji domain protein JMJD6. Demethylation is required for maximal activation of NF-κB. Loss of JMJD6 leads to reduced response, and loss of PRMT1 leads to basal pathway activation with subsequent desensitization to ligands. In human primary cells, variations in the PRMT1/JMJD6 ratio significantly correlate with TRAF6 methylation, basal activation of NF-κB, and magnitude of response to LPS. Reversible arginine methylation of TRAF6 by the opposing effects of PRMT1 and JMJD6 is, therefore, a novel mechanism for regulation of innate immune pathways.
Introduction
Protein arginine methylation is a common posttranslational modification that plays a role in multiple pathways, including cell cycle control, RNA processing, and DNA replication. The protein arginine methyltransferase PRMT1 is responsible for ∼85% of total cellular arginine methylation (1) and catalyzes arginine mono- and dimethylation using S-adenosyl methionine (SAM)3 as a methyl donor. PRMT1 methylates histones; however, many non-histone protein targets are not yet defined. PRMT1 has very broad substrate specificity and preferentially modifies arginines in glycine-arginine-rich motifs. Abnormal function of PRMT1 is closely associated with several types of cancer and cardiovascular disease. Arginine methylation impacts gene transcription and splicing as well as upstream signal transduction including a number of innate immunity pathways (2). Until recently, arginine methylation was thought to be an irreversible modification due to the absence of demethylation enzymes. But PRMT1 is involved in processes such as cell cycle control, where responses are of short duration, and signal turnover is rapid. This suggests a possible dynamic regulation of this modification. Protein arginine demethylases have not been well described, but one of the Jumonji family proteins, JMJD6, has been reported to have arginine demethylase activity (3, 4). JMJD6 is a dioxygenase that can act as both an arginine demethylase and a lysyl hydroxylase. It is required during embryogenesis and is a key regulator of hematopoietic differentiation through its targets U2AF2/U2AF65 (5).
We recently showed that changes in PRMT1 activity are responsible for altered function of the transcription factor FOXO3 in models of liver disease (6). We were thus interested in what other effects might result from these changes in arginine methylation. One such possible methylation target is TNF receptor-associated factor 6 (TRAF6), which has been identified as a potential interaction partner of PRMT1 (7). TRAF6 is a key E3 ubiquitin ligase that mediates the synthesis of Lys-63-linked polyubiquitin chains essential for innate immune signaling downstream of the majority of the toll-like receptors (TLRs). Specific TLRs are expressed in multiple cell types such as monocytes, macrophages, intestinal epithelial cells, and the majority of liver cells including hepatocytes (8). TLRs recognize specific molecular patterns found in a broad range of microbial pathogens such as bacteria and trigger inflammatory and immune responses (9).
In this study we show that TRAF6-dependent TLR pathways are regulated by reversible arginine methylation of TRAF6 in a range of primary and cultured cells. TRAF6 is methylated on arginine residues by PRMT1 and is relatively inactive in its methylated state. After TLR ligand exposure, TRAF6 is demethylated by JMJD6, resulting in its activation. Loss of JMJD6 leads to reduced response to TLR ligands, and loss of PRMT1 leads to both pathway activation in the absence of the ligand and impaired post-ligand response. Basal TRAF6 pathway activation in human liver specimens is determined by the ratio of methylation to demethylation enzymes, and this ratio predicts susceptibility to bacterial infection in patients with cirrhosis.
Experimental Procedures
Cell Culture
Huh7.5 cells (obtained from Dr. Charles Rice) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% FBS, 50 units ml−1 penicillin, and 50 mg ml−1 streptomycin. THP1-LuciaTM NF-κB cells stably expressing NF-κB-inducible luciferase were from InvivoGen and were maintained according to the recommended procedures. Cells were treated where indicated with different TLR ligands (InvivoGen): Pam3CSK4 (TLR1/2 ligand), 100 ng/ml; LPS (TLR4), 10 ng/ml; flagellin (TLR5), 10 ng/ml; FSL-1 (TLR6/2), 1 ng/ml; imiquimod (TLR7), 250 ng/ml. IRAK1/4 inhibitor (catalog #407601) was obtained from Millipore and used at 1 μm. AMI-1 was obtained from EMD4Biosciences and used at 10 μm. Cells were transfected using Lipofectamine LTX transfection reagent (Invitrogen) according to the manufacturer's protocol. Huh7.5 cells were seeded at the density of 5 × 105 cells per well of the 6-well plate and transfected with 1–2 μg of each DNA per well. THP1-LuciaTM NF-κB cells were transfected where indicated with expression plasmids, or siRNA was incubated for 24 h, treated with TLR ligands, and incubated for additional 24 h before luciferase determination using QUANTI-luc reagent (InvivoGen).
Primary Antibodies
Anti-PRMT1 (F339), anti-IκB, anti-NFκB, anti-IRAK1, anti-IRAK3, anti-IRAK4, and anti-ubiquitin (clone P4D1) mouse antibodies were from Cell Signaling. Anti-LaminB (C20) and anti-β-actin were from Santa Cruz. Anti-mono and dimethyl-arginine (clone 7E6), anti-TRAF6 (rabbit), and anti-SAM68 antibodies were from Abcam. Anti-asymmetric-dimethyl-arginine antibodies were from Active Motif. Mouse anti-β-actin, anti-JMJD6, anti-TRAF6, and anti-FLAG (M2) antibodies were from Sigma. Anti-GAPDH was from Ambion. Anti-HA.11 (clone 16B12) was from Covance.
Secondary Antibodies
IRDye 800CW goat anti-mouse IgG and IRDye 680RD goat anti-rabbit IgG were from Li-COR. General HRP-conjugated secondary antibodies were from Southern Biotechnology Associates (Birmingham, AL).
Vectors
pRK5-HA-ubiquitin-Lys-63 plasmid was provided by Ted Dawson via Addgene (Addgene plasmid 17606). p6352 MSCV-CMV-CMV-FLAG-HA-JMJD6 was provided by Peter Howley via Addgene (Addgene plasmid 31358). pDeNy-hTRAF6 plasmid expressing TRAF6 dominant negative was from InvivoGen. pCMV6-TRAF6 and pCMV6-PRMT1 were from Origene. The TRAF6 point mutations and PRMT1 dominant-negative (V82A/L83A/D84A) were generated by site-directed mutagenesis (QuikChange kit, Stratagene).
Luciferase Reporter Assays
Huh7.5 cells were seeded onto 24-well plates. After 24 h, cells were serum-starved for 5–16 h and then cotransfected using Lipofectamine-LTX (Invitrogen) with NF-κB-luc reporter vector (0.2 μg per well) (10), pRL-tk vector (Renilla luciferase reporter, 0.03 μg per well, for normalization of transfection efficiency), and where indicated with p6352 MSCV-CMV-CMV-FLAG-HA-JMJD6, pDeNy-hTRAF6, pCMV6-PRMT1, or pCMV6-TRAF6 vectors (WT or mutants, 0.1 μg per well each). Cells were subsequently incubated for 24 h, treated with TLR ligands, and incubated for an additional 24 h before lysis and luciferase determination. Lysates were used to measure luciferase activity using the Dual luciferase assay kit (Promega) on a single tube Glomax 20/20 luminometer (Promega). Results were expressed as firefly luciferase/Renilla luciferase activity.
Cell Fractionation
Whole cell lysates were prepared from cells that had been washed and harvested by centrifugation in phosphate-buffered saline (PBS), pH 7.5. Cell pellets were resuspended in radioimmune precipitation assay buffer that contained 50 mm Tris, pH 7.5, 150 mm sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1 mm EDTA, and 1% protease and phosphatase inhibitors (Sigma). Lysates were centrifuged at 14,000 rpm for 15 min; supernatants were collected, and protein concentration was measured using the Bio-Rad protein assay kit (Bio-Rad).
Cytosolic and nuclear proteins were prepared as follows. Cells were washed twice with ice-cold PBS resuspended in Buffer A (10 mm HEPES, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 5 mm sodium butyrate, 0.6% CHAPS, protease, and phosphatase inhibitors) and incubated on ice for 30 min. Nuclei were pelleted by centrifugation at 3000 rpm (1000 × g) for 5 min, washed with Buffer A, and collected by centrifugation at 3000 rpm for 5 min. The supernatant constituted the cytosolic fraction. The nuclear pellets were resuspended in Buffer B (20 mm HEPES, pH 7.5, 1.5 mm MgCl2, 0.42 m NaCl, 0.2 mm EDTA, 10% glycerol, 1 mm DTT, protease, and phosphatase inhibitors), sonicated for 12–15 s, and incubated on ice for 30 min. Extracts were then centrifuged at 20,000 × g for 30 min to remove unsolubilized particles.
Immunoprecipitation and Protein Purification
Radioimmune precipitation assay buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 10% glycerol) extracts were precleared using 10 μg/ml preimmune rabbit or mouse IgG (Millipore) for 2 h at 4 °C. 1 μl of protein G magnetic beads (Millipore, LSKMAGG10) per 10 μg of antibody was added, and the sample was incubated for another 1 h at 4 °C. After removal of beads, proteins were immunoprecipitated using 10 μg/ml immunoprecipitation antibody overnight at 4 °C. 1 μl of protein G magnetic beads per 10 μg of antibody was added, and the sample was incubated for 4 h at 4 °C. The beads were magnetically separated and washed 2 times in radioimmune precipitation assay buffer. Beads were then resuspended in 10 μl of a 4× SDS loading buffer (0.25 m Tris, pH 6.8, 40% glycerol, 20% β-mercaptoethanol, 4% SDS) and analyzed by SDS-PAGE. FLAG-TRAF6 and FLAG-PRMT1 were purified using the FLAG® immunoprecipitation kit from Sigma according to the manufacturer's instructions. Proteins were eluted using FLAG peptide.
In Vitro Activity Assays
In vitro methylation assay was performed in a reaction mixture containing purified FLAG-TRAF6 (wt or mutants), 50 μm SAM, 0.5 μg of recombinant human PRMT1 (Origene) in 1× PBS buffer. Reaction mixtures were incubated for 1 h at 37 °C. The in vitro demethylation assay reaction contained 0.2 μg of purified FLAG-TRAF6, 0.5 μg of recombinant human JMJD6 (Origene) in 250 mm HEPES-KOH, pH 8.0, 70 μm Fe(NH4)2(SO4)2, and 10 mm ascorbate acid in the presence or absence of 5 mm α-ketoglutarate. Reaction mixtures were incubated for 1 h at 37 °C. After incubation the methylation or demethylation reaction mixtures were directly used for in vitro ubiquitination assays. The reaction mixtures containing ∼0.02 μg of purified FLAG-TRAF6 (WT or mutants) were supplemented with 0.2 μg of substrate NF-κ-B essential modulator (Origene), recombinant E1, E2, and ubiquitin solution in E3 ligase buffer (auto-ubiquitinylation kit from Enzo Life Sciences) according to the manufacturer's protocol. TRAF6 self-ubiquitination was monitored using 0.2 μg of purified FLAG-TRAF6. The reaction was started by the addition of ATP. Mixtures were incubated for 1 h at 37 °C. The resulting TRAF6 polyubiquitin species were analyzed by Western blotting using anti-ubiquitin antibodies.
Human Specimens
De-identified human liver specimens from normal livers (transplant donors) were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human tissue samples were approved by the Human Subjects Committee of the University of Kansas Medical Center. Subcellular fractions were isolated from frozen specimens by homogenization, passing the sample through a cell strainer (BD Falcon, 40 μm), and further fractionation as described for the cell culture specimens.
Peripheral blood mononuclear cells were isolated as follows. Whole blood was centrifuged for 15 min at 1200 × g with no brake. The buffy coat was then diluted with RPMI, layered over Ficoll, and centrifuged for 45 min at 200 × g with no brake. The peripheral blood mononuclear cell fraction was washed twice with RPMI and twice with PBS and resuspended in MACS buffer (Miltenyi Biotec). CD14+ cells were purified using MACS beads (human CD14) (Miltenyi Biotec, 130-050-201) according to manufacturer's instructions. Cells were differentiated by treatment with 1 × 104 units/ml of M-CSF for 5 days.
Western Blots
Protein extracts (15 μg) were subjected to 10% SDS-polyacrylamide gel electrophoresis, electrophoretically transferred to nitrocellulose membranes (Amersham Biosciences Hybond ECL, GE Healthcare), and blocked in 3% BSA/PBS at room temperature for 1 h. Primary antibodies were incubated overnight at the manufacturer recommended concentrations. Immunoblots were detected with the ECL Plus Western blotting Detection System (Amersham Biosciences) or using near-infrared fluorescence with the ODYSSEY Fc, Dual-Mode Imaging system (Li-COR). Expression levels were evaluated by quantification of relative density of each band normalized to that of the corresponding β-actin or GAPDH band density.
Real Time PCR
RNA was extracted from cultured cells using the RNeasy Mini kit (Qiagen). cDNA was generated with the random primer method using the RNA reverse transcription kit (Applied Biosystems, catalog #4368814). Quantitative real time RT-PCR was performed in a CFX96 real time system (Bio-Rad) using specific sense and antisense primers (PCR array, Bio-Rad) for 40 amplification cycles: 5 s at 95 °C, 30 s at 60 °C. PCR primers used are listed in supplemental Table S1.
siRNA Transfections
Knockdown of PRMT1 and JMJD6 was performed using specific siRNA duplexes (Trilencer-27 27-mer siRNA duplexes) from Origene. Trilencer-27 Universal Scrambled negative control siRNA duplex served as the negative control. SiTran1.0 Transfection Agent (Origene) was used for the transfection after the manufacturer's instructions.
LC MS Analysis
Ten 15-cm dishes of Huh7.5 cells were seeded at 1 × 107 cells per dish and were transiently transfected with 30 μg of TRAF6-FLAG plasmid per dish. Cells were harvested 48 h post-transfection. Extracts were obtained by lysing the cells in radioimmune precipitation lysis buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 10% glycerol, protease, and phosphatase inhibitors). TRAF6-FLAG was purified using a FLAG purification kit from Sigma according to the manufacturer's instructions. TRAF6-FLAG was eluted with FLAG peptide and separated on SDS gel. TRAF6-containing gel segments were digested with endoproteinases trypsin, chymotrypsin, GluC, or AspN. Identification of methyl and dimethyl-Arg on TRAF6 using in-gel digestion with multiple proteases, LC-MS/MS, and database searching was performed by MS Bioworks LLC, Ann Arbor, MI.
Isolation of Mouse Peritoneal Macrophages
Primary peritoneal macrophages were isolated as described previously (11). Male C57BL/6 mice at 5 weeks of age were killed by CO2 asphyxiation. Briefly, 10 ml of sterile PBS was injected into the caudal half of the peritoneal cavity using a 25-gauge needle (beveled side up) followed by shaking the entire body for 10 s. Saline-containing resident peritoneal cells was collected, and cells were plated on uncoated tissue culture plates (Greiner Bio-One, Monroe, NC) and incubated for 60 min at 37 °C. Nonadherent cells were removed by washing five times in warm PBS. Macrophages were maintained in RPMI medium (Invitrogen) containing 10% FBS.
Isolation of Mouse Primary Hepatocytes
Primary mouse hepatocytes were isolated from 5-week-old male C57/B6 mice by a two-step collagenase perfusion method as described previously (12). All perfusion solutions were maintained at 37 °C using a heated water bath, and the perfusion was carried out using a peristaltic pump. After the induction of anesthesia with ketamine, the peritoneal cavity was opened, the inferior vena cava was cannulated with a 24-gauge catheter, and the liver was perfused in situ via the inferior vena cava for 10 min at 37 °C with calcium- and magnesium-free Hanks' balanced salt solution (Hyclone:SH30588.02) followed by perfusion with Hanks' balanced salt solution containing calcium, magnesium (Hyclone, SH30268.01), and 0.025 mg/ml LiberaseTM (Roche Applied Science, 0540112001) until the liver revealed signs of digestion (∼8–10 min). The liver was then excised from the body and placed in a small sterile beaker containing cold calcium- and magnesium-free Hanks' balanced salt solution. The liver was then chopped using a pair of small sterile scissors to release the isolated liver cells. The cell suspension was filtered through nylon gauze (100 μm), centrifuged for 5 min at 50 × g at 4 °C, and then resuspended in fresh cold calcium- and magnesium-free Hanks' balanced salt solution. This was repeated three times to isolate the hepatocyte fraction. Hepatocyte viability was evaluated by the trypan blue exclusion method, and the number of hepatocytes isolated was determined using a hemocytometer.
Enzyme-linked Immunosorbent Assays (ELISA)
ELISAs were carried out as follows. Plastic 96-well microtiter plates (Immuno Maxi Sorb, Nunc) were coated overnight with an excess (0.5 μg) of the first primary antibody. Unbound protein was washed with PBS. Wells were blocked for 2 h with 0.3 ml 3% BSA (Sigma, Cohn Fraction V, essentially fatty acid-free) in PBS. After washing, samples containing the protein of interest (in 50 μl) were added, incubated for 1 h at room temperature, and then washed with PBS. The second primary antibody (0.1 μg in 50 μl) was added, incubated overnight at 4 °C, washed, and then visualized by incubation with secondary antibody conjugated with horseradish peroxidase (The Jackson Laboratory) in the presence of 3% BSA for 1 h followed by reaction with ABTS (Sigma). The reaction was allowed to proceed for 20 min, and then plates were quantitated spectrophotometrically at 410 nm.
Proximity Ligation Assay
Proximity ligation assays (PLA) were carried out using PLA kit (Sigma) according to the manufacturer's instructions. Before detection THP-1 cells were treated with 25 ng/ml PMA, incubated for 48 h, and treated as indicated. After treatment, cells were fixed with 4% paraformaldehyde, washed, permeabilized with 1% Triton in PBS, blocked with supplied PLA blocking buffer, and incubated with primary antibody against TRAF6, methylated arginine, or JMJD6 as indicated. Interactions were visualized using Duolink Brightfield detection reagent (Sigma). The PLA assay omitting one or both primary antibodies was used as a negative control.
Statistics
Results are expressed as the mean ± S.D. Student's t test, paired t test, Spearman's rank correlation, χ2 test, or one-way analysis of variance with Bonferroni post hoc test was used for statistical analyses. Multivariate analysis was performed using SimFit software (W. G. Bardsley, University of Manchester; The Simfit Package). p < 0.05 was considered significant.
Results
TRAF6 Is Arginine-methylated by PRMT1
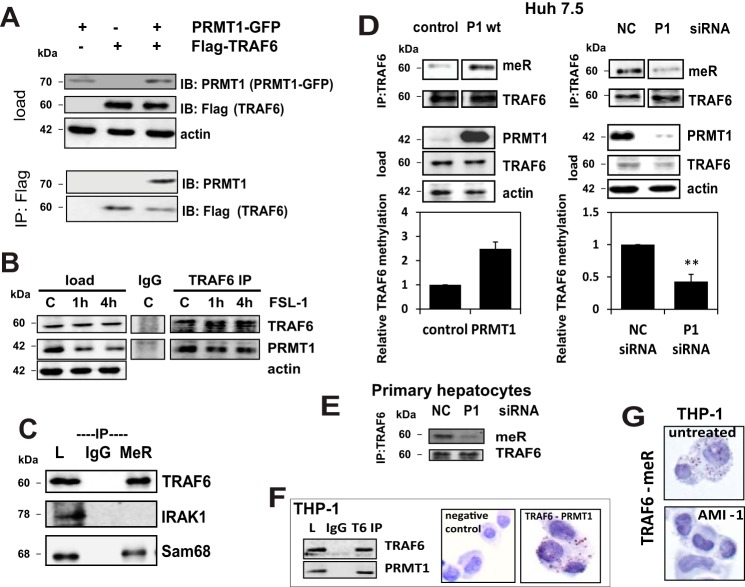
In examining the possible consequences of changes in arginine methylation, we searched for potential interaction partners of PRMT1 in the IntAct Molecular Interaction Database. Among 88 reported proteins, TRAF6 appeared to be a promising target because of its central role in innate immune signaling (interaction_id:EBI-1083602; Ref. 7). To study this possible interaction we first determined if there was a binding interaction between TRAF6 and PRMT1. Fig. 1A demonstrates that overexpressed PRMT1 in Huh7.5 hepatoma cells co-immunoprecipitates with FLAG-TRAF6, indicating binding of these proteins. An identical binding result was obtained for the endogenous proteins (Fig. 1B) both in the presence and absence of a TLR6/2 ligand, FSL-1, suggesting that TRAF6 might be a new substrate for PRMT1. To study this possibility we determined whether TRAF6 was arginine-methylated. Fig. 1C shows the result of immunoprecipitation with a methyl-arginine antibody followed by immunoblotting for TRAF6, or control proteins IRAK1 and SAM68, a protein known to be methylated by PRMT1 (2). Under control conditions we were able to detect methylation of TRAF6 and SAM68, but not IRAK1. Overexpression of PRMT1 increased TRAF6 methylation, and PRMT1 knockdown decreased it in either Huh7.5 cells (Fig. 1D) or primary mouse hepatocytes (Fig. 1E). These effects were statistically significant.
FIGURE 1.
PRMT1 binds and methylates TRAF6. A, Huh7.5 cells were transiently transfected with expression plasmids for PRMT1-GFP and TRAF6-FLAG where indicated. TRAF6-FLAG was immunoprecipitated (IP) using anti-FLAG affinity resin. Immunoprecipitated proteins were blotted (IB) for TRAF6 (anti-FLAG antibody) and PRMT1. B, Western blot analysis of endogenous PRMT1 and TRAF6 co-immunoprecipitated from Huh7.5 cells untreated or treated with FSL-1 (TLR6/2 ligand) for the indicated times using anti-TRAF6 antibody or IgG as a negative control. C, Huh7.5 cells were lysed, and arginine-methylated proteins were immunoprecipitated and analyzed by Western blot for the presence of methylated TRAF6, IRAK1, and SAM68 (positive control) using specific antibodies (L, load; IgG, nonspecific pulldown with mouse IgG; MeR, proteins immunoprecipitated with anti-methyl-arginine antibody). D, TRAF6 was immunoprecipitated and immunoblotted with methyl-arginine antibody from control cells or cells expressing wild type PRMT1 (P1, left) or cells expressing negative control (NC) or PRMT1-specific siRNA (right). Lower panels show the densitometry quantification of MeR/TRAF6 signal ratio from three independent immunoprecipitation experiments. **, p < 0.01. E, TRAF6 immunoprecipitation and methyl-arginine immunoblot of lysates from primary mouse hepatocytes expressing negative control (NC) or PRMT1 specific siRNA. F, Western blot analysis of endogenous PRMT1 and TRAF6 co-immunoprecipitated from THP-1 cells using anti-TRAF6 antibody or IgG as a negative control (left). Shown are Representative images of proximity ligation assays in THP-1 cells (right). TRAF6 interaction with PRMT1 was detected using anti-PRMT1 and anti-TRAF6 antibodies. Negative control, signal in the absence of primary antibodies. G, representative images of proximity ligation assays in THP-1 cells untreated or treated with AMI-1 for 16 h. TRAF6 methylation was detected using the combination of anti-TRAF6 and anti-methyl-arginine antibodies.
Further evidence of TRAF6 methylation and interaction with PRMT1 was obtained in a second cell type, THP-1 monocytes. Fig. 1F, left, shows the interaction of TRAF6 and PRMT1 by immunoprecipitation with an anti-TRAF6 antibody or IgG control followed by immunoblotting for TRAF6 and PRMT1. It demonstrates that PRMT1 co-immunoprecipitates with TRAF6. Additional evidence of TRAF6-PRMT1 interaction in THP-1 cells was obtained by PLA. In this case close proximity of the two tested antibodies produces a positive signal indicated by brown/red dots, which can be readily seen against the blue hematoxylin counterstain but not in the absence of the primary antibodies (Fig. 1F, right).
Direct evidence of TRAF6 arginine methylation in THP-1 cells was obtained by PLA using anti-TRAF6 and anti-methyl-arginine antibodies (Fig. 1G). This showed a positive signal in 97.1% of untreated THP-1 cells. The signal was nearly abolished by treatment with the methylation inhibitor, AMI-1 (1.2% of cells positive, p < 0.001).
We performed LC-MS-MS to directly determine the sites of basal methylation in TRAF6. Overexpressed TRAF6 was purified from untreated cells and subjected to mass spectrometry analysis. We were able to achieve 95% peptide coverage and confirmed the presence of arginine methylation at 12 sites (Table 1). Detailed spectra are shown in the supplemental Figure S1.
TABLE 1.
Identification of methyl-arginine sites in TRAF6 under basal conditions
r denotes modified arginine residue.
| Site | Modification | Sequence | Localization probability (A score) |
|---|---|---|---|
| Arg-78 | Methyl | ICLMALrEAVQTP | 100% (1000.00) |
| Arg-88 | Methyl | QTPCGHrFCKACI | 100% (1000.00) |
| Arg-125 | Methyl | PDNFAKrEILSLM | 100% (1000.00) |
| Arg-167 | Methyl | DCPQCQrPFQKFH | 100% (1000.00) |
| Arg-184 | Methyl | ILKDCPrRQVSCD | 100% (1000.00) |
| Arg-185 | Methyl | LKDCPRrQVSCDN | 100% (1000.00) |
| Arg-224 | Methyl | PDNFAKrEILSLM | 100% (1000.00) |
| Arg-255 | Methyl | CHEKMQrNHLARH | 100% (1000.00) |
| Arg-271 | Methyl | NTQSHMrMLAQAV | 100% (1000.00) |
| Arg-496 | Methyl | DDTLLVrCEVSTR | 100% (190.29) |
| Arg-305 | Dimethyl | IHQLEGrLVRQDH | 100% (1000.00) |
| Arg-443 | Dimethyl | QSEAPVrQNHEEI | 100% (1000.00) |
Arginine Methylation Decreases TRAF6 Signaling Activity
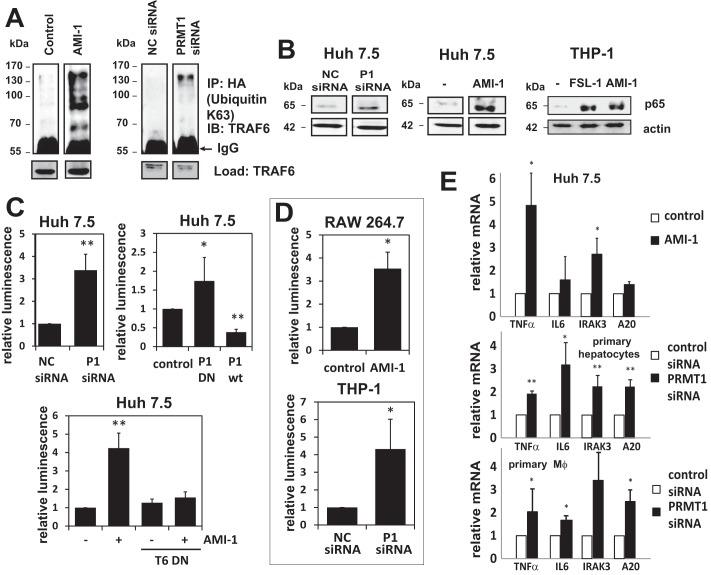
Next we examined the effects of PRMT1 on TRAF6 activity. We first measured TRAF6 self-ubiquitination, as a marker of activation, by immunoprecipitating Lys-63-ubiquitinated proteins from cells overexpressing an HA-ubiquitin-Lys-63 mutant (with all lysines except Lys-63 mutated to arginines) and immunoblotting for TRAF6. Inactivation of PRMT1 either by a small molecule inhibitor of arginine methyl transferases (AMI-1) or by PRMT1 knockdown caused an increase of TRAF6 ubiquitination in the absence of TLR ligand (Fig. 2A). These results suggest that decreased TRAF6 methylation activates TRAF6 in vivo.
FIGURE 2.
PRMT1 activity is required for suppression of the TRAF6-dependent TLR pathway. A, Huh7.5 cells were transfected with HA-ubiquitin-Lys-63 (all lysines were substituted with arginine except Lys-63). Cells were otherwise untreated, pretreated with 10 μm AMI-1, or treated with negative control (NC) or PRMT1-specific siRNA. Lys-63-linked polyubiquitinated proteins were immunoprecipitated (IP) with anti-HA antibody, and the presence of TRAF6 was analyzed by Western blotting (IB). B, nuclear fractions were prepared from Huh7.5 or THP-1 cells treated as indicated with siRNA, the methylation inhibitor AMI-1 (10 μm), or the TLR6/2 ligand FSL-1. Immunoblots for NF-κB p65 are shown. C, NF-κB luciferase reporter assays of Huh7.5 cells expressing negative control (NC) or PRMT1-specific siRNA, expressing either wild type or dominant-negative PRMT1, or cells expressing TRAF6 dominant-negative construct treated with or without 10 μm AMI-1 as indicated. Transfection efficiency was controlled by expression of Renilla luciferase, and data are presented as the mean ± S.D. of relative luminescence. *, p < 0.05; **, p < 0.01. n ≥ 4. D, NF-κB luciferase reporter assays from RAW264.7 cells (control or pretreated with 10 μm AMI-1) or THP-1 cells expressing negative control (NC) or PRMT1-specific siRNA. Data are presented as the mean ± S.D. *p < 0.05. n ≥ 3. E, relative mRNA of NF-κB target genes TNFα, IL6, IRAK3 (IRAK-M), and TNFAIP3 (A20) in Huh7.5 cells, primary mouse hepatocytes, or primary mouse macrophages treated with or without AMI-1 for 16 h. Data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05. n = 3.
To further study the effects of methylation on TRAF6 activity, we knocked down or inhibited PRMT1 in Huh7.5 or THP-1 cells and measured NF-κB activation. PRMT1 inhibition or knockdown increased nuclear translocation of p65 under basal conditions (Fig. 2B). In THP-1 cells the effect was similar to that seen after the addition of a TLR2/6 ligand, FSL-1.
NF-κB reporter luciferase activity was similarly increased by inhibiting PRMT1 either by siRNA, AMI-1, or expressing a dominant-negative form of the protein (Fig. 2C). Overexpression of wt-PRMT1, on the other hand, caused a decrease in the basal level of NF-κB luciferase activity (Fig. 2C). We confirmed that the step at which PRMT1 affects the TLR pathway is TRAF6 as activation of NF-κB luciferase activity by the methylation inhibitor AMI-1 was blocked by the TRAF6 dominant-negative mutant (Fig. 2C). In addition to the effect in Huh7.5 cells, PRMT1 inhibition similarly activated NF-κB activity in the monocytic cell lines Raw264.7 and THP-1 (Fig. 2D).
We further determined the effects of the PRMT1 inhibitor on basal mRNA expression of NF-κB target genes (Fig. 2E). PRMT1 inhibition increased mRNA levels of TNF, IL-6, and IRAK3 in Huh7 cells, primary mouse hepatocytes, and primary mouse peritoneal macrophages (Fig. 2E). These data suggest that PRMT1 is needed to keep the pathway inactive in the absence of the ligand in multiple cell types.
TRAF6 Is Demethylated by the Histone Demethylase JMJD6
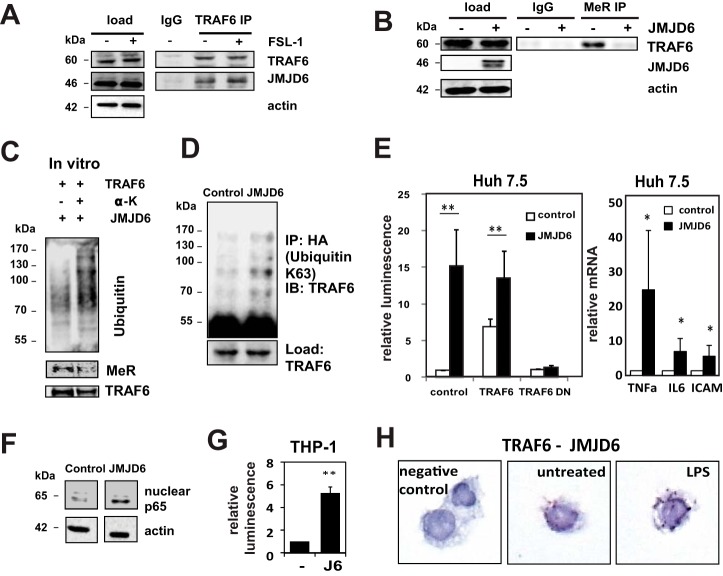
The involvement of arginine methylation in a rapidly responding pathway such as TLR signaling suggests the presence of a specific demethylase enzyme. Recent work (3, 4) has shown that the Jumonji domain-containing protein JMJD6 can demethylate arginine residues. We, therefore, tested whether it was involved in demethylation of TRAF6. First, we tested whether JMJD6 binds to TRAF6. Fig. 3A demonstrates that JMJD6 co-immunoprecipitated with endogenous TRAF6 in the presence or absence of the TLR6/2 ligand FSL-1. JMJD6 overexpression resulted in TRAF6 demethylation (Fig. 3B). JMJD6 enzymatic activity is known to require the presence of α-ketoglutarate (13). Fig. 3C demonstrates that purified JMJD6 is able to demethylate TRAF6 in vitro and increase its self-ubiquitinylation in an α-ketoglutarate-dependent fashion. JMJD6 overexpression increased TRAF6 ubiquitination in vivo (Fig. 3D) similar to the effect of PRMT1 inactivation.
FIGURE 3.
TRAF6 is demethylated and activated by JMJD6. A, Huh7.5 cells were treated with or without FSL-1 (TLR6/2 ligand) for 1 h, and TRAF6 was immunoprecipitated (IP) from cell lysates. Western blot analysis of JMJD6 and TRAF6 is shown. Immunoprecipitation with nonimmune IgG served as the negative control. B, immunoprecipitation with anti-methyl-arginine antibody and immunoblotting for TRAF6 from control cells or cells overexpressing JMJD6 is shown. MeR, proteins immunoprecipitated with anti-methyl-arginine antibody. C, in vitro self-ubiquitinylation of recombinant TRAF6 (0.2 μg per reaction) in the presence of 0.2 μg of JMJD6 with or without its required cofactor, α-ketoglutarate (α-K). Immunoblots are shown for ubiquitin-specific, methyl-arginine-specific, and TRAF6-specific antibodies. D, Huh7.5 cells were transfected with an HA-tagged modified ubiquitin containing Lys-63 as the only lysine with or without JMJD6, immunoprecipitated with anti-HA antibody, and probed for TRAF6 to detect Lys-63-linked poly-ubiquitinated TRAF6. IB, immunoblot. E, left panel, control Huh7.5 cells or cells expressing JMJD6 were co-transfected with wild type or dominant-negative (DN) TRAF6 and NF-κB luciferase reporter assay were performed. Right panel, RNA was isolated from control or JMJD6-transfected cells, and the indicated NF-κB target gene mRNA abundance was determined by RT-qPCR. Data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05. n = 3. ICAM, intercellular adhesion molecule 1. F, Western blot analysis of NF-κB p65 in nuclear fractions of control Huh7.5 cells and cells transfected with JMJD6. G, NF-κB luciferase reporter assay in THP-1 cells, either control cells (−) or cells expressing JMJD6 (J6) (left). Data are presented as the mean ± S.D. **, p < 0.01. n = 3. H, representative images of proximity ligation assays in THP-1 cells untreated or treated with LPS for 30 min (right). TRAF6 interaction with JMJD6 was detected using anti-JMJD6 and anti-TRAF6 antibodies. Negative control, signal in the absence of primary antibodies.
We next examined the effect of JMJD6-induced demethylation on signaling. Overexpression of JMJD6 in Huh7.5 cells strongly activated NF-κB reporter activity, nuclear p65 localization, and mRNA expression of NF-κB-dependent genes (Fig. 3, E and F). This was similar to the effect seen after PRMT1 inhibition. JMJD6-induced stimulation was blocked in the presence of dominant-negative TRAF6 but not wild type TRAF6 (Fig. 3E), demonstrating that activation is at the level of TRAF6. We further confirmed that TRAF6 can be activated by JMJD6 in THP-1 cells where JMJD6 overexpression also activated NF-κB activity (Fig. 3G).
We further studied the interaction between TRAF6 and JMJD6 using PLA (Fig. 3H). We observed positive PLA signals in control cells but not in the absence of the primary antibodies. Interaction persisted after treatment with LPS. Direct binding of TRAF6 and JMJD6, as evidenced by a positive PLA signal, was observed in 79% (76/96) of control cells and 83% (72/87) of cells treated with LPS. These data suggest that JMJD6 demethylates and activates TRAF6.
Arg-88 and Arg-125 Are Involved in Regulation of TRAF6 Activity
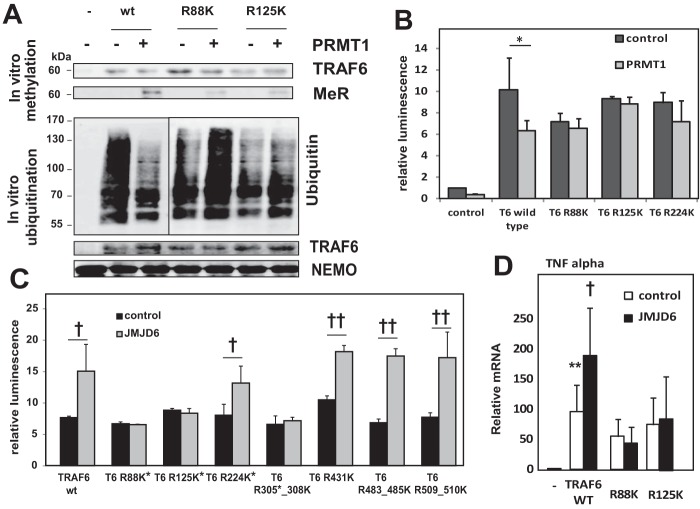
Next we evaluated which methylation sites were important in regulating TRAF6 activity. We mutated multiple arginines to lysine and assessed functional responses in vitro and in vivo. In an in vitro methylation assay, purified PRMT1 and its cofactor SAM were sufficient to methylate wild type TRAF6. R88K and R125K TRAF6 mutants were methylated to a lesser extent than wild type protein (Fig. 4A). These TRAF6 mutants were then used in an in vitro ubiquitination assay measuring the ability of TRAF6 to ubiquitinate the substrate protein NF-κ-B essential modulator. With wild type TRAF6, there was robust ubiquitination in the absence of PRMT1, and this was reduced by the addition of the methyltransferase. Neither R88K nor R125K mutants of TRAF6 showed a decrease of ubiquitin ligase activity in the presence of PRMT1 (Fig. 4A). Similar effects were observed by co-expressing PRMT1 with TRAF6 mutants and detecting TRAF6-dependent NF-κB activation. Wild type TRAF6 induced NF-κB activity, and this was suppressed by PRMT1. In contrast, NF-κB activation by methylation-defective TRAF6 mutant constructs was not suppressed by PRMT1 (Fig. 4B).
FIGURE 4.
Identification of functional TRAF6 methylation sites. A, upper panel, in vitro methylation assay. Wild type or mutant TRAF6 purified from Huh7.5 cells was methylated in vitro by recombinant PRMT1 in the presence of 50 μm SAM and analyzed using anti-TRAF6 and anti-methyl-arginine antibody. Lower panel, in vitro ubiquitination assay. Untreated wild type or mutant TRAF6 or each TRAF6 methylated in vitro by PRMT1 as above was incubated in vitro with recombinant E1, E2, and NF-κ-B essential modulator (NEMO) added as a ubiquitination substrate. Polyubiquitinated NEMO was detected using specific antibody against polyubiquitin chains. MeR, proteins immunoprecipitated with anti-methyl-arginine antibody. B, NF-κB luciferase reporter assays from control cells or cells expressing wild type or mutant TRAF6 co-transfected with or without wild type PRMT1 where indicated. Data are presented as the mean ± S.D. *, p < 0.05, n = 3. C, NF-κB luciferase reporter assays. Control Huh7.5 cells or cells overexpressing JMJD6 were co-transfected with wild type or mutant constructs of TRAF6. *, methylation at this site was confirmed by mass spectrometry. Data are presented as the mean ± S.D. †, p < 0.05; ††, p < 0.01, n ≥ 3. D, relative levels of TNF-α mRNA in Huh7.5 cells transfected with indicated TRAF6 mutants with or without co-expression of JMJD6 as indicated. **, p < 0.01 compared with control; †, p < 0.05 compared with wild type TRAF6 alone. n = 3.
We also examined the effects of these TRAF6 arginine substitutions on JMJD6-mediated signal activation. JMJD6 activated wild type TRAF6 but not R88K, R125K, or R305K/R308K mutants (Fig. 4, C and D). Several other arginine residues such as Arg-224 or Arg-431 were not required for JMJD6 activation.
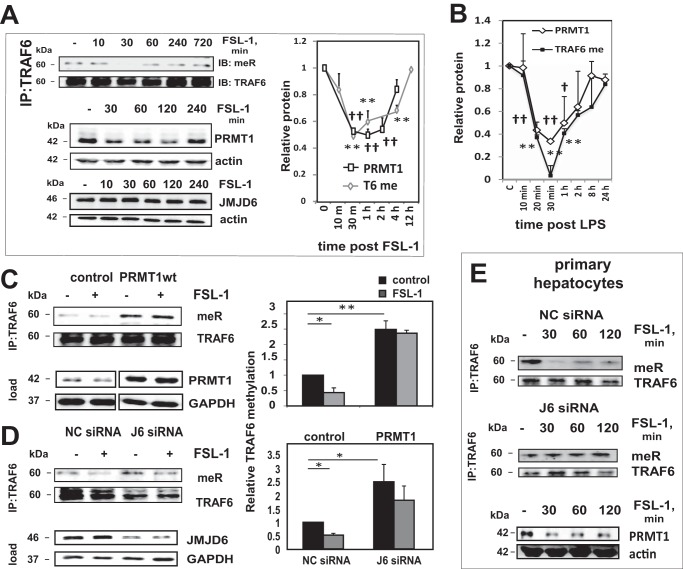
TRAF6 Is Demethylated after TLR Ligand Exposure
To examine the role of TRAF6 methylation in TLR responses, we first determined the arginine methylation status of TRAF6 after pathway activation. Fig. 5A shows results in Huh7.5 cells of immunoprecipitation with TRAF6 antibody followed by immunoblotting for methyl-arginine. Stimulation with FSL-1, a TLR2/6 ligand, induced a loss of TRAF6 methylation by 30 min, which subsequently recovered. We then determined if there were corresponding changes in the levels of the two modifying enzymes, PRMT1 and JMJD6, which might be responsible for changes in methylation. PRMT1 protein level was decreased 2-fold 30 min post-exposure to FSL-1 following a similar time course as was observed for the change in TRAF6 methylation (Fig. 5A). Over this same time period, JMJD6 level remained unchanged.
FIGURE 5.
TRAF6 is demethylated after activation of TLR pathway. A, Western blot analysis of TRAF6 immunoprecipitated (IP) and immunoblotted (IB) with anti-TRAF6 and anti-methyl-arginine antibody (top left) or JMJD6 and PRMT1 protein levels (lower left) in Huh7.5 cells treated with FSL-1 for the indicated times. The right panel shows densitometry quantification of MeR/TRAF6 signal ratio and PRMT1 relative protein levels for three independent experiments. Data are presented as mean ± S.D. **, p < 0.01 (TRAF6 methylation); ††, p < 0.01 (PRMT1) compared with untreated cells. B, relative PRMT1 protein levels and relative TRAF6 methylation in THP-1 cells treated with LPS for the indicated times, measured by ELISA as described under “Experimental Procedures.” Data are presented as the mean ± S.D. **, p < 0.01 (TRAF6 methylation); †, p < 0.05; ††, p < 0.01 (PRMT1) compared with untreated cells. n = 3. C, control cells or cells overexpressing wild type PRMT1 were treated with or without FSL-1 for 30 min, and lysates were immunoprecipitated and immunoblotted with methyl-arginine antibody. Densitometry quantification of MeR/TRAF6 signal ratio is shown for three independent experiments. Data are presented as the mean ± S.D. *, p < 0.05; **, p < 0.01. n ≥ 3. MeR, proteins immunoprecipitated with anti-methyl-arginine antibody. D, cells were transfected with either negative control or JMJD6-specific siRNA and analyzed as in C. Densitometry quantification of MeR/TRAF6 signal ratio is shown for three independent experiments. Data are presented as the mean ± S.D. *, p < 0.05; **, p < 0.01. n ≥ 3. E, top two panels, TRAF6 was immunoprecipitated and immunoblotted with methyl-arginine antibody from primary mouse hepatocytes transfected with negative control (NC) siRNA or JMJD6 (J6)-specific siRNA and treated with FSL-1 ligand for indicated times. Bottom panel, Western blot of PRMT1 protein levels in cells treated with FSL-1 for the indicated times.
We further determined the time course of ligand-induced changes in PRMT1 and TRAF6 methylation in THP-1 monocytes (Fig. 5B). For these studies we used sandwich ELISA assays to measure both PRMT1 level and TRAF6 methylation. This technique produced a more quantitative measure than Western blot and densitometry. In THP-1 cells, both PRMT1 protein levels and TRAF6 methylation decreased rapidly after LPS treatment, reaching nadir at 30 min (Fig. 5B).
The ligand-induced loss of TRAF6 methylation was prevented by either PRMT1 overexpression or JMJD6 knockdown (Fig. 5, C and D), suggesting that loss of PRMT1 in the setting of continued JMJD6 presence was responsible for ligand-induced TRAF6 demethylation. We observed a nearly identical ligand-induced TRAF6 demethylation in primary hepatocytes as well. Demethylation was maximum at 30 min post ligand, corresponding to the loss of PRMT1 protein. As in Huh7.5 cells, demethylation was dependent on the presence of JMJD6 (Fig. 5E). These results demonstrate that TRAF6 is demethylated after TLR ligand exposure in multiple cell types. Because demethylation increases TRAF6 activity, these data suggest that demethylation might be required for TRAF6 activation after ligand exposure.
Impact of TRAF6 Methylation on Ligand-induced TLR Responses
The above data demonstrate that the basal activity of TRAF6 is inhibited by arginine methylation and activated by demethylation. The effect that this might have on TLR ligand-induced signaling is not easy to predict. Demethylation might simply amplify the entire process enhancing ligand responses, but alternatively, basal pathway activation could induce a desensitization phenomenon (14) that might result in increased basal activity but decreased peak ligand responses.
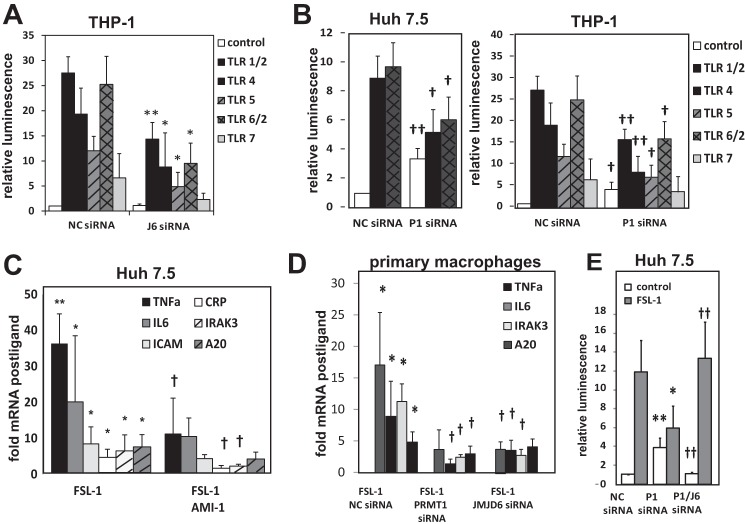
To examine the impact of TRAF6 methylation on ligand responses we tested the effects of increasing or decreasing methylation by reducing the activity of either JMJD6 or PRMT1. Knockdown of JMJD6 by siRNA, which prevented TRAF6 demethylation, had no effect on basal pathway activation but decreased NF-κB activation in response to TLR ligands in both THP-1 and Huh7.5 cells (Fig. 6A). This suggests that a properly timed demethylation response is required for optimal ligand response. Unlike JMJD6 knockdown, knockdown of PRMT1 caused constitutive pathway activation, but it also decreased the NF-B ligand response (Fig. 6B). The PRMT1 inhibitor similarly decreased ligand-induced nuclear translocation of p65 (data not shown). The general observation that PRMT1 inhibition produces basal activation but suppression of ligand responses was also seen for ligand-induced NF-κB target genes mRNA expression as well (Fig. 6, C and D).
FIGURE 6.
Impact of TRAF6 arginine methylation on the ligand-induced TLR response. A, THP-1-Lucia NF-κB reporter monocytes were transfected with negative control siRNA or JMJD6-specific siRNA and treated with ligands for different TLR molecules as indicated. Results of luciferase reporter assays are presented as the mean ± S.D. *, p < 0.05; **, p < 0.01 compared with negative control (NC) siRNA. n ≥ 3. B, luciferase reporter assays in Huh7.5 cells (left panel) or THP-1-Lucia NF-κB reporter monocytes (right panel) transfected with negative control siRNA or PRMT1-specific siRNA. Cells were treated with TLR ligands as indicated. Data are presented as the mean ± S.D. †, p < 0.05; ††, p < 0.01 compared with negative control siRNA. n ≥ 3. C, -fold increase in mRNA levels induced by FSL-1 exposure was determined in Huh7.5 cells treated with or without PRMT1 inhibitor AMI-1 for 16 h where indicated. *, p < 0.05; **, p < 0.01 compared with control; †, p < 0.05 compared with FSL-1 only. ICAM, intercellular adhesion molecule 1. D, FSL-1-induced -fold mRNA changes are shown in primary peritoneal macrophages. Cells were transfected with negative control (NC) siRNA and PRMT1-specific or JMJD6-specific siRNA where indicated. *, p < 0.05, compared with control; †, p < 0.05 compared with NC siRNA, n = 3. E, NF-κB luciferase reporter assays in Huh7.5 cells in the presence or absence of TLR6/2 ligand (1 ng/ml of FSL-1) that were transfected with negative control siRNA, PRMT1-specific siRNA, or both PRMT1- and JMJD6-specific siRNA. Data are presented as the mean ± S.D. *, p < 0.05; **, p < 0.01 compared with NC siRNA; ††, p < 0.01 compared with P1 siRNA. n = 3.
If the loss of TRAF6 methylation was the primary reason for the dual effects of PRMT1 inhibition, we reasoned that this likely required the demethylase activity of JMJD6. To test this possibility we simultaneously knocked down PRMT1 and JMJD6. Simultaneous reduction of JMJD6 completely compensated for the loss of PRMT1 and restored both the low basal and high stimulated NK-κB activity (Fig. 6E). These data show that the activity of TRAF6 is regulated in a dynamic fashion, and its activity depends on the ratio of methylation and demethylation enzymes rather than the amount of either enzyme alone.
Methylation-dependent TLR Responses in Primary Isolated Cells and Human Liver
The above results were obtained in cell culture and in vitro models. To determine if TRAF6 methylation plays a significant role in innate immunity in humans, we conducted a series of experiments in human liver samples and human blood monocyte-derived macrophages (HBDM).
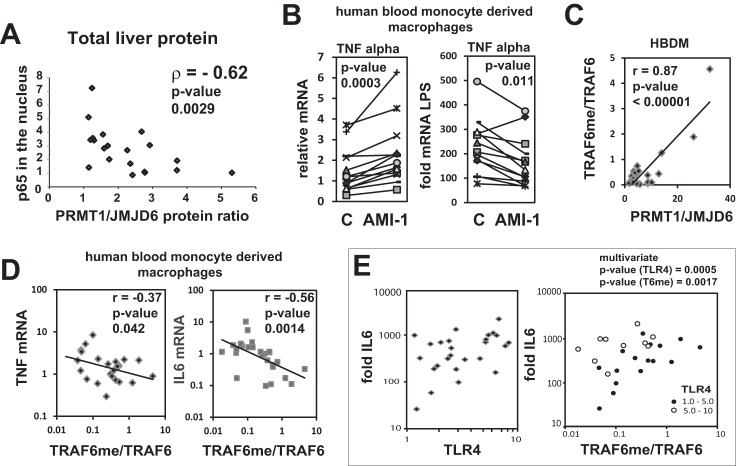
Baseline activation of inflammatory pathways is often related to disease and is one of the reasons for desensitization and poor prognosis for infection clearance (8). To determine if natural variations in PRMT1/JMJD6 ratio can account for variations in basal NF-κB activation in liver, we measured PRMT1/JMJD6 ratios by Western blot in normal human livers and correlated these with nuclear p65, a measure of basal pathway activation (Fig. 6A). In normal liver the ratio of PRMT1/JMJD6 was negatively correlated with baseline nuclear p65 (Fig. 7A, ρ = −0.62, p = 0.003). This suggested that a high degree of basal pathway activation might be attributed to low arginine methylation, similar to that seen in the primary hepatocytes.
FIGURE 7.
TRAF6 methylation correlates with TLR responses in humans. A, cytosolic and nuclear extracts were prepared from normal human liver tissue (liver transplant donor biopsy samples). Western blots were performed for PRMT1 and JMJD6 in cytosolic fractions and for NF-κB p65 in nuclear fractions. Data points represent values for individual liver specimens. n = 20. B, human peripheral blood monocytes were differentiated into macrophages and treated with or without AMI-1 for 16 h. Left panel, relative mRNA of TNFα in individual human macrophage preparations before (C) and after exposure to AMI-1. Right panel, -fold increase in TNFα mRNA 1 h after LPS exposure. Cells were pretreated with AMI-1 as indicated. Individual patient samples are connected by lines, n = 10. C, TRAF6 methylation was measured by ELISA as described under “Experimental Procedures.” Western blots were performed for JMJD6 protein concentration in cytosolic fractions from individual HBDM preparations. PRMT1 was measured using ELISA. n = 26. D, basal mRNA levels of TNFα and IL6 are plotted against relative TRAF6 methylation in individual HBDM preparations. E, -fold increase in IL6 mRNA levels is plotted against relative TLR4 expression or relative TRAF6 methylation in individual HBDM preparations. n = 26. Dependence of LPS-induced mRNA increases on TLR4 expression and TRAF6 methylation (6Tme) were statistically significant by multivariate linear regression, p = 0.0005 and 0.0017, respectively.
Next we looked at whether the PRMT1/JMJD6 ratio defines variations in innate immune response in myeloid cells. For that purpose we used human monocyte-derived macrophages and determined the effect of the methylation inhibitor AMI-1 on basal and LPS-stimulated cytokine mRNA levels. There was considerable variability between macrophage preparations, but in almost every case AMI increased basal levels of TNFα mRNA and reduced the response to LPS (Fig. 6B). We next directly measured TRAF6 methylation in those cells using an ELISA-based assay and measured PRMT1 and JMJD6 protein levels either by ELISA or Western blotting. PRMT1/JMJD6 ratio highly correlated with TRAF6 arginine methylation level (Fig. 6C), suggesting that the relative abundance of these enzymes controls the methylation state of TRAF6 protein in HBDMs.
Similar to the situation in whole liver extracts, low levels of TRAF6 methylation in HBDMs correlated with high basal levels of NF-κB target genes TNFα and interleukin 6 (IL6; Fig. 6D). Response to LPS, measured as -fold change in IL6 mRNA, was dependent on TRAF6 methylation as well as TLR4 expression, and both positive correlations were statistically significant by multivariate analysis (Fig. 6E). Thus, TRAF6 arginine methylation is an important factor defining both basal NF-κB activation and magnitude of response to TLR ligands in human macrophages.
Demethylation-induced TLR Desensitization Is a Result of Up-regulation of IRAK3
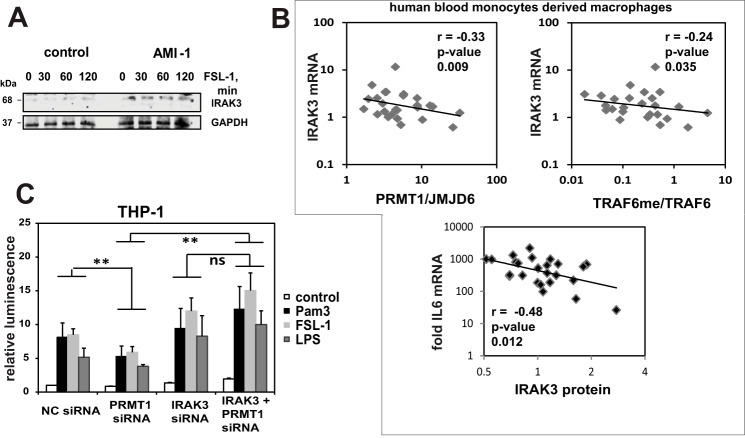
It is well known that innate immune pathway preactivation can lead to tolerance through multiple mechanisms including downstream production of inhibitors such as A20 (15), SOCS-1 (16), sMyD88 (17), or IRAK3, alternatively called IRAK-M (18). To determine if inhibitor accumulation might contribute to the desensitization phenomenon we observed, we looked for accumulation of molecules known to suppress the TLR response. We found that PRMT1 inhibition resulted in a significant increase in protein and mRNA levels of IRAK3 in Huh7.5 cells (Figs. 2E and 8A) consistent with the idea that pathway preactivation and consequent inhibitor accumulation might be responsible for tolerance in cells with reduced PRMT1 activity. We further examined human blood monocyte-derived macrophages and similarly observed a negative correlation between either the PRMT1/JMJD6 ratio or TRAF6 methylation and IRAK3 mRNA levels and observed that IRAK3 protein level negatively correlated with response to LPS (Fig. 7B). This suggests that IRAK3 up-regulation could be a mechanism for why low TRAF6 methylation reduces TLR responses.
FIGURE 8.
IRAK3 is responsible for ligand desensitization under low methylation conditions. A, Western blot analysis of IRAK-3 levels in Huh7.5 cells treated with 10 μm AMI-1 for 16 h with or without 1 ng/ml FSL-1 for the indicated times. B, analysis of human blood derived macrophages. Basal mRNA level of IRAK3 is plotted against PRMT1/JMJD6 (left) or relative TRAF6 methylation (right). -Fold increase in IL6 mRNA after 1 h of LPS treatment is plotted against IRAK3 protein concentration (bottom). Each point represents an individual HBDM preparation, n = 26. C, NF-κB luciferase reporter assays in THP-1 cells that were either untreated (control) or treated with TLR6/2 (FSL-1), TLR4 (LPS) or TLR1/2 (Pam3) ligands as indicated. Cells were first transfected with negative control (NC) siRNA, PRMT1-specific siRNA, IRAK3-specific siRNA, or both IRAK3 and PRMT1 siRNA. Data are presented as the mean ± S.D. **, p < 0.01. n ≥ 3. ns, not significant.
To test this hypothesis we stimulated THP-1 cells with three different TLR ligands and observed the effect of knockdown of PRMT1, IRAK3, or both together on NF-κB luciferase activity. As seen previously, PRMT1 siRNA suppressed the ligand responses. IRAK3 siRNA by itself resulted in a small increase in response to ligands. The addition of IRAK3 siRNA, however, completely abolished the inhibition of ligand response in the presence of PRMT1 siRNA (Fig. 7C). These data suggest that IRAK3 is a main mechanism of desensitization to TLR ligands in cells lacking PRMT1.
Discussion
Arginine methylation is a common posttranslational modification (19) that is important in the regulation of histones, RNA-binding proteins, FOXO transcription factors, estrogen receptor, and many other signaling molecules (20). It has been proposed that arginine methylation is irreversible, but the demonstration that estrogen receptor is rapidly arginine-methylated and -demethylated in response to estrogen (4, 21) suggests that demethylation may be an active process as well. PRMT1 is the major enzyme responsible for asymmetric arginine dimethylation (22).
TRAF6 is a critical E3 ubiquitin ligase required for innate immune signaling (23), and the studies reported here demonstrate that it is regulated by PRMT1-mediated arginine methylation. The overall hypothesis supported by our findings is illustrated in Fig. 9. PRMT1 directly binds to TRAF6, methylating it at multiple sites. Arginine methylation of TRAF6 decreases its ubiquitin ligase activity and serves to suppress basal NF-κB activation. Exposure of cells to TLR ligands activates TRAF6 and transiently reduces PRMT1 without changing JMJD6 levels, thus allowing TRAF6 to be demethylated by JMJD6. Demethylation reaches its maximum at 30 min after ligand exposure. Demethylated TRAF6 is highly active resulting in a maximal burst of NF-κB activation. Once signaling is terminated, PRMT1 is re-accumulated, re-methylating TRAF6.
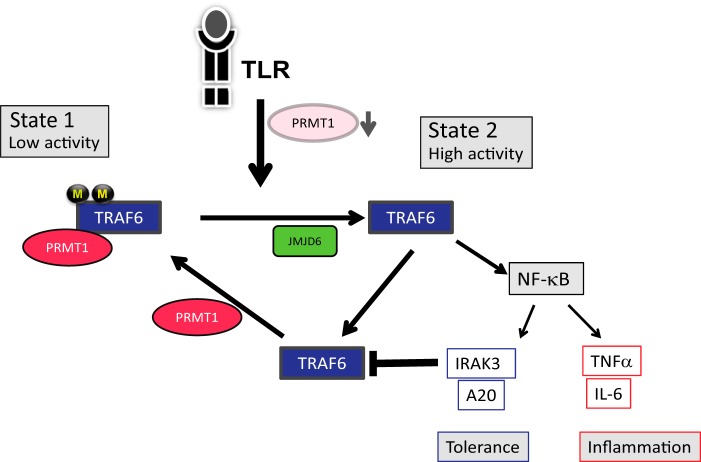
FIGURE 9.
A model of TRAF6 regulation by arginine methylation. In the basal state TRAF6 is associated with PRMT1, methylated, and inactive (State 1). TLR ligands activate TRAF6 and induce PRMT1 degradation. Reduction of PRMT1 levels allows JMJD6 to demethylate TRAF6 and results in the transition to a fully active form (State 2) leading to maximal NF-κB activation. The NF-κB-driven accumulation of inhibitory molecules, such as IRAK-3, eventually produces feedback inhibition reducing pathway activation. This allows the re-accumulation of PRMT1. Restoration of the PRMT1/JMJD6 ratio results in the re-methylation of TRAF6 and the return to the basal, inactive state.
The basal level of TLR pathway activation is determined by the ratio of PRMT1/JMJD6. PRMT1 inhibition or JMJD6 overexpression results in reduced TRAF6 methylation and constitutive pathway activation, whereas PRMT1 overexpression suppresses basal NF-κB activation. An active mechanism to keep basal pathway activation low is well described for multiple pathogen recognition systems (14). Failure to adequately suppress basal activation can produce tolerance to pathogens, and/or inappropriate inflammation (24). In addition to basal activation, inhibition of PRMT1 caused suppression of maximal response to TLR1/2, -2/6, -4, -5, and -7 ligands. This tolerance phenomenon likely results from the accumulation of downstream inhibitory molecules. A number of downstream inhibitors have been described including the inactive kinase IRAK3 (18), the ubiquitin modifying protein A20 (25), and the small heterodimer protein SHP (26). The finding that PRMT1 inhibition increased the level of IRAK3 and that knockdown of IRAK3 prevented tolerance induced by loss of PRMT1 supports the idea that impaired ligand response after baseline activation results from inhibitor accumulation. Our data thus show that PRMT1 is required for keeping the TRAF6-dependent pathway off, and loss of PRMT1 leads to constitutive activation and impaired response to TLR ligands.
The understanding of arginine demethylation remains limited. JMJD6 is an iron and α-ketoglutarate-dependent bifunctional lysine oxygenase and arginine demethylase (3, 13). It demethylates histone arginines in vitro, but its role and specificity as a general protein arginine demethylase is uncertain (27). The studies reported here show that JMJD6 functions to demethylate TRAF6 as part of the TLR response. JMJD6 binds directly to TRAF6 and demethylates it, and overexpression of JMJD6 is sufficient to activate TRAF6 ubiquitin ligase activity and downstream signaling. On the other hand, JMJD6 knockdown increases TRAF6 methylation and impairs the NF-κB responses. The loss of NF-κB activation caused by JMJD6 siRNA could be overcome by simultaneously knocking down PRMT1.
We were able to identify two TRAF6 arginine residues that appear to be required for regulation. Substitution of either Arg-88 or Arg-125 with lysine abolished the ability of JMJD6 to stimulate NF-κB activation. Mutation of these same arginines also prevented the ability of PRMT1 overexpression to decrease activity (Fig. 4).
Taken together, these data demonstrate that JMJD6 activates the TRAF6-dependent TLR signaling pathway and this activation is caused by direct JMJD6 demethylation of TRAF6. Recently, Poulard et al. (4) demonstrated that JMJD6 is able to demethylate estrogen receptor-α, raising the possibility that JMJD6 is a multispecific arginine demethylase involved in dynamic signaling processes.
The mechanisms that generate the specificity of ligand-induced methylation events are not fully understood. PRMT1-dependent modifications are involved in multiple pathways, and the enzyme has a broad spectrum of recognition motifs (1, 22). Overexpression of PRMT1 or reducing PRMT1 activity causes global increases or decreases in protein asymmetric dimethyl-arginine (6). The specificity of arginine methylation signaling might thus reside with the demethylation enzymes such as JMJD6. Our data suggest that JMJD6 has a role in TLR pathway activation both in hepatocytes and in macrophages. JMJD6 knock-out mice have been shown to have decreased macrophage cytokine production in response to LPS (28), and JMJD6-deficient macrophages have impaired cytokine production in response to LPS (29). Our results suggest that this could be due to a lack of TRAF6 demethylation.
The loss of TRAF6 methylation 30 min after stimulation by TLR ligands correlates with an ∼50% decrease in PRMT1 level at the same time, whereas JMJD6 level was unchanged. Although we cannot exclude the possibility of changes in JMJD6 activity or localization, the decrease in total PRMT1 protein was sufficient to explain ligand-induced demethylation. Although we looked for evidence of PRMT1 ubiquitination before the loss of the protein, we were not able to find this. Understanding the mechanisms responsible for the loss of PRMT1 will thus require further studies.
An important finding from our studies is that the basal TLR pathway activation was determined by the ratio of the methylation/demethylation enzymes and not simply by either process alone. Although PRMT1 knock-down activated, and JMJD6 knock down inhibited basal activation, the simultaneous knockdown of both resulted in no effect. These data suggest that the basal activity of the TLR pathway is determined by a state of TRAF6 “methylation balance” that is determined by the ratio of PRMT1/JMJD6. High PRMT1/JMJD6 ratios should result in basal inactivation, and normal ligand responses and low ratios should result in elevated basal activity but impaired response to bacterial antigens.
We examined the relevance of these findings to humans by studying PRMT1/JMJD6 ratios in liver and blood monocyte-derived macrophages. Our data show that variations in the PRMT1/JMJD6 ratio may account for some of the variability of TLR responses. In normal liver there was a correlation between PRMT1/JMJD6 ratio and basal TLR pathway activation as measured by nuclear p65 abundance. In macrophages, the PRMT1/JMJD6 ratios define TRAF6 methylation levels, which in turn correlate with both basal activation and magnitude of the response to LPS.
In conclusion, the current study has demonstrated a novel mechanism for TRAF6 regulation showing that arginine methylation of the protein is necessary to keep MyD88-dependent TLR pathways inactive and prevent tolerance. We further identified JMJD6 as a selective TRAF6 demethylase involved in this pathway. The methylation state of TRAF6 is dynamically regulated by the ratio of PRMT1/JMJD6, and transient loss of PRMT1 is responsible for promoting demethylation and maximal response to ligand. Further understanding of dynamic changes in protein arginine methylation may provide opportunities for manipulating innate immune responses in both infectious and autoimmune diseases.
Author Contributions
I. T. and S. W. designed experiments, analyzed data, and wrote the paper. I. T., S. K., A. A., and M. T. V. performed the experiments. A. N., B. R., and K. D. provided materials. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
The human liver specimens used in this study were derived from samples provided by the University of Kansas Liver Center Tissue Bank. We thank Drs. Jameson Forster, Sean Kumer, Tim Schmitt, and Bashar Abdulkarim for assistance in obtaining these specimens. We thank Drs. Stan Lemon, Kui Li, Wen-Xing Ding, and Luciano DiTacchio for critical comments on an early version of this manuscript.
This work was supported, in whole or in part, by National Institute on Alcohol Abuse and Alcoholism Grant AA012863 and National Institutes of Health COBRE Grant P20 RR021940 (Center for Research Resources). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1 and Table S1.
- SAM
- S-adenosyl methionine
- TLR
- toll-like receptor
- PLA
- proximity ligation assay
- TRAF6
- TNF receptor-associated factor 6
- HBDM
- human blood monocyte-derived macrophage.
References
- 1. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 [DOI] [PubMed] [Google Scholar]
- 2. Bedford M. T., Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang B., Chen Y., Zhao Y., Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 4. Poulard C., Rambaud J., Hussein N., Corbo L., Le Romancer M. (2014) JMJD6 regulates ERα methylation on arginine. PLoS ONE 9, e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boeckel J. N., Guarani V., Koyanagi M., Roexe T., Lengeling A., Schermuly R. T., Gellert P., Braun T., Zeiher A., Dimmeler S. (2011) Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. U.S.A. 108, 3276–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tikhanovich I., Kuravi S., Campbell R. V., Kharbanda K. K., Artigues A., Villar M. T., Weinman S. A. (2014) Regulation of FOXO3 by phosphorylation and methylation in hepatitis C virus infection and alcohol exposure. Hepatology 59, 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakamoto N., Kanai T. (2014) Role of toll-like receptors in immune activation and tolerance in the liver. Front. Immunol. 5, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drexler S. K., Foxwell B. M. (2010) The role of toll-like receptors in chronic inflammation. Int. J. Biochem. Cell Biol. 42, 506–518 [DOI] [PubMed] [Google Scholar]
- 10. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 11. Davies J. Q., Gordon S. (2005) Isolation and culture of murine macrophages. Methods Mol. Biol. 290, 91–103 [DOI] [PubMed] [Google Scholar]
- 12. Skvorak K. J., Paul H. S., Dorko K., Marongiu F., Ellis E., Chace D., Ferguson C., Gibson K. M., Homanics G. E., Strom S. C. (2009) Hepatocyte transplantation improves phenotype and extends survival in a murine model of intermediate maple syrup urine disease. Mol. Ther. 17, 1266–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Webby C. J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M. L., Schmitz C., Butler D. S., Yates J. R. 3rd, Delahunty C. M., Hahn P., Lengeling A., Mann M., Proudfoot N. J., Schofield C. J., Böttger A. (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 [DOI] [PubMed] [Google Scholar]
- 14. Kondo T., Kawai T., Akira S. (2012) Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 33, 449–458 [DOI] [PubMed] [Google Scholar]
- 15. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 16. Nakagawa R., Naka T., Tsutsui H., Fujimoto M., Kimura A., Abe T., Seki E., Sato S., Takeuchi O., Takeda K., Akira S., Yamanishi K., Kawase I., Nakanishi K., Kishimoto T. (2002) SOCS-1 participates in negative regulation of LPS responses. Immunity 17, 677–687 [DOI] [PubMed] [Google Scholar]
- 17. Janssens S., Burns K., Tschopp J., Beyaert R. (2002) Regulation of interleukin-1- and lipopolysaccharide-induced NF-κB activation by alternative splicing of MyD88. Curr. Biol. 12, 467–471 [DOI] [PubMed] [Google Scholar]
- 18. Kobayashi K., Hernandez L. D., Galán J. E., Janeway C. A. Jr., Medzhitov R., Flavell R. A. (2002) IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110, 191–202 [DOI] [PubMed] [Google Scholar]
- 19. Bedford M. T., Richard S. (2005) Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 20. Yang Y., Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 [DOI] [PubMed] [Google Scholar]
- 21. Le Romancer M., Treilleux I., Leconte N., Robin-Lespinasse Y., Sentis S., Bouchekioua-Bouzaghou K., Goddard S., Gobert-Gosse S., Corbo L. (2008) Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol. Cell 31, 212–221 [DOI] [PubMed] [Google Scholar]
- 22. Nicholson T. B., Chen T., Richard S. (2009) The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol. Res. 60, 466–474 [DOI] [PubMed] [Google Scholar]
- 23. Choi Y. (2005) Role of TRAF6 in the immune system. Adv. Exp. Med. Biol. 560, 77–82 [DOI] [PubMed] [Google Scholar]
- 24. Anwar M. A., Basith S., Choi S. (2013) Negative regulatory approaches to the attenuation of Toll-like receptor signaling. Exp. Mol. Med. 45, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 26. Yuk J. M., Shin D. M., Lee H. M., Kim J. J., Kim S. W., Jin H. S., Yang C. S., Park K. A., Chanda D., Kim D. K., Huang S. M., Lee S. K., Lee C. H., Kim J. M., Song C. H., Lee S. Y., Hur G. M., Moore D. D., Choi H. S., Jo E. K. (2011) The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat. Immunol. 12, 742–751 [DOI] [PubMed] [Google Scholar]
- 27. Hahn P., Wegener I., Burrells A., Böse J., Wolf A., Erck C., Butler D., Schofield C. J., Böttger A., Lengeling A. (2010) Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS ONE 5, e13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zakharova L., Dadsetan S., Fomina A. F. (2009) Endogenous Jmjd6 gene product is expressed at the cell surface and regulates phagocytosis in immature monocyte-like activated THP-1 cells. J. Cell Physiol. 221, 84–91 [DOI] [PubMed] [Google Scholar]
- 29. Böse J., Gruber A. D., Helming L., Schiebe S., Wegener I., Hafner M., Beales M., Köntgen F., Lengeling A. (2004) The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.