Background: Rab small GTPases are membrane trafficking proteins in eukaryotes.
Results: Comprehensive knockdown screening identified six Rab isoforms that are involved in regulating Golgi morphology in HeLa-S3 cells.
Conclusion: Five of the six Rabs, including Rab2A and Rab2B, non-redundantly regulate Golgi morphology. A Rab2B-specific effector, GARI-L4, also regulates it.
Significance: This is the first study to systematically analyze all human Rabs in the Golgi.
Keywords: G protein, Golgi, membrane trafficking, organelle, Rab, small GTPase, small interfering RNA (siRNA), yeast two-hybrid
Abstract
Rab small GTPases are crucial regulators of the membrane traffic that maintains organelle identity and morphology. Several Rab isoforms are present in the Golgi, and it has been suggested that they regulate the compacted morphology of the Golgi in mammalian cells. However, the functional relationships among the Golgi-resident Rabs, e.g. whether they are functionally redundant or different, are poorly understood. In this study, we used specific siRNAs to perform genome-wide screening for human Rabs that are involved in Golgi morphology in HeLa-S3 cells. The results showed that knockdown of any one of the six Rab isoforms (Rab1A/1B/2A/2B/6B/8A) induced fragmentation of the Golgi in HeLa-S3 cells and that its phenotype was rescued by re-expression of their respective siRNA-resistant construct. We then performed systematic knockdown-rescue experiments in relation to each of the six Rabs. Interestingly, with the exception of the Rab8A knockdown, the Golgi fragmentation phenotype induced by knockdown of a single Rab isoform, e.g. Rab2B, was efficiently rescued by re-expression of its siRNA-resistant Rab alone, not by any of the other five Rabs, e.g. Rab2A, which is highly homologous to Rab2B, indicating that these Rab isoforms non-redundantly regulate Golgi morphology possibly through interaction with isoform-specific effector molecules. In addition, we identified Golgi-associated Rab2B interactor-like 4 (GARI-L4) as a novel Golgi-resident Rab2B-specific binding protein whose knockdown also induced fragmentation of the Golgi. Our findings suggest that the compacted Golgi morphology of mammalian cells is finely tuned by multiple sets of Rab (or Rab-effector complexes) that for the most part function independently.
Introduction
The Rab-type small GTPases constitute the largest family of membrane trafficking proteins that are conserved in all eukaryotic cells (for reviews, see Refs. 1–4). They function as molecular switches that when activated by binding to GTP drive transport of vesicular carriers from donor organelles to acceptor organelles. Thus, the proper functions of Rab small GTPases are thought to be important for maintenance of organelle identity and morphology (3, 4). A representative example is the role of Rab proteins in the morphology and functions of the Golgi, which is closely connected to the endoplasmic reticulum and trans-Golgi network via membrane trafficking (5–9). A number of mammalian Rab isoforms have been reported to be present in the Golgi, including Rab1 in the cis-Golgi, Rab33B in the medial Golgi, and Rab6 in the trans-Golgi, and functional ablation of certain Golgi-resident Rabs, e.g. Rab1, Rab2, Rab6, Rab8, Rab18, Rab29, Rab30, and Rab41/43, either by RNA interference-mediated knockdown or by overexpression of its GTPase-activating protein (or of a dominant negative form of Rab), has been shown to cause a change in the morphology of the Golgi from compacted (“compacted Golgi” located just near the nucleus) to dispersed (“fragmented Golgi” located throughout the cytoplasm) (10–17). Thus, membrane trafficking within the Golgi or between the Golgi and other organelles appears to be important for maintenance of the identity and morphology of the Golgi (9).
In addition to their membrane trafficking roles, Rab small GTPases may be structurally involved in the stacked cisternal structures of the Golgi of mammalian cells because Golgi matrix proteins, e.g. golgin family members, are known to bind a variety of Golgi Rabs mainly through their coiled-coil domains (18–24). More specifically, GCC185 contains multiple Rab-binding sites and interacts with different Rabs, including with Rab1 (cis-Golgi), Rab33B (medial Golgi), and Rab6 (trans-Golgi) (24), and that may contribute to the stacking of the Golgi cisternae. Although it is now widely believed that functions of Rab small GTPases are indispensable for the compacted Golgi morphology of mammalian cells (5–9), several important questions regarding the functional diversity and redundancy of Rab small GTPases in the Golgi remain unanswered, e.g. how many of the ∼60 mammalian Rab isoforms are involved in Golgi identity/morphology, and is there any functional redundancy of the Rabs (e.g. Rab2A and Rab2B) in maintaining the compacted Golgi morphology?
To answer several of the remaining unanswered questions, in this study, we comprehensively screened for Rabs that regulate the morphology of the Golgi of HeLa-S3 cells by knocking down each of the 62 known human classical Rabs. The results showed that at least six Rab isoforms, Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, and Rab8A, are independently required for a compacted Golgi and that knockdown of any one of the Rab isoforms alone induced fragmentation of the Golgi. We also identified Golgi-associated Rab2B interactor-like 4 (GARI-L4)2 as a novel specific Rab2B-binding protein that is also required for the Golgi morphology of HeLa-S3 cells. We discuss the functional specialization of Rab small GTPases in the Golgi based on our findings.
Experimental Procedures
Antibodies
Rabbit polyclonal antibodies against Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, Rab8A-C (amino acids 162–207), and Rab29 were generated by using purified glutathione S-transferase (GST)-tagged Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, Rab8A-C, and Rab29, respectively, as the antigen (25), and they were affinity-purified with antigen-immobilized beads as described previously (26). The specificity of each anti-Rab antibody was evaluated by probing recombinant FLAG-tagged Rab proteins expressed in COS-7 cells as described previously (27) (see Fig. 4A). The following other antibodies were obtained commercially: horseradish peroxidase (HRP)-conjugated anti-FLAG tag (M2) mouse monoclonal antibody, anti-FLAG tag antibody-conjugated agarose (Sigma-Aldrich), HRP-conjugated anti-T7 tag mouse monoclonal antibody (Merck Biosciences Novagen), anti-GM130 mouse monoclonal antibody (BD Biosciences), anti-GFP rabbit polyclonal antibody (Medical and Biological Laboratories, Co., Ltd., Nagoya, Japan), and anti-β-actin mouse monoclonal antibody (Applied Biological Materials, Richmond, British Columbia, Canada).
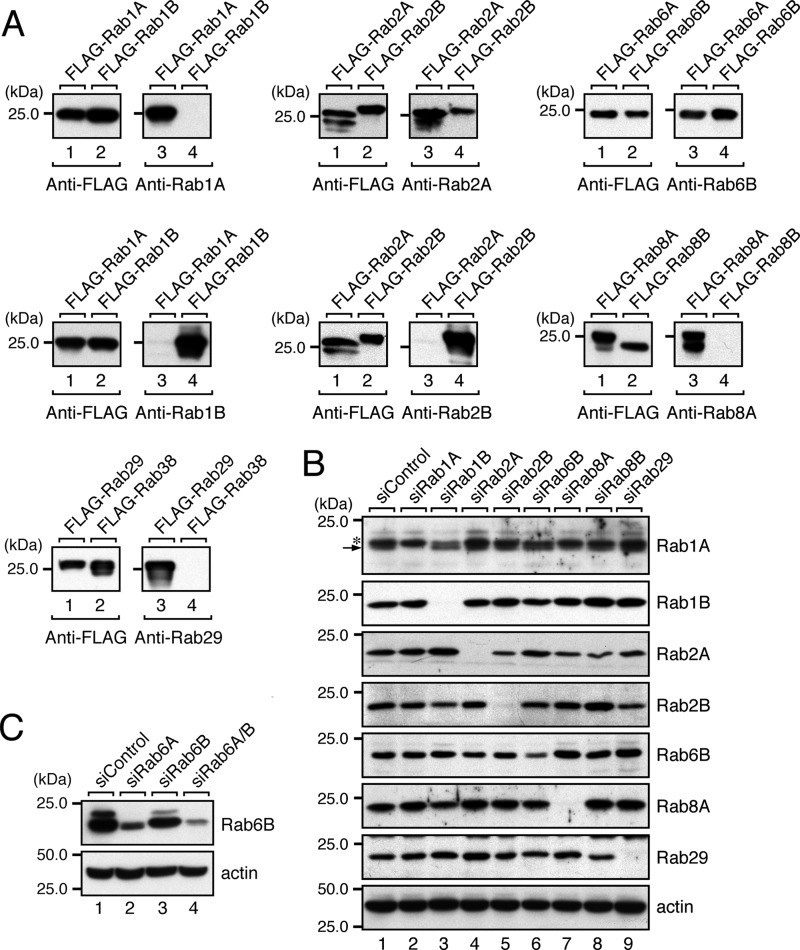
FIGURE 4.
Knockdown of candidate Rabs in HeLa-S3 cells as revealed by immunoblotting with specific antibodies. A, specificity of the antibodies used in this study. The specificity of each antibody was evaluated by using recombinant FLAG-tagged mouse Rab proteins expressed in COS-7 cells. The specificity of anti-Rab1A antibody was evaluated by analyzing COS-7 cell lysates expressing FLAG-Rab1A and FLAG-Rab1B by 10% SDS-PAGE followed by immunoblotting with HRP-conjugated anti-FLAG tag antibody (left panel) and anti-Rab1A antibody (right panel). All of the antibodies except anti-Rab2A antibody and anti-Rab6B antibody specifically recognized a single Rab isoform, whereas the anti-Rab2A antibody weakly recognized Rab2B, and the anti-Rab6B antibody recognized both Rab6A and Rab6B equally. B, knockdown of candidate Rabs in HeLa-S3 cells with specific siRNAs. Total cell lysates of HeLa-S3 cells that had been knocked down by each Rab siRNA were analyzed by 10% SDS-PAGE followed by immunoblotting with the antibodies indicated. A longer SDS-PAGE gel than usual (14 cm long instead of 8.5 cm long) was used to detect Rab1A by separating Rab1A (arrow) from the upper nonspecific bands (asterisk). All of the Rabs except Rab6B were almost completely knocked down by siRNA treatment. Because our anti-Rab6B antibody also recognized Rab6A (see A), we used partially Rab6A-knocked down samples only in the Rab6B panel (see “Experimental Procedures” for details). The residual signals in lane 6 (fifth panel) most likely correspond to endogenous Rab6A. C, double knockdown of Rab6A and Rab6B in HeLa-S3 cells with specific siRNAs. Note that knockdown of Rab6A alone dramatically decreased the anti-Rab6B immunoreactive bands without affecting the Golgi morphology (see Fig. 2A). In addition, double knockdown of Rab6A/B in HeLa-S3 cells did not further increase the rate of Golgi fragmentation (data not shown). The positions of the molecular mass markers (in kDa) are shown on the left in A–C.
cDNA Cloning and Plasmid Construction
cDNAs of the 62 classical Rabs (listed in Table 1) (28) were amplified from Marathon-Ready human brain and/or testis cDNA (BD Biosciences) by PCR with specific oligonucleotides as described previously (29). The sequence information for the oligonucleotides used in this study is available from the authors on request. Purified PCR products were directly inserted into the pGEM-T Easy vector (Promega, Madison, WI) and verified with an automated sequencer. The cDNAs were excised from the pGEM-T Easy vector with appropriate restriction enzymes and then subcloned into the pEGFP-C1 vector (Clontech-Takara Bio Inc.). Unless otherwise specified, the Rabs used in this study were from humans, and mouse Rabs are abbreviated as mRabs throughout (e.g. in Fig. 6). cDNAs of mouse and human GARI-L4 were similarly amplified by PCR and directly inserted into the pGEM-T Easy vector. After verification of their sequences, human and mouse GARI-L4 cDNAs (gene IDs 149647 and 619288, respectively) were subcloned into the pEGFP-C1 vector, pmStr-C1 vector (30), and/or pEF-T7 tag expression vector (29). The sequences of the oligonucleotides for GARI-L4 are also available from the authors on request.
TABLE 1.
Target sequences of siRNAs toward all human Rabs
| Name | Gene ID | Site | siRNA target sequence |
|---|---|---|---|
| Rab1A | 5861 | st3 | CCACAAAGAAAGTAGTAGA |
| st4 | GGAAAGATTTCGAACAATC | ||
| Rab1B | 81876 | st2 | CCAAGAAGGTGGTGGACAA |
| st3 | TGACGTCACTGACCAGGAA | ||
| Rab2A | 5862 | st1 | TGAGGCAAATGGCATTAAA |
| st2 | TGACCTTACTATTGGTGTA | ||
| Rab2B | 84932 | st1 | ACAGTCAATTTCAACATCA |
| st3 | GTTCCAACATGGTTATCAT | ||
| Rab3A | 5864 | st2 | AGGACAACATTAACGTCAA |
| Rab3B | 5865 | st3 | CCAATGAAGAGTCCTTCAA |
| Rab3C | 115827 | st4 | CAGTTGGGATCGATTTCAA |
| Rab3D | 9545 | st1 | GGACGAACGTGTTGTGCCT |
| Rab4A | 5867 | st2 | GGACCTGGATGCAGATCGT |
| Rab4B | 53916 | st1 | GCCCCAACATCGTGGTCAT |
| Rab5A | 5868 | st1 | AGGCCGACCTAGCAAATAA |
| Rab5B | 5869 | st2 | AGACAGCTATGAACGTGAA |
| Rab5C | 5878 | st2 | ACGAAATCTTCATGGCAAT |
| Rab6A | 5870 | st3 | TCATCATGCTAGTAGGAAA |
| Rab6B | 51560 | st1 | AGACGGACCTGGCTGATAA |
| st3 | CCATTGGGATTGACTTCTT | ||
| Rab6C | 84084 | st2 | CCTTTTCCCTTCATTAATA |
| Rab6D/41a | 347517 | st2 | GGAGCGCTTTCACAGCCTA |
| Rab7/7Aa | 7879 | st2 | GGAGCTGACTTTCTGACCA |
| Rab8A | 4218 | st1 | CCATAGGAATTGACTTTAA |
| st2 | TCATGCTGGTCTACGACAT | ||
| Rab8B | 51762 | st1 | TGACAAAACTCAACAGAAA |
| Rab9A | 9367 | st1 | GGAAGCGGTTCGAAGAGTT |
| Rab9B | 51209 | st1 | GCAGGGTCTTCGTGCTGTT |
| Rab10 | 10890 | st2 | GGAATAGACTTCAAGATCA |
| Rab11A | 8766 | st2 | GCATCCAGGTTGATGGAAA |
| Rab11B | 9230 | st1 | GCAACATCGTCATCATGCT |
| Rab12 | 201475 | st3 | AGGAATGAGTTGTCCAATA |
| Rab13 | 5872 | st1 | TGAGAAATCTTTCGAGAAT |
| Rab14 | 51552 | st1 | ATGGCTTATTGTTCCTCGA |
| Rab15 | 376267 | st2 | CCTCAACATTAAAGAGTCA |
| Rab17 | 64284 | st2 | GGAAGGATTCCTTCCTCAA |
| Rab18 | 22931 | st4 | TCCAGAACTTGCAGCAACA |
| Rab19 | 401409 | st2 | TCTGCCAAGGAGTCAAAGA |
| Rab20 | 55647 | st3 | GGCCGTCACACACAGTGGA |
| Rab21 | 23011 | st1 | GGAACTCTTTCTTGACCTT |
| Rab22A | 57403 | st2 | GAAGAGACATTTTCAACAT |
| Rab22B/31a | 11031 | st1 | AGTGCGACCTCTCAGATAT |
| Rab23 | 51715 | st2 | GAACATCAGTGAAAGAAGA |
| Rab24 | 53917 | st1 | GAGGAGGGCTGCCAAATCT |
| Rab25 | 57111 | st2 | CCAATCTACTCTCCCGATT |
| Rab26 | 25837 | st1 | GGCATTGACTTCCGGAACA |
| Rab27A | 5873 | st1 | CCAGTGTACTTTACCAATA |
| Rab27B | 5874 | st2 | GCAAATGCTTATTGTGAAA |
| Rab28 | 9364 | st2 | AGGCAGATATTGTAAACTA |
| Rab29 | 8934 | st1 | TGAGAGTCCTCATTGAAAA |
| Rab30 | 27314 | st1 | GCAACAAGGTCATCACTGT |
| Rab32 | 10981 | st3 | CCAAAGCTTTCCTAATGAA |
| Rab33A | 9363 | st1 | AAAGCATGGTCGAGCATTA |
| Rab33B | 83452 | st1 | AGAGCATGGTTCAGCACTA |
| Rab34 | 83871 | st3 | TGCATTGCATCAACCTACT |
| Rab35 | 11021 | st2 | GCAGTTTACTGTTGCGTTT |
| Rab36 | 9609 | st3 | GCCCCAGCTTTCACAGCCA |
| Rab37 | 326624 | st2 | CATGTTTCCTGATCCAATT |
| Rab38 | 23682 | st3 | AGCACATACTTGCAAATGA |
| Rab39A | 54734 | st3 | CCGACGATCTTTTGAACAT |
| Rab39B | 116442 | st4 | GAGAGGAGATGTTTGTGCT |
| Rab40A | 142684 | st2 | GCCTCTGCAAAGTGGAGAT |
| Rab40ALb | 282808 | ||
| Rab40B | 10966 | st1 | CGGCATTGATCGATGGATT |
| Rab40C | 57799 | st1 | AGAACTGCATGACCTTCTT |
| Rab41/43a | 339122 | st1 | CCATGAAGACGCTGGAGAT |
| Rab42/7Ba | 338382 | st1 | GTAGGGCTCTGTCGAGGTA |
| Rab43/42a | 115273 | st1 | CCAGGTCCTTTTACCGGAA |
a The nomenclature of the human Rabs in this study is as described in Itoh et al. (28). The human Rab6D, Rab7, Rab22B, Rab41, Rab42, and Rab43 in this study have been registered as Rab41, Rab7A, Rab31, Rab43, Rab7B, and Rab42, respectively, in the NCBI (The National Center for Biotechnology Information) database, which is available at www.ncbi.nlm.nih.gov/gene/.
b Because of the high sequence identity between Rab40A and Rab40AL, Rab40A siRNA st1 was also able to knock down Rab40AL efficiently.
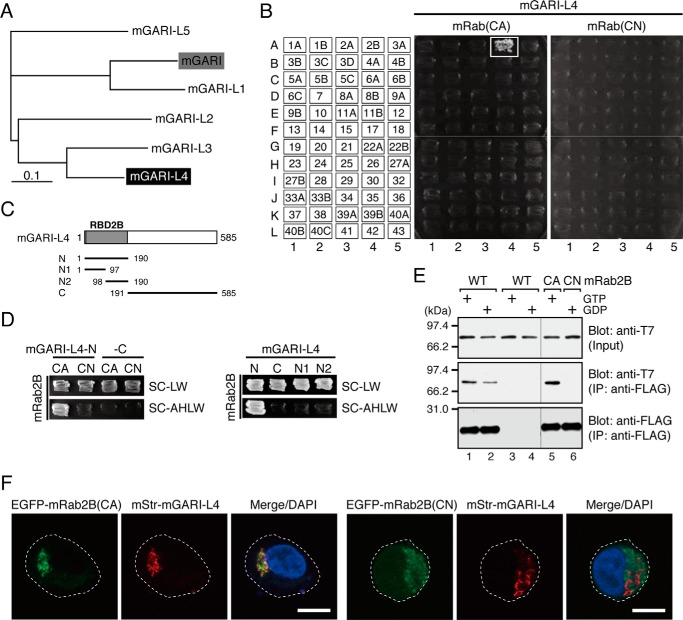
FIGURE 6.
GARI-L4 is a novel Rab2B-specific binding protein in the Golgi. A, phylogenetic relationships between mouse GARI (shaded box) and its related proteins, GARI-L1–5. GARI-L4, on which we focused in this study, is shown against a black background. The phylogenetic tree was drawn by using the ClustalW program set at the default parameters. B, specific interaction between GARI-L4 and Rab2B(CA) as revealed by yeast two-hybrid assays. The assays were performed as described previously (22, 33, 34). Yeast cells containing pGAD-C1-GARI-L4 and pGBD-C1-Rabs(CA/CN)ΔCys (34) were streaked on synthetic complete medium lacking adenine, histidine, leucine, and tryptophan (selection medium) and incubated at 30 °C for 1 week. A positive patch is represented by a white box. C, schematic representation of the truncated mutants of mouse GARI-L4 used in this study. The N-terminal 190 amino acids of GARI-L4 showed high similarity to the N-terminal domain of GARI (22) and were found to function as a Rab-binding domain specific for Rab2B (RBD2B) (see D). D, mapping of the site responsible for Rab2B binding in mouse GARI-L4. Interaction between GARI-L4 mutants (in C) and Rab2B(CA/CN) was assessed by yeast two-hybrid assays as described in B. Yeast cells containing pGAD-C1-GARI-L4 mutants and pGBD-C1-Rab2B(CA/CN)ΔCys (34) were streaked on synthetic complete medium lacking leucine and tryptophan (SC-LW) and synthetic complete medium lacking adenine, histidine, leucine, and tryptophan (selection medium; SC-AHLW) and incubated at 30 °C for 1 day and 1 week, respectively. E, GTP-dependent interaction between T7-tagged mouse GARI-L4 and FLAG-tagged mouse Rab2B (WT/CA/CN) in cultured mammalian cells. Co-immunoprecipitation assays in COS-7 cells were performed as described previously (29, 37). F, co-localization of mouse GARI-L4 with Rab2B(CA), but not with Rab2B(CN), in HeLa-S3 cells. HeLa-S3 cells were co-transfected with pmStr-C1-mGARI-L4 and pEGFP-C1-mRab2B(CA or CN) and then analyzed by confocal fluorescence microscopy. Scale bars, 20 μm.
Small interfering RNA (siRNA)-resistant (SR) forms of human Rabs (named RabSR) were prepared by conventional or two-step PCR techniques essentially as described previously (31, 32). At least five nucleotides in the target sequence of each siRNA were changed without altering the amino acids. The resultant human RabSR cDNAs were subcloned into the pEGFP-C1 vector. Sequence information regarding the mutant oligonucleotides for site-directed mutagenesis is also available from the authors on request.
A series of deletion mutants of mouse GARI-L4, i.e. N (amino acids 1–190), N1 (amino acids 1–97), N2 (amino acids 98–190), and C (191–585), was prepared by conventional PCR techniques (see Fig. 6C). Each cDNA fragment of mouse GARI-L4 was subcloned into the pGAD-C1 vector (33). All other plasmids, including pEF-FLAG-mRab2B, pEF-FLAG-mRab2B(Q65L) (constitutively active (CA) form), pEF-FLAG-mRab2B(S20N) (constitutively negative (CN) form), and pGBD-C1-mRab bait vectors, were prepared as described previously (22, 34, 35).
Cell Cultures and Transfections
HeLa-S3 cells were obtained from the RIKEN BioResource Center (Ibaraki, Japan). HeLa-S3 cells and COS-7 cells were cultured at 37 °C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin G, and 100 μg/ml streptomycin under 5% CO2. One day after plating, plasmids and/or siRNAs were transfected into cultured cells by using Lipofectamine LTX, Lipofectamine 2000, or Lipofectamine RNAiMAX (Life Technologies), each according to the manufacturer's instructions.
RNA Interference
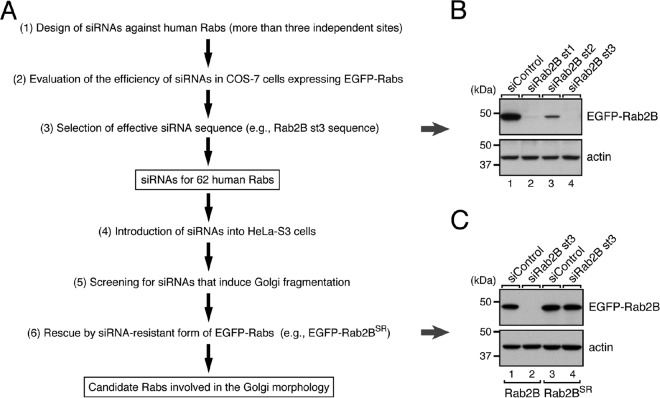
At least three different siRNAs against each human Rab and human GARI-L4 (site (st) 1 target sequence, 5′-GAGCCAACCTTACTACAAA-3′; st2 target sequence, 5′-GACGGTTGGTTCTATGACA-3′) were chemically synthesized by Nippon Gene Co., Ltd. (Toyama, Japan). The knockdown efficiency of the siRNAs against each Rab was evaluated by co-expressing siRNA and pEGFP-C1-Rab (or pEGFP-C1-RabSR) in COS-7 cells, and one or more effective siRNAs for each Rab was selected. Knockdown of endogenous Rab proteins in HeLa-S3 cells (see Fig. 4B) was achieved by transfecting 2 pmol of siRNAs into HeLa-S3 cells in a 6-cm dish. Three days after transfection, the cells were harvested, and Rab knockdown efficiency was evaluated by immunoblotting with specific antibodies. Because the anti-Rab6B antibody used in this study recognized both Rab6A and Rab6B, knockdown of endogenous Rab6B was evaluated by using 0.5 pmol of Rab6A siRNA (e.g. 1.5 pmol of Rab6B siRNA + 0.5 pmol of Rab6A siRNA; total, 2 pmol) to partially reduce the Rab6A protein level (Fig. 4B).
Immunofluorescence Analysis
HeLa-S3 cells were cultured on glass-bottomed dishes (35-mm dish; MatTek Corp., Ashland, MA) and transfected with siRNAs and/or plasmids as described above. Three days after transfection, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, and then stained with anti-GM130 antibody (1:1000 dilution) and DAPI as described previously (36). The stained cells were examined for immunofluorescence signals with a confocal laser-scanning fluorescence microscope (Fluoview 1000, Olympus, Tokyo, Japan), and the images were processed with Adobe Photoshop software (CS5). More than 50 cells were analyzed to obtain the results of the Golgi fragmentation assays shown in Fig. 2A. The results of the knockdown-rescue experiments in Figs. 3B and 5 were obtained by analyzing 30 cells in one experiment and independently repeating the experiments three times. The results are expressed as the means and S.D. of the data obtained in three independent experiments. The statistical analyses were performed by using Student's unpaired t test or Dunnett's test, and p values <0.05 were considered statistically significant (*, p < 0.05; **, p < 0.01).
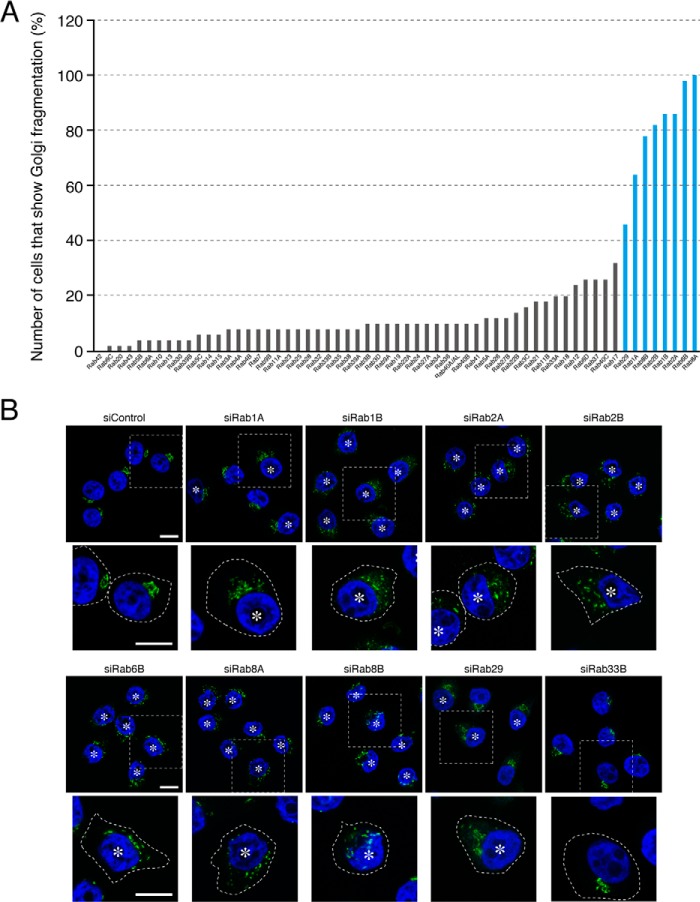
FIGURE 2.
Effect of knockdown of each human Rab isoform on the morphology of the Golgi in HeLa-S3 cells. A, screening for Rabs whose knockdown induced fragmentation of the Golgi. Rabs whose knockdown resulted in Golgi fragmentation in more than 40% of the siRNA-transfected cells were considered candidates for Rabs involved in the morphology of the Golgi (blue bars). B, typical images of Rab knockdown HeLa-S3 cells. HeLa-S3 cells were transfected with each Rab siRNA or control siRNA. Three days after transfection, the cells were fixed and stained with anti-GM130 antibody (a Golgi marker; green) and DAPI (a nucleus marker; blue). The asterisks indicate the cells that exhibited Golgi fragmentation. High magnification views of the boxed areas are shown in the lower panels, and the cells are outlined with a broken line. Scale bars, 20 μm.
FIGURE 3.
Functional rescue of the Golgi fragmentation of Rab knockdown cells by re-expressing the respective Rab isoform. A, typical images of Rab knockdown cells re-expressing a respective RabSR mutant (simply designated as Rab in the figure). HeLa-S3 cells were transfected with each Rab siRNA together with pEGFP-C1 or pEGFP-C1-RabSR. Three days after transfection, the cells were fixed and stained with anti-GM130 antibody (a Golgi marker; red) and DAPI (a nucleus marker; blue). EGFP-expressing cells and EGFP-Rab-expressing cells were identified by green fluorescence. Scale bars, 20 μm. B, quantification of the Golgi fragmentation shown in A. The bars represent the means and S.D. (error bars) of data from three independent experiments. *, p < 0.05; **, p < 0.01, Student's unpaired t test. NS, not significant. Note that the Golgi fragmentation phenotype induced by each Rab siRNA was significantly rescued by re-expression of the respective Rab isoform, although the rescue effect of Rab8ASR in Rab8A knockdown cells was relatively weak. Approximately 30–40% of the Rab1A/1B/2A/2B/6B/8ASR-expressing cells still exhibited the Golgi fragmentation phenotype presumably because of the low co-transfection efficiency of the siRNAs and plasmids.
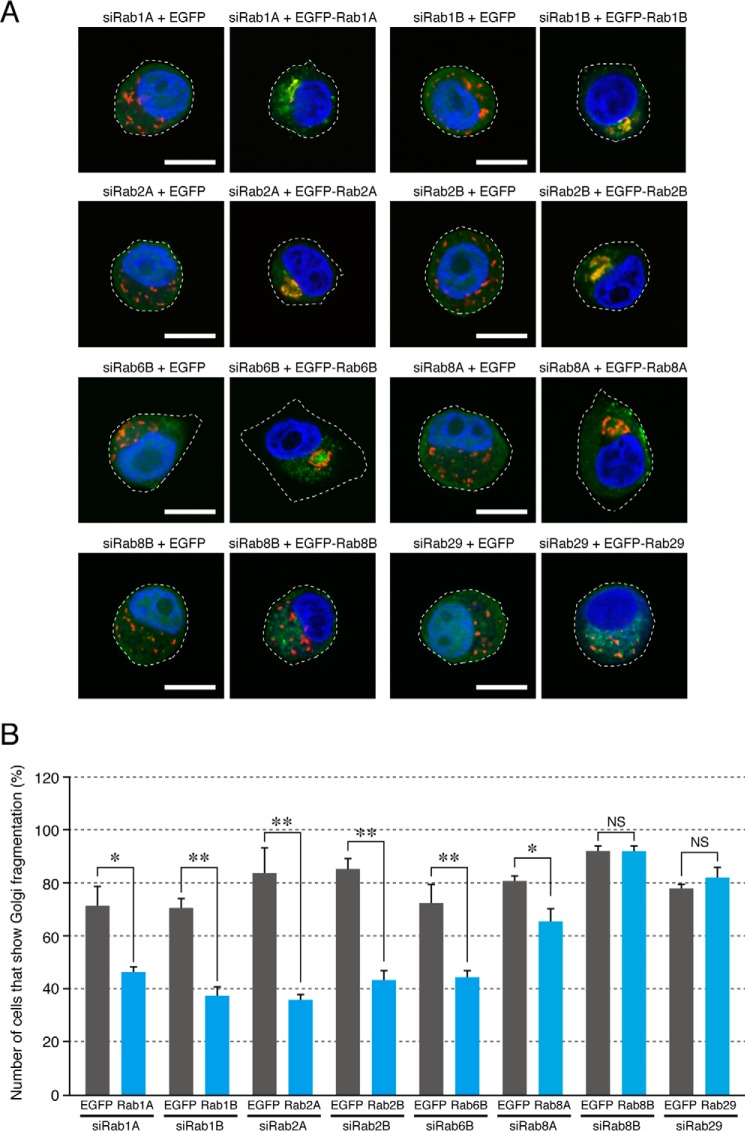
FIGURE 5.
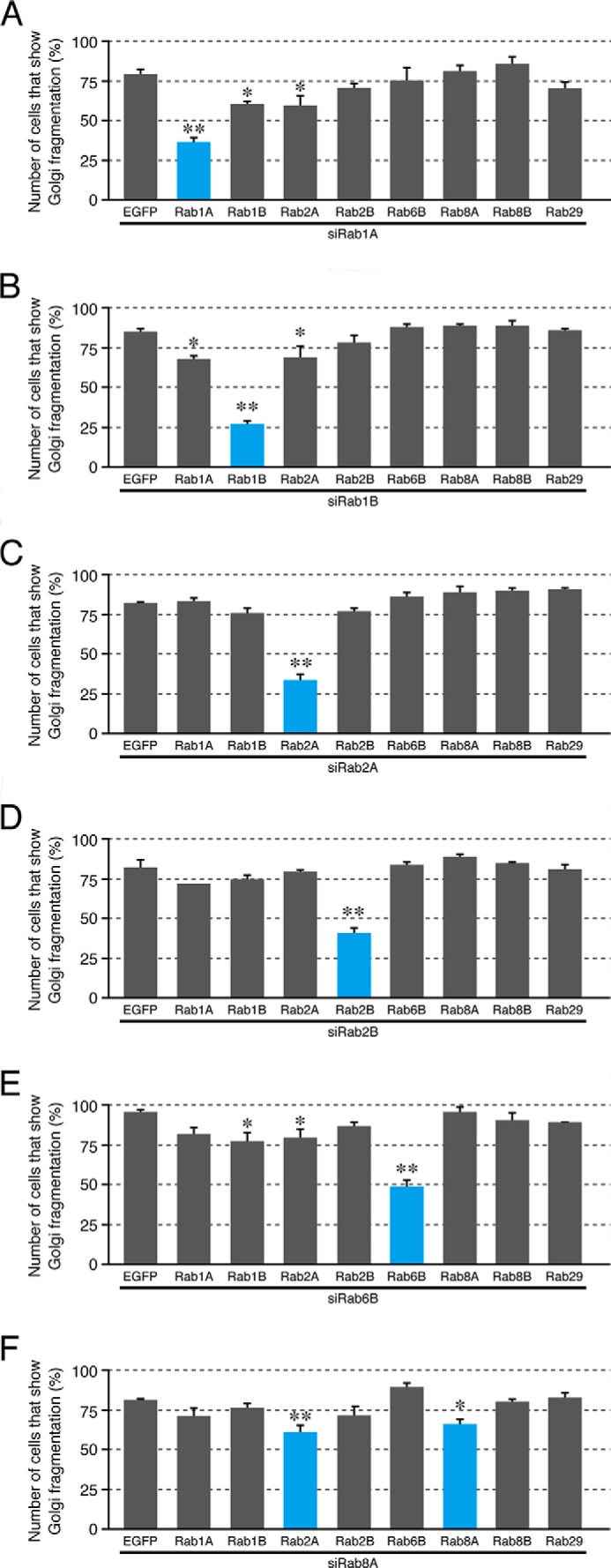
Non-redundant roles of Rab1A, Rab1B, Rab2A, Rab2B, and Rab6B in the compacted Golgi morphology. HeLa-S3 cells were transfected with Rab1A siRNA (A), Rab1B siRNA (B), Rab2A siRNA (C), Rab2B siRNA (D), Rab6B siRNA (E), and Rab8A siRNA (F) together with pEGFP-C1 or pEGFP-C1-Rab1A/1B/2A/2B/6B/8A/8B/29 (or their RabSR mutant). Three days after transfection, the cells were fixed and stained with anti-GM130 antibody and DAPI. EGFP-expressing cells and EGFP-Rab-expressing cells were identified by green fluorescence, and their Golgi morphology was analyzed. The bars represent the means and S.D. (error bars) of data from three independent experiments. *, p < 0.05; **, p < 0.01, Dunnett's test. Note that the Golgi fragmentation phenotype induced by knockdown of Rab1A, Rab1B, Rab2A, Rab2B, and Rab6B was most effectively rescued by re-expression of Rab1ASR, Rab1BSR, Rab2ASR, Rab2BSR, and Rab6BSR, respectively (blue bars). Approximately 30–40% of the Rab1A/1B/2A/2B/6BSR-expressing cells (blue bars) still exhibited the Golgi fragmentation phenotype presumably because of the low co-transfection efficiency of the siRNAs and plasmids.
Miscellaneous Procedures
Co-immunoprecipitation assays in COS-7 cells to evaluate the interaction between T7-tagged mouse GARI-L4 and FLAG-tagged mouse Rab2B and immunoblotting were performed essentially as described previously (29, 37). All of the procedures used to perform yeast two-hybrid assays have been described elsewhere (22, 33, 34).
Results
Genome-wide Screening for Rabs That Regulate the Morphology of the Golgi in HeLa-S3 Cells
We used specific siRNAs against 62 different human Rabs to perform knockdown experiments as a means of comprehensively screening for Rabs that regulate the morphology of the Golgi by the following procedures (Fig. 1A and Table 1). First, we designed and synthesized siRNAs for at least three different sites in each Rab. The knockdown efficiency of each siRNA was evaluated by co-expressing EGFP-Rabs together with their respective siRNAs in COS-7 cells, and the effective siRNAs were selected (Fig. 1B). Each of the effective siRNAs was then introduced into HeLa-S3 cells, and their effect on Golgi morphology was evaluated by immunostaining with anti-GM130 antibody (a Golgi marker). The results of the screening showed that knockdown of any one of the eight Rab isoforms, i.e. Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, Rab8A, Rab8B, and Rab29, affected the morphology of the Golgi in HeLa-S3 cells (Fig. 2A, blue bars). In the control cells, GM130-positive signals were concentrated at one side of the nucleus (so-called compacted Golgi (Fig. 2B, far left panel in the top row), whereas knockdown of Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, Rab8A, Rab8B, or Rab29 induced “fragmentation of the Golgi,” and the fragmented Golgi appeared to be dispersed throughout the cytoplasm (Fig. 2B, asterisks; more than 40% of the cells exhibited the Golgi fragmentation phenotype). Most of these Rabs are known to be associated with the cis-Golgi (Rab1A, Rab1B, and Rab2A) or trans-Golgi/trans-Golgi network (Rab6B, Rab8A, Rab8B, and Rab29), and knockdown of some of them (e.g. Rab1A, Rab1B, and Rab29) had previously been shown to induce Golgi fragmentation (10–13, 17), thereby validating our screening procedures.
FIGURE 1.
Screening strategy for candidates for Rabs that regulate Golgi morphology in this study. A, a flowchart of the screening procedure to identify human Rabs that are involved in the morphology of the Golgi in HeLa-S3 cells. B, selection of siRNAs effective against Rab2B. COS-7 cells were transfected with pEGFP-C1-Rab2B together with Rab2B siRNAs. Three days after transfection, the cells were harvested, and their cell lysates were analyzed by 10% SDS-PAGE followed by immunoblotting with HRP-conjugated anti-GFP antibody (upper panel) and anti-actin antibody (lower panel). C, generation and validation of an SR form of Rab2B. COS-7 cells were transfected with pEGFP-C1-Rab2B (lanes 1 and 2) or pEGFP-C1-Rab2BSR (lanes 3 and 4) together with control siRNA (lanes 1 and 3) or Rab2B siRNA st3 (lanes 2 and 4). Expression and siRNA resistance of Rab2BSR were evaluated as described in B. The positions of the molecular mass markers (in kDa) are shown on the left in B and C.
To exclude the possibility that the Golgi fragmentation phenotype induced by siRNAs against these eight Rabs was attributable to an off-target effect, we attempted to confirm our results by two independent approaches. In our first approach, we performed knockdown-rescue experiments after producing an siRNA-resistant form of each Rab and verifying its siRNA resistance in COS-7 cells (see Fig. 1C). As anticipated, re-expression of EGFP-tagged Rab1ASR, Rab1BSR, Rab2ASR, Rab2BSR, Rab6BSR, or Rab8ASR in the respective Rab knockdown cells significantly decreased the proportion of cells that exhibited the Golgi fragmentation phenotype (Fig. 3, A and B). Re-expression of Rab8BSR or Rab29SR in the respective knockdown cells, however, did not restore the compacted Golgi at all (Fig. 3, A and B) despite the fact that involvement of Rab8 and Rab29 in Golgi morphology had been reported by other groups (15, 17). The Rab8B and Rab29 siRNAs used in this study may also reduce the expression level of some other unrelated molecule(s) that is required for the compacted Golgi morphology. Alternatively, the N-terminal EGFP tagging of Rab8B and Rab29 may distort their function. Be that as it may, however, because of the absence of any rescue effect by Rab8BSR or Rab29SR, we decided not to include these two Rabs in our subsequent analysis. In our second approach, we tested a different siRNA site in each of the six remaining Rabs, i.e. Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, and Rab8A, and the results confirmed that the same phenotype was observed in relation to two independent siRNA sites (more than 40% of the siRNA-treated cells exhibited the Golgi fragmentation phenotype; data not shown). These results indicated that the Golgi fragmentation phenotype that had been observed was not attributable to an off-target effect of the siRNAs.
Because the A isoform and B isoform of Rab1, Rab2, Rab6, and Rab8 are highly homologous to each other, our results showing that knockdown of only a single Rab isoform, i.e. Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, and Rab8A, is sufficient to induce the Golgi fragmentation phenotype (Fig. 2) is somewhat surprising. We also noted that knockdown of Rab6B, but not of Rab6A, induced the phenotype, suggesting the presence of a difference in function between these two Rab6 isoforms in the Golgi. Although we tried to design the siRNA sequences we used in this study to be specific for each human Rab isoform, we could not completely rule out the possibility that some of the siRNAs (e.g. Rab1A siRNA) targeted other closely related Rabs (e.g. Rab1B) despite the presence of several mismatches (e.g. four mismatches were found between the Rab1A siRNA target sequence and the corresponding Rab1B sequence). To investigate this possibility, we generated a specific antibody against each Rab isoform and confirmed its specificity by immunoblotting with recombinant FLAG-tagged Rab proteins expressed in COS-7 cells (27) (Fig. 4A). The results showed that the antibodies against Rab1A, Rab1B, Rab2B, Rab8A, and Rab29 specifically recognized a single Rab isoform of the respective Rab. However, the anti-Rab2A antibody weakly recognized Rab2B, and the anti-Rab6B antibody recognized both A and B isoforms of Rab6 equally. We then used these antibodies to assess the knockdown of each Rab in siRNA-treated cells at the protein level. As anticipated, each siRNA caused a dramatic reduction of its respective single Rab isoform; e.g. Rab1B siRNA specifically reduced the Rab1B protein level, and other Rab siRNAs did not affect the Rab1B protein level (Fig. 4B, second panel, lane 3). The reduction in Rab6B protein level was not very evident even after Rab6B siRNA treatment, but this result was most unlikely attributable to insufficient knockdown of endogenous Rab6B for the following reason. Because our anti-Rab6B antibody recognized both Rab6A and Rab6B (Fig. 4A), the residual band signals presumably corresponded to Rab6A, and in actual fact, hardly any anti-Rab6B-positive immunoreactive bands were detected after double knockdown of Rab6A and Rab6B with Rab6A siRNA and Rab6B siRNA (Fig. 4C). These results led us to hypothesize that two closely related, phylogenetically similar Rab isoforms, i.e. an A isoform and a B isoform of Rab1, Rab2, Rab6, and Rab8, have distinct functions at least in the Golgi.
Rab1A, Rab1B, Rab2A, Rab2B, and Rab6B Non-redundantly Regulate the Golgi Morphology
Next, we performed systematic knockdown-rescue experiments to determine whether the six Rabs, i.e. Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, and Rab8A, are involved in Golgi morphology redundantly or differently. In brief, we attempted to rescue the Golgi fragmentation phenotype induced by knockdown of each Rab by expressing one of these six Rabs. As shown in Fig. 5A, the Golgi fragmentation phenotype induced by Rab1A knockdown was most effectively rescued by re-expression of Rab1A itself, whereas expression of the other Rabs, including Rab1B, had little or no significant rescuing effect on Golgi morphology. Similar results were obtained for Rab1B, Rab2A, Rab2B, and Rab6B: the compacted Golgi morphology was restored by re-expression of the Rab that had been knocked down with its specific siRNA (Fig. 5, B–E). The Golgi fragmentation phenotype induced by Rab8A knockdown, however, was rescued by re-expression of either Rab2A or Rab8A, although their rescue effect was very weak (Fig. 5F), and expression of Rab8B or Rab29, which we identified by the initial knockdown screening (Fig. 2A), failed to rescue the Golgi fragmentation phenotype induced by knockdown of Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, or Rab8A (Fig. 5). These results taken together indicated that the five Rabs identified here (Rab1A/1B/2A/2B/6B) are likely to regulate the compacted Golgi morphology in a non-redundant manner at least in HeLa-S3 cells.
GARI-L4 Is a Novel Rab2B-specific Binding Protein
The fact that the five Rabs identified here non-redundantly regulate compacted Golgi morphology led us to hypothesize that they exert their function(s) through interaction with a single isoform-specific effector molecule. We especially focused on one of the Golgi morphology-regulating Rabs, Rab2B, because nothing had been published in the literature about its function, including about its function in Golgi morphology, and because its effector molecule(s) had not been characterized. In a previous study in which we conducted comprehensive screening for Rab effectors, we identified Golgi-associated Rab2B interactor (GARI) as a Rab2B-specific binding protein whose function was unknown (22). We also identified five GARI homologues, named GARI-L1–5, in silico (22) (Fig. 6A) and suggested that they also function as Rab2B effectors.
By using previously established yeast two-hybrid Rab panels (22, 34), we succeeded in showing that GARI-L4, a previously uncharacterized protein, specifically interacts with a constitutively active form of Rab2B (mimics the GTP-bound form) but not with its constitutively negative form (mimics the GDP-bound form) (Fig. 6B). The GTP-dependent interaction between GARI-L4 and Rab2B was also confirmed by co-immunoprecipitation assays in cultured mammalian cells (Fig. 6E), and the results of the immunofluorescence analysis also supported our finding: GARI-L4 was well co-localized with Rab2B(CA) in the Golgi, whereas virtually no co-localization between Rab2B(CN) and GARI-L4 was observed in HeLa-S3 cells (Fig. 6F).
We then produced a series of truncated mutants of GARI-L4 to identify the Rab2B-binding site of GARI-L4 (Fig. 6C). The results of the yeast two-hybrid assays showed that the N-terminal 190 amino acids of GARI-L4 (GARI-L4-N in Fig. 6C), which are well conserved between GARI and GARI-like proteins, are required for specific GTP-Rab2B binding (Fig. 6D, left panels), whereas the remaining C-terminal domain of GARI-L4 (GARI-L4-C in Fig. 6C), which exhibits no clear sequence similarity between GARI and GARI-like proteins, failed to interact with Rab2B (Fig. 6D, left panels). Because further truncation of GARI-L4-N (GARI-L4-N1 and -N2 in Fig. 6C) completely impaired Rab2B binding activity (Fig. 6D, right panels), the N-terminal 190 amino acids of GARI-L4 are likely to be a minimal GTP-Rab2B-binding domain (and have therefore been designated as Rab-binding domain specific for Rab2B (RBD2B) here).
GARI-L4 Is Required for the Compacted Golgi Morphology in HeLa-S3 Cells
Finally, postulating that if GARI-L4 actually functions as a Rab2B-specific effector in the regulation of Golgi morphology then knockdown of GARI-L4 in HeLa-S3 cells should phenocopy the Rab2B deficiency, i.e. fragmentation of the Golgi, we investigated the involvement of GARI-L4 in the compacted Golgi morphology by means of RNA interference technology. We prepared two independent siRNAs against human GARI-L4, both of which effectively knocked down expression of recombinant GARI-L4 (Fig. 7A), and transfected them into HeLa-S3 cells. As anticipated, HeLa-S3 cells that had been treated with GARI-L4 siRNA st1/st2 often exhibited Golgi fragmentation, the same as occurred after Rab2B knockdown (Fig. 7, B and C). We therefore concluded that GARI-L4 as well as Rab2B is required for the compacted Golgi morphology in HeLa-S3 cells.
FIGURE 7.
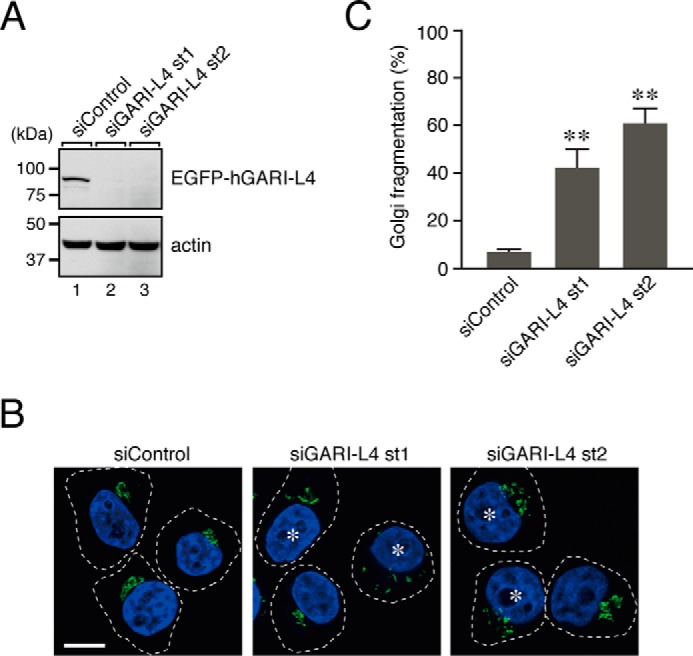
Knockdown of GARI-L4 in HeLa-S3 cells induced the Golgi fragmentation phenotype, the same as Rab2B knockdown did. A, knockdown efficiency of siRNAs against human GARI-L4 (hGARI-L4) as assessed in COS-7 cells. COS-7 cells were co-transfected with GARI-L4 siRNA st1/st2 and pEGFP-C1-hGARI-L4, and their knockdown effect was evaluated by immunoblotting with anti-GFP antibody (upper panel) and anti-actin antibody (lower panel). The positions of the molecular mass markers (in kDa) are shown on the left. B, typical images of GARI-L4 knockdown HeLa-S3 cells. HeLa-S3 cells were transfected with GARI-L4 siRNA or control siRNA. Three days after transfection, the cells were fixed and stained with anti-GM130 antibody (a Golgi marker; green) and DAPI (a nucleus marker; blue). The asterisks indicate the cells that exhibited Golgi fragmentation. Scale bar, 20 μm. C, quantification of the Golgi fragmentation shown in B. The bars represent the means and S.D. (error bars) of data from three independent experiments. **, p < 0.01, Dunnett's test.
Discussion
Because functional ablation of certain Rab isoforms either by knockdown or overexpression of their dominant negative mutants has been found to induce fragmentation of the Golgi in mammalian cells (10–17), Rab proteins are generally thought to be key regulators of Golgi morphology in mammalian cells (5–9). However, the functional relationships among these presumed Golgi morphology-regulating Rabs or the involvement of additional Rab isoforms in the Golgi morphology of mammalian cells had never been investigated. In the present study, we performed systematic genome-wide screening for Rabs that are involved in the compacted Golgi morphology of HeLa-S3 cells (Fig. 1) and succeeded in identifying six candidate Rabs, Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, and Rab8A, whose knockdown caused fragmentation of the Golgi (Figs. 2 and 3). We then performed systematic knockdown-rescue experiments in regard to each of these six Rabs and demonstrated that they independently regulate the morphology of the Golgi in HeLa-S3 cells (Fig. 5). It should be noted that the functions of two phylogenetically related Rabs, i.e. Rab1A and Rab1B (91.7% amino acid identity), Rab2A and Rab2B (85.8% amino acid identity), Rab6A and Rab6B (90.9% amino acid identity), and Rab8A and Rab8B (84.4% amino acid identity), are not interchangeable in terms of their function in relation to compacted Golgi morphology. Because the intracellular distribution of Rab1A and Rab1B, of Rab2A and Rab2B, of Rab6A and Rab6B, or of Rab8A and Rab8B in HeLa-S3 cells was indistinguishable at least at the fluorescence microscopy level (data not shown), different functions of the two Rab isoforms are unlikely to be attributable to their different subcellular localizations in HeLa-S3 cells.
Mammalian Rabs have been classified into ∼40 subfamilies (Rab1–43 in humans and mice) (28, 38), and members in the same subfamily were originally thought to have a redundant function(s) at least in part. Actually, four members of the Rab3 subfamily (Rab3A/B/C/D) are known to have redundant roles in mouse survival (39), and two members of the Rab8 subfamily (Rab8A/B) have compensatory roles in the apical transport of epithelial cells (40). Recent accumulating evidence, however, indicates that the A and B isoforms of Rabs have different functions in certain membrane trafficking events and different subcellular localizations even within the same cell type (41–47). One good example is the Rab27 subfamily in which Rab27A and Rab27B differently regulate the exocytosis of certain types of lysosome-related organelles, e.g. azurophilic granules in neutrophils (41) and secretory granules in mast cells (42), and the exosome secretion pathway (43). More specifically, Rab27A and Rab27B have been suggested to regulate exosome secretion through interaction with different effector molecules, Slp4-a and Slac2-b, respectively (43). Therefore, it is highly possible that the Golgi morphology-regulating Rabs identified here also recruit single-isoform-specific effector molecules to regulate the compacted Golgi morphology in HeLa-S3 cells rather than binding to the same or multiple Rab effector molecules, e.g. golgins (18–24). Actually, it has been reported that SKIP (SifA and kinesin-interacting protein) (48), GARI (22), and TRIP8b (49) are a Rab1A-specific binding protein, Rab2B-specific binding protein, and Rab8B-specific binding protein, respectively, although their involvement in Golgi morphology has never been investigated. In this study, we identified GARI-L4 as a novel GTP-Rab2B-specific binding protein (Fig. 6) that has no sequence similarity to the known Rab-binding proteins in the Golgi, e.g. GCC185, which binds 14 Rabs, including Rab2A/B (24). In addition, we found that knockdown of GARI-L4 also caused fragmentation of the Golgi (Fig. 7), the same as Rab2B knockdown did. These results indicated that GARI-L4 is likely to function as a Rab2B-specific effector that regulates the compacted Golgi morphology of HeLa-S3 cells. It would be interesting to determine whether the Golgi fragmentation phenotype induced by knockdown of other Rabs can be reversed by co-expression of both Rab2B and GARI-L4. In our preliminary experiments, co-expression of Rab2B and GARI-L4 in Rab1B knockdown cells did not restore compacted Golgi morphology (data not shown). Further studies are now underway in our laboratory.
Because we systematically knocked down each Rab isoform in HeLa-S3 cells, we cannot rule out the possibility that some members of the same Rab subfamily redundantly regulate compacted Golgi morphology. One of the candidate Rab subfamilies for involvement in the morphology of the Golgi was the Rab33 subfamily because both Rab33A and Rab33B are present in the Golgi (47, 50). However, simultaneous knockdown of both Rab33A and Rab33B had no effect on the Golgi morphology of HeLa-S3 cells (data not shown). Similarly, knockdown of all members of the Rab3, Rab4, and Rab5 subfamilies had no clear effect on the Golgi morphology of HeLa-S3 cells (data not shown), suggesting that members of the same Rab subfamily may not have redundant roles in the regulation of Golgi morphology at least not in HeLa-S3 cells. At any rate, further systematic combinational knockdown studies will be necessary to determine whether Rab isoforms (especially comparing different Rab subfamilies) have redundant functions in the regulation of Golgi morphology. During the course of preparing this manuscript, Galea et al. (51) reported the results of a systematic analysis of “58 human Rabs” (but not all Rabs) in the Golgi-to-endoplasmic reticulum retrograde trafficking in HeLa cells. Importantly, combinational knockdown of Rabs in their study revealed functional cooperation between different subfamilies of Rabs, although they did not analyze functional redundancy between A and B isoforms of Rabs and single isoform-specific effectors at all. Thus, two independent studies, both of which focused on the Golgi, are different, and their findings may complement each other in making it possible to understand the mechanism of regulation of Golgi morphology in mammalian cells.
In summary, we comprehensively analyzed all known human Rab isoforms with regard to a possible role in Golgi morphology and found that six Rab isoforms, i.e. Rab1A, Rab1B, Rab2A, Rab2B, Rab6B, and Rab8A, independently regulate the Golgi morphology of HeLa-S3 cells possibly through interaction with a single isoform-specific Rab effector molecule, including GARI-L4. We speculate that each of these six Rabs controls a specific step or specific type of membrane traffic in the Golgi that cannot be compensated for by other Rab isoforms. Further investigation of the precise membrane traffic pathways mediated by these six Rabs should provide new insights into the mechanism of the maintenance of the compacted Golgi morphology of mammalian cells.
Author Contributions
M. A. designed and performed experiments. M. F. designed, performed, and analyzed the experiments and wrote the paper.
Acknowledgments
We thank Hotaka Kobayashi, Takahide Matsui, Koutaro Ishibashi, and Daisuke Omote for help in the initial screening for Rabs and in the analysis of GARI-L4 and members of the Fukuda laboratory for valuable discussions.
This work was supported in part by Grants-in-aid for Scientific Research 26111501, 15H04367, and 15H01198 from the Ministry of Education, Culture, Sports, and Technology of Japan (to M. F.). The authors declare that they have no conflicts of interest with the contents of this article.
- GARI-L
- Golgi-associated Rab2B interactor-like
- CA
- constitutively active
- CN
- constitutively negative
- EGFP
- enhanced green fluorescent protein
- GARI
- Golgi-associated Rab2B interactor
- mRab
- mouse Rab
- mStr
- monomeric Strawberry
- SR
- siRNA-resistant
- st
- site.
References
- 1. Fukuda M. (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65, 2801–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 3. Pfeffer S. R. (2013) Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barr F. A. (2013) Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfeffer S. R. (2010) How the Golgi works: a cisternal progenitor model. Proc. Natl. Acad. Sci. U.S.A. 107, 19614–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfeffer S. R. (2012) Rab GTPase localization and Rab cascades in Golgi transport. Biochem. Soc. Trans. 40, 1373–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mizuno-Yamasaki E., Rivera-Molina F., Novick P. (2012) GTPase networks in membrane traffic. Annu. Rev. Biochem. 81, 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papanikou E., Glick B. S. (2014) Golgi compartmentation and identity. Curr. Opin. Cell Biol. 29, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu S., Storrie B. (2015) How Rab proteins determine Golgi structure. Int. Rev. Cell Mol. Biol. 315, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilson B. S., Nuoffer C., Meinkoth J. L., McCaffery M., Feramisco J. R., Balch W. E., Farquhar M. G. (1994) A Rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J. Cell Biol. 125, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haas A. K., Yoshimura S., Stephens D. J., Preisinger C., Fuchs E., Barr F. A. (2007) Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 120, 2997–3010 [DOI] [PubMed] [Google Scholar]
- 12. Dejgaard S. Y., Murshid A., Erman A., Kizilay O., Verbich D., Lodge R., Dejgaard K., Ly-Hartig T. B., Pepperkok R., Simpson J. C., Presley J. F. (2008) Rab18 and Rab43 have key roles in ER-Golgi trafficking. J. Cell Sci. 121, 2768–2781 [DOI] [PubMed] [Google Scholar]
- 13. Zenner H. L., Yoshimura S., Barr F. A., Crump C. M. (2011) Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J. Virol. 85, 8012–8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly E. E., Giordano F., Horgan C. P., Jollivet F., Raposo G., McCaffrey M. W. (2012) Rab30 is required for the morphological integrity of the Golgi apparatus. Biol. Cell 104, 84–101 [DOI] [PubMed] [Google Scholar]
- 15. Rendón W. O., Martínez-Alonso E., Tomás M., Martínez-Martínez N., Martínez-Menárguez J. A. (2013) Golgi fragmentation is Rab and SNARE dependent in cellular models of Parkinson's disease. Histochem. Cell Biol. 139, 671–684 [DOI] [PubMed] [Google Scholar]
- 16. Majeed W., Liu S., Storrie B. (2014) Distinct sets of Rab6 effectors contribute to ZW10- and COG-dependent Golgi homeostasis. Traffic 15, 630–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S., Ma Z., Xu X., Wang Z., Sun L., Zhou Y., Lin X., Hong W., Wang T. (2014) A role of Rab29 in the integrity of the trans-Golgi network and retrograde trafficking of mannose-6-phosphate receptor. PLoS One 9, e96242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barr F. A. (1999) A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol. 9, 381–384 [DOI] [PubMed] [Google Scholar]
- 19. Moyer B. D., Allan B. B., Balch W. E. (2001) Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic 2, 268–276 [DOI] [PubMed] [Google Scholar]
- 20. Weide T., Bayer M., Köster M., Siebrasse J. P., Peters R., Barnekow A. (2001) The Golgi matrix protein GM130: a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2, 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Short B., Preisinger C., Körner R., Kopajtich R., Byron O., Barr F. A. (2001) A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukuda M., Kanno E., Ishibashi K., Itoh T. (2008) Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics 7, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 23. Sinka R., Gillingham A. K., Kondylis V., Munro S. (2008) Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J. Cell Biol. 183, 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayes G. L., Brown F. C., Haas A. K., Nottingham R. M., Barr F. A., Pfeffer S. R. (2009) Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol. Biol. Cell 20, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Itoh T., Fujita N., Kanno E., Yamamoto A., Yoshimori T., Fukuda M. (2008) Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell 19, 2916–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukuda M., Mikoshiba K. (1999) A novel alternatively spliced variant of synaptotagmin VI lacking a transmembrane domain: implications for distinct functions of the two isoforms. J. Biol. Chem. 274, 31428–31434 [DOI] [PubMed] [Google Scholar]
- 27. Imai A., Yoshie S., Nashida T., Shimomura H., Fukuda M. (2004) The small GTPase Rab27B regulates amylase release from rat parotid acinar cells. J. Cell Sci. 117, 1945–1953 [DOI] [PubMed] [Google Scholar]
- 28. Itoh T., Satoh M., Kanno E., Fukuda M. (2006) Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells 11, 1023–1037 [DOI] [PubMed] [Google Scholar]
- 29. Fukuda M., Kanno E., Mikoshiba K. (1999) Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J. Biol. Chem. 274, 31421–31427 [DOI] [PubMed] [Google Scholar]
- 30. Ohbayashi N., Maruta Y., Ishida M., Fukuda M. (2012) Melanoregulin regulates retrograde melanosome transport through interaction with the RILP-p150Glued complex in melanocytes. J. Cell Sci. 125, 1508–1518 [DOI] [PubMed] [Google Scholar]
- 31. Fukuda M., Kojima T., Aruga J., Niinobe M., Mikoshiba K. (1995) Functional diversity of C2 domains of synaptotagmin family: mutational analysis of inositol high polyphosphate binding domain. J. Biol. Chem. 270, 26523–26527 [DOI] [PubMed] [Google Scholar]
- 32. Tamura K., Ohbayashi N., Ishibashi K., Fukuda M. (2011) Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J. Biol. Chem. 286, 7507–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukuda M., Kobayashi H., Ishibashi K., Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170 [DOI] [PubMed] [Google Scholar]
- 35. Fukuda M. (2003) Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2: identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J. Biol. Chem. 278, 15373–15380 [DOI] [PubMed] [Google Scholar]
- 36. Kuroda T. S., Itoh T., Fukuda M. (2005) Functional analysis of Slac2-a/melaonophilin as a linker protein between Rab27A and myosin Va in melanosome transport. Methods Enzymol. 403, 419–431 [DOI] [PubMed] [Google Scholar]
- 37. Fukuda M., Kanno E. (2005) Analysis of the role of Rab27 effector Slp4-a/granuphilin-a in dense-core vesicle exocytosis. Methods Enzymol. 403, 445–457 [DOI] [PubMed] [Google Scholar]
- 38. Diekmann Y., Seixas E., Gouw M., Tavares-Cadete F., Seabra M. C., Pereira-Leal J. B. (2011) Thousands of rab GTPases for the cell biologist. PLoS Comput. Biol. 7, e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schlüter O. M., Schmitz F., Jahn R., Rosenmund C., Südhof T. C. (2004) A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 24, 6629–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato T., Iwano T., Kunii M., Matsuda S., Mizuguchi R., Jung Y., Hagiwara H., Yoshihara Y., Yuzaki M., Harada R., Harada A. (2014) Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J. Cell Sci. 127, 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson J. L., Brzezinska A. A., Tolmachova T., Munafo D. B., Ellis B. A., Seabra M. C., Hong H., Catz S. D. (2010) Rab27a and Rab27b regulate neutrophil azurophilic granule exocytosis and NADPH oxidase activity by independent mechanisms. Traffic 11, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh R. K., Mizuno K., Wasmeier C., Wavre-Shapton S. T., Recchi C., Catz S. D., Futter C., Tolmachova T., Hume A. N., Seabra M. C. (2013) Distinct and opposing roles for Rab27a/Mlph/MyoVa and Rab27b/Munc13-4 in mast cell secretion. FEBS J. 280, 892–903 [DOI] [PubMed] [Google Scholar]
- 43. Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M. C., Darchen F., Amigorena S., Moita L. F., Thery C. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 [DOI] [PubMed] [Google Scholar]
- 44. Ishida M., Ohbayashi N., Maruta Y., Ebata Y., Fukuda M. (2012) Functional involvement of Rab1A in microtubule-dependent anterograde melanosome transport in melanocytes. J. Cell Sci. 125, 5177–5187 [DOI] [PubMed] [Google Scholar]
- 45. Chen P. I., Kong C., Su X., Stahl P. D. (2009) Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J. Biol. Chem. 284, 30328–30338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lapierre L. A., Dorn M. C., Zimmerman C. F., Navarre J., Burnette J. O., Goldenring J. R. (2003) Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp. Cell Res. 290, 322–331 [DOI] [PubMed] [Google Scholar]
- 47. Ishibashi K., Uemura T., Waguri S., Fukuda M. (2012) Atg16L1, an essential factor for canonical autophagy, participates in hormone secretion from PC12 cells independently of autophagic activity. Mol. Biol. Cell 23, 3193–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishida M., Ohbayashi N., Fukuda M. (2015) Rab1A regulates anterograde melanosome transport by recruiting kinesin-1 to melanosomes through interaction with SKIP. Sci. Rep. 5, 8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen S., Liang M. C., Chia J. N., Ngsee J. K., Ting A. E. (2001) Rab8b and its interacting partner TRIP8b are involved in regulated secretion in AtT20 cells. J. Biol. Chem. 276, 13209–13216 [DOI] [PubMed] [Google Scholar]
- 50. Zheng J. Y., Koda T., Fujiwara T., Kishi M., Ikehara Y., Kakinuma M. (1998) A novel Rab GTPase, Rab33B, is ubiquitously expressed and localized to the medial Golgi cisternae. J. Cell Sci. 111, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 51. Galea G., Bexiga M. G., Panarella A., O'Neill E. D., Simpson J. C. (2015) A high content screening microscopy approach to dissect the role of Rab proteins in Golgi-to-ER retrograde trafficking. J. Cell Sci. 128, 2339–2349 [DOI] [PubMed] [Google Scholar]