Background: CD300 family molecules regulate innate immune responses.
Results: We identified and characterized a novel immunoreceptor, CD300H.
Conclusion: CD300H mediates neutrophil chemoattractant production.
Significance: CD300H may play an important role in innate immunity.
Keywords: chemokine, dendritic cell, monocyte, neutrophil, single-nucleotide polymorphism (SNP)
Abstract
Recruitment of circulating monocytes and neutrophils to infection sites is essential for host defense against infections. Here, we identified a previously unannotated gene that encodes an immunoglobulin-like receptor, designated CD300H, which is located in the CD300 gene cluster. CD300H has a short cytoplasmic tail and associates with the signaling adaptor proteins, DAP12 and DAP10. CD300H is expressed on CD16+ monocytes and myeloid dendritic cells. Ligation of CD300H on CD16+ monocytes and myeloid dendritic cells with anti-CD300H monoclonal antibody induced the production of neutrophil chemoattractants. Interestingly, CD300H expression varied among healthy subjects, who could be classified into two groups according to “positive” and “negative” expression. Genomic sequence analysis revealed a single-nucleotide substitution (rs905709 (G→A)) at a splice donor site on intron 1 on either one or both alleles. The International HapMap Project database has demonstrated that homozygosity for the A allele of single nucleotide polymorphism (SNP) rs905709 (“negative” expression) is highly frequent in Han Chinese in Beijing, Japanese in Tokyo, and Europeans (A/A genotype frequencies 0.349, 0.167, and 0.138, respectively) but extremely rare in Sub-Saharan African populations. Together, these results suggest that CD300H may play an important role in innate immunity, at least in populations that carry the G/G or G/A genotype of CD300H.
Introduction
Recruitment of blood leukocytes to sites of infection is essential for host defense against infection. Circulating monocytes (Mo)2 and neutrophils are especially important effectors in the initiation of inflammatory responses to microbes (1). Mo in the human peripheral blood are divided into subsets on the basis of their cell surface expression of CD14 and CD16 (2). A major subset with a phenotype of CD14+CD16− is referred to as classical or inflammatory Mo (iMo); these are similar to Ly6ChiCCR2hi Mo in mice (3). The CD16+ Mo subset can further be divided into two subsets, namely CD14+CD16+ and CD14dimCD16+, on the basis of CD14 expression (4). CD14dimCD16+ Mo resemble Ly6ClowCx3CR1hi Mo in mice and are referred to as patrolling Mo (pMo) (5–7). pMo crawl along the luminal side of the vascular endothelium in the steady state. CD14+CD16+ Mo are referred to as intermediate monocytes (intMo) and are found at low frequency (∼5% of blood monocytes) (8) in the peripheral blood; they represent a continuous transition from iMo (CD14+CD16–) to pMo (CD14dimCD16+) (7, 9). iMo produce TNF-α, IL-6, and IL-1 in response to bacteria-associated signals. In contrast, pMo respond poorly to bacterial components but potently to viruses and nucleic acids; they thus selectively detect viral infection and injury and produce inflammatory cytokines (4). In mice, Ly6Clow pMo provide immune surveillance and rapidly extravasate before neutrophils in response to Listeria monocytogenes infection (5). In addition, activation of intravascular Ly6Clow pMo is responsible for neutrophil recruitment via TLR7-dependent CXCL1 production (6). However, the precise mechanism by which pMo produce chemokines, particularly in humans, remains unclear.
CD300 family molecules are type 1 immunoreceptors belonging to the immunoglobulin superfamily and are encoded by seven genes on human chromosome 17 and nine genes on mouse chromosome 11 (10, 11). They are expressed on myeloid lineage cells, including monocytes-macrophages, granulocytes, dendritic cells, and mast cells, suggesting that they play an important role in innate immunity. CD300A (also named MAIR-I (12), LMIR1 (13), and CLM-8 (14) in mice) and CD300LF (MAIR-V (15, 16), LMIR3 (17–20), and CLM-1 (21) in mice) mediate inhibitory signals via the immunoreceptor tyrosine-based inhibitory motif in their cytoplasmic regions. By contrast, CD300LB (LMIR5 (22)), CD300C, CD300LD (MAIR-IV (23), LMIR4 (17, 24), CLM-5 (25)), and CD300E have short cytoplasmic tails with no signaling motifs. However, CD300LB and CD300E each contain a positively charged lysine residue, whereas CD300C and CD300LD contain a negatively charged glutamic acid in their transmembrane domains (10) (11). These receptors noncovalently associate with membrane-bound signaling adaptor proteins, including DNAX adaptor protein 12 (DAP12), DAP10, and the γ chain of the Fc receptor for IgE (FcϵRIγ) through interaction with a negatively charged amino acid (aspartic acid) in the transmembrane domain of the adaptors and thus transmit activating signals (10, 11). DAP12 and FcϵRIγ contain an immunoreceptor tyrosine-based activation motif in their cytoplasmic regions, whereas DAP10 contains a YXXM motif, a potential Src homology 2 domain-binding site for the p85 regulatory subunit of the PI3K. Among the seven genes encoding human CD300 molecules, those encoding CD300A, CD300LB, CD300C, CD300E, CD300LF, and CD300LG each have mouse homologs, as determined from their functional, molecular, and genetic characteristics (10, 11). However, the mouse counterpart of human CD300LD and the human counterpart of mouse MAIR-II (CLM-4/LMIR2) and MAIR-VI (CLM-3/LIMR7) are unclear.
Here, we identified a previously unannotated gene encoding a novel molecule, designated CD300H, in the CD300 family gene cluster on human chromosome 17. We demonstrate that CD300H is expressed on CD14+CD16+ intMo and CD14dimCD16+ pMo and associates with DAP12 or DAP10. Upon cross-linking of CD300H on CD16+ Mo and CD11c+ dendritic cells (DCs), it mediates an activating signal for the production of neutrophil chemoattractants.
Experimental Procedures
Cloning of Human CD300H cDNA
Full-length cDNA of human CD300H was isolated from human CD14+ monocyte-derived cDNA by reverse transcription-polymerase chain reaction (RT-PCR) using CD300H-specific primers (5′-ATGACCCAGAGGGCTGGGGC-3′ and 5′-TCATGACTCTGTCCAAGGAG-3′).
Generation of Fc Fusion Protein
Fusion proteins of the entire extracellular domains of CD300H, CD300A, and CD300C with the Fc portion of human IgG1 (CD300H-Fc, CD300A-Fc, and CD300C-Fc, respectively) were generated as described previously (26).
Antibodies and Flow Cytometry Analyses
Monoclonal antibody (mAb) against CD300H (TX93; mouse IgG2a) was generated in our laboratory by immunizing mice with CD300H-Fc (12); mAb against CD300C (TX47; mouse IgG1) was generated by immunizing mice with BW5147 transfectant expressing CD300C, as described previously (12). mAb against CD300A and CD300C (TX49; mouse IgG1) was generated in our laboratory, as described previously (27). mAbs specific to human CD3 (clone UCHT1), CD14 (clone M5E2), CD56 (clone B159), CD11c (clone B-ly6), CD16 (clone 3G8), HLA-DR (clone G46-6), CD40 (clone 5C3), CD80 (clone BB1), and CD86 (clone 2331) were purchased from BD Biosciences (San Jose, CA). mAbs specific to human CD123 (clone AC145), CD203c (clone FR3-16A11), and CD304 (clone AD5-17F6) were purchased from Miltenyi Biotec (Auburn, CA). Flow cytometry analyses were performed with a FACS Fortessa flow cytometer (BD Biosciences). FlowJo software (Tree Star, Ashland, OR) was used for data analyses.
Phylogenic Analysis
The protein sequences of human CD300 family molecules were aligned by using the ClustalW program (28). NJplot was used to construct tree diagrams (29).
Cells and Transfectants
Murine T cell lines 2B4 and BW5147, human monocytic cell lines THP-1 and U937, and human embryonic kidney cell line 293T were used; they were cultured in RPMI 1640 medium containing 5% fetal calf serum (FCS) except for 293T cells. 293T cells were cultured in DMEM containing 10% FBS. Blood samples were collected from healthy donors at the University of Tsukuba under an institutional review board-approved protocol. Written informed consent was obtained from the donors. Human peripheral blood mononuclear cells (PBMCs) were isolated from whole blood with LymphoPrep (Axis-Shield, Oslo, Norway) in accordance with the manufacturer's protocol. CD16+ Mo were isolated from PBMCs by using the MACS cell separation system (Miltenyi Biotec); NK cells were depleted from the PBMCs by using anti-CD56 MicroBeads, and then CD16+ Mo were purified from the NK cell-depleted PBMCs by using anti-CD16 MicroBeads. The purity of the CD16+ Mo was more than 85%, as determined by flow cytometry. CD16− Mo were isolated from PBMCs by using the MACS cell separation system (Miltenyi Biotec); CD16+ cells were depleted from the PBMCs by using anti-CD16 MicroBeads, and then CD16− Mo were purified from the CD16+ cell-depleted PBMCs by using anti-CD14 MicroBeads. The purity of the CD16− Mo was more than 90%, as determined by flow cytometry. CD11c+ DCs were isolated from PBMCs by using the Myeloid dendritic cell isolation kit (Miltenyi Biotec). The purity of the CD11c+ DCs was more than 95%, as determined by flow cytometry. 2B4, U937, and BW5147 transfectants stably expressing CD300H, CD300A, or CD300C tagged with FLAG or HA at the N terminus were established, as described previously (12). 293T transfectant transiently expressing FLAG-tagged CD300Hs was established by using Lipofectamine 2000 (Invitrogen).
SNP Genotyping
Genomic DNA was extracted and purified from peripheral blood leukocyte using the QuickGene 610 L system (Fujifilm, Tokyo, Japan). Polymorphism was analyzed by genomic PCR with primer pair 5′-AAACCCAGAAGAGGCCAGAG-3′ and 5′-GGTTTGGACCTTGACTGTGC-3′ and subsequent direct sequencing with the Big Dye Termination Kit (Applied Biosystems, Foster City, CA).
Biochemical Analyses
Cells were lysed in lysis buffer containing 1% Nonidet P-40 (Sigma-Aldrich) or digitonin (Calbiochem, Darmstadt, Germany) supplemented with protease inhibitors, as described previously (26). Cell lysates were immunoprecipitated with control Ig, anti-FLAG mAb (M2, Sigma-Aldrich), anti-HA (3F10, Roche Applied Science, Mannheim, Germany), anti-DAP12 polyclonal antibody (FL-113, Santa Cruz Biotechnology, Inc.), anti-DAP10 (H-2, Santa Cruz Biotechnology), or anti-FcϵRIγ-chain polyclonal antibody (06-727, Millipore, Billerica, MA). Immunoprecipitates were resolved by SDS-PAGE, transferred onto polyvinylidine difluoride membranes by electroblotting, and then immunoblotted with anti-FLAG polyclonal antibody (F7425, Sigma-Aldrich), anti-HA (3F10, Roche Applied Science), anti-DAP12 (FL-113, Santa Cruz Biotechnology), anti-DAP10 (H-2, Santa Cruz Biotechnology), or anti-FcϵRIγ chain (T2040, United States Biological) followed by horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (eB182, eBioscience, San Diego, CA), anti-mouse IgG (eB144, eBioscience), or anti-rat IgG (NA935, GE Healthcare UK Ltd., Buckinghamshire, UK).
Stimulation of CD300H for Cytokine and Chemokine Production
CD16+ Mo (5 × 104 or 1 × 105/well), CD16− Mo, and CD11c+ DCs (5 × 104/well) were plated into 96-well flat-bottom plates precoated with 10 μg/ml F(ab′)2 fragments of either anti-CD300H (TX93) or control mouse IgG and cultured for 4 or 24 h at 37 °C in 5% CO2. TNF-α concentration in the culture supernatants was measured with ELISA kits purchased from BD Biosciences; concentrations of IL-12p70, TNF-α, IL-10, IL-6, IL-1β, and IL-8 in the culture supernatants were measured with a BDTM cytometric bead array (BD Biosciences); and the mRNAs of IL6, CXCL1, CXCL2, CXCL5, and CXCL8 were quantified by real-time quantitative PCR. The sequences of the specific primers were as follows: IL-6 forward, 5′-GATGAGTACAAAAGTCCTGATCCA-3′; IL-6 reverse, 5′-CTGCAGCCACTGGTTCTGT-3′; CXCL1 forward, 5′-TCCTGCATCCCCCATAGTTA-3′; CXCL-1 reverse, 5′-CTTCAGGAACAGCCACCAGT-3′; CXCL2 forward, 5′-CTTGTCTCAACCCCGCATCG-3′; CXCL2 reverse, 5′-TCCTTCAGGAACAGCCACCA-3′; CXCL5 forward, 5′-TTCGCCATAGGCCCACAGT-3′; CXCL5 reverse, 5′-TTTCCATGCGTGCTCATTTCTC-3′; CXCL8 forward, 5′-AGACAGCAGAGCACACAAGC-3′; CXCL8 reverse, 5′-CACAGTGAGATGGTTCCTTCC-3′.
cDNA Synthesis and RT-PCR
Total RNA was extracted with Isogen reagent (Nippon Gene, Tokyo, Japan), and cDNA was synthesized by using a High-Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA). Real-time RT-PCR was performed with SYBR Green master mix (Applied Biosystems). Expression of each target gene was normalized against that of GAPDH (primer sequences 5′-CTTCACCACCATGGAGAAGGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′).
Neutrophil Migration Assay
Neutrophils were isolated from whole blood with Polymorphprep (Axis-Shield) in accordance with the manufacturer's protocol. Supernatant obtained from TX93-stimulated CD16+ Mo was added to the lower compartment of 96-well Transwell plates (pore size 3.0 μm; Corning, Inc.) at a total volume of 235 μl/well. Neutrophils (1 × 105) were placed in the upper compartment at a total volume of 75 μl, and then the plates were incubated for 30 min at 37 °C in 5% CO2. Cells in the lower compartment were collected and counted by using a Guava easyCyte Mini flow cytometer (Millipore).
Statistical Analyses
The unpaired Student's t test was used for statistical analyses. p values of <0.05 were considered statistically significant. All statistical analyses were carried out using GraphPad Prism version 5.0c software (GraphPad Software, San Diego, CA).
Results
Cloning of the Gene Encoding Human CD300H
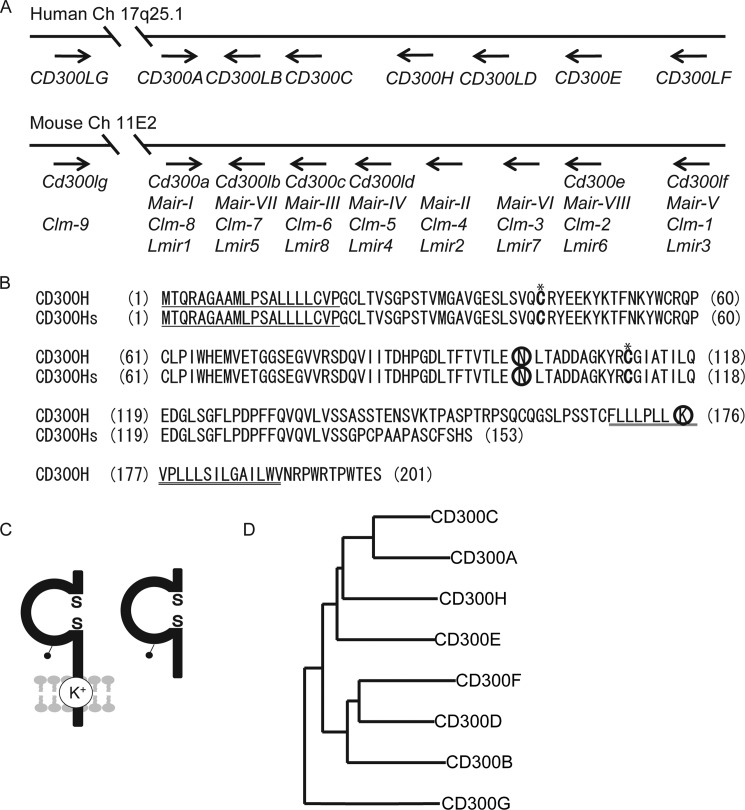
In our search for a human homolog of MAIR-II in the National Center for Biotechnology Information database, we found a previously unannotated gene located in the human CD300 gene cluster on chromosome 17 (Fig. 1A). We isolated full-length cDNA from human CD14+ Mo-derived cDNA by RT-PCR. Sequence analysis revealed that an open reading frame of 603 bp encoded a type I transmembrane receptor, which contained a 20-amino acid (aa) leader sequence, a 148-aa extracellular region containing an Ig-like domain, a 23-aa transmembrane region, and a 10-aa cytoplasmic domain with no signaling motif (Fig. 1B). The Ig-like domain contained a potential N-glycosylation site. The transmembrane domain contained a positively charged lysine residue (Fig. 1C). Because this homolog was the eighth member of the human CD300 family of receptors, we termed it CD300H. We also identified a splicing variant that lacked a transmembrane domain; we called this variant CD300Hs (Fig. 1, B and C). A database search demonstrated that CD300H had 44% amino acid identity with mouse MAIR-II/CLM-4/LMIR2 and CD300c/CLM-6, but it had lower rates of identity with CD300e/CLM-2/LMIR6 (42%), CLM-3/LMIR7 (35%), CD300ld/CLM-5/LMIR4 (40%), CD300lb/CLM-7/LMIR5 (35%), and CD300lg/CLM-9 (35%) among the non-immunoreceptor tyrosine-based inhibitory motif-bearing receptors. The gene encoding CD300H was located between CD300C and CD300LD on human chromosome 17q25.1 (Fig. 1A). We analyzed the degree of identity of the extracellular Ig domain of CD300H with the sequences of CD300 family members and constructed a molecular phylogenetic tree by using ClustalW2 phylogeny. The Ig-like domains of CD300A and CD300C had the greatest identity (53%) with that of CD300H; other members of the family had lower identity levels (Fig. 1D).
FIGURE 1.
Molecular characteristics of CD300H. A, localization on mouse and human CD300 family gene maps. Mouse and human family genes are mapped on chromosomes 11 and 17, respectively. B, amino acid sequences of CD300H and CD300Hs. Numbers in parentheses (left), aa positions; underlining, putative leader and transmembrane sequences; circles, potential N-linked glycosylation sites in the extracellular domain and the charged aa residue in the transmembrane region; *, cysteine residues potentially involved in disulfide bonding of the immunoglobulin-like domains. C, schematic diagram of CD300H (left) and CD300Hs (right) proteins. The pair of cysteine residues in the extracellular portion is potentially able to participate in intrachain disulfide bonding for the formation of Ig-like domains. D, phylogenetic tree showing the relationships among the Ig-like domain amino acid sequences of human CD300 molecules. Each neighbor-joining phylogenetic tree was generated by aligning the translated sequences of the open reading frames for CD300 proteins by using the ClustalW algorithm.
Expression of CD300H
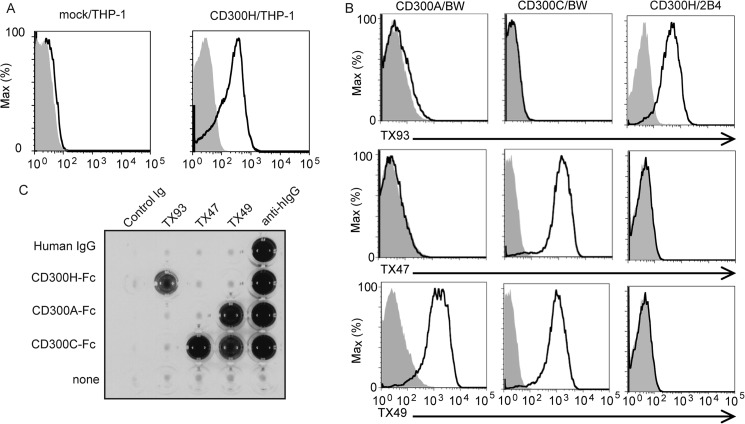
To investigate the cellular distribution of CD300H, we generated a mAb against CD300H (clone TX93) and analyzed its specificity. TX47 and TX49 were previously generated mAbs against CD300C and against CD300A and CD300C, respectively (Fig. 2, B and C) (27). Although CD300H had a high level of identity with CD300A and CD300C, TX93 mAb bound specifically to CD300H-expressing THP-1 and 2B4 transfectants but not to CD300A- or CD300C-expressing BW5147 transfectants (Fig. 2, A and B). In addition, TX93 mAb bound to CD300H-Fc fusion protein but not to CD300A-Fc or CD300C-Fc protein (Fig. 2C). These results indicated that TX93 mAb specifically recognized CD300H.
FIGURE 2.
Characterization of the anti-CD300H monoclonal antibody, TX93. A, flow cytometry of THP-1 cells or THP-1 transfectants expressing CD300H (5 × 105 cells/experiment) were stained with biotinylated control mouse immunoglobulin (shaded) or anti-CD300H (TX93) (thick lines) followed by allophycocyanin-conjugated streptavidin. B, flow cytometry of BW5147 transfectants expressing CD300A or CD300C or 2B4 transfectants expressing CD300H (5 × 105 cells/experiment) were stained with control mouse immunoglobulin (shaded), TX93, anti-CD300C (TX47), or anti-CD300A and CD300C (TX49) (thick lines), followed by phycoerythrin-conjugated goat F(ab′)2 fragment anti-mouse IgG. C, microtiter plates coated with control human IgG, CD300H-Fc, CD300A-Fc, or CD300C-Fc were incubated with control mouse IgG, TX93, TX47, or TX49, and binding was analyzed by ELISA. Data are representative of two independent experiments.
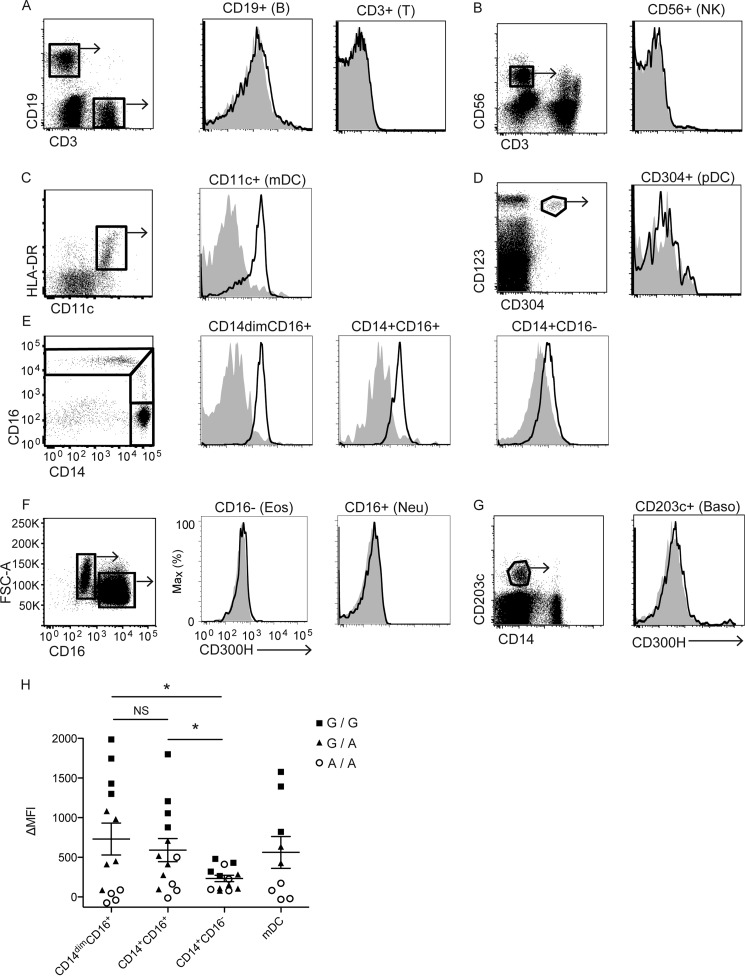
Flow cytometry analyses of human peripheral blood cells demonstrated that CD300H was expressed on myeloid lineage cells, including CD16+ Mo (Fig. 3, E and H) and myeloid DCs (n = 9; Fig. 3, C and H) but not on plasmacytoid DCs (n = 3; Fig. 3D). By contrast, CD300H was not expressed on lymphocytes (T cells, B cells, and NK cells) (n = 3 for each; Fig. 3, A and B) or granulocytes (eosinophils, neutrophils, and basophils) (n = 3 for each; Fig. 3, F and G). CD16+ Mo can be divided into two subsets (namely CD14dimCD16+ and CD14+CD16+ Mo) (4) on the basis of their CD14 expression; mean fluorescence intensity (MFI) of CD300H expression was comparable between these subsets (p = 0.5816, n = 13 for each; Fig. 3, E and H). Compared with CD14dimCD16+ and CD14+CD16+ Mo, CD16− Mo (n = 13) exhibited a significantly lower level of CD300H expression (p = 0.0234 and p = 0.0270, respectively; Fig. 3, E and H).
FIGURE 3.
Expression of human CD300H protein. A–G, flow cytometry of peripheral blood mononuclear cells (5 × 105 cells/experiment) from healthy donors. Cells were stained with specific antibodies to distinguish different cell subsets and simultaneously stained with biotinylated control mouse immunoglobulin (shaded) or anti-CD300H (TX93) (thick lines), followed by phosphatidylethanolamine/Cy-7 streptavidin. PI-positive dead cells were excluded. Cells in outlined areas were gated and analyzed for CD300H expression. Cells in C are lineage (CD3, CD19, CD56, CD14)−. Cells in E are lineage (CD3, CD19, CD56)− HLA-DR+. HLA-DR, human leukocyte antigen-DR; FSC-A, forward scatter-area; B, B cell; T, T cell; NK, natural killer cell; mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell; Eos, eosinophil; Neu, neutrophil; Baso, basophil. Data are representative of at least three independent experiments. H, the MFI of CD300H expression on each cell type from each individual carrying A/A (circles), G/A (triangles), or G/G (squares) genotype is shown. ΔMFI, MFI of control stained with isotype was subtracted from MFI of CD300H stained with anti-CD300H (TX93). *, p < 0.05; NS, not significant.
Individual Differences in CD300H Expression
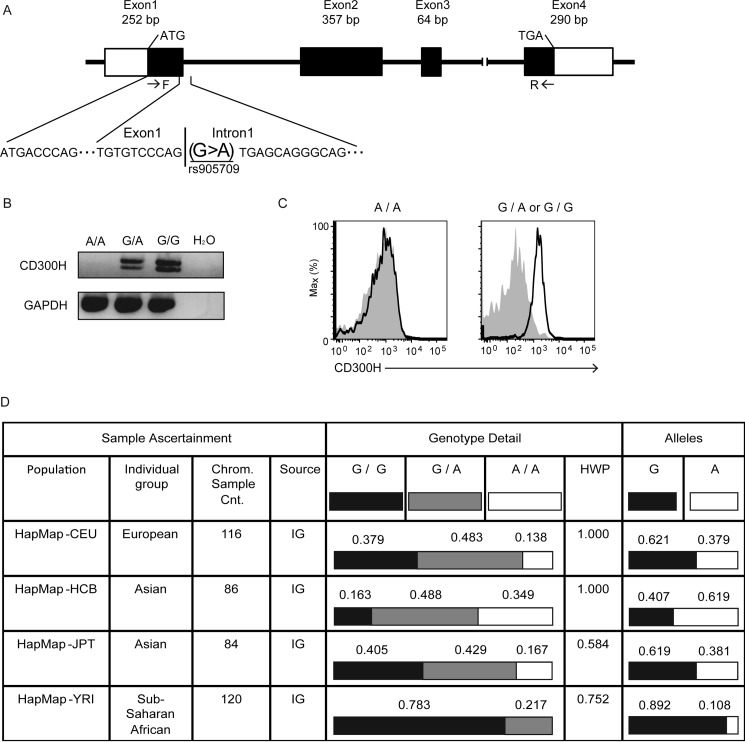
Genomic DNA database analysis demonstrated that CD300H consisted of four exons (Fig. 4A), but the gene encoding CD300Hs lacked exon 3, which encoded the transmembrane region. Interestingly, we observed that 4 of the 13 individuals examined in Fig. 3 showed no detectable expression of CD300H on CD16+ Mo (Fig. 3H, open circles). Genomic DNA analyses of these individuals demonstrated a single-nucleotide substitution (rs905709; G→A) of CD300H, which abolished the intron 1 donor splice site (Fig. 4A). This can lead to loss of CD300H transcript expression through the action of nonsense-mediated mRNA decay machinery (30). Indeed, persons carrying the rs905709 A/A genotype showed a loss of CD300H transcripts (Fig. 4B) and protein (Figs. 3H and 4C). In contrast, CD16+ Mo derived from subjects with a G/A or G/G genotype expressed the CD300H transcript (Fig. 4B) and protein on their cell surfaces (Figs. 3H and 4C). The A/A genotype frequency of rs905709 differed significantly in the different ethnic populations and is observed at high frequency in Han Chinese in Beijing, China (HCB), Japanese in Tokyo, Japan (JPT), and CEPH (Utah Residents with Northern and Western European Ancestry, CEU) populations (A/A genotype frequencies 0.349, 0.167, and 0.138, respectively). However, it is extremely rare in Yoruba in Ibadan, Nigeria (YRI) (Fig. 4D) (31).
FIGURE 4.
A single-nucleotide polymorphism, rs905709 (allele A), at the splice donor site on intron 1 of CD300H is responsible for the loss of CD300H expression. A, schematic structure of the exon-intron of CD300H. F and R, sites of primers. Peripheral blood mononuclear cells derived from healthy donors carrying the A/A, G/A, or G/G genotype were subjected to RT-PCR analysis for the expression of CD300H and GAPDH transcripts (B) or were analyzed for surface expression of CD300H on CD14dimCD16+ Mo by flow cytometry analysis (C), as described in the legend to Fig. 3. Data are representative of two independent experiments. D, table from the National Center for Biotechnology Information SNP Web site showing the population diversity of SNP rs905709 in Utah residents with ancestry from Northern and Western Europe (CEU), Han Chinese in Beijing (HCB), Japanese in Tokyo (JPT), and Yoruba in Ibadan, Nigeria (YRI) populations. HWP, Hardy-Weinberg probability; Chrom. Sample Cnt, chromosome sample count; IG, individual genotype.
CD300H Associates with DAP12 and DAP10 in U937 Cells
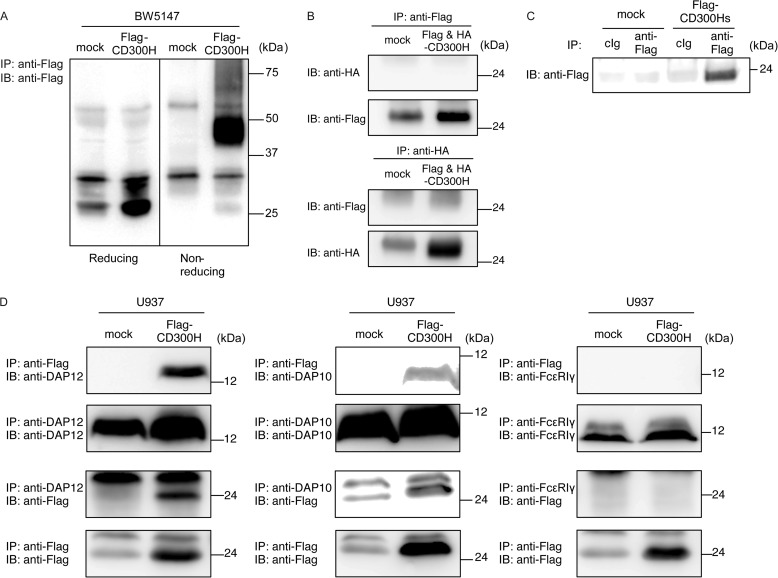
To analyze the biochemical characteristics of CD300H, we generated a BW5147 transfectant stably expressing FLAG-tagged CD300H at the N terminus. Immunoprecipitation of CD300H protein from lysates of BW5147 transfectant and subsequent immunoblotting with anti-FLAG antibody revealed that human CD300H had a molecular mass of ∼25 kDa under reducing conditions and ∼50 kDa under non-reducing conditions (Fig. 5A), suggesting that CD300H forms dimer formation. To determine whether CD300H forms homodimeric or heterodimeric structure, we generated BW5147 transfectant simultaneously expressing FLAG-tagged and hemagglutinin (HA)-tagged CD300H and performed a co-immunoprecipitation assay. We found that HA-tagged CD300H was not co-immunoprecipitated with FLAG-tagged CD300H and vice versa (Fig. 5B), suggesting that CD300H forms heterodimeric structure with an undetermined molecule with a molecular mass of ∼25 kDa expressed in BW5147 cells.
FIGURE 5.
Biochemical analyses of CD300H. A, BW5147 cells and transfectants expressing FLAG-tagged CD300H (5 × 106 cells/experiment) were lysed in 1% Nonidet P-40 buffer, immunoprecipitated (IP) with anti-FLAG, and immunoblotted (IB) with anti-FLAG. B, BW5147 transfectants simultaneously expressing FLAG-CD300H and HA-CD300H were lysed in digitonin buffer, immunoprecipitated with anti-HA or anti-FLAG, and immunoblotted with anti-HA or anti-FLAG in reducing conditions. C, culture supernatant from 293T cells transiently expressing FLAG-tagged CD300Hs or mock were immunoprecipitated with anti-FLAG and immunoblotted with anti-FLAG. D, U937 cells and transfectants stably expressing FLAG-tagged CD300H were lysed in digitonin buffer, immunoprecipitated with anti-FLAG, anti-DAP12, anti-DAP10, or anti-FcϵRIγ, and immunoblotted with anti-FLAG, anti-DAP12, anti-DAP10, or anti-FcϵRIγ. Data are representative of two independent experiments.
Next, we analyzed to determine whether CD300Hs is a secreted form of CD300H. We performed immunoblot analysis of culture supernatant of 293T transfectant transiently expressing CD300Hs tagged with FLAG at the N terminus and found that CD300Hs was a secreted form of CD300H (Fig. 5C).
CD300H possesses a lysine residue in the transmembrane region, suggesting that CD300H associates with DAP12, DAP10, and/or FcϵRIγ chain. We found that FLAG-tagged CD300H was co-immunoprecipitated with endogenous DAP12 and DAP10, and vice versa, but not with FcϵRIγ chain, in U937 cells (Fig. 5D). This result indicates that, unlike mouse MAIR-II, CD300H associates with DAP12 but not with the FcϵRIγ chain in U937 cells.
CD300H Transduces an Activating Signal in CD16+ Mo
Our findings that CD300H was expressed on CD16+ Mo and was associated with DAP12 and DAP10 in U937 cell line suggested that CD300H was an activating receptor on CD16+ Mo. Human CD16+ Mo consists of two distinct subsets. CD14dimCD16+ Mo resemble murine Ly6Clow Mo (5), which are designated pMo and produce chemokines. On the other hand, CD14+CD16+ Mo are designated as intMo (32), main producers of inflammatory cytokines, such as TNF-α and IL-1β (33). Therefore, we investigated whether CD300H mediated signaling for inflammatory cytokine and chemokine production in CD16+ Mo. We sorted CD16+ Mo from the PBMCs of healthy donors with the G/G (CD300H+) or A/A genotype (CD300H−) and then stimulated them with plate-coated F(ab′)2 fragments of TX93 mAb or control Ig (Fig. 6, A and B). Stimulation of CD300H+CD16+ Mo with TX93 mAb induced significantly more TNF-α and IL-6 production than did stimulation with control Ig. In contrast, stimulation with TX93 mAb did not increase cytokine production by CD300H−CD16+ Mo compared with stimulation with control Ig (Fig. 6, B and C). Thus, CD300H transduces an activating signal for cytokine production in CD16+ Mo. A recent study has demonstrated that murine Ly6Clow Mo recruit neutrophils in a TLR7-dependent manner to mediate focal necrosis of endothelial cells (6). CD16+ Mo produce chemoattractants for neutrophils. To examine whether CD300H is involved in this process, CD16+ Mo were stimulated with plate-coated F(ab′)2 fragments of TX93 mAb or control Ig for 24 h; the culture supernatant was then plated in the lower compartment of a Transwell, and neutrophils derived from the peripheral blood were added to the upper compartment. In the case of CD300H+CD16+ Mo but not CD300H−CD16+ Mo, the average number of neutrophils that migrated from the upper to the lower compartment upon TX93 mAb stimulation was significantly greater than that with control Ig-stimulated culture supernatant (∼1.3-fold increase; p < 0.05) (Fig. 6D). These results suggested that CD300H mediated a signal for the production of a neutrophil chemoattractant. Indeed, the expression of CXCL8, CXCL1, CXCL2, and CXCL5, which encode chemokines for neutrophil attraction, was significantly greater in CD300H+ Mo, but not CD300H− Mo, than in the controls after stimulation with plate-coated F(ab′)2 fragments of TX93 mAb (Fig. 6E).
FIGURE 6.
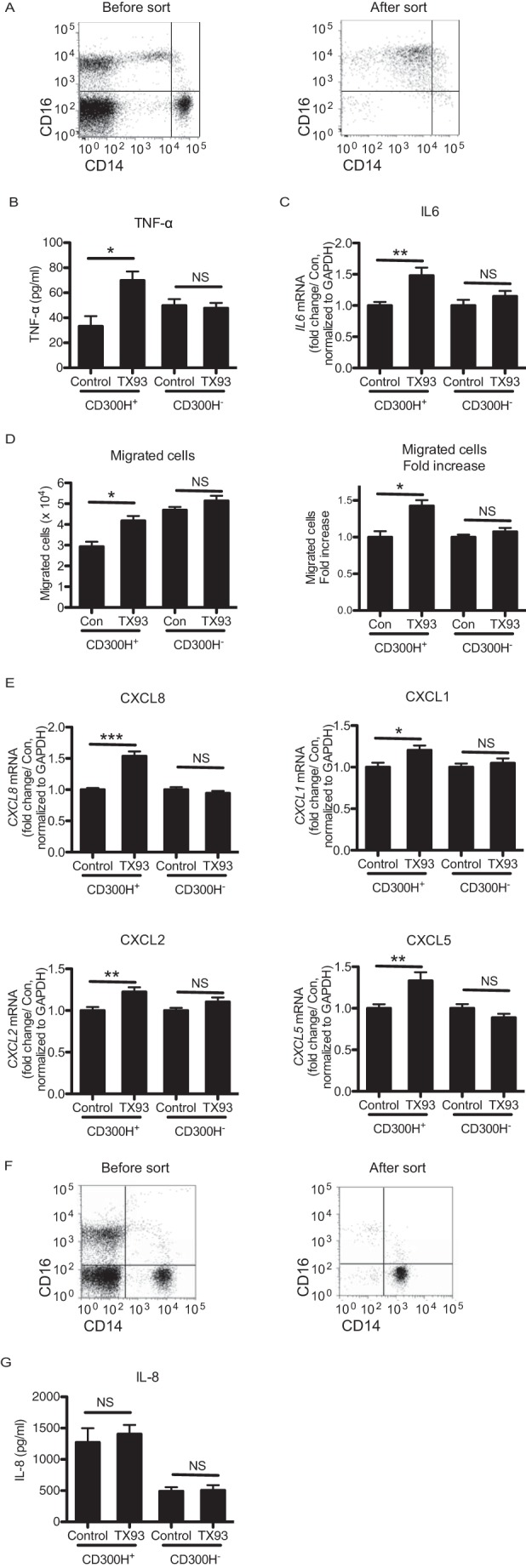
CD300H ligation in CD16+ Mo induces cytokine and chemokine secretion. CD16+ Mo (A–E) and CD16− Mo (F and G) were isolated from peripheral blood mononuclear cells by using the MACS system. Their purity was then analyzed by using flow cytometry (A and F). MACS-isolated Mo from G/G (CD300H+) or A/A (CD300H−) genotype donors were stimulated with plate-coated F(ab′)2 fragment of TX93 or control Ig for 24 h (B, D, and G) or 4 h (C and E). Culture supernatant was analyzed by ELISA (B) or subjected to neutrophil migration assay (D) or cytometric bead array (G). Cells were analyzed for IL-6 (C) and chemokine (E) expression by using quantitative polymerase chain reaction. In D, culture supernatant was transferred to the lower compartment of transwells, and neutrophils were placed in the upper compartment. This was followed by a 30-min incubation. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant. All data are representative of three independent experiments. Data in C and E are shown as mean ± S.D. (error bars) of four (CD300H+) and three (CD300H−) different donors.
CD300H Had No Effect on IL-8 Production in CD16− Mo
Because CD16− Mo also expresses CD300H (Fig. 3E), we analyzed the CD300H activity on CD16−CD14+ classical Mo sorted from the peripheral blood from donors with the G/G (CD300H+) or A/A genotype (CD300H−). We analyzed the production of cytokines, including CXCL8 (IL-8), IL-12p70, TNF-α, IL-10, IL-6, and IL-1β, after stimulation with anti-CD300H mAb by using cytometric bead array. However, we could not detect a significant amount of cytokines except for IL-8 and observed no significant differences in IL-8 production between stimulation with control antibody and anti-CD300 mAb (Fig. 6, F and G). This might be the result of the low level of CD300H expression on CD16− Mo compared with CD16+ Mo.
CD300H Transduces an Activating Signal in CD11c+ DC
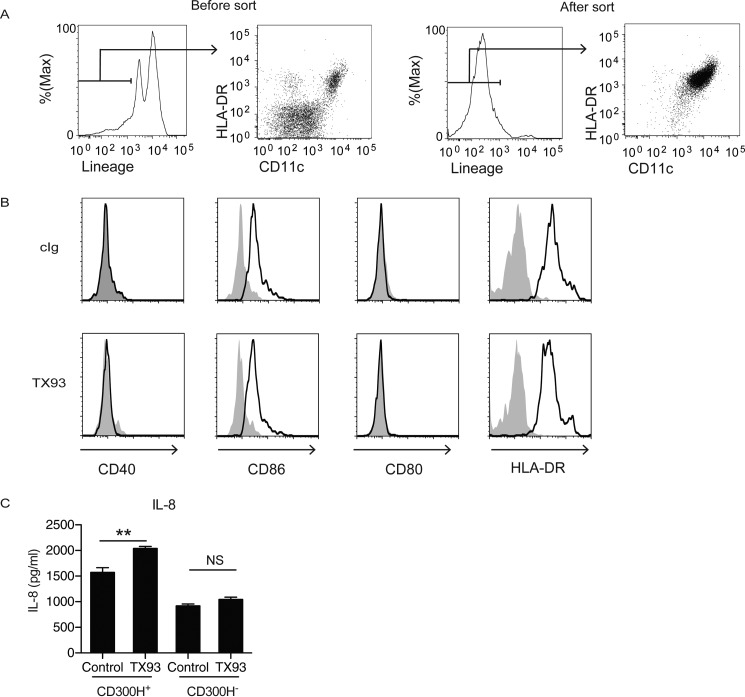
CD11c+ DCs also express CD300H at a level comparable with CD16+ Mo. We addressed whether CD300H transduces activating signal in CD11c+ DCs. We purified CD11c+ cells by the MACS sorting system (Fig. 7A), stimulated them with plate-coated F(ab′)2 fragment of TX93 mAb, and analyzed surface expression of co-stimulatory molecule and cytokine production by flow cytometry. We found that TX93 stimulation did not enhance the expression level of co-stimulatory molecules, such as CD40, CD86, CD80, and HLA-DR, on the G/G genotype (CD300H+)-derived CD11c+ DCs (Fig. 7B). However, CD11c+ DCs from G/G genotype, but not A/A genotype (CD300H−), produced IL-8 upon CD300H stimulation (Fig. 7C). These results indicate that CD300H induced IL-8 production by CD16+ Mo and CD11c+ DCs and may contribute to neutrophil attraction.
FIGURE 7.
CD300H ligation in CD11c+ DCs induces IL-8 production. CD11c+ DCs were isolated from peripheral blood mononuclear cells by using the MACS system. A, the purity was analyzed by using flow cytometry. MACS-isolated DCs from G/G (CD300H+) or A/A (CD300H−) genotype donors were stimulated with plate-coated F(ab′)2 fragment of TX93 or control Ig for 24 h. B, cells were analyzed for expression of co-stimulatory molecules by flow cytometry. C, culture supernatant was analyzed by cytometric bead array. Lineage: CD3, CD19, CD56, CD14. **, p < 0.01; NS, not significant. Data are representative of three independent experiments. Error bars, S.D.
Discussion
We identified and characterized a new member of the human CD300 family of immunoreceptors, which we designated CD300H. It is expressed on CD14+CD16+ intMo and CD14dimCD16+ pMo in the peripheral blood and transduces an activating signal for the production of inflammatory cytokines (TNF-α and IL-6) and chemokines (CXCL1, CXCL2, CXCL5, and IL-8) by CD16+ Mo. In addition, CD300H induces IL-8 production by CD11c+ DC. Importantly, we demonstrated that CD300H mediates neutrophil recruitment through the production of chemoattractants for those cells, suggesting that CD300H plays an important role in host immunity against bacterial and viral infections.
Despite recent progress in human Mo studies, the physiological roles of CD16+ Mo have not been fully elucidated. One recent report described a healthy family with CD16+ Mo deficiency in the blood (34), suggesting that that the function of CD16+ Mo is redundant. In accordance with this idea, we also found a subpopulation of healthy people whose CD16+ Mo did not express CD300H, suggesting that CD300H on CD16+ Mo is functionally redundant. However, because CD16+ pMo are implicated in inflammatory diseases, such as arthritis and atherosclerosis (35–37), or autoimmune diseases, such as systemic lupus erythematosus (4), deficiency of CD16+ pMo or CD300H on CD16+ pMo might be potentially advantageous, rather than disadvantageous, for maintaining homeostasis. The genetic mutation known as CCR5-Δ32 is a well studied example of an advantageous allele with a well characterized geographic distribution. This mutation plays an important role in human immunodeficiency virus (HIV)-1 resistance because it prevents functional expression of the CCR5 chemokine receptor, the entry receptor for HIV-1 (38). The mutation is found principally in Europe and Western Asia, where average frequencies are ∼10%, but not in Western and Central Africa (38). The mutant allele, which can be regarded as a natural knockout in humans, is not accompanied by an obvious phenotype in homozygous individuals, although CCR5 has a protective role against L. monocytogenes infection in mice (39). Additional studies are required to determine the functional role of CD300H in immunity and inflammation.
CD300H has a splicing variant, CD300Hs, which lacks the transmembrane domain. We found that CD300Hs is a secreted form of CD300H. Recent findings demonstrated that activating immunoglobulin-like receptors CD300b and triggering receptor expressed on myeloid cells (TREM-1) also produce soluble forms sCD300b and sTREM-1, respectively, by proteolytic cleavage (40, 41). Whereas sCD300b amplifies LPS-induced inflammation (40, 42), sTREM-1 attenuates excessive inflammatory response by counterregulating TREM-1 (41), probably by acting as a decoy receptor, as observed in the TNF-α system (43). Therefore, expression of CD300Hs may modulate CD300H-mediated inflammatory responses in vivo.
Our studies suggested that CD300H may form a heterodimeric structure in mouse T cell lymphoma, BW5147. Recent evidence demonstrated that CD300C forms either a homodimer or heterodimer with all CD300 family members. Although these dimer formations require Ig-like domains, CD300C did not bind other Ig-like receptors, such as TREM-1 and CD28 (44) (45). The Ig-like domain of CD300H has 53% homology to that of CD300C, suggesting that CD300H may also form heterodimer with mouse CD300 family members expressed on the BW5147 cell line. Because we did not detect surface expression of mouse CD300a, MAIR-II, and CD300lf on BW5147 cells by flow cytometry using TX41 (12), TX52 (46), and TX70 (15) mAbs, respectively (data not shown), CD300H may interact with mouse CD300 family members other than these Ig-like receptors. Unlike CD300C, CD300H has a positively charged lysine residue in its transmembrane region and interacts with DAP12 and DAP10. Thus, CD300H may be able to mediate an activating signal upon ligand binding. However, the heterodimer of CD300H with other CD300 receptors may positively or negatively modulate the signaling.
To elucidate the physiological roles of CD300H on CD16+ pMo in vivo, the counterpart of CD300H in mice needs to be identified. Although we identified CD300H in our search for a human homolog of MAIR-II, there are several differences between MAIR-II and CD300H. First, MAIR-II inhibits B-cell receptor-mediated signaling in B cells (47), whereas CD300H expression on human peripheral blood B cells was not detected by flow cytometry using TX93 mAb (Fig. 3). Second, MAIR-II is a unique receptor that binds to either DAP12 or FcϵRIγ; however, CD300H did not show an association with FcϵRIγ in U937 cells. Third, CD300H is located adjacent to CD300C on human chromosome 17, whereas Cd300ld/MAIR-IV/LMIR4/Clm-5, but not MAIR-II, is located adjacent to Cd300c on mouse chromosome 11. Recent studies have demonstrated that CD300 family receptors recognize lipids, such as phosphatidylserine, ceramide, and lipoprotein, as ligands that mediate signals in mast cells and macrophages via these receptors (48–50). We and others reported that human CD300A binds to phosphatidylserine and phosphatidylethanolamine (11, 27, 50). Because CD300H has high amino acid identity with CD300A, we assessed whether CD300H also binds to phosphatidylserine and phosphatidylethanolamine by dot blot analyses using a lipid-bound membrane. However, CD300H bound to neither phosphatidylserine nor phosphatidylethanolamine (data not shown). Future studies are required for identification of a ligand for CD300H to understand the function of CD300H in vitro and in vivo.
Author Contributions
K. N. conducted the experiments and analyzed the data. S. T.-H. designed the experiments, analyzed the data, and wrote the paper. E. N. analyzed the data. A. S. supervised the overall project and wrote the paper.
Acknowledgments
We thank N. Akaboshi and K. Hayashi for technical assistance, Jun Ohashi (University of Tokyo) for helpful discussions, S. Mitsuishi and Y. Nomura for secretarial assistance, and the members of the laboratory for discussions.
This work was supported in part by grants provided by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- Mo
- monocytes
- iMo
- inflammatory Mo
- pMo
- patrolling Mo
- intMo
- intermediate Mo
- DC
- dendritic cell
- PBMC
- peripheral blood mononuclear cell
- aa
- amino acid(s)
- MFI
- mean fluorescence intensity.
References
- 1. Soehnlein O., Lindbom L. (2010) Phagocyte partnership during the onset and resolution of inflammation. Nat. Rev. Immunol. 10, 427–439 [DOI] [PubMed] [Google Scholar]
- 2. Shi C., Pamer E. G. (2011) Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon S., Taylor P. R. (2005) Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 [DOI] [PubMed] [Google Scholar]
- 4. Cros J., Cagnard N., Woollard K., Patey N., Zhang S. Y., Senechal B., Puel A., Biswas S. K., Moshous D., Picard C., Jais J. P., D'Cruz D., Casanova J. L., Trouillet C., Geissmann F. (2010) Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 33, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auffray C., Fogg D., Garfa M., Elain G., Join-Lambert O., Kayal S., Sarnacki S., Cumano A., Lauvau G., Geissmann F. (2007) Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666–670 [DOI] [PubMed] [Google Scholar]
- 6. Carlin L. M., Stamatiades E. G., Auffray C., Hanna R. N., Glover L., Vizcay-Barrena G., Hedrick C. C., Cook H. T., Diebold S., Geissmann F. (2013) Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell 153, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ziegler-Heitbrock L., Ancuta P., Crowe S., Dalod M., Grau V., Hart D. N., Leenen P. J., Liu Y. J., MacPherson G., Randolph G. J., Scherberich J., Schmitz J., Shortman K., Sozzani S., Strobl H., Zembala M., Austyn J. M., Lutz M. B. (2010) Nomenclature of monocytes and dendritic cells in blood. Blood 116, e74–e80 [DOI] [PubMed] [Google Scholar]
- 8. Wong K. L., Yeap W. H., Tai J. J., Ong S. M., Dang T. M., Wong S. C. (2012) The three human monocyte subsets: implications for health and disease. Immunol. Res. 53, 41–57 [DOI] [PubMed] [Google Scholar]
- 9. Wong K. L., Tai J. J., Wong W. C., Han H., Sem X., Yeap W. H., Kourilsky P., Wong S. C. (2011) Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118, e16–e31 [DOI] [PubMed] [Google Scholar]
- 10. Shibuya A., Nakahashi-Oda C., Tahara-Hanaoka S. (2009) Regulation of immune responses by the activating and inhibitory myeloid-associated immunoglobulin-like receptors (MAIR) (CD300). Immune Netw. 9, 41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borrego F. (2013) The CD300 molecules: an emerging family of regulators of the immune system. Blood 121, 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yotsumoto K., Okoshi Y., Shibuya K., Yamazaki S., Tahara-Hanaoka S., Honda S., Osawa M., Kuroiwa A., Matsuda Y., Tenen D. G., Iwama A., Nakauchi H., Shibuya A. (2003) Paired activating and inhibitory immunoglobulin-like receptors, MAIR-I and MAIR-II, regulate mast cell and macrophage activation. J. Exp. Med. 198, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumagai H., Oki T., Tamitsu K., Feng S. Z., Ono M., Nakajima H., Bao Y. C., Kawakami Y., Nagayoshi K., Copeland N. G., Gilbert D. J., Jenkins N. A., Kawakami T., Kitamura T. (2003) Identification and characterization of a new pair of immunoglobulin-like receptors LMIR1 and 2 derived from murine bone marrow-derived mast cells. Biochem. Biophys. Res. Commun. 307, 719–729 [DOI] [PubMed] [Google Scholar]
- 14. Moshkovits I., Shik D., Itan M., Karo-Atar D., Bernshtein B., Hershko A. Y., van Lookeren Campagne M., Munitz A. (2014) CMRF35-like molecule 1 (CLM-1) regulates eosinophil homeostasis by suppressing cellular chemotaxis. Mucosal Immunol. 7, 292–303 [DOI] [PubMed] [Google Scholar]
- 15. Can I., Tahara-Hanaoka S., Hitomi K., Nakano T., Nakahashi-Oda C., Kurita N., Honda S., Shibuya K., Shibuya A. (2008) Caspase-independent cell death by CD300LF (MAIR-V), an inhibitory immunoglobulin-like receptor on myeloid cells. J. Immunol. 180, 207–213 [DOI] [PubMed] [Google Scholar]
- 16. Can I., Tahara-Hanaoka S., Shibuya A. (2008) Expression of a splicing isoform of MAIR-V (CD300LF), an inhibitory immunoglobulin-like receptor on myeloid cells. Hybridoma 27, 59–61 [DOI] [PubMed] [Google Scholar]
- 17. Izawa K., Kitaura J., Yamanishi Y., Matsuoka T., Oki T., Shibata F., Kumagai H., Nakajima H., Maeda-Yamamoto M., Hauchins J. P., Tybulewicz V. L., Takai T., Kitamura T. (2007) Functional analysis of activating receptor LMIR4 as a counterpart of inhibitory receptor LMIR3. J. Biol. Chem. 282, 17997–18008 [DOI] [PubMed] [Google Scholar]
- 18. Izawa K., Kitaura J., Yamanishi Y., Matsuoka T., Kaitani A., Sugiuchi M., Takahashi M., Maehara A., Enomoto Y., Oki T., Takai T., Kitamura T. (2009) An activating and inhibitory signal from an inhibitory receptor LMIR3/CLM-1: LMIR3 augments lipopolysaccharide response through association with FcRγ in mast cells. J. Immunol. 183, 925–936 [DOI] [PubMed] [Google Scholar]
- 19. Izawa K., Yamanishi Y., Maehara A., Takahashi M., Isobe M., Ito S., Kaitani A., Matsukawa T., Matsuoka T., Nakahara F., Oki T., Kiyonari H., Abe T., Okumura K., Kitamura T., Kitaura J. (2012) The receptor LMIR3 negatively regulates mast cell activation and allergic responses by binding to extracellular ceramide. Immunity 37, 827–839 [DOI] [PubMed] [Google Scholar]
- 20. Izawa K., Isobe M., Matsukawa T., Ito S., Maehara A., Takahashi M., Yamanishi Y., Kaitani A., Oki T., Okumura K., Kitamura T., Kitaura J. (2014) Sphingomyelin and ceramide are physiological ligands for human LMIR3/CD300f, inhibiting FcepsilonRI-mediated mast cell activation. J. Allergy Clin. Immunol. 133, 270–273 [DOI] [PubMed] [Google Scholar]
- 21. Chung D. H., Humphrey M. B., Nakamura M. C., Ginzinger D. G., Seaman W. E., Daws M. R. (2003) CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclast formation. J. Immunol. 171, 6541–6548 [DOI] [PubMed] [Google Scholar]
- 22. Yamanishi Y., Kitaura J., Izawa K., Matsuoka T., Oki T., Lu Y., Shibata F., Yamazaki S., Kumagai H., Nakajima H., Maeda-Yamamoto M., Tybulewicz V. L., Takai T., Kitamura T. (2008) Analysis of mouse LMIR5/CLM-7 as an activating receptor: differential regulation of LMIR5/CLM-7 in mouse versus human cells. Blood 111, 688–698 [DOI] [PubMed] [Google Scholar]
- 23. Nakano T., Tahara-Hanaoka S., Nakahashi C., Can I., Totsuka N., Honda S., Shibuya K., Shibuya A. (2008) Activation of neutrophils by a novel triggering immunoglobulin-like receptor MAIR-IV. Mol. Immunol. 45, 289–294 [DOI] [PubMed] [Google Scholar]
- 24. Enomoto Y., Yamanishi Y., Izawa K., Kaitani A., Takahashi M., Maehara A., Oki T., Takamatsu R., Kajikawa M., Takai T., Kitamura T., Kitaura J. (2010) Characterization of leukocyte mono-immunoglobulin-like receptor 7 (LMIR7)/CLM-3 as an activating receptor: its similarities to and differences from LMIR4/CLM-5. J. Biol. Chem. 285, 35274–35283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujimoto M., Takatsu H., Ohno H. (2006) CMRF-35-like molecule-5 constitutes novel paired receptors, with CMRF-35-like molecule-1, to transduce activation signal upon association with FcRγ. Int. Immunol. 18, 1499–1508 [DOI] [PubMed] [Google Scholar]
- 26. Tahara-Hanaoka S., Shibuya K., Onoda Y., Zhang H., Yamazaki S., Miyamoto A., Honda S., Lanier L. L., Shibuya A. (2004) Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 16, 533–538 [DOI] [PubMed] [Google Scholar]
- 27. Nakahashi-Oda C., Tahara-Hanaoka S., Honda S., Shibuya K., Shibuya A. (2012) Identification of phosphatidylserine as a ligand for the CD300a immunoreceptor. Biochem. Biophys. Res. Commun. 417, 646–650 [DOI] [PubMed] [Google Scholar]
- 28. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 29. Perrière G., Gouy M. (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369 [DOI] [PubMed] [Google Scholar]
- 30. Rebbapragada I., Lykke-Andersen J. (2009) Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 21, 394–402 [DOI] [PubMed] [Google Scholar]
- 31. International HapMap Consortium (2003) The International HapMap Project. Nature 426, 789–796 [DOI] [PubMed] [Google Scholar]
- 32. Ziegler-Heitbrock L. (2014) Monocyte subsets in man and other species. Cell Immunol. 289, 135–139 [DOI] [PubMed] [Google Scholar]
- 33. Belge K. U., Dayyani F., Horelt A., Siedlar M., Frankenberger M., Frankenberger B., Espevik T., Ziegler-Heitbrock L. (2002) The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J. Immunol. 168, 3536–3542 [DOI] [PubMed] [Google Scholar]
- 34. Frankenberger M., Ekici A. B., Angstwurm M. W., Hoffmann H., Hofer T. P., Heimbeck I., Meyer P., Lohse P., Wjst M., Häussinger K., Reis A., Ziegler-Heitbrock L. (2013) A defect of CD16-positive monocytes can occur without disease. Immunobiology 218, 169–174 [DOI] [PubMed] [Google Scholar]
- 35. Iwahashi M., Yamamura M., Aita T., Okamoto A., Ueno A., Ogawa N., Akashi S., Miyake K., Godowski P. J., Makino H. (2004) Expression of Toll-like receptor 2 on CD16+ blood monocytes and synovial tissue macrophages in rheumatoid arthritis. Arthritis Rheum. 50, 1457–1467 [DOI] [PubMed] [Google Scholar]
- 36. Schlitt A., Heine G. H., Blankenberg S., Espinola-Klein C., Dopheide J. F., Bickel C., Lackner K. J., Iz M., Meyer J., Darius H., Rupprecht H. J. (2004) CD14+CD16+ monocytes in coronary artery disease and their relationship to serum TNF-α levels. Thromb. Haemost. 92, 419–424 [DOI] [PubMed] [Google Scholar]
- 37. Ziegler-Heitbrock L. (2007) The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J. Leukoc. Biol. 81, 584–592 [DOI] [PubMed] [Google Scholar]
- 38. Samson M., Libert F., Doranz B. J., Rucker J., Liesnard C., Farber C. M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C., Muyldermans G., Verhofstede C., Burtonboy G., Georges M., Imai T., Rana S., Yi Y., Smyth R. J., Collman R. G., Doms R. W., Vassart G., Parmentier M. (1996) Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 [DOI] [PubMed] [Google Scholar]
- 39. Zhou Y., Kurihara T., Ryseck R. P., Yang Y., Ryan C., Loy J., Warr G., Bravo R. (1998) Impaired macrophage function and enhanced T cell-dependent immune response in mice lacking CCR5, the mouse homologue of the major HIV-1 coreceptor. J. Immunol. 160, 4018–4025 [PubMed] [Google Scholar]
- 40. Yamanishi Y., Takahashi M., Izawa K., Isobe M., Ito S., Tsuchiya A., Maehara A., Kaitani A., Uchida T., Togami K., Enomoto Y., Nakahara F., Oki T., Kajikawa M., Kurihara H., Kitamura T., Kitaura J. (2012) A soluble form of LMIR5/CD300b amplifies lipopolysaccharide-induced lethal inflammation in sepsis. J. Immunol. 189, 1773–1779 [DOI] [PubMed] [Google Scholar]
- 41. Gibot S., Kolopp-Sarda M. N., Béné M. C., Bollaert P. E., Lozniewski A., Mory F., Levy B., Faure G. C. (2004) A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J. Exp. Med. 200, 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Phongsisay V., Iizasa E., Hara H., Yamasaki S. (2014) LMIR5 extracellular domain activates myeloid cells through toll-like receptor 4. Mol. Immunol. 62, 169–177 [DOI] [PubMed] [Google Scholar]
- 43. Lantz M., Gullberg U., Nilsson E., Olsson I. (1990) Characterization in vitro of a human tumor necrosis factor-binding protein: a soluble form of a tumor necrosis factor receptor. J. Clin. Invest. 86, 1396–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Comas-Casellas E., Martínez-Barriocanal Á., Miró F., Ejarque-Ortiz A., Schwartz S. Jr., Martín M., Sayós J. (2012) Cloning and characterization of CD300d, a novel member of the human CD300 family of immune receptors. J. Biol. Chem. 287, 9682–9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martínez-Barriocanal A., Comas-Casellas E., Schwartz S. Jr., Martín M., Sayós J. (2010) CD300 heterocomplexes, a new and family-restricted mechanism for myeloid cell signaling regulation. J. Biol. Chem. 285, 41781–41794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakahashi C., Tahara-Hanaoka S., Totsuka N., Okoshi Y., Takai T., Ohkohchi N., Honda S., Shibuya K., Shibuya A. (2007) Dual assemblies of an activating immune receptor, MAIR-II, with ITAM-bearing adapters DAP12 and FcRγ chain on peritoneal macrophages. J. Immunol. 178, 765–770 [DOI] [PubMed] [Google Scholar]
- 47. Nakano-Yokomizo T., Tahara-Hanaoka S., Nakahashi-Oda C., Nabekura T., Tchao N. K., Kadosaki M., Totsuka N., Kurita N., Nakamagoe K., Tamaoka A., Takai T., Yasui T., Kikutani H., Honda S., Shibuya K., Lanier L. L., Shibuya A. (2011) The immunoreceptor adapter protein DAP12 suppresses B lymphocyte-driven adaptive immune responses. J. Exp. Med. 208, 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cannon J. P., O'Driscoll M., Litman G. W. (2012) Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics 64, 39–47 [DOI] [PubMed] [Google Scholar]
- 49. Nakahashi-Oda C., Tahara-Hanaoka S., Shoji M., Okoshi Y., Nakano-Yokomizo T., Ohkohchi N., Yasui T., Kikutani H., Honda S., Shibuya K., Nagata S., Shibuya A. (2012) Apoptotic cells suppress mast cell inflammatory responses via the CD300a immunoreceptor. J. Exp. Med. 209, 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simhadri V. R., Andersen J. F., Calvo E., Choi S. C., Coligan J. E., Borrego F. (2012) Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood 119, 2799–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]