Abstract
Receptor tyrosine kinases (RTKs) are a family of ligand-binding cell surface receptors that regulate a wide range of essential cellular activities, including proliferation, differentiation, cell-cycle progression, survival and apoptosis. As such, these proteins play an important role during development and throughout life; germline mutations in genes encoding RTKs cause several developmental syndromes, while somatic alterations contribute to the pathogenesis of many aggressive cancers. This creates an interesting paradigm in which mutation timing, type and location in a gene leads to different cell signaling and biological responses, and ultimately phenotypic outcomes. In this review, we highlight the roles of RTKs in developmental disorders and cancer. The multifaceted roles of these receptors, their genetic signatures and their signaling during developmental morphogenesis and oncogenesis are discussed. Additionally, we propose that comparative analysis of RTK mutations responsible for developmental syndromes may shed light on those driving tumorigenesis.
Introduction
Phosphorylation is a reversible post-translational modification, which controls the activity and localization of many proteins, and is dynamically regulated by kinases and phosphatases. Protein kinases catalyze the transfer of a phosphate from ATP to threonine, serine and tyrosine residues of specific target proteins. To date, >520 protein kinases have been identified, ∼90 of which are tyrosine kinases (1,2). Receptor tyrosine kinases (RTKs) are a subclass of tyrosine kinases which are involved in mediating intercellular communication and orchestrating a wide range of complex biological functions (3). Genetic studies have demonstrated a role for RTK signaling in congenital and acquired human disease (1,4). These studies highlight a paradigm in which the timing, type and location of mutations in RTKs, whether in germline or somatic tissue, dictates diverse phenotypic effects. Here we review germline and somatic mutations in RTKs and discuss the consequences of aberrant signaling during developmental morphogenesis and oncogenesis.
Receptor Tyrosine Kinases
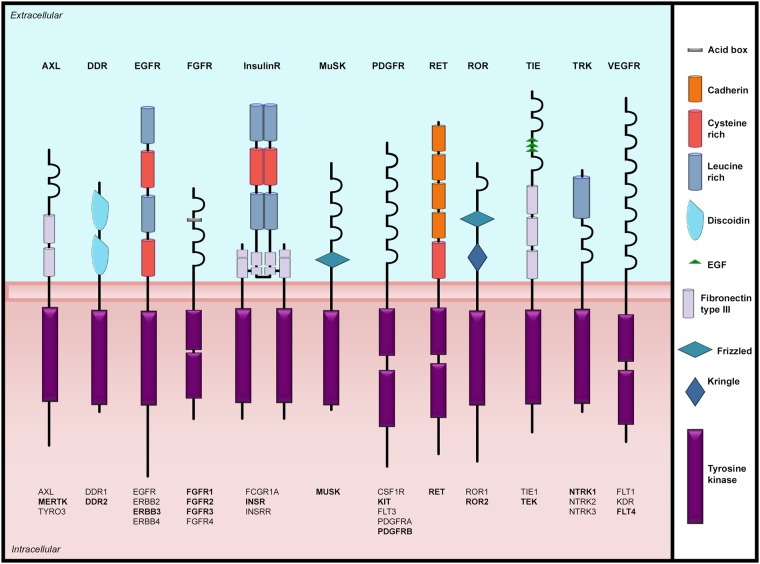
There are 58 RTKs identified to date, which can be subdivided into 20 subfamilies, all of which share a basic structure consisting of an extracellular ligand-binding domain linked to an intracellular protein kinase core via a single-pass transmembrane domain (Fig. 1) (2,3). RTK activation is a complex biological process and has been reviewed elsewhere (3). Briefly, canonical RTKs function by binding their specific ligand to induce dimerization and conformational changes. Additionally, there is a subset of RTKs that exist as oligomers even in the absence of the activating ligand; these oligomers are then activated by ligand binding which induces conformational changes (3). For both mechanisms, ligand activation leads to trans-autophosphorylation of tyrosine residues in the dimer/oligomer and activation of RTK catalytic activity (5). In an undimerized/inactive state the catalytic activity of the RTKs is inhibited by intramolecular interactions, which are released following activation by phosphorylation of the tyrosine kinase domain (TKD) (3,6). RTK phosphorylation occurs in two phases, intramolecular followed by intermolecular (3,6). In the first phase, trans-autophosphorylation between the dimer pair destabilizes cis-inhibition, permitting RTK catalytic activity (3,6). Autophosphorylation of the TKD continues creating phosphotyrosine-based binding sites. In the second phase, phosphotyrosine recognition motif containing cytoplasmic signaling proteins are recruited (3,6). Once bound, these proteins are activated by phosphorylation to initiate intracellular signaling for cell proliferation, growth, survival, apoptosis, differentiation, morphogenesis, cell-cycle progression, migration and autophagy (3,6). In addition to dimerization and intramolecular phosphorylation, RTK signaling is also regulated by positive and negative feedback mechanisms, tissue-specific splicing, RTK post-translational modifications, endocytosis and ligand availability (3,7–9). Together, these mechanisms prevent unwanted protein kinase activation, and enable temporal and tissue-specific kinase activity.
Figure 1.
Receptor tyrosine kinase families involved in human developmental disorders. Schematic representation of the RTKs with all family members listed below each receptor. Receptors involved in developmental disease are indicated in bold. Structural domains are marked according to the key.
Kinasopathies
Germline mutations disrupting RTK signaling pathways have been identified as the cause of a number of congenital malformation syndromes; we refer to this collection of disorders as the ‘developmental receptor tyrosine kinasopathies’ (DRTKs). To date, at least 35 DRTKs have been described in the Online Mendelian Inheritance in Man (OMIM) database, caused by mutations in 15 RTK genes (Table 1) (12). Interestingly, skeletal abnormalities are over-represented in DRTKs; mutations in DDR2, FGFR1, FGFR2, FGFR3 and ROR2 are associated with over 20 clinically distinct skeletal dysplasias. Specific mutations in these genes during embryogenesis cause defects in osteoblast/osteoclast/chondrocyte proliferation, growth, differentiation and apoptosis resulting in abnormal bone morphogenesis (13–16). Other affected systems include the nervous and endocrine systems. For example, a mutation in ERBB3 has been associated with lethal congenital contractural syndrome type 2 (17), and a number of insulin resistance conditions are caused by dysregulated INSR signaling, including familial hyperinsulinemic hypoglycemia (18) and Donohue (19) and Rabson–Mendenhall syndromes (20).
Table 1.
Examples of DRTKs
| Gene family | Gene symbol | OMIM number | Developmental disease (OMIM number) | Inheritance | Suspected mechanism | Disease mutation and COSMIC mutation overlap (known drivers are indicated in bold)a |
|---|---|---|---|---|---|---|
| AXL | MERTK | 604 705 | Retinitis pigmentosa 38 (613 862) | AR | LOF | |
| DDR | DDR2 | 191 311 | Short limb-hand spondylometaepiphyseal dysplasia (271 665) | AR | LOF | E113K, R752C |
| EGFR | ERBB3 | 190 151 | Lethal congenital contractural syndrome 2 (607 598) | AR | LOF | |
| FGFR | FGFR1 | 136 350 | Hartsfield syndrome (615 465) | AD/AR | ? | |
| Hypogonadotrophic hypogonadism 2 (147 950) | AD | LOF | R250W, A343V, G703S, V795I | |||
| Trigoncephaly (190 440) | AD | ? | ||||
| Pfeiffer syndrome (101 600) | AD | GOF | ||||
| Osteoglophonic dysplasia (166 250) | AD | GOF | ||||
| FGFR2 | 176 943 | Antley–Bixler syndrome (207 410) | AD | ? | W290C | |
| Apert syndrome (101 200) | AD | GOF | S252W, P253R | |||
| Beare–Stevenson cutis gyrata syndrome (123 790) | AD | GOF | S372C, Y375C | |||
| Bent bone dysplasia syndrome (614 592) | AD | ? | ||||
| Crouzon syndrome (123 500) | AD | GOF | S267P, W290R, D549H, R678G | |||
| Jackson–Weiss syndrome (123 150) | AD | GOF | ||||
| LADD syndrome (149 730) | AD | LOF | A648T | |||
| Pfeiffer syndrome (101 600) | AD | GOF | W290C | |||
| FGFR3 | 134 934 | Achondroplasia (100 800) | AD | GOF | G380R | |
| Severe achondroplasia with developmental delay and acanthosis nigricans (187 600) | AD | GOF | K650M | |||
| Crouzon syndrome with acanthosis nigricans (612 247) | AD? | GOF | A391E | |||
| Hypochondroplasia (146 000) | AD | GOF | N540S, K650N, K650T, K650Q | |||
| LADD syndrome (149 730) | AD | LOF | ||||
| Muenke craniosynostosis (602 849) | AD | GOF | ||||
| Thanatophoric dysplasia I (187 600) | AD | GOF | R248C, S249C, G370C, S371C, Y373C, K650M | |||
| Thanatophoric dysplasia II (187 601) | AD | GOF | K650E | |||
| INSR | INSR | 147 670 | Donohue syndrome (246 200) | AR | LOF | R924 |
| Rabson–Mendenhall syndrome (262 190) | AR | LOF | ||||
| MUSK | MUSK | 601 296 | Myasthenic syndrome type 9 (616 325) | AR | LOF | V790M |
| Fetal akinesia deformation sequence (208 150) | AR | LOF | ||||
| PDGFR | KIT | 164 920 | Piebaldism (172 800) | AD | LOF | W557, F584L, G664R, R796G |
| PDGFRß | 173 410 | Infantile myofibromatosis (228 550) | AD | ? | ||
| RET | RET | 164 761 | Hirschsprung disease (142 623) | AD | LOF | R77C, V145G, V202M, R231H, T278N, R330Q, R330W, R360W, A373V, E480K, R844W, G894S, R912Q, E921K, M980T |
| Multiple endocrine neoplasia 2B (162 300) | AD | GOF | M918T | |||
| ROR | ROR2 | 602 337 | Brachydactyly type B1 (113 000) | AD | GOF | |
| Robinow syndrome (268 310) | AR | LOF | ||||
| TIE | TEK | 600 221 | Multiple cutaneous and mucosal venous malformations (600 195) | AD/somatic | GOF | R849W |
| TRK | NTRK1 | 191 315 | Insensitivity to pain, congenital, with anhidrosis (256 800) | AR | LOF | |
| VEGFR | FLT4 | 136 352 | Hereditary lymphedema type IA (153 100) | AD | LOF | G1024E, R1041Q, R1041W, R1114L, P1137L |
To evaluate the range of causative mutations in DRTKs, we conducted a review of coding pathogenic variants reported for each of the associated disorders (Table 1); of the more than 500 disease-associated mutations reported in the Human Gene Mutation Database (HGMD) (21) and ClinVar (22) over two-thirds are missense mutations. Notably, both loss-of-function and gain-of-function mutations are observed, the latter being more common. For some of the dominant disorders, the mutations cluster exclusively in a given functional domain. For example, all 38 reported mutations causing autosomal dominant hereditary lymphedema type 1A are localized within the two intracellular TKDs of FLT4 and have been shown to reduce receptor activation, suggesting a domain-dependent mechanism for this disorder (21–23). Extreme clustering is observed in achondroplasia, the most frequent form of skeletal dysplasia with short stature; the Gly380Arg mutation in FGFR3 is observed in ∼97% of patients (24). Clearly, for a subset of the DRTKs, the type and location of the mutation has a very specific impact on the phenotypic outcome.
Phenotypic heterogeneity in DRTKs
Receptor tyrosine kinases possess intricate mechanisms to direct quantitatively and qualitatively distinct cell-type specific responses in precise developmental windows; these can include involvement of an accessory molecule, as well as differences in receptor and ligand expression levels and splice isoforms (3,25,26). This perhaps explains how mutations in some RTK genes cause multiple developmental syndromes (Table 1). For example, gain-of-function mutations in the extracellular immunoglobulin domain of FGFR1 are associated with Pfeiffer syndrome (27) and osteoglophonic dysplasia (28), while loss-of-function mutations in both the extracellular domain and TKD of the protein cause hypogonadotrophic hypogonadism (29). Similarly, gain-of-function mutations in FGFR3 cause achondroplasia, severe achondroplasia with acanthosis nigricans, Crouzon syndrome with acanthosis nigricans, hypochondroplasia, Muenke craniosynostosis and thanatophoric dysplasia type I and II (Table 1). Interestingly, specific substitutions of FGFR3 at Lys650, have been reported to cause hypochondroplasia with Lys650Asn/Gln mutations, TDII with Lys650Glu and TD1 or severe achondroplasia with acanthosis nigricans with Lys650Met, demonstrating that the nature and severity of the disease can be influenced by the specific change in a single amino acid (30). Overall, the range of clinical disease resulting from mutations in each RTK is likely a balance of many factors such as the location and type of the mutation, the function of the given kinase isoform, and the mutation's impact on receptor integrity and kinase activity in the context of the individual's genetic background. These observations emphasize the complexity of phenotype–genotype associations in the RTKs and further investigation will be required to more fully understand these intricacies.
Genetic heterogeneity in DRTKs
Just like different mutations in the same RTKs can cause very different diseases, mutations in different RTKs can cause the same disease. This genetic heterogeneity is widely observed in the FGFR family. For example, heterozygous mutations in either FGFR2 or FGFR3 cause Crouzon syndrome, FGFR1 and FGFR2 cause Pfeiffer syndrome and FGFR2 and FGFR3 cause lacrimo-auriculo-dento-digital (LADD) syndrome (Table 1). Additionally, mutations in signaling components which dysregulate RTK pathways can contribute to genetic heterogeneity. For example, a subset of LADD syndrome is caused by heterozygous mutations in FGF10, a FGFR ligand that interacts with FGFR2 (31,32). LADD-associated mutations in any of FGFR2, FGFR3 or FGF10 result in reduced downstream signaling and this developmental disorder (31,33). Overall, given the extensive and complex regulatory circuits for RTK signaling, there are often many molecules within a given pathway that can result in a similar phenotype.
Somatic mutations in receptor tyrosine kinasopathies and developmental syndromes
Germline mutations underlie the majority of the DRTKs recognized to date (Table 1). However, there is an inherent bias for identification of germline mutations as they are readily detected in DNA extracted from blood, the DNA source used in most gene discovery studies. Somatic mutations are more challenging to identify and require high degree of clinical suspicion, access to appropriate patient tissue samples, and analysis by deep sequencing. Therefore, it is possible that a number of developmental disorders caused by somatic mutations in RTKs have yet to be identified. These disorders may present as a milder or atypical form of a known disease, or as a novel condition. Mutations in FGFR3 highlight this interesting paradigm; the Arg248Cys substitution typically results in TD1, though an individual with somatic mosaicism for that same substitution was reported with atypical features of achondroplasia at 2 years of age (34). Remarkably, this same FGFR3 Arg248Cys substitution was identified in epidermal nevi (OMIM 162 900) and was absent from adjacent normal skin in a small number of individuals (35). The contrast between the skeletal and epidermal phenotypes with this mutation is striking and likely reflects the timing and the point in embryonic lineage where the mutation arose. It is quite probable that as deep sequencing becomes more widely available novel developmental syndromes will be identified that are secondary to somatic mutations in RTKs.
Cancer predisposition and the DRTKs
Several RTKs have been recognized to contribute to both cancer and developmental syndromes (Table 1). This raises the question of whether patients with DRTKs, especially those with mutations also implicated in tumorigenesis, would be predisposed to certain types of cancer. Multiple endocrine neoplasia type IIB (MEN2B), caused by recurrent germline mutations in RET, is characterized by early aggressive medullary thyroid cancer, pheochromocytoma, mucosal neuromas and a Marfanoid body habitus with dysmorphic facies (36). MEN2B is, to our knowledge, the only example of a DRTK with an inherited predisposition to cancer. There is no clear evidence to support that patients with other DRTKs are at increased risk for cancer, though some cases have been reported. For example, two patients with Apert syndrome with germline Pro253Arg mutations in FGFR2 were reported with cancer, one with early-onset low-grade papillary carcinoma of the bladder (37), and the other with an ovarian dysgerminoma (38). The Pro253Arg mutation has been reported in endometrial carcinomas and has been demonstrated to be oncogenic (39). Notably, many of the DRTKs are characterized by a shortened lifespan, which would preclude cancer formation (e.g. TD1 is associated with mortality in the neonatal period). So while RTKs have a clear and emerging role in cancer pathology, it seems that with the exception of MEN2B, cancer is not prevalent in known DRTKs; the explanation for this apparent discordance is unknown but may reflect the timing and specific cellular environment of the mutation.
RTKs and Cancer
In contrast to the ordered proliferation and differentiation of development, cancer represents an accumulation of genetic and epigenetic changes resulting in a disregard for the constraints of differentiation, proliferation, programed cell death and localization. By the time cancers reach an advanced state, genomic instability often results in hundreds of mutations, which can be categorized as either ‘driver’ mutations, those conferring a selective growth advantage to cells and are instrumental in cancer initiation or progression, or ‘passenger’ mutations, which do not contribute to oncogenesis (40,41). A number of the driver mutations identified occur in genes involved in key developmental pathways, such as gastrulation, angiogenesis and patterning, and contribute to specific malignant phenotypes (41,42).
Protein kinases, including RTKs, are one of the most frequently mutated gene families implicated in cancer, which has prompted numerous studies on their role in cancer pathogenesis [reviewed in (1,4,43)]. There are four main mechanisms of RTK dysregulation in human cancers: genomic rearrangements, autocrine activation, overexpression and gain- or loss-of-function mutations (1,3). Unchecked RTK signaling can disrupt the balance between cell growth, cell-cycle progression and apoptosis and when coupled with factors such as timing, location, duration and strength of dysregulated RTK signaling, may sensitize cells to oncogenic transformation or trigger RTK-induced oncogenesis (3,41).
Driver mutations in RTK genes
High-throughput DNA sequencing of tumor tissues has begun to shed light on the complex genomic landscape of human cancers. Initiatives such as the Catalogue Of Somatic Mutations In Cancer (COSMIC) database archive genetic sequence with the ultimate goal of elucidating the molecular determinants of cancer (44). Discovery and functional characterization of driver mutations is changing the understanding of cancer formation and progression, and providing opportunities for targeted treatments (41,43). Distinguishing passenger from driver mutations is a central challenge in cancer genome analysis. Statistical approaches may be able to identify candidate cancer genes, but are not always able to predict the tumorigenic potential of individual mutations (43). For example, activating mutations in the RTK FLT3 have been recognized to cause a common class of acute myeloid leukemia (AML) (45). Subsequent high-throughput FLT3 sequencing in a cohort of AML patients followed by functional characterization for each identified variant confirmed constitutively active kinase activity in a subset of mutations (driver mutations) but revealed that many were likely passenger mutations (46). Computationally, these passenger mutations could not be distinguished from driver mutations, highlighting the need for functional validation studies and novel strategies to identify driver mutations (46).
Kinases, Kinasopathies and Cancer: Two Sides of the Same Coin
Mutations in RTKs contribute to the pathogenesis of both cancer and developmental syndromes (4,43). Notably, it has been recognized that some mutations which cause DRTKs are also drivers in somatic cancer (Table 1) (10,11). For example, the Gly380Arg mutation in FGFR3 that causes achondroplasia has been demonstrated to be a driver mutation in bladder cancer (10,47). Similarly, somatic mutations in FGFR2 were present in 12% of endometrial carcinomas (39), two of which (Ser252Trp, Pro253Arg) have been identified as driver mutations and are identical to germline mutations reported in Apert and Crouzon syndromes (10,39,48).
We set out to investigate the overlapping subset of mutations that cause DRTKs and have also been identified in tumor tissue in COSMIC. Interestingly, mutations shared between developmental syndromes and cancer were either seen in a very small or very large number of tumor samples. For example, of the FGFR3 gain-of-function mutations reported to cause TDI, six are in COSMIC and are found in 3105 samples; five of these mutations (Arg248Cys, Ser249Cys, Gly370Cys, Ser371Cys and Tyr373Cys) showed a high distribution in urinary tract neoplasia, while one mutation (Lys650Met) was found more frequently in skin cancer (44). FGFR variants that cause DRTKs have been shown to be oncogenic in several tumor types (10). For instance, Lys650Glu (TDII), a recognized driver mutation in FGFR3 that results in constitutively elevated kinase activity, has been identified in spermatocytic seminoma (49), bladder carcinoma (50), and multiple myeloma (51). Looking for overlap between known pathogenic mutations that cause developmental syndromes and are present in tumor tissue may provide information on whether that mutation has a significant impact on protein function, and thus has potential to contribute to cancer.
Conclusions
RTK signaling is tightly regulated from embryogenesis throughout life; disruptions in these pathways cause developmental disorders and malignancies. Analysis of the mutations that cause DRTKs reveals complex genotype–phenotype patterns and highlights the intricacies of RTK signaling during development. A subset of the mutations that are responsible for DRTKs also play a role in tumorigenesis. The identification of driver mutations amongst the plethora of somatic cancer variation is advantageous as it facilitates development and use of targeted therapeutics. Currently, there are several clinically available small molecule inhibitors and monoclonal antibodies against specific RTKs (52,53) that are dysregulated in human cancers. For example, a number of c-Met small molecule inhibitors have been developed, a few of which are currently in clinical trials with promising results (e.g. INC280 and foretinib) (54). In the future, it is anticipated that many small molecule inhibitors will be generated for an array of targets. As we move into the age of personalized medicine, information from an individual's germline or cancer genome, combined with an understanding of RTK dynamics and mutational effects, will facilitate improved development of targeted therapeutics for these patients.
Conflict of Interest statement. None declared.
Funding
The author's work is supported by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec and Children′s Hospital of Eastern Ontario Foundation. L.M.M. is supported by a scholarship from the Canadian Institutes of Health Research (CIHR) and Consortium National de Formation en Santé.
References
- 1.Blume-Jensen P., Hunter T. (2001) Oncogenic kinase signalling. Nature, 411, 355–365. [DOI] [PubMed] [Google Scholar]
- 2.Robinson D.R., Wu Y.M., Lin S.F. (2000) The protein tyrosine kinase family of the human genome. Oncogene, 19, 5548–5557. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M.A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell, 141, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahiry P., Torkamani A., Schork N.J., Hegele R.A. (2010) Kinase mutations in human disease: interpreting genotype-phenotype relationships. Nat. Rev. Genet., 11, 60–74. [DOI] [PubMed] [Google Scholar]
- 5.Heldin C.H. (1995) Dimerization of cell surface receptors in signal transduction. Cell, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger J. (2000) Cell signaling by receptor tyrosine kinases. Cell, 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 7.Casaletto J.B., McClatchey A.I. (2012) Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer, 12, 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P.P., Dikic I. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol., 5, 461–466. [DOI] [PubMed] [Google Scholar]
- 9.Adrain C., Freeman M. (2014) Regulation of receptor tyrosine kinase ligand processing. Cold Spring Harb. Perspect. Biol., 6, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greulich H., Pollock P.M. (2011) Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol. Med., 17, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jhiang S.M. (2000) The RET proto-oncogene in human cancers. Oncogene, 19, 5590–5597. [DOI] [PubMed] [Google Scholar]
- 12.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. (2015) OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res., 43, D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labrador J.P., Azcoitia V., Tuckermann J., Lin C., Olaso E., Manes S., Bruckner K., Goergen J.L., Lemke G., Yancopoulos G. et al. (2001) The collagen receptor DDR2 regulates proliferation and its elimination leads to dwarfism. EMBO Reports, 2, 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ornitz D.M., Marie P.J. (2002) FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev., 16, 1446–1465. [DOI] [PubMed] [Google Scholar]
- 15.Billiard J., Way D.S., Seestaller-Wehr L.M., Moran R.A., Mangine A., Bodine P.V. (2005) The orphan receptor tyrosine kinase Ror2 modulates canonical Wnt signaling in osteoblastic cells. Mol. Endocrinol., 19, 90–101. [DOI] [PubMed] [Google Scholar]
- 16.Maeda K., Kobayashi Y., Udagawa N., Uehara S., Ishihara A., Mizoguchi T., Kikuchi Y., Takada I., Kato S., Kani S. et al. (2012) Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat. Med., 18, 405–412. [DOI] [PubMed] [Google Scholar]
- 17.Narkis G., Ofir R., Manor E., Landau D., Elbedour K., Birk O.S. (2007) Lethal congenital contractural syndrome type 2 (LCCS2) is caused by a mutation in ERBB3 (Her3), a modulator of the phosphatidylinositol-3-kinase/Akt pathway. Am. J. Hum. Genet., 81, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hojlund K., Hansen T., Lajer M., Henriksen J.E., Levin K., Lindholm J., Pedersen O., Beck-Nielsen H. (2004) A novel syndrome of autosomal-dominant hyperinsulinemic hypoglycemia linked to a mutation in the human insulin receptor gene. Diabetes, 53, 1592–1598. [DOI] [PubMed] [Google Scholar]
- 19.Psiachou H., Mitton S., Alaghband-Zadeh J., Hone J., Taylor S.I., Sinclair L. (1993) Leprechaunism and homozygous nonsense mutation in the insulin receptor gene. Lancet, 342, 924. [DOI] [PubMed] [Google Scholar]
- 20.Kadowaki T., Kadowaki H., Rechler M.M., Serrano-Rios M., Roth J., Gorden P., Taylor S.I. (1990) Five mutant alleles of the insulin receptor gene in patients with genetic forms of insulin resistance. J. Clin. Invest., 86, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. (2014) The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet., 133, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res., 42, D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karkkainen M.J., Ferrell R.E., Lawrence E.C., Kimak M.A., Levinson K.L., McTigue M.A., Alitalo K., Finegold D.N. (2000) Missense mutations interfere with VEGFR-3 signalling in primary lymphoedema. Nat. Genet., 25, 153–159. [DOI] [PubMed] [Google Scholar]
- 24.Bellus G.A., Hefferon T.W., De Ortiz Luna R.I., Hecht J.T., Horton W.A., Machado M., Kaitila I., McIntosh L., Francomano C.A. (1995) Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet., 56, 367–373. [PMC free article] [PubMed] [Google Scholar]
- 25.Beenken A., Mohammadi M. (2009) The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov., 8, 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li A.W., Murphy P.R. (2000) Expression of alternatively spliced FGF-2 antisense RNA transcripts in the central nervous system: regulation of FGF-2 mRNA translation. Mol. Cell. Endocrinol., 162, 69–78. [DOI] [PubMed] [Google Scholar]
- 27.Schell U., Hehr A., Feldman G.J., Robin N.H., Zackai E.H., de Die-Smulders C., Viskochil D.H., Stewart J.M., Wolff G., Ohashi H. et al. (1995) Mutations in FGFR1 and FGFR2 cause familial and sporadic Pfeiffer syndrome. Hum. Mol. Genet., 4, 323–328. [DOI] [PubMed] [Google Scholar]
- 28.White K.E., Cabral J.M., Davis S.I., Fishburn T., Evans W.E., Ichikawa S., Fields J., Yu X., Shaw N.J., McLellan N.J. et al. (2005) Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am. J. Hum. Genet., 76, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitteloud N., Acierno J.S. Jr., Meysing A., Eliseenkova A.V., Ma J., Ibrahimi O.A., Metzger D.L., Hayes F.J., Dwyer A.A., Hughes V.A. et al. (2006) Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc. Natl. Acad. Sci. U. S. A., 103, 6281–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellus G.A., Spector E.B., Speiser P.W., Weaver C.A., Garber A.T., Bryke C.R., Israel J., Rosengren S.S., Webster M.K., Donoghue D.J. et al. (2000) Distinct missense mutations of the FGFR3 lys650 codon modulate receptor kinase activation and the severity of the skeletal dysplasia phenotype. Am. J. Hum. Genet., 67, 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shams I., Rohmann E., Eswarakumar V.P., Lew E.D., Yuzawa S., Wollnik B., Schlessinger J., Lax I. (2007) Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the fibroblast growth factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol. Cell. Biol., 27, 6903–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohmann E., Brunner H.G., Kayserili H., Uyguner O., Nurnberg G., Lew E.D., Dobbie A., Eswarakumar V.P., Uzumcu A., Ulubil-Emeroglu M. et al. (2006) Mutations in different components of FGF signaling in LADD syndrome. Nat. Genet., 38, 414–417. [DOI] [PubMed] [Google Scholar]
- 33.Lew E.D., Bae J.H., Rohmann E., Wollnik B., Schlessinger J. (2007) Structural basis for reduced FGFR2 activity in LADD syndrome: Implications for FGFR autoinhibition and activation. Proc. Natl. Acad. Sci. U. S. A., 104, 19802–19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takagi M., Kaneko-Schmitt S., Suzumori N., Nishimura G., Hasegawa T. (2012) Atypical achondroplasia due to somatic mosaicism for the common thanatophoric dysplasia mutation R248C. Am. J. Med. Genet. A, 158A, 247–250. [DOI] [PubMed] [Google Scholar]
- 35.Hafner C., van Oers J.M., Vogt T., Landthaler M., Stoehr R., Blaszyk H., Hofstaedter F., Zwarthoff E.C., Hartmann A. (2006) Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J. Clin. Invest., 116, 2201–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison P.J., Nevin N.C. (1996) Multiple endocrine neoplasia type 2B (mucosal neuroma syndrome, Wagenmann-Froboese syndrome). J. Med. Genet., 33, 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreou A., Lamy A., Layet V., Cailliez D., Gobet F., Pfister C., Menard M., Frebourg T. (2006) Early-onset low-grade papillary carcinoma of the bladder associated with Apert syndrome and a germline FGFR2 mutation (Pro253Arg). Am. J. Med. Genet. A, 140, 2245–2247. [DOI] [PubMed] [Google Scholar]
- 38.Rouzier C., Soler C., Hofman P., Brennetot C., Bieth E., Pedeutour F. (2008) Ovarian dysgerminoma and Apert syndrome. Pediatr. Blood Cancer, 50, 696–698. [DOI] [PubMed] [Google Scholar]
- 39.Dutt A., Salvesen H.B., Chen T.H., Ramos A.H., Onofrio R.C., Hatton C., Nicoletti R., Winckler W., Grewal R., Hanna M. et al. (2008) Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc. Natl. Acad. Sci. U. S. A., 105, 8713–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Negrini S., Gorgoulis V.G., Halazonetis T.D. (2010) Genomic instability—--an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol., 11, 220–228. [DOI] [PubMed] [Google Scholar]
- 41.Pon J.R., Marra M.A. (2015) Driver and passenger mutations in cancer. Annu. Rev. Pathol., 10, 25–50. [DOI] [PubMed] [Google Scholar]
- 42.Bellacosa A. (2013) Developmental disease and cancer: biological and clinical overlaps. Am. J. Med. Genet. A, 161A, 2788–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torkamani A., Verkhivker G., Schork N.J. (2009) Cancer driver mutations in protein kinase genes. Cancer Lett., 281, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forbes S.A., Tang G., Bindal N., Bamford S., Dawson E., Cole C., Kok C.Y., Jia M., Ewing R., Menzies A. et al. (2010) COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res., 38, D652–D657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reindl C., Bagrintseva K., Vempati S., Schnittger S., Ellwart J.W., Wenig K., Hopfner K.P., Hiddemann W., Spiekermann K. (2006) Point mutations in the juxtamembrane domain of FLT3 define a new class of activating mutations in AML. Blood, 107, 3700–3707. [DOI] [PubMed] [Google Scholar]
- 46.Frohling S., Scholl C., Levine R.L., Loriaux M., Boggon T.J., Bernard O.A., Berger R., Dohner H., Dohner K., Ebert B.L. et al. (2007) Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell, 12, 501–513. [DOI] [PubMed] [Google Scholar]
- 47.van Rhijn B.W., van Tilborg A.A., Lurkin I., Bonaventure J., de Vries A., Thiery J.P., van der Kwast T.H., Zwarthoff E.C., Radvanyi F. (2002) Novel fibroblast growth factor receptor 3 (FGFR3) mutations in bladder cancer previously identified in non-lethal skeletal disorders. Eur. J. Hum. Genet., 10, 819–824. [DOI] [PubMed] [Google Scholar]
- 48.Pollock P.M., Gartside M.G., Dejeza L.C., Powell M.A., Mallon M.A., Davies H., Mohammadi M., Futreal P.A., Stratton M.R., Trent J.M. et al. (2007) Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene, 26, 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goriely A., Hansen R.M., Taylor I.B., Olesen I.A., Jacobsen G.K., McGowan S.J., Pfeifer S.P., McVean G.A., Rajpert-De Meyts E., Wilkie A.O. (2009) Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat. Genet., 41, 1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cappellen D., De Oliveira C., Ricol D., de Medina S., Bourdin J., Sastre-Garau X., Chopin D., Thiery J.P., Radvanyi F. (1999) Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet., 23, 18–20. [DOI] [PubMed] [Google Scholar]
- 51.Chesi M., Brents L.A., Ely S.A., Bais C., Robbiani D.F., Mesri E.A., Kuehl W.M., Bergsagel P.L. (2001) Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood, 97, 729–736. [DOI] [PubMed] [Google Scholar]
- 52.Gupta S., El-Rayes B.F. (2008) Small molecule tyrosine kinase inhibitors in pancreatic cancer. Biologics, 2, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esteva F.J. (2004) Monoclonal antibodies, small molecules, and vaccines in the treatment of breast cancer. Oncologist, 9(Suppl 3), 4–9. [DOI] [PubMed] [Google Scholar]
- 54.Qi X.S., Guo X.Z., Han G.H., Li H.Y., Chen J. (2015) MET inhibitors for treatment of advanced hepatocellular carcinoma: a review. World J. Gastroenterol., 21, 5445–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]