Abstract
Genetic variants, including single-nucleotide variants (SNVs) and copy number variants (CNVs), in the non-coding regions of the human genome can play an important role in human traits and complex diseases. Most of the genome-wide association study (GWAS) signals map to non-coding regions and potentially point to non-coding variants, whereas their functional interpretation is challenging. In this review, we discuss the human non-coding variants and their contributions to human diseases in the following four parts. (i) Functional annotations of non-coding SNPs mapped by GWAS: we discuss recent progress revealing some of the molecular mechanisms for GWAS signals affecting gene function. (ii) Technical progress in interpretation of non-coding variants: we briefly describe some of the technologies for functional annotations of non-coding variants, including the methods for genome-wide mapping of chromatin interaction, computational tools for functional predictions and the new genome editing technologies useful for dissecting potential functional consequences of non-coding variants. (iii) Non-coding CNVs in human diseases: we review our emerging understanding the role of non-coding CNVs in human disease. (iv) Compound inheritance of large genomic deletions and non-coding variants: compound inheritance at a locus consisting of coding variants plus non-coding ones is described.
Introduction
Targeted locus resequencing and whole exome sequencing (WES) in the coding regions of the human genome have revealed a plethora of causative mutations or variants associated with human disease. The possible pathogenic roles of various non-synonymous variants, including missense, nonsense, frameshift and other types of mutations, can also be predicted by multiple computational tools based on the conserved sequence of protein-coding genes and amino acid changes predicted by the genetic code. Furthermore, the Encyclopedia of DNA Elements (ENCODE) project has provided large data sets of experimental evidence regarding the function of non-coding regulatory elements and gained new insights into the nature of transcription, chromatin structure and histone modification in the human genome (1). However, the ENCODE is not the Rosetta stone equivalent to the genetic code (1,2). Instead, the ENCODE will enable us to further investigate the major but non-coding portions in the human genome and glean insights into how these non-coding variants potentially affect biological functions.
Accumulated evidence suggests that genetic variants in the non-coding but functional elements can be a cause for missing heritability. Notably, most of the single-nucleotide variants/polymorphisms (SNVs/SNPs) suggested by genome-wide association studies (GWAS) that provide statistical evidence for increased risk of complex diseases have been mapped to non-coding regions. The risk variants revealed by GWAS may be linked with a neighboring causative coding variant, but an alternative and more likely explanation is that functionally relevant genetic variants reside in non-coding regions with functional consequences on nearby genes. Accumulating evidence has shown that the GWAS variants are significantly enriched in the functional non-coding regions such as enhancer elements, DNase hypersensitivity regions and chromatin marks (3–5). In addition, large-scale structural variants (SVs), including genomic copy number variants (CNVs) and copy number neutral SVs such as inversions, often encompass more than one gene. In previous studies on the functions of CNVs, much effort has been expended on studying dosage-sensitive gene(s) in the CNV regions. However, the important role of non-coding regions (including intergenic sequence, non-coding elements within a protein-coding gene and non-coding RNAs such as microRNAs and lncRNAs—long non-coding RNAs) in biological functions and human diseases demand more attention.
In this review, we discuss non-coding variants, the potential effects of such variants on gene function and experimental approaches to assay functional effects of non-coding variants. We briefly describe (i) functional non-coding SNVs revealed by GWAS, (ii) recent advances in functional assays and annotation of non-coding variants, (iii) pathogenic CNVs in non-coding regions and (iv) non-coding CNVs/SNVs in compound inheritance. Illustrative examples are provided from studies of the molecular mechanisms of disease. These will provide further insight into the genetic mechanisms underlying both Mendelian and common complex diseases and other traits.
Functional Annotations of Non-coding SNPs Mapped by GWAS
In the post-GWAS era, an obstacle to understanding genetic mechanisms for common/complex disease and indeed a challenging task has been to uncover the perturbed biological functions implicated in the GWAS signals. A recent review has suggested the following approaches for this purpose: fine mapping in the GWAS loci to identify the functional variants responsible for traits and diseases, functional annotations using computational tools and in vivo/in vitro experimental assays in cells and model organisms (6). More details have been provided in the study by Edwards et al. (6). Here we review some new progress in annotating non-coding SNPs mapping to the GWAS identified loci.
Non-coding variants in enhancers are one of the major candidates for functional interpretation of GWAS loci. For example, the NOS1AP gene on human chromosome 1q has been long known to be associated with variability of QT interval and cardiac repolarization (7), whereas the underlying mechanism was unclear. A recent study utilized high-coverage resequencing and regional association for fine mapping in the GWAS locus for QT interval variation, which identified 210 common risk variants (8). Notably, all of them are non-coding. Further enhancer/suppressor analysis of 12 selected variants located in cardiac phenotype associated DNaseI hypersensitivity sites assisted in the identification of an upstream enhancer variant (rs7539120) associated with QT interval (8). This variant can affect cardiac function by increasing NOS1AP transcript expression in cardiomyocyte-intercalated discs and increase risk of cardiac arrhythmias (8). Similar evidence for functional enhancer SNPs has also been observed at many other loci, including the intronic enhancer SNPs at the MEIS1 gene associated with restless legs syndrome (9) and at the BCL11A gene associated with fetal hemoglobin levels (10), the intergenic enhancer SNP upstream to the MYB gene that is a critical regulator of erythroid development and fetal hemoglobin levels (11), and the recessive mutations in a distal enhancer located 25 kb downstream of PTF1A that is associated with isolated pancreatic agenesis (12). Furthermore, systematic investigations on the functions of non-coding variants in enhancer elements can be conducted using massively parallel reporter assays (MPRA), the technologies derived from high-throughput sequencing, in cell models or even in vivo in model organisms (13,14).
Notably, when investigating the functions of hypothesized enhancer variants, their targets cannot be limited to protein-coding genes. Instead, non-coding genes such as microRNAs (miRNAs) (15) and lncRNAs (16) must be considered. A recent study on the schizophrenia-associated locus at 1p21.3 identified a rare enhancer SNP (chr1:98515539A>T, hg19) with increased risk (15). The chromatin conformation capture assay showed that this risk allele has no obvious influence on the neighboring genes such as DPYD, but can reduce the expression of non-coding genes MIR137/MIR2682 (15).
In some instances, such functional variants are located in either the 5′ or 3′ untranslated region (UTR) of the disease-associated genes. Utilizing the methods of fine mapping, conditional analyses and exome array genotyping in >8000 individuals from the ‘Metabolic Syndrome in Men’ cohort (17), a recent study identified the association of rs11603334 (a SNP located in the 5′ UTR of ARAP1) with fasting proinsulin and type 2 diabetes (18). The allele-specific expression assay in human pancreatic islet samples showed that the risk allele of rs11603334 can upregulate gene expression of ARAP1 by 2-fold, which is also supported by the observation of decreased binding of pancreatic beta cell transcriptional regulators PAX6 and PAX4 to the rs11603334 risk allele and its corresponding increased promoter activity (18).
In the case of hypertriglyceridemia-associated APOA5, the 3′ UTR SNP rs2266788 was predicted to create a potential miRNA binding site for liver-expressed miR-485-5p (19). Luciferase reporter assays in both HEK293T cells with a miR485-5p precursor and in HuH-7 cells with endogenously expressed miR-485-5p suggested that the mutant allele of rs2266788 is involved in the miR-485-5p-mediated downregulation of APOA5 (19).
Despite the above progress, the identification and functional annotations of causal variants in the non-coding regions remain a challenge. This can result from the limitation of fine mapping methods, the existence of functional elements beyond ENCODE and our current state of knowledge, and the failure of in vitro assays in mimicking in vivo conditions.
Technical Progress in Interpretation of Non-coding Variants
Genome interaction
Functional non-coding variants can affect cis- or trans-regulatory elements, suggesting potential genome interaction that can be investigated by various genome technologies. Chromosome Conformation Capture (3C) enables the study of chromatin looping and genome architecture in three dimensions; 3C has been used for functional annotation of the schizophrenia-associated loci suggested by a genome-wide association study (20,21). The schizophrenia-associated SNPs identified by GWAS are located in the predicted enhancers downstream of CACNA1C. They map to where the 3C assay in human dorsolateral prefrontal cortex and induced pluripotent stem cells (iPSCs)-derived neurons has revealed promoter–enhancer interaction through chromosome loops (21).
Further adaptations were made based on the original 3C technology to derive 4C, 5C and HiC. Recently, a new technology named Chromatin Interaction Analysis by Paired-End Tag sequencing (ChIA-PET), which allows the detection of long-range interactions at regions bound by a target protein of interest, has been employed in generating a genome-wide interaction map of regulatory elements in human cells (22).
The 4C-seq (Circularized Chromosome Conformation Capture combined with high-throughput sequencing) method can achieve the resolution of ∼1 kb, which is useful to map enhancer–promoter interactions. The 4C-seq method has also been used to investigate the chromosome-wide effect of the CNVs associated with Williams–Beuren Syndrome (23). Chromatin looping and long-range interactions have been observed between the Williams–Beuren Syndrome critical region and its flanking genes.
However, the 4C-seq technology can only investigate a single region of interest at a time (24). Accordingly, a high-throughput method called Capture-C, which combines oligonucleotide capture technology (OCT), 3C and high-throughput sequencing, has been developed to analyze interactions between cis-regulatory elements at hundreds of selected loci at high resolution in a single assay (25).
Recently, the in situ Hi-C technology, in which DNA–DNA proximity ligation is performed in intact nuclei, has been developed to comprehensively map genome-wide chromatin interactions (26). The new three-dimensional map of the human genome at the resolution of 1 kb can clearly reveal domain structure, compartmentalization and thousands of chromatin loops that frequently link promoters and enhancers (26).
The above genome technologies to discern chromosome interactions will expedite functional annotations of the risk SNPs and CNVs associated with complex diseases.
Regulatory quantitative trait loci
One of the major roles of functional non-coding variants is to regulate gene expression. Although it is still challenging to identify the causal variants and uncover their molecular mechanisms in gene regulation, quantitative trait loci (QTL)-mapping approaches in cell lines or tissue samples can be adopted as a first step toward functional interpretation of non-coding variants (27). Various regulatory QTLs, including the classic expression QTL (eQTL), have been comprehensively discussed in a recent review (27).
In addition to eQTL, alternative splicing and its associated splicing QTL (sQTL) are another important aspect for functional annotation of non-coding variants (28). Using whole blood samples from the Framingham Heart Study, a recent genome-wide study identified >572k cis-sQTLs in 2650 unique genes (28). In addition, these sQTLs were significantly enriched for the risk SNPs revealed in previous GWAS summarized and annotated by the National Human Genome Research Institute (http://www.genome.gov/gwastudies/). Furthermore, 4.5% of the GWAS SNPs have been identified as cis-sQTLs but not cis-eQTLs, which indicates that sQTLs could be independent factors from QTL in interpreting the functional mechanism underlying GWAS loci (28). Notably, these sQTLs only explain a small portion of the GWAS signals. Therefore, further experimental investigation using additional cell and tissue types may identify more sQTLs for functional interpretation.
Recent studies in the angle of DNA replication also reveal the roles of non-coding variants in controlling DNA replication timing (29) and suggest their indirect connections with gene expression regulation (30). Based on the sequencing data of the 1000 Genomes Project and newly developed computational methods, researchers have observed inter-individual variations of replication timing in hundreds of loci throughout the human genome, at 16 of which the inherited genetic variants are associated with the replication timing variation (30). These replication timing QTLs (rtQTL) can be affected in a cis manner by local genetic variants and also be involved in the long-range regulation of gene expression (30).
Computational functional predictions for non-coding variants
Non-synonymous variants and their effects on protein functions can be predicted by computational methods based on quantifying constraint on the affected residue from conserved protein sequences. However, such an approach cannot be applied to non-coding variants. Therefore, alternative types of computational methods utilizing various genomic and epigenomic annotations have been developed, such as the tool named Genome-Wide Annotation of VAriants (GWAVA) that can support prioritization of non-coding variants (31). The regulatory mutations from the Human Gene Mutation database (HGMD) (32) were used as the disease variant set for training data, and common SNPs of the 1000 Genomes Project (33) were selected as the control variant set for GWAVA to identify annotations useful for discriminating functional non-coding variants from benign variants (31).
The Combined Annotation-Dependent Depletion (CADD) is another approach to prioritize deleterious and pathogenic variants by the ‘C score’ derived from integrating multiple annotations including conservation metrics, regulatory information, transcript information and protein-level scores (34). The ability of CADD to measure deleteriousness of human genetic variants has been achieved by training a support vector machine to contrast the annotations of fixed derived alleles in the human genomes (i.e. less deleterious) with those of simulated data sets that have more deleterious variants. Notably, CADD can also be applied to short insertions and deletions (indels) in the non-coding regions.
Genome editing and stem cell models
The above computational methods could provide functional predictions for non-coding variants; however, further functional (especially in vivo) assays are important experimental steps to investigate the molecular mechanism of non-coding variants in human disease. A classic approach to study functional consequences of human disease-associated SNV has been to engineer variation into mouse models. An analogous approach applicable to large CNVs associated with genomic disorders, such as the Smith–Magenis deletion syndrome and Potocki–Lupski duplication syndrome (35), has been of proven utility for elucidating functional evidence for the role of dosage-sensitive genes and mirror traits (35–38).
In the past few years, the renovation and development of the CRISPR/Cas technology (39) enables the quick and accurate genome editing in various model organism for both SNPs and small indels. Recently, the CRISPR/Cas methods have also been successfully utilized for engineering large CNVs and inversions at the megabase level in mice (40).
However, evolutionary conservation is still the prerequisite for functional assays of non-coding variants using genome editing technologies in model organisms. When the non-coding variants of interest are not conserved in mouse, it has also been feasible to directly obtain the human genetic variant of interest using the stem cell technologies, such as patient-derived iPSCs, for further in vitro functional assays (41,42).
Non-coding CNVs in Human Diseases
CNVs, whose functional effects were elucidated by studies of genomic disorders, play an important role in human Mendelian traits and complex diseases (43–46). These large pathogenic CNVs frequently affect multiple genes or truncate a specific functional gene, which can consequently lead to clinical conditions via the molecular mechanisms of gene dosage effect (haploinsufficiency and triplosensitivity), gene interruption, gene fusion and other effects on gene function including non-coding variant effects (47). Evidence documented during the past few years supports that CNVs in the non-coding regions can also be pathogenic by a position effect mechanism (47).
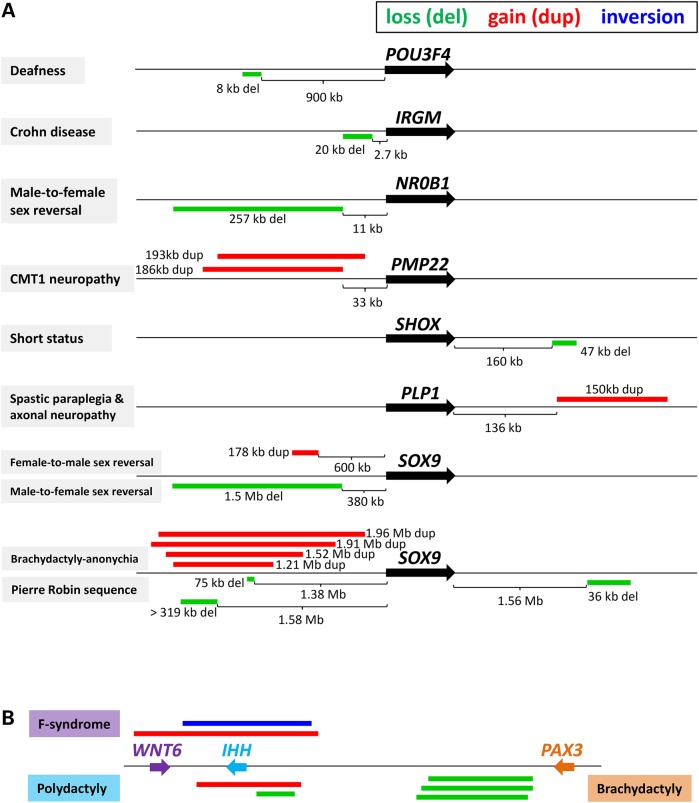
Functional CNVs (both deletions and duplications) have been frequently identified in upstream non-coding regions of disease-associated genes (Fig. 1), including POU3F4 (48), IRGM (49), NR0B1 (50) and PMP22 (51,52). In some instances, the CNVs in the non-coding regions downstream to a candidate gene can also be pathogenic by perturbing gene regulation; for example, the 150 kb duplications downstream to the PLP1 gene is associated with spastic paraplegia type 2 with axonal peripheral neuropathy (54). In addition, a recurrent 47 kb deletion in the downstream region of SHOX, involving a previously uncharacterized SHOX enhancer, has been identified in 15% of patients with Leri–Weill dyschondrosteosis (a skeletal dysplasia characterized by short stature) and 2% of subjects with idiopathic short stature in the Spanish population (53).
Figure 1.
(A) Functional CNVs in the gene-flanking regions are associated with human diseases (not to scale). References: POU3F4 (48), IRGM (49), NR0B1 (50), PMP22 (51,52), SHOX (53), PLP1 (54) and SOX9 (55–58). (B) Various CNVs/SVs in the genomic region involving WNT6, IHH and PAX3 genes can cause different phenotypes of limb malformations (59). Deletion (del) is shown in green bar, duplication (dup) in red and inversion in blue.
Intriguingly, deletions and duplications in either upstream or downstream regions of a specific gene can result in different clinical phenotypes as shown at the SOX9 locus (Fig. 1). A 1.5 Mb de novo deletion, 380 kb upstream to SOX9, has been identified in a 46,XY patient with male-to-female sex reversal (55). In contrast, upstream duplications of SOX9 with variable sizes have been observed to cause female-to-male sex reversal (56). Notably, three deletions in the long-range (>1 Mb in linear distance) flanking regions of SOX9 (two upstream and one downstream to SOX9) are associated with Pierre Robin sequence, a developmental defect sometimes associated with cleft palate (57). This suggests the existence of very long-range cis-regulatory elements in the flanking regions of SOX9 (57). Interestingly, genomic duplications of such cis-regulatory elements of SOX9 can also lead to brachydactyly-anonychia in Cooks syndrome (58).
The variable phenotypes manifested by overlapping CNVs upstream to the SOX9 gene may reflect the different cis-regulatory elements located in the non-overlapping regions (60). Alternatively, the inconsistent phenotypes may also suggest potential position effects and misexpression of these genes outside of the CNV regions. For example, the employment of chromosome conformation capture (4C-seq) technology in the Williams–Beuren Syndrome critical region (WBSCR) showed that the WBSCR deletion can cause chromatin conformation changes outside of the deletion region and disrupt the expression of the genes flanking WBSCR (23). Similarly, RNA sequencing of lymphoblast lines from carriers of 16p11.2 CNVs also showed altered expression of the genes outside the CNV region (61). Recently, a more complex example has been shown in the locus spanning WNT6/IHH/EPHA4/PAX3 genes, in which deletion, duplication and inversion SVs can misregulate their flanking genes and result in variable phenotypes of limb malformations (59). A pathogenicity model of SV-rearranged structure of neighboring topologically associated domains (TADs, the regulatory units within which enhancers and promoters can interact) and their associated enhancer and boundary elements was proposed (59).
Besides the functional CNVs involving the non-coding regulatory elements of protein-coding genes, pathogenic CNVs can also affect non-coding genes, such as lncRNAs (62). It was observed that 16q24.1 deletions can cause a lethal lung developmental disorder, alveolar capillary dysplasia with misalignment of pulmonary veins. The smallest overlapping region of these 16q24.1 deletions is 75 kb, involving lung-specific lncRNA genes that were suggested to contribute to long-range regulation of FOXF1, a protein-coding gene in which point mutations can result in the same lung disease (62).
Compound Inheritance of Large Genomic Deletions and Non-coding Variants
Utilizing genome-wide CNV detection technologies, genetic researchers have identified many large genomic CNVs as the causative genetic factors of human diseases (63). Accordingly, more and more pathogenic loci have been incorporated into molecular diagnostic panels (64,65). However, in some rare instances, these pathogenic CNVs can also be found in human populations at a very low frequency or transmitted from unaffected parents. These variable phenotypes suggested the possible existence of some modifying genetic factors in addition to the large genomic CNVs. Recent progress has shown that the SNPs and CNVs in non-coding regions can be the genetic modifiers.
Frequent 1q21.1 deletion in the human genome has been observed in the rare thrombocytopenia with absent radii (TAR) syndrome (66). This deletion in the TAR patients can be inherited from unaffected parents, suggesting an autosomal recessive inheritance (66). Intriguingly, low-frequency regulatory SNPs of RBM8A, the TAR syndrome-associated gene in 1q21.1, were identified as the second recessive allele to downregulate the expression of RBM8A (67). Such an autosomal recessive combination of one regulatory SNP of RBM8A and a 1q21.1 deletion CNV can explain the majority (51/53) of the affected patients with TAR syndrome (67).
Non-coding CNVs can also be the second allele at a locus consistent with a recessive model. In a recent study on Burn–McKeown syndrome, large 18q23 deletions were identified as the first recessive alleles in 4 out of 11 patients (68). The smallest region of overlap shared by these deletions affect TXNL4A, the gene involved in Burn–McKeown syndrome (68). Interestingly, a low-frequency 34 bp deletion of the TXNL4A gene promoter regions was shared by all the above four cases with large 18q23 deletions. Dual luciferase assays showed that this promoter deletion can dramatically reduce TXNL4A expression (68).
In the above two loci, RBM8A and TXNL4A, the non-coding pathogenic variations are suggested to be recessive (67,68), consistent with compound inheritance of two autosomal recessive alleles. However, more complexity has been recently revealed in the TBX6 locus of human chromosome 16p11.2. Compound inheritance was demonstrated for a rare null allele and a common hypomorphic allele in association with a common/complex trait—scoliosis (69). This compound inheritance model accounted for 8–11% of congenital scoliosis in the Chinese population studied (69).
Deletion CNVs of 16p11.2 are associated with various complex disease phenotypes, including autism (70,71), obesity (72,73), Müllerian aplasia (74) and congenital scoliosis (69,75–77). This 16p11.2 deletion is recurrent and mediated by non-allelic homologous recombination occurring between two large flanking human genomic low-copy repeats; therefore, the deletion sizes and gene contents are consistent between deletion carriers. How did the same genomic deletion cause different clinical conditions? In addition, the inter-individual phenotypic variability has also been observed between the deletion carriers in the same family (76). Therefore, the popular mechanism of haploinsufficiency cannot completely explain the clinical disorders associated with 16p11.2 deletion. Instead, based on the genetic model of autosomal recessive inheritance, some groups tried to find the additional functional mutations in the coding genes within the 16p11.2 deletion regions to explain the variable phenotypes of scoliosis between deletion carriers, but no non-synonymous mutations were identified in the candidate gene TBX6 in 16p11.2 (69,75–77). Intriguingly, a common haplotype of TBX6 has been identified in the deletion carriers with congenital scoliosis (69). In vitro functional assays suggest that two non-coding SNPs, which define the above scoliosis risk haplotype, can moderately downregulate the gene expression level of TBX6, representing a hypomorphic allele of TBX6 (69). These findings of a contributing CNV and a functional SNP, with a rare plus a common variant, fit more with a Clan Genomics model, in which the unique combinations of rare variants characteristic of a recent family lineage can have a causative role in disease, than a dichotomous rare Mendelian disease versus common/complex disease genetic model (78).
Compound inheritance for a bleeding diathesis was shown for Sotos syndrome patients due to recurrent deletion CNV involving both NSD1 and FXII in association with a non-coding functional SNP at the FXII locus; both genes are encompassed within the recurrent CNV associated with Sotos syndrome (79). Subjects bearing the functional SNP associated with lower expression were documented to have the lowest FXII levels (79). Thus, variability of expression in a deletion CNV can result from non-coding variants on the remaining allele. Common functional SNVs may be more likely to contribute to variability of clinical expression of microdeletion syndromes than rare variant alleles in coding sequences (80).
Conclusions
Human genetic mutations/variants in the coding regions have been long known as important genetic contributors to Mendelian and complex diseases. Accordingly, the genome technologies of WES (81–83) and genome-wide microarrays (64,65) have been widely used for gene discovery and for molecular diagnostics of pathogenic SNVs and CNVs for human diseases, especially congenital disorders. However, recent progress in genetic and genomic studies has identified functional genetic variants in non-coding regions of the human genome. Therefore, whole genome sequencing (WGS) could be informative and promising for the future clinical practice (84,85) not only because of its ability to characterize non-coding functional variants but also because it enables robust identification of SV, including CNV and copy number neutral SV such as inversions and translocations. This research trend also demands the technical and methodological innovations to assist in functional interpretation and data mining from millions of non-coding variants that can be found in a single human genome (84).
Funding
This work was supported in part by National Basic Research Program of China (2012CB944600 and 2011CBA00401 to F.Z.), National Natural Science Foundation of China (81222014 and 31171210 to F.Z.), Shu Guang Project (12SG08 to F.Z.), National Program for Support of Top-notch Young Professionals (to F.Z.), US National Institute of Neurological Disorders and Stroke (R01NS058529 to J.R.L.), the National Institute of General Medical Sciences (R01GM106373 to J.R.L.), the National Institute of Child Health and Development (U54HD083092) Intellectual and Developmental Disabilities Research Center, and the National Human Genome Research Institute/National Heart Blood Lung Institute jointly funded Baylor Hopkins Center for Mendelian Genomics (U54HG006542 to J.R.L.).
Acknowledgements
We thank Professors Eric Boerwinkle and Bo Wen for their critical comments.
Conflict of Interest statement. J.R.L. holds stock ownership in 23andMe, Inc., and Lasergen, Inc., is a paid consultant for Regeneron Pharmaceuticals and is a co-inventor on multiple United States and European patents related to molecular diagnostics. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Baylor Miraca Genetics Laboratories.
References
- 1.The Encode Project Consortium. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graur D., Zheng Y., Price N., Azevedo R.B., Zufall R.A., Elhaik E. (2013) On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol., 5, 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahonen T., Saltevo J., Laakso M., Kautiainen H., Kumpusalo E., Vanhala M. (2009) Gender differences relating to metabolic syndrome and proinflammation in Finnish subjects with elevated blood pressure. Mediators Inflamm., 2009, 959281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degner J.F., Pai A.A., Pique-Regi R., Veyrieras J.B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., Crawford G.E. et al. (2012) DNase I sensitivity QTLs are a major determinant of human expression variation. Nature, 482, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. (2013) Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet., 45, 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards S.L., Beesley J., French J.D., Dunning A.M. (2013) Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet., 93, 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arking D.E., Pfeufer A., Post W., Kao W.H., Newton-Cheh C., Ikeda M., West K., Kashuk C., Akyol M., Perz S. et al. (2006) A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat. Genet., 38, 644–651. [DOI] [PubMed] [Google Scholar]
- 8.Kapoor A., Sekar R.B., Hansen N.F., Fox-Talbot K., Morley M., Pihur V., Chatterjee S., Brandimarto J., Moravec C.S., Pulit S.L. et al. (2014) An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am. J. Hum. Genet., 94, 854–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spieler D., Kaffe M., Knauf F., Bessa J., Tena J.J., Giesert F., Schormair B., Tilch E., Lee H., Horsch M. et al. (2014) Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res., 24, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer D.E., Kamran S.C., Lessard S., Xu J., Fujiwara Y., Lin C., Shao Z., Canver M.C., Smith E.C., Pinello L. et al. (2013) An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science, 342, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadhouders R., Aktuna S., Thongjuea S., Aghajanirefah A., Pourfarzad F., van Ijcken W., Lenhard B., Rooks H., Best S., Menzel S. et al. (2014) HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest., 124, 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weedon M.N., Cebola I., Patch A.M., Flanagan S.E., De Franco E., Caswell R., Rodriguez-Segui S.A., Shaw-Smith C., Cho C.H., Lango Allen H. et al. (2014) Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet., 46, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., Lee C., Andrie J.M., Lee S.I., Cooper G.M. et al. (2012) Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol., 30, 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G. Jr, Kinney J.B. et al. (2012) Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol., 30, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan J., Shi J., Fiorentino A., Leites C., Chen X., Moy W., Chen J., Alexandrov B.S., Usheva A., He D. et al. (2014) A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. Am. J. Hum. Genet., 95, 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhartiya D., Jalali S., Ghosh S., Scaria V. (2014) Distinct patterns of genetic variations in potential functional elements in long noncoding RNAs. Hum. Mutat., 35, 192–201. [DOI] [PubMed] [Google Scholar]
- 17.Stancakova A., Javorsky M., Kuulasmaa T., Haffner S.M., Kuusisto J., Laakso M. (2009) Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes, 58, 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulzer J.R., Stitzel M.L., Morken M.A., Huyghe J.R., Fuchsberger C., Kuusisto J., Laakso M., Boehnke M., Collins F.S., Mohlke K.L. (2014) A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am. J. Hum. Genet., 94, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caussy C., Charriere S., Marcais C., Di Filippo M., Sassolas A., Delay M., Euthine V., Jalabert A., Lefai E., Rome S. et al. (2014) An APOA5 3′ UTR variant associated with plasma triglycerides triggers APOA5 downregulation by creating a functional miR-485-5p binding site. Am. J. Hum. Genet., 94, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ripke S., O'Dushlaine C., Chambert K., Moran J.L., Kahler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M. et al. (2013) Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet., 45, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roussos P., Mitchell A.C., Voloudakis G., Fullard J.F., Pothula V.M., Tsang J., Stahl E.A., Georgakopoulos A., Ruderfer D.M., Charney A. et al. (2014) A role for noncoding variation in schizophrenia. Cell Rep., 9, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidari N., Phanstiel D.H., He C., Grubert F., Jahanbani F., Kasowski M., Zhang M.Q., Snyder M.P. (2014) Genome-wide map of regulatory interactions in the human genome. Genome Res., 24, 1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gheldof N., Witwicki R.M., Migliavacca E., Leleu M., Didelot G., Harewood L., Rougemont J., Reymond A. (2013) Structural variation-associated expression changes are paralleled by chromatin architecture modifications. PLoS One, 8, e79973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Werken H.J., Landan G., Holwerda S.J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B.A. et al. (2012) Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods, 9, 969–972. [DOI] [PubMed] [Google Scholar]
- 25.Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R., Higgs D.R. (2014) Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet., 46, 205–212. [DOI] [PubMed] [Google Scholar]
- 26.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pai A.A., Pritchard J.K., Gilad Y. (2015) The genetic and mechanistic basis for variation in gene regulation. PLoS Genet., 11, e1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Joehanes R., Chen B.H., Huan T., Ying S., Munson P.J., Johnson A.D., Levy D., O'Donnell C.J. (2015) Identification of common genetic variants controlling transcript isoform variation in human whole blood. Nat. Genet., 47, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhind N., Gilbert D.M. (2013) DNA replication timing. Cold Spring Harb. Perspect. Biol., 5, a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koren A., Handsaker R.E., Kamitaki N., Karlic R., Ghosh S., Polak P., Eggan K., McCarroll S.A. (2014) Genetic variation in human DNA replication timing. Cell, 159, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie G.R., Dunham I., Zeggini E., Flicek P. (2014) Functional annotation of noncoding sequence variants. Nat. Methods, 11, 294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenson P.D., Mort M., Ball E.V., Howells K., Phillips A.D., Thomas N.S., Cooper D.N. (2009) The Human Gene Mutation Database: 2008 update. Genome Med., 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The 1000 Genomes Project Consortium Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricard G., Molina J., Chrast J., Gu W., Gheldof N., Pradervand S., Schutz F., Young J.I., Lupski J.R., Reymond A. et al. (2010) Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol., 8, e1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walz K., Paylor R., Yan J., Bi W., Lupski J.R. (2006) Rai1 duplication causes physical and behavioral phenotypes in a mouse model of dup(17)(p11.2p11.2). J. Clin. Invest., 116, 3035–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacaria M., Spencer C., Gu W., Paylor R., Lupski J.R. (2012) Enriched rearing improves behavioral responses of an animal model for CNV-based autistic-like traits. Hum. Mol. Genet., 21, 3083–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacaria M., Saha P., Potocki L., Bi W., Yan J., Girirajan S., Burns B., Elsea S., Walz K., Chan L. et al. (2012) A duplication CNV that conveys traits reciprocal to metabolic syndrome and protects against diet-induced obesity in mice and men. PLoS Genet., 8, e1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shalem O., Sanjana N.E., Zhang F. (2015) High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet., 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraft K., Geuer S., Will A.J., Chan W.L., Paliou C., Borschiwer M., Harabula I., Wittler L., Franke M., Ibrahim D.M. et al. (2015) Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Rep., 10, 833–839. [DOI] [PubMed] [Google Scholar]
- 41.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M. et al. (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron, 80, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sareen D., O'Rourke J.G., Meera P., Muhammad A.K., Grant S., Simpkinson M., Bell S., Carmona S., Ornelas L., Sahabian A. et al. (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med., 5, 208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupski J.R. (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet., 14, 417–422. [DOI] [PubMed] [Google Scholar]
- 44.Lee C., Morton C.C. (2008) Structural genomic variation and personalized medicine. N. Engl. J. Med., 358, 740–741. [DOI] [PubMed] [Google Scholar]
- 45.Lupski J.R. (2009) Genomic disorders ten years on. Genome Med., 1, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lupski J.R. (2015) Structural variation mutagenesis of the human genome: impact on disease and evolution. Environ. Mol. Mutagen., 56, doi:10.1002/em.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lupski J.R., Stankiewicz P. (2005) Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet., 1, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Kok Y.J., Vossenaar E.R., Cremers C.W., Dahl N., Laporte J., Hu L.J., Lacombe D., Fischel-Ghodsian N., Friedman R.A., Parnes L.S. et al. (1996) Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum. Mol. Genet., 5, 1229–1235. [DOI] [PubMed] [Google Scholar]
- 49.McCarroll S.A., Huett A., Kuballa P., Chilewski S.D., Landry A., Goyette P., Zody M.C., Hall J.L., Brant S.R., Cho J.H. et al. (2008) Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat. Genet., 40, 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyk M., Berg J.S., Pursley A., Curtis F.K., Fernandez B.A., Bien-Willner G.A., Lupski J.R., Cheung S.W., Stankiewicz P. (2007) Male-to-female sex reversal associated with an approximately 250 kb deletion upstream of NR0B1 (DAX1). Hum. Genet., 122, 63–70. [DOI] [PubMed] [Google Scholar]
- 51.Zhang F., Seeman P., Liu P., Weterman M.A., Gonzaga-Jauregui C., Towne C.F., Batish S.D., De Vriendt E., De Jonghe P., Rautenstrauss B. et al. (2010) Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am. J. Hum. Genet., 86, 892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weterman M.A., van Ruissen F., de Wissel M., Bordewijk L., Samijn J.P., van der Pol W.L., Meggouh F., Baas F. (2010) Copy number variation upstream of PMP22 in Charcot-Marie-Tooth disease. Eur. J. Hum. Genet., 18, 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benito-Sanz S., Royo J.L., Barroso E., Paumard-Hernandez B., Barreda-Bonis A.C., Liu P., Gracia R., Lupski J.R., Campos-Barros A., Gomez-Skarmeta J.L. et al. (2012) Identification of the first recurrent PAR1 deletion in Leri-Weill dyschondrosteosis and idiopathic short stature reveals the presence of a novel SHOX enhancer. J. Med. Genet., 49, 442–450. [DOI] [PubMed] [Google Scholar]
- 54.Lee J.A., Madrid R.E., Sperle K., Ritterson C.M., Hobson G.M., Garbern J., Lupski J.R., Inoue K. (2006) Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann. Neurol., 59, 398–403. [DOI] [PubMed] [Google Scholar]
- 55.Pop R., Conz C., Lindenberg K.S., Blesson S., Schmalenberger B., Briault S., Pfeifer D., Scherer G. (2004) Screening of the 1 Mb SOX9 5′ control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J. Med. Genet., 41, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox J.J., Willatt L., Homfray T., Woods C.G. (2011) A SOX9 duplication and familial 46,XX developmental testicular disorder. N. Engl. J. Med., 364, 91–93. [DOI] [PubMed] [Google Scholar]
- 57.Benko S., Fantes J.A., Amiel J., Kleinjan D.J., Thomas S., Ramsay J., Jamshidi N., Essafi A., Heaney S., Gordon C.T. et al. (2009) Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet., 41, 359–364. [DOI] [PubMed] [Google Scholar]
- 58.Kurth I., Klopocki E., Stricker S., van Oosterwijk J., Vanek S., Altmann J., Santos H.G., van Harssel J.J., de Ravel T., Wilkie A.O. et al. (2009) Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat. Genet., 41, 862–863. [DOI] [PubMed] [Google Scholar]
- 59.Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell, 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bien-Willner G.A., Stankiewicz P., Lupski J.R. (2007) SOX9cre1, a cis-acting regulatory element located 1.1 Mb upstream of SOX9, mediates its enhancement through the SHH pathway. Hum. Mol. Genet., 16, 1143–1156. [DOI] [PubMed] [Google Scholar]
- 61.Blumenthal I., Ragavendran A., Erdin S., Klei L., Sugathan A., Guide J.R., Manavalan P., Zhou J.Q., Wheeler V.C., Levin J.Z. et al. (2014) Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am. J. Hum. Genet., 94, 870–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szafranski P., Dharmadhikari A.V., Brosens E., Gurha P., Kolodziejska K.E., Zhishuo O., Dittwald P., Majewski T., Mohan K.N., Chen B. et al. (2013) Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res., 23, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F., Gu W., Hurles M.E., Lupski J.R. (2009) Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet., 10, 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reddy U.M., Page G.P., Saade G.R., Silver R.M., Thorsten V.R., Parker C.B., Pinar H., Willinger M., Stoll B.J., Heim-Hall J. et al. (2012) Karyotype versus microarray testing for genetic abnormalities after stillbirth. N. Engl. J. Med., 367, 2185–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wapner R.J., Martin C.L., Levy B., Ballif B.C., Eng C.M., Zachary J.M., Savage M., Platt L.D., Saltzman D., Grobman W.A. et al. (2012) Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med., 367, 2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klopocki E., Schulze H., Strauss G., Ott C.E., Hall J., Trotier F., Fleischhauer S., Greenhalgh L., Newbury-Ecob R.A., Neumann L.M. et al. (2007) Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am. J. Hum. Genet., 80, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albers C.A., Paul D.S., Schulze H., Freson K., Stephens J.C., Smethurst P.A., Jolley J.D., Cvejic A., Kostadima M., Bertone P. et al. (2012) Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet., 44, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wieczorek D., Newman W.G., Wieland T., Berulava T., Kaffe M., Falkenstein D., Beetz C., Graf E., Schwarzmayr T., Douzgou S. et al. (2014) Compound heterozygosity of low-frequency promoter deletions and rare loss-of-function mutations in TXNL4A causes Burn-McKeown syndrome. Am. J. Hum. Genet., 95, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. et al. (2015) TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med., 372, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T. et al. (2008) Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med., 358, 667–675. [DOI] [PubMed] [Google Scholar]
- 71.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H. Jr, Dobyns W.B. et al. (2008) Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet., 17, 628–638. [DOI] [PubMed] [Google Scholar]
- 72.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S. et al. (2010) A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature, 463, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. et al. (2011) Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature, 478, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandbacka M., Laivuori H., Freitas E., Halttunen M., Jokimaa V., Morin-Papunen L., Rosenberg C., Aittomaki K. (2013) TBX6, LHX1 and copy number variations in the complex genetics of Mullerian aplasia. Orphanet. J. Rare Dis., 8, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimojima K., Inoue T., Fujii Y., Ohno K., Yamamoto T. (2009) A familial 593-kb microdeletion of 16p11.2 associated with mental retardation and hemivertebrae. Eur. J. Med. Genet., 52, 433–435. [DOI] [PubMed] [Google Scholar]
- 76.Shen Y., Chen X., Wang L., Guo J., Shen J., An Y., Zhu H., Zhu Y., Xin R., Bao Y. et al. (2011) Intra-family phenotypic heterogeneity of 16p11.2 deletion carriers in a three-generation Chinese family. Am. J. Med. Genet. B Neuropsychiatr. Genet., 156, 225–232. [DOI] [PubMed] [Google Scholar]
- 77.Al-Kateb H., Khanna G., Filges I., Hauser N., Grange D.K., Shen J., Smyser C.D., Kulkarni S., Shinawi M. (2014) Scoliosis and vertebral anomalies: additional abnormal phenotypes associated with chromosome 16p11.2 rearrangement. Am. J. Med. Genet. A, 164A, 1118–1126. [DOI] [PubMed] [Google Scholar]
- 78.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. (2011) Clan genomics and the complex architecture of human disease. Cell, 147, 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurotaki N., Shen J.J., Touyama M., Kondoh T., Visser R., Ozaki T., Nishimoto J., Shiihara T., Uetake K., Makita Y. et al. (2005) Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet. Med., 7, 479–483. [DOI] [PubMed] [Google Scholar]
- 80.Shiow L.R., Paris K., Akana M.C., Cyster J.G., Sorensen R.U., Puck J.M. (2009) Severe combined immunodeficiency (SCID) and attention deficit hyperactivity disorder (ADHD) associated with a Coronin-1A mutation and a chromosome 16p11.2 deletion. Clin. Immunol., 131, 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. et al. (2013) Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med., 369, 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. et al. (2014) Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T. et al. (2015) The genetic basis of Mendelian phenotypes: discoveries, challenges, and ppportunities. Am. J. Hum. Genet., 97, doi:10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lupski J.R., Reid J.G., Gonzaga-Jauregui C., Rio Deiros D., Chen D.C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D.A. et al. (2010) Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med., 362, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor J.C., Martin H.C., Lise S., Broxholme J., Cazier J.B., Rimmer A., Kanapin A., Lunter G., Fiddy S., Allan C. et al. (2015) Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat. Genet., 47, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]