Abstract
Key message
A QTL for thrips resistance on pepper chromosome 6 was identified and validated. This QTL affects thrips larval development and explains 50 % of the variation.
Abstract
Thrips is one of the most damaging pests in pepper (Capsicum). Resistance to thrips was identified in Capsicum annuum. This study was aimed at the elucidation of the genetic background of thrips resistance in Capsicum through QTL mapping. The QTL analysis was carried out for Frankliniella occidentalis resistance in an F2 population consisting of 196 plants derived from an interspecific cross between the highly resistant C. annuum AC 1979 as female parent and the highly susceptible C. chinense 4661 as male parent. Fifty-seven SSR, 109 AFLP, and 5 SNP markers were used to construct a genetic map with a total length of 1636 cM. Damage caused by larvae and the survival of first and second instar larval stages observed in a no-choice test were used as parameters of resistance. Interval mapping detected one QTL for each of these parameters, all co-localizing near the same marker on chromosome 6. Use of this marker as co-factor in a multiple-QTL mapping analysis failed to uncover any additional QTLs. This QTL explained about 50 % of the genetic variation, and the resistance allele of this QTL was inherited from the resistant parent. Thrips resistance was not linked to trichome density.
Introduction
Pepper (Capsicum) production worldwide is constrained by thrips as one of the most damaging pests (Siemonsma and Piluek 1994). Adult thrips are about 1 mm long, and females are usually a bit larger than males. They belong to the insect order Thysanoptera. There are at least 16 species of thrips that attack Capsicum (Talekar 1991; Capinera 2001). Among these is Frankliniella occidentalis, which is the major species found on pepper in Europe (Tommasini and Maini 1995), and it has recently been found in Asia as well (Zhang et al. 2007). Thrips cause direct damage by feeding on pepper fruits, flowers, and leaves (Welter et al. 1990; Tommasini and Maini 1995; Shipp et al. 1998). Feeding of thrips on leaves may affect leaf size and carbon allocation in the plant (Welter et al. 1990; Shipp et al. 1998), reduce photosynthetic capacity (Tommasini and Maini 1995), and eventually reduce yield (Steiner 1990; Welter et al. 1990). Thrips feeding on pepper fruit causes bronzing and silvering of the fruit skin, thereby reducing market quality (Shipp et al. 1998). Thrips can also transmit several viruses, one of the most important being Tomato Spotted Wilt Virus (TSWV) (Ulman et al. 1992). This virus is acquired during the first and early second larval stage when there is a temporary association between mid-gut, visceral muscles, and salivary glands (Moritz et al. 2004). After that, the virus is transferred into a plant with the saliva of a feeding adult (Jones 2005).
Thrips management and control practices include chemical treatments, biological control, and integrated pest management. Unfortunately, they do not completely solve the problems caused by thrips (Reitz et al. 2003; Weintraub 2007). Thrips-resistant varieties would increase the effectiveness of thrips control. Resistance to thrips may also delay and reduce the transmission of viruses as was shown by Maris et al. (2003) for TSWV. Several pepper accessions have been found to carry resistance to thrips which may be exploited further to breed thrips-resistant varieties (Fery and Schalk 1991; Maris et al. 2003; Maharijaya et al. 2011, 2012).
Molecular marker linkage maps have been constructed for several Capsicum populations (Minamiyama et al. 2006; Yi et al. 2006; Barchi et al. 2007; Lee et al. 2009; Wu et al. 2009). These have been used to detect quantitative trait loci (QTLs) for plant development and fruit characteristics (Palloix et al. 2009; Borovsky and Paran 2011) and for resistance against pathogens, including anthracnose (Colletotrichum spp.) (Voorrips et al. 2004), Phytophthora capsici (Thabuis et al. 2004), and powdery mildew (Lefebvre et al. 2003). For resistance to thrips in pepper, a QTL has been identified by Syngenta Biotechnology Inc. on chromosome 5 (Linders et al. 2010). In other crops, QTLs for resistance to thrips were detected in cowpea (Muchero et al. 2010), potato (Galvez et al. 2005), common bean (Frei et al. 2005), and cabbage (Loptien 2013).
In earlier papers, we described the identification and characterisation of several sources of thrips resistance. We developed test methods to evaluate plant resistance to thrips and showed that these in vitro tests (detached leaf assays) correlate well with greenhouse tests based on damage scores (Maharijaya et al. 2011). The effect of resistance in pepper on thrips reproduction and development was studied using three highly resistant, three medium resistant, and three susceptible accessions. Resistance factors in leaves of resistant pepper accessions were shown to have significant effects, mostly on the larval stages. These factors completely blocked the development of L1 larvae to the L2 stage on the resistant accessions (Maharijaya et al. 2012).
Our current study was aimed at elucidating the genetic basis of the resistance to thrips that we identified earlier in Capsicum annuum AC 1979 (Maharijaya et al. 2011, 2012) through a QTL mapping approach. Since the resistant parent of our population was the same as the one used by Linders et al. (2010) we compared the mapping results. The presence of trichomes has been implicated in resistance against the thrips Scirtothrips dorsalis (Yadwad et al. 2008) in pepper, therefore we included this trait in our study as well.
Materials and methods
Plant material
A mapping population consisting of 196 F2 plants was developed from a cross between C. annuum AC 1979 as female parent and C. chinense 4661 as male parent. The parents were chosen based on evaluation results for resistance against two thrips species, F. occidentalis and Thrips parvispinus using several different resistance tests (Maharijaya et al. 2011, 2012). Capsicum annuum AC 1979 was highly resistant while C. chinense 4661 was very susceptible in these tests. Apart from the contrasting resistance levels the parents also differ in leaf characteristics (Wahyuni et al. 2011) including trichome density: C. annuum AC 1979 does not have trichomes while C. chinense 4661 is densely covered with trichomes. Both accessions were obtained from the Centre for Genetic Resources, the Netherlands. An interspecific cross was used to obtain a sufficient level of polymorphism in the F2 population.
The F2 population was grown together with two first-generation inbred lines, obtained by self-pollination of the two parental plants, and with cuttings of the F1 in a glasshouse at Wageningen University and Research Centre, the Netherlands. From 4 F2 plants that were heterozygous for marker Isotig18917-234 (close to the QTL maximum) F3 lines were obtained by selfing. A total of 41 F3 plants were used in this study.
The plants were maintained in a glasshouse at 25 °C, 16/8 h day/night without pesticide application. Pests were controlled biologically using predator organisms according to standard Dutch pepper cultivation practices. Seeds were sown in rockwool plugs in trays; seedlings were transplanted onto rockwool 5 weeks after germination. The F3 plants were grown under the same conditions but in a later year.
Thrips
A F. occidentalis population was collected from thrips-infested Arabidopsis thaliana plants in a greenhouse of Wageningen UR (Wageningen, the Netherlands). After confirmation of the collected thrips as F. occidentalis a population was developed and maintained by rearing female thrips on small cucumber fruits in a climate chamber at 25 °C, 16/8 h day/night. Thrips larvae (L1 stage) were obtained by allowing thrips to lay eggs in small cucumber fruits for 1 day, after which the adult thrips were brushed off and fruits were kept at 25 °C for 4 days, when the larvae emerged (Mollema et al. 1993). The number of synchronized larvae was sufficient to infest a complete replication of the resistance test in 1 day.
Resistance test
Five newly emerged F. occidentalis L1 larvae were placed on a single fresh fully opened leaf that was placed with the abaxial side downwards in a sterile 50 × 9 mm petri dish with lid (BD Falcon®). We used the third to sixth fully opened leaf counting from the top of the stem, taken from plants between 6 and 9 weeks after transplanting. Leaves and larvae were incubated in a climate chamber at 25 °C, 16 h light, and 70 % RH.
Damage caused by larvae was scored after 2 days using a visual scale ranging from 0 (no damage) to 3 (severe damage) as described in Maharijaya et al. (2011). Development of L1 larvae into the L2 stage was assessed by counting the number of L2 larvae and dividing this by the total number of larvae placed on the leaf. The transition from larval stage L1 to L2 was determined by the presence of skin tissue that remained on the leaf disk after molting, which can be seen under a stereo microscope. Development of L2 larvae was assessed by counting the number of pre-pupae divided by the original number of L1 larvae. Pre-pupae can be recognized by the presence of short wing sheaths. Leaves were replaced by fresh ones every 3 days until all larvae had died or reached the pre-pupa stage; this required incubation and observation up to 8 days. These two parameters are referred to as “survival to L2” and “survival to pre-pupa,” respectively.
For the F2 population experiment, each replication consisted of one petri dish per F2 plant, three dishes for each parental inbred, and two dishes of the F1. The complete F2 test consisted of five replications. The F3 lines experiment consisted of four replications, each with one dish per F3 plant. In both tests, each replication was started on a single day with approximately 1 week intervals.
Trichome density
Trichome density was scored according to a visual scale: 0 (<50 cm−2), 1 (50–100 cm−2), 2 (100–200 cm−2), and 3 (>200 cm−2) at the region near the veins and midrib on the abaxial side of a fully developed leaf. Observations of trichome density were done at three different plant stages: early vegetative stage (3 weeks after transplanting), vegetative stage (6 weeks after transplanting), and reproductive stage (9 weeks after transplanting). Observations were performed on the third to sixth fully expanded leaf, counting from the top of the stem.
Statistical analysis
F2 population experiment
Means for each F2 plant, the parental inbreds and the F1 were obtained by ANOVA analysis with the five replications of the resistance test as blocks, after transforming the fraction survival to L2 and pre-pupa stages as y = arcsine (sqrt(x)) in order to stabilize variances. Pearson correlation coefficients were calculated for the three parameters observed in the resistance test and leaf trichome densities, based on the means of F2 individuals. Broad-sense heritability was estimated for all parameters according to Allard (1999) using the formula: Heritability (h2) = (σ2F2 − (σ2F1 + σ2PR + σ2PS)/3)/(σ2F2), where σ2F2, σ2F1, σ2PR, and σ2PS are the variances of the F2, F1, resistant, and susceptible parent, respectively.
F3 lines experiment
Like in the F2 experiment, means for each F3 plant were obtained by ANOVA analysis with the four replications of the resistance test as blocks, after transforming the fraction survival to L2 and pre-pupa stages as y = arcsine (sqrt(x)) in order to stabilize variances. The means per plant were treated as response variable in a linear regression model with F3 line and marker score (0 = homozygous annuum allele, 1 = heterozygous, and 2 = homozygous chinense allele) as regressors.
Molecular markers and linkage map
The KingFisher® (www.thermo.com) device was used with the AGOWA mag® Maxi DNA Isolation Kit (www.agowa.de) for isolating genomic DNA of the F2 and F3 individuals, F1, and parents. AFLP (Amplified Fragment Length Polymorphism) markers as described by Vos et al. (1995) were detected using combinations of EcoRI and MseI or PstI and MseI primers with two selective nucleotides for PstI and three selective nucleotides for EcoRI. The pre-amplification primers were E01, P00, and M02. Fifteen primers combination were used: P17-M39, P17-M32, P14-M50, P14-M49, P14-M48, P14-M41, P11-M61, P11-M48, E38-M49, E36-M48, E35-M58, E35-M49, E35-M48, E34-M48, and E32-M49 [primer sequences as in Keygene (2004)]. The PstI and EcoRI primers were labeled with fluorescent dyes IRD700 and IRD 800 (Li-Cor, Lincoln, USA). The AFLP products were separated and visualized on a 6 % denaturing polyacrylamide gel using a Li-Cor® sequencer. AFLP data were scored using Quantar software (Keygene®). Polymorphic bands were scored co-dominantly when there was a distinct difference in intensities between putatively homozygous and heterozygous bands.
Fifty-seven primer pairs were used to amplify simple sequence repeat (SSR) markers, which were used to assign the linkage groups obtained to pepper chromosomes based on published maps (Yi et al. 2006; Lee et al. 2009; Wu et al. 2009) and an unpublished map from INRA (Institut National de La Recherche Agronomique, France; personal communication, Dr. A. Palloix) (Table 1). The PCR mix for SSR markers contained 5 µl of 50 ng genomic DNA, 0.25 µl 1 M each of forward and reverse primer, 0.4 µl dNTP, 1 µl LC Green® (Idaho Technology), 0.1 µL Phire™ Hot Start DNA Polymerase (Finnzymes®), 2 µL buffer, and 5 µl MQ. The solution was overlaid with 20 µL of mineral oil. The thermal cycling condition were set as follows: incubation at 94 °C for 2 min, 40 cycles of 94 °C for 60 s, 60 °C for 60 s, 72 °C for 60 s, followed by 5 min 72 °C extension, and hold at 4 °C. The PCR products were analyzed with the LightScanner® system (Idaho Technology) using melting temperature from 60 to 95 °C at the default melting rate (0.1 °C s−1). LightScanner® analysis software was used to normalize the curves and to score them as heterozygote or one of the two homozygotes. In cases where the heterozygote patterns could not be well discriminated from one of the homozygotes the marker was scored dominantly. Four SNP markers [LM_2001, LM_2002, LM_2004, LM_2006, developed by Linders et al. (2010)] were used as reference for the position of the QTL for thrips resistance identified chromosome 5 by this group. One SNP marker (Isotig18917-234, personal communication, Dr. A. Palloix) close to the maximum of the thrips resistance QTL on chromosome 6 identified in the present study was used as reference for this QTL. The PCR protocol, visualization, and scoring methods for these markers were the same as those for SSR primers. PCR primers for the 57 SSR and 5 SNP markers are listed in Table 1.
Table 1.
List of chromosome assignments and primers for SSR and SNP markers
| Markers | Chr.a | Forward primer (5′–3′) | Reverse primer (5′–3′) | |
|---|---|---|---|---|
| 1 | Epms 725 | 1 | TTGAATCGTTGAAGCCCATT | ATCTGAAGCTGGGCTCCTTT |
| 2 | Hpms 1-41 | 1 | GGGTATCATCCGTTGAAAGTTAGG | CAAGAGGTATCACAACATGAGAGG |
| 3 | Hpms 1-281 | 1 | TGAGGCAGTGGTATGGTCTGC | CCCGAGTTCGTCTGCCAATAG |
| 4 | Gpms 169 | 2 | TCGAACAAATGGGTCATGTG | GATGAGGGTCCTGTGCTACC |
| 5 | Gpms 37 | 2 | ATTTGTATATTATTTCTTGGCCTTG | TGAACTACCCAATTCCAGCC |
| 6 | Hpms E073 | 3 | TTATTCAGGCCCACTTATCGAA | CAGCAGCCAAATTCTTGATTTC |
| 7 | Hpms E008 | 3 | CCCCTTAACTTTTAATTCTAGATCTGC | TCGTTGTTCCTCCATCACCTCA |
| 8 | Gpms 198 | 3 | AGCTTTAGACAGTGTCTGCGTG | TGATGATAAATTGCCTTCCG |
| 9 | Epms 386 | 3 | ACGCCAAGAAAATCATCTCC | CCATTGCTGAAGAAAATGGG |
| 10 | Hpms E122 | 3 | GCAATGGCTCAGGTCTCCATCT | TGTCGCCCTTTAATGCAAAACC |
| 11 | Gpms 93 | 3 | ATCCTTGGCGTATTTTGCAC | TTCACTTTGCACACAGGCTT |
| 12 | HpmsAT2 14 | 4 | TTTAGGGTTTCCAACTCTTCTTCC | CTAACCCCACCAAGCAAAACAC |
| 13 | Hpms 1-165 | 4 | GGCTATTTCCGACAAACCCTCAG | CCATTGGTGTTTTCACTGTTGTG |
| 14 | Hpms E099 | 4 | CAATCATTGCCACCTTATTTTTGC | TCACAAGGGGTTGATGGAAATG |
| 15 | Hpms E055 | 4 | GGCCGCTTAAAGTTGTTCAAGG | TGTGGCTAGCGGTGTTATGCAC |
| 16 | Hpms E049 | 4 | CACTCCAACAGCAGCAGCAAAC | CCTTGCCGATGTTGAAGCTTTT |
| 17 | Hpms E085 | 4 | TGCCCAAATATCAGTCAAGCTCA | TGGTTGTTGTTCTCATGGTGGTG |
| 18 | Hpms E111 | 4 | CCATCATTTCTCCCCAATTCCA | GAGAGCAGAAGAAGGGGTGGTG |
| 19 | Hpms E116 | 5 | CATCTCTCCGTTGAATCTATTTCC | ACGGTCATCCATTAGAACCGTA |
| 20 | Hpms 2-45 | 5 | CGAAAGGTAGTTTTGGGCCTTTG | TGGGCCCAATATGCTTAAGAGC |
| 21 | Gpms 165 | 5 | TGAACAATAATAATTGACAGGACAG | AGCCTCGCAGTTTGTTCTTAC |
| 22 | Hpms 2-23 | 5 | CCCTCGGCTCAGGATAAATACC | CCCAGACTCCCACTTTGTG |
| 23 | Hpms E015 | 5 | TTGTGAGGGTTTGACACTGGGA | CCGAGCTCGATGAGGATGAACT |
| 24 | Hpms E014 | 6 | CTTTGGAACATTTCTTTGGGGG | GCGGACGTAGCAGTAGGTTTGG |
| 25 | Hpms E088 | 6 | GCAAATGGTTCCCTAAACTGCTT | GCTCTCCGTTTCCGATGTGATT |
| 26 | Hpms E078 | 6 | TTTGTGAAGAAGCAACCGGTGA | TGTGAGGAAGAAAGTGCGAAGG |
| 27 | Hpms 1-5 | 6 | CCAAACGAACCGATGAACACTC | GACAATGTTGAAAAAGGTGGAAGAC |
| 28 | HpmsAT2-20 | 6 | TGCACTGTCTTGTGTTAAAATGACG | AAAATTGCACAAATATGGCTGCTG |
| 29 | HpmsE113 | 6 | CCCTAAAGCTCGAGAAATTGAAGC | GAATGCTGTTGCTGGGGTTGTT |
| 30 | Epms 376 | 6 | ACCCACCTTCATCAACAACC | ATTTGTGGCTTTTCGAAACG |
| 31 | Hpms E068 | 7 | TGTTCCTTTTGTTGTTACCTTTTG | CGTCTAGGAATGGAAGAAGAGC |
| 32 | Hpms E057 | 7 | ACCCACTCCCTCTCCTCTTTGG | GCAGTGGAAAAACAGTCCTGTGG |
| 33 | Hpms 1-227 | 7 | CGTGGCTTCAAGTATGGACTGC | GGGGCGGAACTTTTCTTATCC |
| 34 | Epms 342 | 8 | CTGGTAGTTGCAAGAGTAGATCG | ATGATCTTTGACGACGAGGG |
| 35 | Hpms E115 | 1/8 | TCATCTCATAGCCTGCCCCCTA | CCACTTGAAGAAGCCATGACCA |
| 36 | Hpms 1-148 | 1/8 | GGCGGAGAAGAACTAGACGATTAGC | CCACCCATTCCACATAGACG |
| 37 | Hpms E004 | 1/8 | TGGGAAGAGAAATTGTGAAAGCA | CAATGCCAACAATGGCATCCTA |
| 38 | Epms 310 | 8 | TGGGAAGAGAAATTGTGAAAGC | AGGAAACATGGTTCAATGCC |
| 39 | Gpms 194 | 9 | AGGTGGCAGTTGAGGCTAAG | GTTCTAGGTCTTTGCCCTGG |
| 40 | Hpms 1-3 | 9 | TGGGAAATAGGATGCGCTAAACC | AACTTTAAGACTCAAAATCCATAACC |
| 41 | Hpms E051 | 9 | TGGCCAGCTTCACACAGAGGTA | TGTCACAATATTGGAGGCCAGAA |
| 42 | Epms 419 | 9 | TTCAGGTGCAGGTATCATCG | GGGTACTTGTCCATTTATCCAG |
| 43 | Hpms E143 | 9 | CCATTCAGCTAGGGTTCAGTCCA | CGACCAAATCGAATCTTCGTGA |
| 44 | Hpms E013 | 10 | GCGCCAAGTGAGTTGAATTGAT | CACCAATCCGCTTGCTGTTGTA |
| 45 | Hpms E059 | 10 | GCAAGGACGCAGTCGTTAGACA | CCGCCTGTGCTGAATTGTTTAG |
| 46 | Hpms 2-21 | 10 | TTTTTCAATTGATGCATGACCGATA | CATGTCATTTTGTCATTGATTTGG |
| 47 | Hpms E065 | 10 | TGAAATAGGCCAATCCCTTTGC | ATTCCCTGGGATTCCTGCATTA |
| 48 | Hpms E031 | 10 | CCCTAAATCAACCCCAAATTCAA | CCCCCATTACCTGACTGCAAAA |
| 49 | Hpms E096 | 10 | CGGGTCAAACAAAAACCGAAGT | GCTTGTGGTTGAGCTCGCTCTT |
| 50 | Gpms 159 | 10 | AAGAACATGAGGAACTTTAACCATG | TTCACCCTTCTCCGACTCC |
| 51 | Epms 561 | 11 | ATTGGACTTCAAATTTGGCC | AAACCAAAATCAGCATTAAAATATAAAC |
| 52 | Epms 410 | 11 | GGAAACTAAACACACTTTCTCTCTC | ACTGGACGCCAGTTTGATTC |
| 53 | Epms 391 | 11 | TTTCTTCTCTGGCCCTTTTG | ACGCCTATTGCGAATTTCAG |
| 54 | Hpms 2-2 | 11 | GCAAGGATGCTTAGTTGGGTGTC | TCCCAAAATTACCTTGCAGCAC |
| 55 | Hpms E094 | 12 | CCAGTTGAGAGCTGCTGCAAAA | CACCAACAAAACAAAGGCCACA |
| 56 | Hpms E128 | 12 | TGGATCCCAAAAGACTCAGAACA | TATTTCCCTCAGTCGAGGTCGT |
| 57 | Hpms E064 | 12 | CCCTCCTTTTACCTCGTCAAAAA | ATGCCAAGGAGCAATGAGAACC |
| 58 | LM_2001 | 5 | CTTTGGAGGTAGCGGTATG | CAACAAACGAACCACAATG |
| 59 | LM_2002 | 5 | CCCGTTTACAAGCAAAGAG | GACCCCTGAAGAACCTCTC |
| 60 | LM_2004 | 5 | TGTAGGATTACAAGAACATTATCG | GCGAGCTATTACACCGAAG |
| 61 | LM_2006 | 5 | TCGGCCTGACTAGTATTGAC | CGGGTACCAGATGTAGGG |
| 62 | Isotig18917-234 | 6 | ACTAGTAAGAGCAGGGGTG | TCAATAGATCCAAATGCAGATTGAAC |
Putative chromosome positions and primer sequences of the Hpms markers are based on Lee et al. (2009) and Yi et al. (2006); of the Gpms and Epms markers on an unpublished Institut National de La Recherche Agronomique (INRA, France) map (personal communication, Dr. A. Palloix) and Wu et al. (2009); of the LM markers on Linders et al. (2010); of SNP marker Isotig18917-234 were communicated by Dr A. Palloix
aPutative chromosome position
A linkage map was constructed using JoinMap 4.1 software (Van Ooijen 2006). Markers with more than about 40 missing values were discarded. Groups of markers of a more or less constant composition over a range of LOD values were used as a starting point to create linkage groups. Where multiple linkage groups were found with SSR markers known to reside on the same pepper chromosome an attempt was made to combine the markers into one linkage group. Mapping within linkage groups was carried out with the regression algorithm and a maximum jump level of 5. The final result was obtained by deleting markers that did not fit well as judged by the nearest neighbor stress or the mean Chi square contribution.
QTL mapping
Potential QTLs for damage, larval survival, and trichome density were identified using the MapQTL 6.0 package (Van Ooijen 2009). First, interval mapping analysis was performed to find regions with potential QTL effects. Second, co-dominant markers in these regions were used as co-factors in multiple-QTL mapping (MQM). Significance thresholds of log of odds (LOD) corresponding to a genome-wide confidence level of P < 0.05 were determined for each trait using the permutation test of MapQTL 6.0 with 1000 iterations. The QTL graphs were prepared with MapChart 2.3 (Voorrips 2002).
Results
Thrips resistance in the F2 population
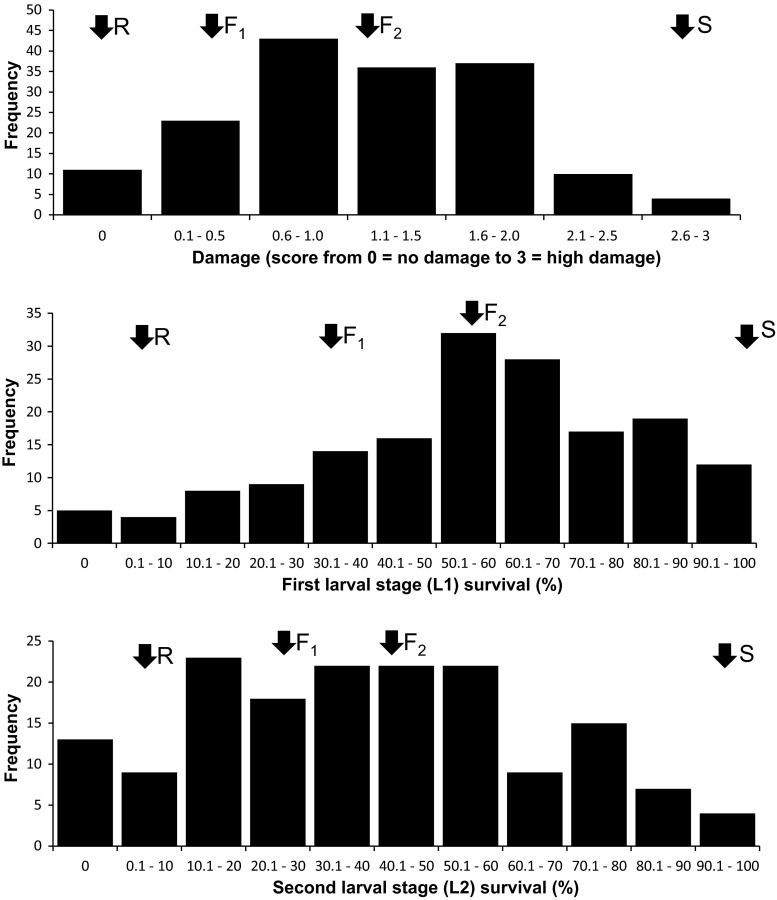
The F2 population showed a continuous variation for damage caused by larvae, for survival from L1 to L2, and for survival to pre-pupa (Fig. 1). Frequency distributions of damage and survival to pre-pupa were skewed toward the resistant parent, while for survival to L2 it was skewed toward the susceptible parent. In all replicates of the resistant parent, the damage was 0 and the survival to L2 and survival to pre-pupa was very low, while all replicates of the susceptible parent exhibited significant feeding damage and very high rates of survival to L2 and to pre-pupa. The broad-sense heritability of all parameters scored in the laboratory tests with F. occidentalis was high (Table 2).
Fig. 1.
Frequency distributions for overall damage caused by first instar larva, survival to L2 (second larval stage), and survival to pre-pupa in F2 population from a cross between resistant and susceptible accessions of pepper. Arrows indicate the approximate means of the resistant parent (R), susceptible parent (S), F1 and F2 population
Table 2.
Values of resistance-related traits for parents, F1 and F2 plants after infestation with newly emerged L1 larvae of Frankliniella occidentalis
| Damageb | Survival to Lc2 | Survival to pre-pupad | |
|---|---|---|---|
| Resistant parent | 0.00 ± 0.00a | 0.20 ± 0.12 | 0.20 ± 0.12 |
| Susceptible parent | 2.73 ± 0.04 | 1.57 ± 0.00 | 1.36 ± 0.20 |
| F1 | 0.40 ± 0.28 | 0.60 ± 0.06 | 0.55 ± 0.12 |
| F2 | 1.16 ± 0.69 | 0.88 ± 0.38 | 0.66 ± 0.38 |
| Heritabilitye | 0.94 | 0.96 | 0.93 |
aMean ± standard deviation
bScore of relative damage caused by L1 larvae of F. occidentalis at 2 days after infestation: 0 (no damage) to 3 (severe damage)
carcsine(sqrt(x)) transformation of fraction of L1 larvae that survived to L2 stage
darcsine(sqrt(x)) transformation of fraction of L1 larvae that survived to pre-pupa stage
eBroad sense heritability
Damage caused by larvae, survival of L1 to L2 and survival to pre-pupa were highly correlated with coefficients 0.68–0.78 (P < 0.001). None of the parameters scored in the resistance tests were significantly correlated with trichome density (Table 3).
Table 3.
Spearman rank correlation coefficients and significance between all parameters scored in the F2 population
| Survival to La2 | Survival to pre-pupab | Leaf trichome density | |||
|---|---|---|---|---|---|
| Early vegetative | Late vegetative | Reproductive | |||
| Damage caused by larva | 0.68* | 0.72* | 0.13 | 0.11 | 0.12 |
| Survival to La2 | 0.78* | 0.10 | 0.09 | 0.09 | |
| Survival to pre-pupab | 0.08 | 0.09 | 0.09 | ||
| Leaf trichome density | |||||
| Early vegetative | 0.86* | 0.71* | |||
| Late vegetative | 0.83* | ||||
Leaf trichome density was measured in three life stages of the plant
* Indicates significance P < 0.001
aarcsine(sqrt(x)) of fraction of L1 larvae that survived to L2 stage
barcsine(sqrt(x)) of fraction of L1 larvae that survived to pre-pupa stage
Linkage map
A linkage map was constructed consisting of 22 linkage groups. The linkage groups varied in length between 16.5 and 197.8 cM, with a total length of 1636.2 cM. The total map included 171 markers (57 SSR, 109 AFLP, and 5 SNP), of which 88 (51.5 %) were scored co-dominantly.
Linkage groups were assigned to pepper chromosomes based on SSR anchor markers. Seven chromosomes (1, 2, 6, 7, 8, 9, and 11) had only one linkage group assigned, while the other five had two or in one case (chromosome 3) three linkage groups assigned. Four linkage groups consisting of a total of 20 AFLPs and spanning 205.1 cM could not be assigned to chromosomes. Four markers (LM_2001, LM_2002, LM_2004, and LM_2006) described by Linders et al. (2010) as mapping to chromosome 5 were confirmed to map on that chromosome.
QTL mapping
Interval mapping of damage, survival to L2, and survival to pre-pupa all resulted in the detection of the same, highly significant QTL on chromosome 6 (P06, Fig. 2). MQM mapping using the marker nearest to the top of the three LOD profiles (Hpms078) as co-factor failed to reveal any additional QTLs. In particular, no QTL signal was found on chromosome 5 at the four markers mentioned by Linders et al. (2010) to target a QTL for thrips resistance (Fig. 2). The LOD scores at marker Hpms078 were 20.6, 24.3, and 18.8, with an explained phenotypic variance of 43.9, 49.4, and 41.1 % for damage, survival of L1 to L2, and survival to pre-pupa, respectively (Table 4); a LOD threshold of 3.6 corresponding to a genome-wide P = 0.05 was estimated by a 1000-fold permutation test for all three traits. Since the heritabilities of damage, survival to L2 and survival to pre-pupa were 0.94, 0.96, and 0.93 (Table 2), the QTL explained 46.7, 51.5, and 44.2 %, respectively, of the genetic variance in the F2 for the three traits. The resistance allele of this QTL was inherited from the resistant parent. The dominance effect of the QTL was small in comparison with the additive effect, with susceptibility being partially dominant over resistance (Table 4).
Fig. 2.
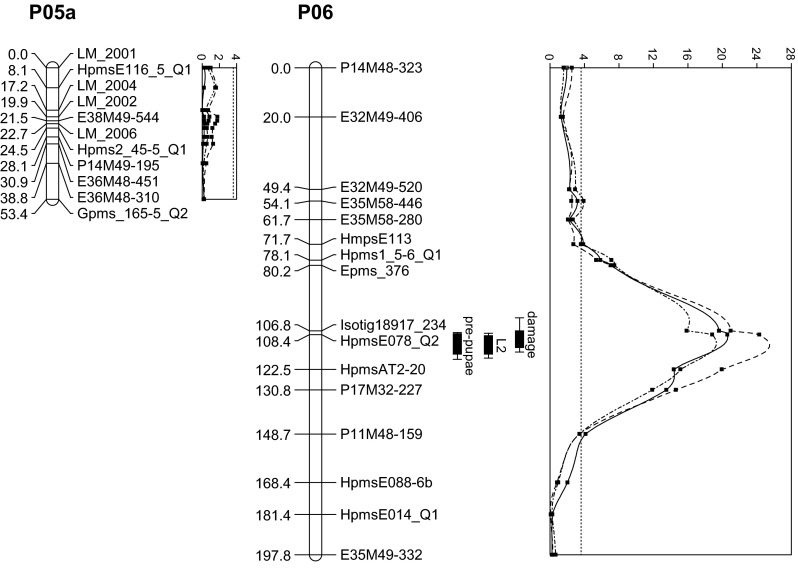
LOD profiles and 1-LOD and 2-LOD support intervals for resistance QTLs on chromosomes 5 and 6. Solid, dashed, and dot-dashed lines represent the profiles for damage, survival to L2, and survival to pre-pupae, respectively, after inoculation with newly emerged L1 larvae of F. occidentalis. The dotted line at LOD 3.6 represents the LOD threshold. On chromosome 5 no QTLs were detected for these traits
Table 4.
QTL effects for resistance-related traits after inoculation with F. occidentalis and for leaf trichome density
| Traits | Marker at QTL peak | Chromosome | Positiona | LODb | Additive effectc | Dominance effect | % Expl.d |
|---|---|---|---|---|---|---|---|
| Damage | HpmsE078 | P06 | 108.4 | 20.6 | −0.66 | 0.10 | 43.9 |
| Survival to Le2 | HpmsE078 | P06 | 108.4 | 24.3 | −0.37 | 0.09 | 49.4 |
| Survival to pre-pupaf | HpmsE078 | P06 | 108.4 | 18.8 | −0.34 | 0.10 | 41.1 |
| Trichome density early vegetativeg | HpmsE031 | P10b | 40.5 | 15.4 | −0.63 | 0.14 | 30.4 |
| Trichome density late vegetativeg | HpmsE031 | P10b | 40.5 | 21.7 | −0.69 | 0.26 | 39.9 |
| Trichome density reproductiveg | HpmsE031 | P10b | 40.5 | 27.5 | −0.74 | 0.30 | 47.5 |
aPosition of the QTL, in cM, referred to the linkage group
bLogarithm of the odds (LOD); for all six traits a threshold of 3.6, corresponding to a genome-wide confidence level of 0.05, was estimated from permutation tests
cNegative values indicate that C. annuum alleles result in lower genotypic values than C. chinense alleles
dPercentage of phenotypic variance explained by each QTL
earcsine(sqrt(x)) of fraction of L1 larvae that survived to L2 stage
farcsine(sqrt(x)) of fraction of L1 larvae that survived to pre-pupa stage
gbased on a visual scale: 0 (<50 cm−2), 1 (50–100/cm−2), 2 (100–200 cm−2), and 3 (>200 cm−2) at the region near to the veins and midrib on the abaxial leaf surface of fully developed leaves at three different plant stages: early vegetative stage (3 weeks after planting), vegetative stage (6 weeks after planting), and reproduction stage (9 weeks after planting)
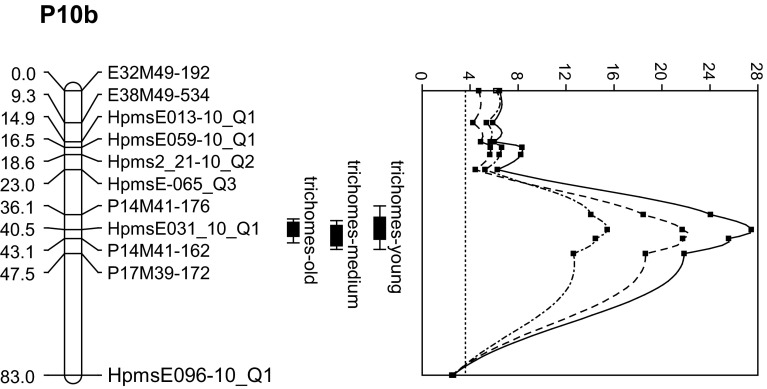
For each of the three leaf ages in which observations were made for leaf trichome density a highly significant QTL was detected on chromosome 10 (Fig. 3). The LOD scores for the detected QTL at all leaf ages were above the LOD score corresponding to a genome-wide confidence level of 95 %, which was 3.6 as determined by permutation test with 1000 iterations. The peak of the LOD profile for leaves at early vegetative, vegetative, and reproductive stage was near marker HpmsE031; at this marker, 30.4, 39.9, and 47.5 % of the variance of the F2 plant means were explained by the QTL, respectively. Use of HpmsE031 as co-factor in MQM analysis failed to uncover any additional QTLs.
Fig. 3.
LOD profiles and 1-LOD and 2-LOD support intervals for QTL for trichome density on chromosome 10. Solid, dashes, and dot-dashed lines represent the trichome density at early vegetative, late vegetative, and reproductive stage, respectively. The dotted line at LOD 3.6 represents the LOD threshold
Confirmation of the resistance QTL on chromosome 6 in F3 lines
The effect of the QTL was validated in a set of four F3 lines, each originating from an F2 plant heterozygous at the SNP marker Isotig18917-234, close to the QTL maximum (Table 5). Overall regression analysis showed significant effects of the marker on all three parameters: damage score (P = 0.013), survival to L2 stage (P = 0.011), and survival to pre-pupa stage (P = 0.012). The effects of the individual F3 lines were not significantly different from each other at P = 0.05, except marginally between lines 40 and 43 for survival to L2 stage (P = 0.043).
Table 5.
Mean scores for three resistance-related traits in the F3-line experiment, averaged per line and per genotype for marker Isotig18917-234
| Trait | Damage | Survival to La2 | Survival to pre-pupaa | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypeb | RR | RS | SS | Total | RR | RS | SS | Total | RR | RS | SS | Total |
| F3-line | ||||||||||||
| 40 | 0.99 (7)c | 0.75 (5) | 2.05 (5) | 1.23 (17) | 0.26 (7) | 0.09 (5) | 0.85 (5) | 0.37 (17) | 0.17 (7) | 0.07 (5) | 0.71 (5) | 0.27 (17) |
| 43 | 2.00 (1) | 2.00 (7) | 2.03 (3) | 2.01 (11) | 0.67 (1) | 0.80 (7) | 0.80 (3) | 0.79 (11) | 0.50 (1) | 0.60 (7) | 0.67 (3) | 0.61 (11) |
| 134 | 0.50 (1) | 2.07 (5) | (0) | 1.81 (6) | 0.12 (1) | 0.59 (5) | (0) | 0.50 (6) | 0.01 (1) | 0.39 (5) | (0) | 0.30 (6) |
| 211 | 1.06 (4) | 1.30 (2) | 3.00 (1) | 1.41 (7) | 0.30 (4) | 0.31 (2) | 0.97 (1) | 0.42 (7) | 0.16 (4) | 0.11 (2) | 0.89 (1) | 0.24 (7) |
| Total | 1.05 (13) | 1.61 (19) | 2.15 (9) | 1.55 (41) | 0.29 (13) | 0.50 (19) | 0.85 (9) | 0.52 (41) | 0.17 (13) | 0.33 (19) | 0.72 (9) | 0.36 (41) |
aThe means for survival to L2 and pre-pupa stages were calculated on the transformed scale and back-transformed to the original scale
bThe genotypes for marker Isotig18917-234 are coded as RR: homozygous for the resistant (C. annuum) parent allele, RS: heterozygous, SS: homozygous for the susceptible (C. chinense) parent allele
cNumbers in brackets are the numbers of plants per category
Discussion
Resistance test
The high heritabilities found for thrips resistance as measured by damage, survival to L2, and survival to pre-pupa indicate that variation due to environmental factors was minor relative to genetic effects. This was achieved using a climate room with controlled environmental conditions and a thrips rearing that supplied us with large quantities of uniform and synchronized larvae. This is an important advantage for genetic studies in comparison with greenhouse or field tests. In previous work (Maharijaya et al. 2011), we have shown that the resistance estimated from the laboratory test corresponds well with that estimated from greenhouse and field tests.
The high correlations between damage caused by larvae, survival to L2, and survival to pre-pupa indicate that differences in tolerance (i.e., the ability of the plant to restrict symptom development in spite of the presence and activities of the pest) do not play an important role in this case. The low number of larvae that survived on resistant plants shows that the mechanism of pepper defense against thrips larvae is based on antibiosis (Horber 1980). It has been reported before that resistance in pepper blocks larval development of F. occidentalis in pepper (Maris et al. 2004; Maharijaya et al. 2012).
Trichome density is not related to thrips resistance in pepper
No correlation was found between trichome density and any of the resistance parameters in our study with F. occidentalis. Also the resistant parent of our mapping population was glabrous, while the susceptible parent carried trichomes. This contrasts with an earlier finding that trichomes are associated with resistance to a different thrips species (S. dorsalis) in pepper (Yadwad et al. 2008). This difference might be caused by the difference in thrips species, but also by the fact that Yadwad et al. (2008) rated the resistance based on damage caused by adult thrips in a preference test, whereas we used a no-choice test with larvae. Further, the significant correlations of thrips resistance and trichome density found by Yadwad et al. (2008) were F2 population specific. For only four out of seven F2 populations, each consisting of 60 plants, they found a significant correlation of resistance against thrips with trichome density at the mature pepper stage (R = 0.27–0.48), and no correlation was found for any of those seven populations at flowering stage.
Linkage map
Twenty-two linkage groups were constructed, for twelve chromosomes in the haploid pepper genome. The mapping of SSR markers in our linkage map was consistent with that in previous populations (Minamiyama et al. 2006; Yi et al. 2006; Barchi et al. 2007; Wu et al. 2009). The total length of our linkage map was 1630 cM which is comparable to the maps published by these authors. Although in several cases we still have more than one linkage group per chromosome, it is likely that our map covers most of the pepper genome.
One major QTL for thrips resistance on chromosome 6 of pepper
Since the three parameters of resistance in our test: damage, survival to L2, and survival to pre-pupa were highly correlated (Table 3), it is not surprising that the QTLs found for those three parameters co-localize near the same markers on chromosome 6. Only one QTL was detected for all three parameters, even when using a MQM approach. This QTL explained about 50 % of the genetic variation for the three parameters, leaving the other half unexplained. Since most of the genome is covered by our linkage map the missing genetic effect cannot be caused by other major QTLs, as these would have been detected by the MQM mapping. Therefore it is likely that several QTLs with small effects are segregating in this population as well. In principle the presence of other QTL might be deduced from differences in the average level of resistance of the four F3 lines. We did not detect any significant F3-line effects, but this may be due to the limited size of each line and the fact that each segregated for the major QTL.
While the major QTL has a small dominance effect with susceptibility partially dominant over resistance, the mean of the F2 population is near to the midparent value and the F1 is more resistant than the midparent, which suggests that the residual genetic effects are (partially) dominant for resistance.
The major QTL described by Linders et al. (2010) on chromosome 5 was not detected in our study, in spite of the fact that we included several markers linked to it. Likewise they gave no hint of a possible resistance QTL on chromosome 6. As they used the same resistant parent as we did (C. annuum AC 1979), but a different susceptible parent, this suggests that at least two major factors are involved in the resistance present in the shared parent, but that in both mapping populations only one of these segregated. If this is true, our susceptible parent contains the resistant allele of the QTL on chromosome 5. As this parent is indeed highly susceptible (Maharijaya et al. 2011, 2012), the chromosome 5 QTL then does not provide any resistance in absence of the resistance allele on chromosome 6 QTL, and the reverse is also likely to be the case.
It is less likely that the two QTL are specific to certain subpopulations of F. occidentalis since the resistance donor was even resistant to two different thrips species (F. occidentalis and T. parvispinus). An alternative hypothesis is that the different QTL are due to different test methods, as the assay of Linders et al. (2010) involved a period of 3–4 weeks of population development in a choice situation, whereas we studied survival and development of individual larvae in a no-choice situation. However, in earlier experiments (Maharijaya et al. 2011), we observed no antixenosis, so the choice vs non-choice contrast is unlikely to be an explanation for the discrepancy; and if the QTL on chromosome 6 was segregating in their population it would have had a large impact on population development and most likely have been detected.
A highly significant QTL for trichome density was detected on chromosome 10. As expected from the absence of correlation between trichome density and resistance parameters, this QTL was not linked with the QTL for resistance. Our QTL for trichome density was found at the same position as the QTL found by Kim et al. (2010).
The QTL on chromosome 6 is an important factor affecting thrips resistance in pepper, which implies that pepper breeders can benefit from the introgression of this QTL. As the source of resistance is an accession of C. annuum, which is the dominant pepper crop species, it may be assumed that the introgression of this region to other C. annuum will be straightforward.
Author contribution statement
AM, BV, and REV conceived the project. AM performed most of the practical work and was the main author of the manuscript. BV and REV supervised the work on a daily basis and contributed extensively to the manuscript. REV also produced the linkage map and performed the QTL analyses. RGFV and AP contributed to the writing of the manuscript. GSB assisted with the insect rearing and KP assisted with the marker assays.
Acknowledgments
The research was financially supported by the Royal Netherlands Academy of Arts and Sciences in the framework of the Scientific Programme Indonesia (SPIN). We thank Song Yanru for her work on the F3 lines.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Allard RW. Principles of plant breeding. 2. New York: John Willey & Sons Inc.; 1999. [Google Scholar]
- Barchi L, Bonnet J, Boudet C, Signoret P, Nagy I, Lanteri S, Palloix A, Lefebvre V. A high-resolution, intraspecific linkage map of pepper (Capsicum annuum L.) and selection of reduced recombinant inbred line subsets for fast mapping. Genome. 2007;50:51–60. doi: 10.1139/g06-140. [DOI] [PubMed] [Google Scholar]
- Borovsky Y, Paran I. Characterization of fs10.1, a major QTL controlling fruit elongation in Capsicum. Theor Appl Genet. 2011;123:657–665. doi: 10.1007/s00122-011-1615-7. [DOI] [PubMed] [Google Scholar]
- Capinera JL. In: Order thysanoptera-thrips: Handbook of vegetable pests. Capinera JL, editor. San Diego: Academic Press; 2001. pp. 535–550. [Google Scholar]
- Fery RL, Schalk JM. Resistance in pepper (Capsicum annuum L.) to western flower thrips [Frankliniella occidentalis (Pergande)] HortScience. 1991;26:1073–1074. [Google Scholar]
- Frei A, Blair MW, Cardona C, Beebe SE, Gu H, Dorn S. QTL mapping of resistance to Thrips palmi karny in common bean. Crop Sci. 2005;45:379–387. doi: 10.2135/cropsci2005.0379. [DOI] [Google Scholar]
- Galvez HF, Fernandez EC, Hautea DM. Molecular mapping of resistance to thrips in potato. Philipp Agric Sci. 2005;88:268–280. [Google Scholar]
- Horber E. In: Types and classification of resistance: breeding plants resistant to insects. Maxwell F, Jennings P, editors. New York: Wiley; 1980. pp. 15–21. [Google Scholar]
- Jones DR. Plant viruses transmitted by thrips. Eur J Plant Pathol. 2005;113:119–157. doi: 10.1007/s10658-005-2334-1. [DOI] [Google Scholar]
- Keygene NV (2004) Nomenclature of AFLP primer enzyme combinations. http://wheat.pw.usda.gov/ggpages/keygeneAFLPs.html. Accessed 3 Apr 2012
- Kim HJ, Han J-H, Kwon J-K, Park M, Kim B-D, Choi D. Fine mapping of pepper trichome locus 1 controlling trichome formation in Capsicum annuum L. CM334. Theor Appl Genet. 2010;120:1099–1106. doi: 10.1007/s00122-009-1237-5. [DOI] [PubMed] [Google Scholar]
- Lee H-R, Bae I-H, Park S-W, Kim H-J, Min W-K, Han J-H, Kim K-T, Kim B-D. Construction of an integrated pepper map using RFLP, SSR, CAPS, AFLP, WRKY, rRAMP, and BAC end sequences. Mol Cells. 2009;27:21–37. doi: 10.1007/s10059-009-0002-6. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Daubèze AM, Rouppe van der Voort J, Peleman J, Bardin M, Palloix A. QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor Appl Genet. 2003;107:661–666. doi: 10.1007/s00122-003-1307-z. [DOI] [PubMed] [Google Scholar]
- Linders EGA, Nicolet JLME, Van Wijk HJ (2010) Insect resistant plant, Vol. PCT/EP2008/055374: Syngenta Participations AG United States
- Loptien HM (2013) Thrips resistant cabbage, Vol. EP2653029A1: Rijk Zwaan Zaadteelt en Zaanhandel B.V., The Netherlands
- Maharijaya A, Vosman B, Steenhuis-Broers G, Harpenas A, Purwito A, Visser RGF, Voorrips RE. Screening of pepper accessions for resistance against two thrips species (Frankliniella occidentalis and Thrips parvispinus) Euphytica. 2011;177:401–410. doi: 10.1007/s10681-010-0277-x. [DOI] [Google Scholar]
- Maharijaya A, Vosman B, Verstappen F, Steenhuis-Broers G, Mumm R, Purwito A, Visser RG, Voorrips RE. Resistance factors in pepper inhibit larval development of thrips (Frankliniella occidentalis) Entomol Exp Appl. 2012;145:62–71. doi: 10.1111/j.1570-7458.2012.01304.x. [DOI] [Google Scholar]
- Maris PC, Joosten NN, Goldbach RW, Peters D. Restricted spread of tomato spotted wilt virus in thrips-resistant pepper. Phytopathology. 2003;93:1223–1227. doi: 10.1094/PHYTO.2003.93.10.1223. [DOI] [PubMed] [Google Scholar]
- Maris PC, Joosten NN, Goldbach RW, Peters D. Decreased preference and reproduction, and increased mortality of Frankliniella occidentalis on thrips-resistant pepper plants. Entomol Exp Appl. 2004;113:149–155. doi: 10.1111/j.0013-8703.2004.00220.x. [DOI] [Google Scholar]
- Minamiyama Y, Tsuro M, Hirai M. An SSR-based linkage map of Capsicum annuum. Mol Breed. 2006;18:157–169. doi: 10.1007/s11032-006-9024-3. [DOI] [Google Scholar]
- Mollema C, Steenhuis MM, Inggamer H, Sona C. Evaluating the resistance to Frankliniella occidentalis in cucumber: methods, genotypic variation and effects upon thrips biology. Bull IOBC/WPRS. 1993;16(5):77–83. [Google Scholar]
- Moritz G, Kumm S, Mound L. Tospovirus transmission depends on thrips ontogeny. Virus Res. 2004;100:143–149. doi: 10.1016/j.virusres.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Muchero W, Ehlers J, Roberts P. QTL analysis for resistance to foliar damage caused by Thrips tabaci and Frankliniella schultzei (Thysanoptera: Thripidae) feeding in cowpea [Vigna unguiculata (L.) Walp.] Mol Breed. 2010;25:47–56. doi: 10.1007/s11032-009-9307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palloix A, Barchi L, Lefebvre V, Sage-Palloix AM, Lanteri S. QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor Appl Genet. 2009;118:1157–1171. doi: 10.1007/s00122-009-0970-0. [DOI] [PubMed] [Google Scholar]
- Reitz S, Yearby EL, Funderburk JE, Stavisky J, Momol MT, Olson SM. Integrated management tactics for Frankliniella thrips (Thysanoptera: Thripidae) in field-grown pepper. J Econ Entomol. 2003;96:1201–1214. doi: 10.1093/jee/96.4.1201. [DOI] [PubMed] [Google Scholar]
- Shipp J, Hao X, Papadopoulos A, Binns M. Impact of Western Flower Thrips (Thysanoptera: Thripidae) on growth, photosynthesis and productivity of greenhouse sweet pepper. Sci Hortic. 1998;78:87–102. doi: 10.1016/S0304-4238(97)00130-1. [DOI] [Google Scholar]
- Siemonsma JS, Piluek K (eds) (1994) Vegetables. Plant resources of South-East Asia. Pudoc Scientific Publisher, Wageningen
- Steiner MY. Determining population characteristics and sampling procedures for the Western Flower Thrips (Thysanoptera, Thripidae) and the Predatory Mite Amblyseius-Cucumeris (Acari, Phytoseiidae) on greenhouse cucumber. Environ Entomol. 1990;19:1605–1613. doi: 10.1093/ee/19.5.1605. [DOI] [Google Scholar]
- Talekar N. Thrips on pepper: AVRDC’s research strategy. Bangkok: Regional Consulation Workshop; 1991. [Google Scholar]
- Thabuis A, Palloix A, Servin B, Daubèze AM, Signoret P, Hospital F, Lefebvre V. Marker-assisted introgression of Phytophthora capsici resistance QTL alleles into a bell pepper line: validation of additive and epistatic effects. Mol Breed. 2004;14:9–20. doi: 10.1023/B:MOLB.0000037991.38278.82. [DOI] [Google Scholar]
- Tommasini M, Maini S. Frankliniella occidentalis and other thrips harmful to vegetable and ornamental crops in Europe. In: Van Lenteren J, Loomans AJM, Tommasini MG, Maini S, Ruidavets J, editors. Biological control of thrips pests. Wageningen: Wageningen University Papers; 1995. pp. 1–42. [Google Scholar]
- Ulman DE, Cho JJ, Mau RFL, Hunter WB, Westcot DM, Suter DM. Thrips-tomato spotted wilt virus interactions: morphological, behavioural and cellular components influencing thrips transmission. Adv Dis Vector Res. 1992;9:196–240. [Google Scholar]
- Van Ooijen JW. JoinMap 4, software for the calculation of genetic linkage maps in experimental population. Wageningen: Plant Research International BV and Kayzma BV; 2006. [Google Scholar]
- Van Ooijen JW. MapQTL6, software for the mapping of quantitative trait loci in experimental population of diploid species. Kyazma BV: Wageningen; 2009. [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Voorrips RE, Finkers R, Sanjaya L, Groenwold R. QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annuum and C. chinense. Theor Appl Genet. 2004;109:1275–1282. doi: 10.1007/s00122-004-1738-1. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Lee TVD, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucl Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahyuni Y, Ballester A-R, Sudarmonowati E, Bino RJ, Bovy AG. Metabolite biodiversity in pepper (Capsicum) fruits of thirty-two diverse accessions: variation in health-related compounds and implications for breeding. Phytochemistry. 2011;72:1358–1370. doi: 10.1016/j.phytochem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Weintraub PG. Integrated control of pests in tropical and subtropical sweet pepper production. Pest Manag Sci. 2007;63:753–760. doi: 10.1002/ps.1366. [DOI] [PubMed] [Google Scholar]
- Welter SC, Rosenheim JA, Johnson MW, Mau RFL, Gusukumaminuto LR. Effects of Thrips-Palmi and Western Flower Thrips (Thysanoptera, Thripidae) on the yield, growth, and carbon allocation pattern in cucumbers. J Econ Entomol. 1990;83:2092–2101. doi: 10.1093/jee/83.5.2092. [DOI] [Google Scholar]
- Wu F, Eannetta N, Xu Y, Durrett R, Mazourek M, Jahn M, Tanksley S. A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet. 2009;118:1279–1293. doi: 10.1007/s00122-009-0980-y. [DOI] [PubMed] [Google Scholar]
- Yadwad A, Sridevi O, Salimath PM. Leaf trichomes-based resistance in chilli (Capsicum annuum) to thrips and mites. Indian J Agric Sci. 2008;78:518–521. [Google Scholar]
- Yi G, Lee J, Lee S, Choi D, Kim B-D. Exploitation of pepper EST–SSRs and an SSR-based linkage map. Theor Appl Genet. 2006;114:113–130. doi: 10.1007/s00122-006-0415-y. [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Wu Q, Li XF, Zhang YJ, Xu BY, Zhu GR. Life history of Western Flower Thrips, Frankliniella occidentalis (Thysanoptera, Thripidae), on five different vegetable leaves. J Appl Entomol. 2007;131:347–354. doi: 10.1111/j.1439-0418.2007.01186.x. [DOI] [Google Scholar]