Abstract
Proximal spinal muscular atrophy (SMA) is the most frequent cause of hereditary infant mortality. SMA is an autosomal recessive neuromuscular disorder that results from the loss of the Survival Motor Neuron 1 (SMN1) gene and retention of the SMN2 gene. The SMN2 gene produces an insufficient amount of full-length SMN protein that results in loss of motor neurons in the spinal cord and subsequent muscle paralysis. Previously we have shown that overexpression of human SMN in neurons in the SMA mouse ameliorates the SMA phenotype while overexpression of human SMN in skeletal muscle had no effect. Using Cre recombinase, here we show that either deletion or replacement of Smn in motor neurons (ChAT-Cre) significantly alters the functional output of the motor unit as measured with compound muscle action potential and motor unit number estimation. However ChAT-Cre alone did not alter the survival of SMA mice by replacement and did not appreciably affect survival when used to deplete SMN. However replacement of Smn in both neurons and glia in addition to the motor neuron (Nestin-Cre and ChAT-Cre) resulted in the greatest improvement in survival of the mouse and in some instances complete rescue was achieved. These findings demonstrate that high expression of SMN in the motor neuron is both necessary and sufficient for proper function of the motor unit. Furthermore, in the mouse high expression of SMN in neurons and glia, in addition to motor neurons, has a major impact on survival.
Introduction
The motor neuron disorder proximal 5q SMA is the most frequent hereditary cause of death in infants (1). The loss of motor neuron function results in paralysis of muscles and respiratory failure (2). SMA is caused by the loss of function of SMN1 and retention of SMN2 (3,4). Both genes encode the SMN protein, however the SMN2 gene produces only a small amount of full-length transcript. The truncated SMN protein lacking the amino acids encoded by exon 7 does not oligomerize efficiently and is rapidly degraded (5–7). The ubiquitously expressed SMN protein plays an essential role in snRNP assembly and splicing in all cell types (8). Yet reduced levels of SMN protein result in the selective loss of motor neurons and impairment of the neuromuscular junction (NMJ) formation and maturation (9–11). Over expression of SMN has been shown to completely ameliorate the SMA phenotype in mice (12,13). Several promising therapies that increase the amount of full-length SMN protein including gene therapy, oligonucleotide therapy and small molecules have been developed (14–23). Yet any therapy if administered at the wrong time or in the wrong tissue will not be effective. Previously, others have studied the temporal requirement for SMN expression in the mouse (24,25). It is critical for the advancement of therapies in SMA to understand the spatial requirement of SMN for proper function of the motor neuron. Previously we reported that over expression of SMN in neurons completely ameliorates the SMA phenotype while over expression in muscle had no effect (13). Other studies have suggested the importance of high levels of SMN in neurons by either deleting or replacing Smn and then examining neuromuscular junction (NMJ) physiology and morphology (26–31). While NMJ morphology was improved there was no substantial increase in survival of the SMA mouse in these studies.
Here we present a comprehensive study of the requirement of SMN in the motor neuron, neurons and glia with both elimination of Smn using a floxed exon 7 allele (SmnF7) and replacement of Smn using a hybrid Smn exon 7 allele (SmnINV) in the SMNΔ7 mouse model of SMA (24,32). We assayed the function of the motor unit using clinically relevant electrophysiological measurements. In these experiments SMN2 and the SMNΔ7 transgene provide the ubiquitous low level of SMN protein required for survival of all cells and tissues (12,33). We found that the reduction of SMN from all neurons and glia resulted in loss of function of the motor unit and death of the mouse. Conversely replacement of SMN in neurons and glia corrected the functional output of the motor unit and survival of the mouse. Interestingly, replacement of Smn in just the motor neuron did not alter the survival of the SMA mouse yet the functional deficit of the motor unit was fully restored. Furthermore replacement of SMN in neurons and glial cells was sufficient to rescue both survival of the mouse and the function of the motor unit in the SMNΔ7 mouse model.
Results
Specific deletion and replacement of SMN upon Cre-mediated recombination in neural tissue
In order to study the requirement of SMN in the nervous system we selectively eliminated and replaced SMN expression using the Cre/lox system. We performed these experiments with multiple Cre drivers and floxed Smn alleles in the SMNΔ7 SMA mouse. The SMNΔ7 model contains two copies of the human SMN2 transgene, is homozygous for the SMNΔ7 transgene and has a disruption in the mouse Smn allele (Smn−/−; SMN2+/+; SMNΔ7+/+) (33). We conducted these experiments in the presence of low levels of SMN expression from the SMN2 and SMNΔ7 transgenes because the complete absence of SMN from any tissue is lethal (32,34,35). SMN expression is required in all cell types for survival due to SMN's ubiquitous essential role in snRNP assembly. Thus we were able to specifically examine the effect of decreasing or increasing SMN levels in the nervous system while SMN2 (and SMNΔ7) provided the required low level of SMN protein necessary for general cellular development and viability.
To eliminate Smn expression we used the previously characterized Smn allele containing a floxed Smn exon 7 (SmnF7) (32). Conversely, to replace Smn expression we used an Smn allele (SmnINV) containing a hybrid genomic cassette consisting of an inverted Smn exon 7 fused to human SMN exons 7–8 and flanked by lox66 and lox71 sites (24,36). Upon Cre-mediated recombination the entire cassette is inverted so that SMN is expressed from a mouse Smn exon 1–7/human SMN2 exon 8 hybrid allele (SmnRe) (24). These Smn alleles are diagrammed in Figure 1A. The sequence of the SmnINV genomic cassette also contains a portion of adjacent intronic regions of the mouse Smn and human SMN alleles (38).
Figure 1.

Diagram of Floxed Smn alleles and crosses used in this study. (A) The disrupted Smn allele is referred to as SmnKO (37). The Floxed exon 7 Smn allele is referred to as SmnF7 before recombination (32). After recombination when exon 7 is deleted the allele is called SmnD7. For the replacement of Smn, the allele termed SmnINV, contains human Smn exon 7 with flanking regions of intron followed by an inverted copy of mouse Smn exon 7 with flanking regions of intron (24,38). The two copies of exon 7 were cloned in between lox66 and a lox71 sites. Upon activation of recombination by a Cre driver the area between the lox sites is inverted resulting in a functional Smn exon 7 followed by an inverted SMN exon 7. After recombination this allele is referred to as SmnRe. In all crosses the SmnINV or SmnF7 alleles were crossed to mice that were heterozygous for the SmnKO allele and contained a Cre driver. Thus all affected animals reported here are SmnD7/KO or SmnRe/KO after recombination. (B) Mice containing a Cre driver and heterozygous for the Smn knockout allele (SmnKO) were crossed to mice homozygous for the floxed exon7 SmnF7 allele. Upon recombination the floxed exon7 Smn allele is referred to as SmnD7. Similarly, mice that contain a Cre driver and are heterozygous for the Smn knockout allele (SmnKO) were crossed to mice heterozygous for the floxed exon7 SmnINV allele. Mice homozygous for the SmnINV allele are not viable beyond 21 days of age. Upon recombination the floxed exon7 Smn allele is referred to as SmnRe. Note that the progeny of these crosses only contain one deletion or replacement Smn allele over the SmnKO knockout allele. This is to ensure maximal efficiency of the Cre driver on just one target allele. Moreover, the progeny of these crosses were only used for data analysis and not for breeding to eliminate the possibility of germ line recombination.
For full interpretation of the SMN requirement in nerve and muscle we chose to perform both deletion and replacement of Smn exon 7 with several Cre drivers. To eliminate the possibility of germ line recombination all floxed alleles were maintained separately from mice containing Cre drivers. The progeny of floxed alleles crossed to Cre drivers were only used for study and never for subsequent breeding. The mouse crosses used in this study to generate deletion of Smn (SmnD7) and replacement of Smn (SmnRe) are diagramed in Figure 1B. Littermates with a WT copy of Smn and the Cre driver (Cre+; SmnD7/WT or Cre+; SmnRe/WT) were used as corresponding controls.
Before we selectively eliminated or replaced Smn in neuronal tissue we first determined the integrity of each Smn allele with the ubiquitous Sox2-Cre driver. Upon Sox2-Cre-mediated deletion of Smn exon 7 (SmnD7), the mice developed an SMA-like phenotype and survived on average 13.8 ± 1.3 days (n = 9) (38). Conversely, as expected, ubiquitous Sox2-Cre recombination of the SmnINV allele resulted in mice that lived beyond one year of age with no phenotype (n = 10) (38). In this study, the control mice without Cre are phenotypically similar to SMNΔ7 mice with a comparable lifespan (33). Once we established that the Smn alleles were fully functional we crossed the SmnF7and the SmnINV alleles to five different Cre drivers (Nestin-Cre, ChAT-Cre, Gad2-Cre, rSyn1-Cre and SYN1-iCre) listed in Table 2 (39–46). The primers used to genotype the Smn alleles and the Cre lines are listed in Table 3.
Table 2.
Floxed alleles used in this study
| Jax number | Strain name | Recombination |
Result upon Cre recombination | References | |
|---|---|---|---|---|---|
| Before | After | ||||
| 006138 | Smn1tm1Jme/J | SmnF7 | SmnD7 | Deletion of Smn exon 7 | (32) |
| 007249 | Smn1tm3(SMN2/Smn1)Mrph/J | SmnINV | SmnRe | Replacement of Smn exon 7 | (24) |
| 007914 | Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | <STOP>tdTom | tdTom | RFP expression | (47) |
Table 3.
Primer sequences used for genotyping
| Transgene/Allele | Forward primer | Reverse primer |
|---|---|---|
| SmnF7 | AGA AGG AAA GTG CTC ACA TAC AAA TT | TGT CTA TAA TCC TCA TGC TAT GGA G |
| SmnINV | ACT TCT TAA TTT GTA TGT GAG CAC T | CGC TTC ACA TTC CAG ATC TGT CTG |
| SmnRe | ATT TAA GGA ATG TGA GCA CCT TCC | CGC TTC ACA TTC CAG ATC TGT CTG |
| Cre | CCT GTT TTG CAC GTT CAC CG | ATG CTT CTG TCC GTT TGC CG |
| Nes-Cre | TCT CAA GAT TCT CTC AGA AAA TCA CC | GTT GCT GGA TAG TTT TTA CTG CCA G |
| ChAT-Cre | CTC ATC TGT GGA GTT TGC AGA AGC | GAA AGA CCC CTA GGA ATG CTC |
| SYN1-iCre | CAG GGC CTT CTC CAC ACC AGC | CTG GCT GTG AAG ACC ATC |
| HB9:GFP | GTC GAG CTG GAC GGC GAC GT | CTG CAC GCT GCC GTC CTC GA |
| tdTomato | ACG CTG ATC TAC AAG GTG AAG ATG C | CAT TAA AGC AGC GTA TCC ACA TAG CG |
| Rosa26 | AAG GGA GCT GCA GTG GAG TA | CCG AAA ATC TGT GGG AAG TC |
Characterization of Cre driver using tdTomato-RFP expression
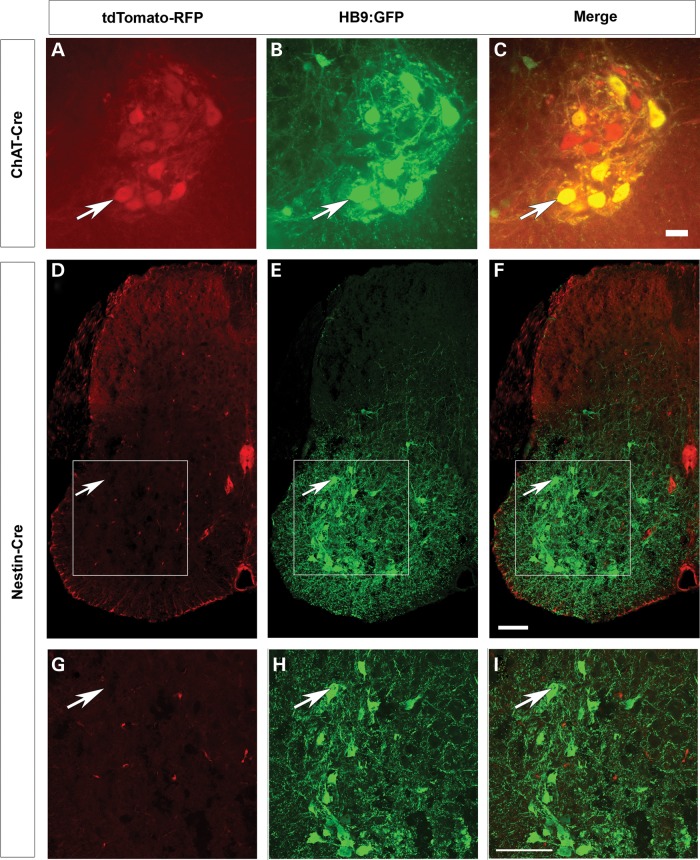
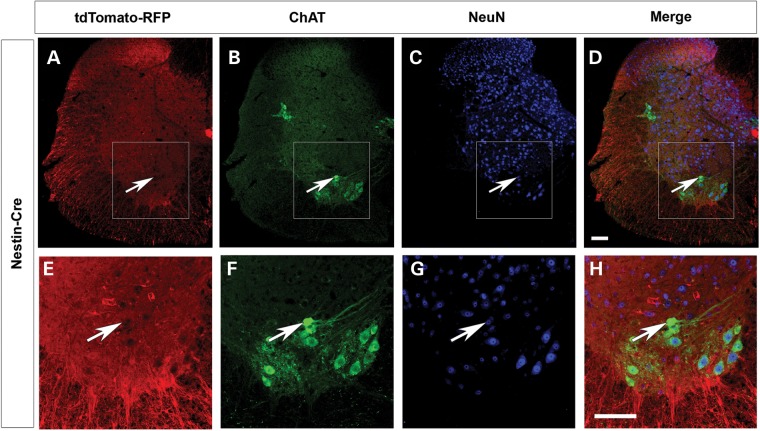
While in these experiments SMN2 provides a low level of SMN in all tissues, any additional deletion or replacement of Smn due to ectopic expression of the Cre driver could contribute to the overall weight and survival of the mouse. Thus it is important to ascertain the expression of any Cre line outside of the tissue of interest. To fully characterize the expression of each Cre driver, we used the tdTomato-RFP reporter line (47). When a Cre line is crossed to tdTomato-reporter line the floxed stop cassette preceding RFP is removed allowing RFP expression irreversibly in the cells in which the Cre driver is expressed. Thus we can identify the tissues in which recombination has occurred by the presence of RFP immunofluorescence in those cells. The mice also contained the HB9:GFP transgene to identify the motor neurons (48). Expression of ChAT-Cre was isolated to the large motor neurons of the ventral horn (Fig. 2A) and colocalized with expression of HB9:GFP (Fig. 2B and C). As expected certain motor neurons did not express HB9:GFP but did express ChAT-Cre (Fig. 2C) (49,50). No additional RFP expression was found in any other tissue including skeletal muscle, heart, lung, pancreas, kidney, adrenal gland, liver and spleen (data not shown). Nestin-Cre expression was found throughout the white matter of the spinal cord; however surprisingly, minimal RFP expression was detected in the motor neurons themselves (Fig. 2D–I). The absence of Nestin-Cre expression is demonstrated by the lack of colocalization of tdTomato-RFP in the motor neuron upon staining with choline acetyltransferase (ChAT) antibody (Fig. 3). Additionally we examined the organs of ten different Nestin-Cre mice and identified limited expression in other tissues (Supplementary Material, Fig. S1). In all 10 mice patches of RFP positive blood vessels and connective tissue were found. In skeletal muscle four of the ten mice studied displayed small patches of weak RFP expression (Supplementary Material, Fig. S1A–C). The expression in muscle was very limited in area with only a few fibers showing expression, and this expression was far less intense than expression driven by the Myf5-Cre muscle driver (Supplementary Material, Fig. S1A–C) (38). In six of the ten mice examined skeletal muscle was completely negative for RFP. One of ten mice displayed RFP expression in the heart (Supplementary Material, Fig. S1D–F). Lung was weakly positive for RFP in more than half of the mice examined, but bronchioles were always negative (Supplementary Material, Fig. S1G–I). Pancreas expression was found in most animals in the acinar cells, however the islet cells responsible for the endocrine activity of pancreas and the pancreatic duct were always negative in all samples (Supplementary Material, Fig. S1J–L). Most kidney sections were positive for RFP in the nephrons, tubules and glomeruli (Supplementary Material, Fig. S1M–O). More than half of the mice studied had scattered expression in the medulla of the adrenal gland (Supplementary Material, Fig. S1P–R). No RFP expression was detected in the liver or spleen (data not shown). In summary, Nestin-Cre expression was consistently identified in the spinal cord, blood vessels, acinar cells of the pancreas and kidney nephrons. Most of the ten mice studied had no expression in skeletal muscle, heart, lung, adrenal gland, liver and spleen.
Figure 2.
Nestin-Cre and ChAT-Cre expression in the spinal cord. Mice containing ChAT-Cre (A–C) or Nestin-Cre (D–I) were crossed to tdTomato-RFP reporter line where mice contained HB9:GFP transgene to label the motor neurons (B, E and H). (A–C) ChAT-Cre is heavily expressed in all motor neurons (arrow). Scale bar: 100 µm (D–I) Nestin-Cre is expressed throughout the white matter but very weakly in motor neurons (arrow). (G–I) Higher magnification inset image shown in the white box in images D–F. Representative images from ChAT-Cre mice (n = 3) and Nestin-Cre mice (n = 10) at P12. Scale bar: 100 µm in all panels.
Figure 3.
The expression of the Nestin-Cre driver in the spinal cord. (A) Nestin-Cre pattern of expression was determined by crossing mice to the tdTomato-RFP reporter line (red). (B) ChAT staining marks the motor neurons and (C) NeuN staining identifies the nuclei in spinal cord sections of P10 mice. (D) The merged image reveals a lack of Nestin-Cre expression in some motor neurons. The white arrow indicates a hole where tdTomato-RFP is not present. (E–H) Insets from the white boxes in A–D clearly show the absence of RFP in some motor neurons. Scale bars = 100 µm.
To give an overall representation of the expression pattern of Nestin-Cre and ChAT-Cre together we visualized tdTomato-RFP at P2 in a whole-body sagittal section (Supplementary Material, Fig. S2). The brain and spinal cord reveal the most intense staining while the connective tissue found at the periphery of the gut and the lungs was weakly positive. Most notably however is that the skeletal muscle is completely negative when tdTomato is driven by Nestin-Cre and ChAT-Cre.
Survival and weight of mice are improved upon replacement of Smn in neural tissue
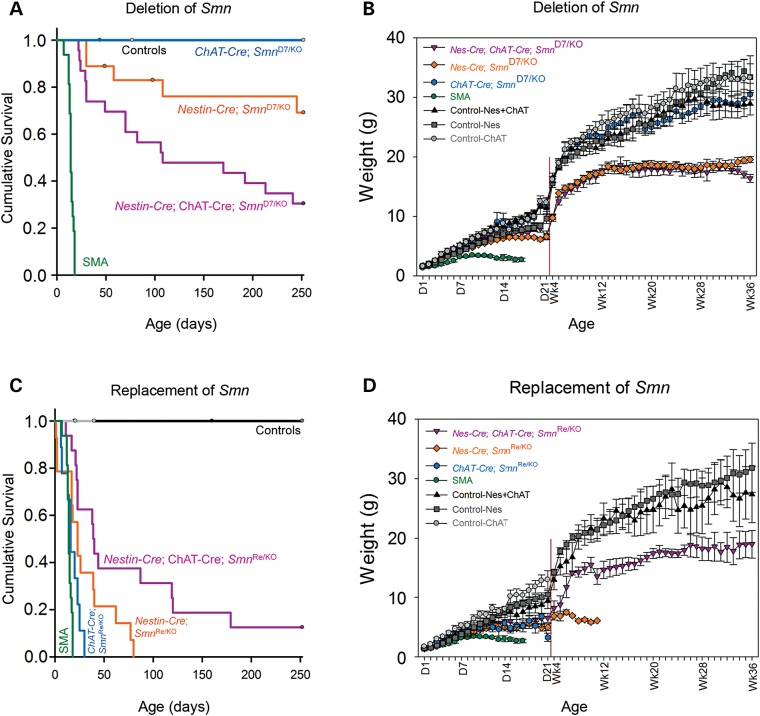
We monitored both survival and weight in the mice to study the impact of deletion or replacement of Smn. Upon deletion of Smn from the motor neurons we expected to generate an SMA-like phenotype where the mice survive approximately 14 days. Instead we found that upon deletion of Smn with ChAT-Cre 100% of the mice survived past 250 days and displayed no apparent SMA-like weakness (n = 7 total mice) (Fig. 4A). Deletion of Smn in most neurons and glia with Nestin-Cre resulted in 79% of mice living longer than 100 days (n = 18 total mice) with a tremor of the forepaws and clasp of the hindlimbs at the time of weaning (∼25 days) (Fig. 4A). Noting that Nestin-Cre had weak expression in the motor neurons we chose to delete Smn with both Nestin-Cre + ChAT-Cre, to ensure that recombination occurred in all neurons and glia. This resulted in only 50% of the mice surviving past 100 days of age (n = 23 total mice) (Fig. 4A). All mice displayed severe tremor and weakness illustrated by a full hindlimb clasp. The mice that did survive past 100 days weighed half as much as control mice, similar to the Nestin-Cre deletion mice (Fig. 4B). Indeed the phenotype of the mice with deletion of Smn with both Nestin-Cre + ChAT-Cre was clearly severe with significant weight loss and weakness.
Figure 4.
Survival and weight of SMA mice are improved upon replacement of Smn with neuron-specific Cre drivers. (A) Upon deletion of Smn from the motor neurons with ChAT-Cre 100% of the mice survived past 250 days with no SMA-like behavioral phenotype (mean survival >250 days, n = 7 total mice, P < 0.001 versus SMA). Deletion of Smn in all neurons and glia with Nestin-Cre resulted in 79% of mice living longer than 100 days (mean survival 205.4 ± 22.7 days, n = 18 total mice, P < 0.001 versus SMA). Deleting Smn with both Nestin-Cre + ChAT-Cre resulted in only 50% of the mice surviving past 100 days of age (mean survival 140.1 ± 20.5 days, n = 23 total mice, P < 0.001 versus SMA). SMA mice lived 14.3 ± 0.7 days, n = 16 mice. (B) At 36 weeks of age there was no significant difference between the weight of the ChAT-Cre deletion mice and controls (30.5 ± 1.8 g versus 33.3 ± 3.7 g). Nestin-Cre deletion mice were 19.6 ± 0.5 g and Nestin-Cre + ChAT-Cre deletion mice were 16.4 ± 0.7 g, which is approximately 50% smaller than control mice. (C) Replacement of Smn with the ChAT-Cre driver resulted in mice that lived 17.3 ± 2.7 days (n = 9, P = 0.3 versus SMA). Replacement of Smn with Nestin-Cre further increased survival to a mean of 30.5 ± 7.0 days (n = 14, P = 0.02 versus SMA). Finally replacement of Smn with both Nestin-Cre + ChAT-Cre increased survival so that 30% of the mice lived longer than 100 days (mean survival 80.4 ± 21.6 days, n = 16 total mice, P < 0.0001 versus SMA). (D) ChAT-Cre replacement mice weighed 5.4 ± 1.0 g at P16 while Nestin-Cre replacement mice weighted 4.9 ± 5.0 g. SMA mice at P16 weighed on average 2.7 ± 0.4 g. Nestin-Cre + ChAT-Cre replacement mice that survived past 100 days weighted approximately 50% less than controls at 36 weeks of age.
To study the opposite effect of replacing Smn in motor neurons we crossed mice containing SmnINV replacement allele to ChAT-Cre mice. We found that high expression of SMN in motor neurons alone only increased survival of the mouse from 14 days to a mean of 17.3 ± 2.7 days (n = 9) (Fig. 4C). Replacement of Smn with Nestin-Cre increased survival to a mean of 30.5 ± 7.0 days (n = 14) (Fig. 4C). The mice weighed 40% less than controls and displayed mild tail necrosis (Fig. 4D). Finally replacement of Smn with both Nestin-Cre + ChAT-Cre increased survival to a mean of 80.4 ± 21.6 days (n = 16) and now 38% of the mice lived longer than 100 days (Fig. 4C). Interestingly these mice displayed no weakness or clasping of the hindlimbs, and thus it appeared that the primary neuromuscular defects were corrected. Distal limb swelling, slight tail necrosis and occasionally ear necrosis were observed only in mice that survived past 100 days. The weights of the Nestin-Cre + ChAT-Cre mice were also improved over that of the Nestin-Cre Smn replacement mice, although they were still approximately 50% smaller than the control mice (Fig. 4D). Thus replacement of Smn in motor neurons and glia was sufficient to rescue both the SMA phenotype and survival of the mouse.
Muscle fiber size is decreased upon deletion of Smn in neural tissue
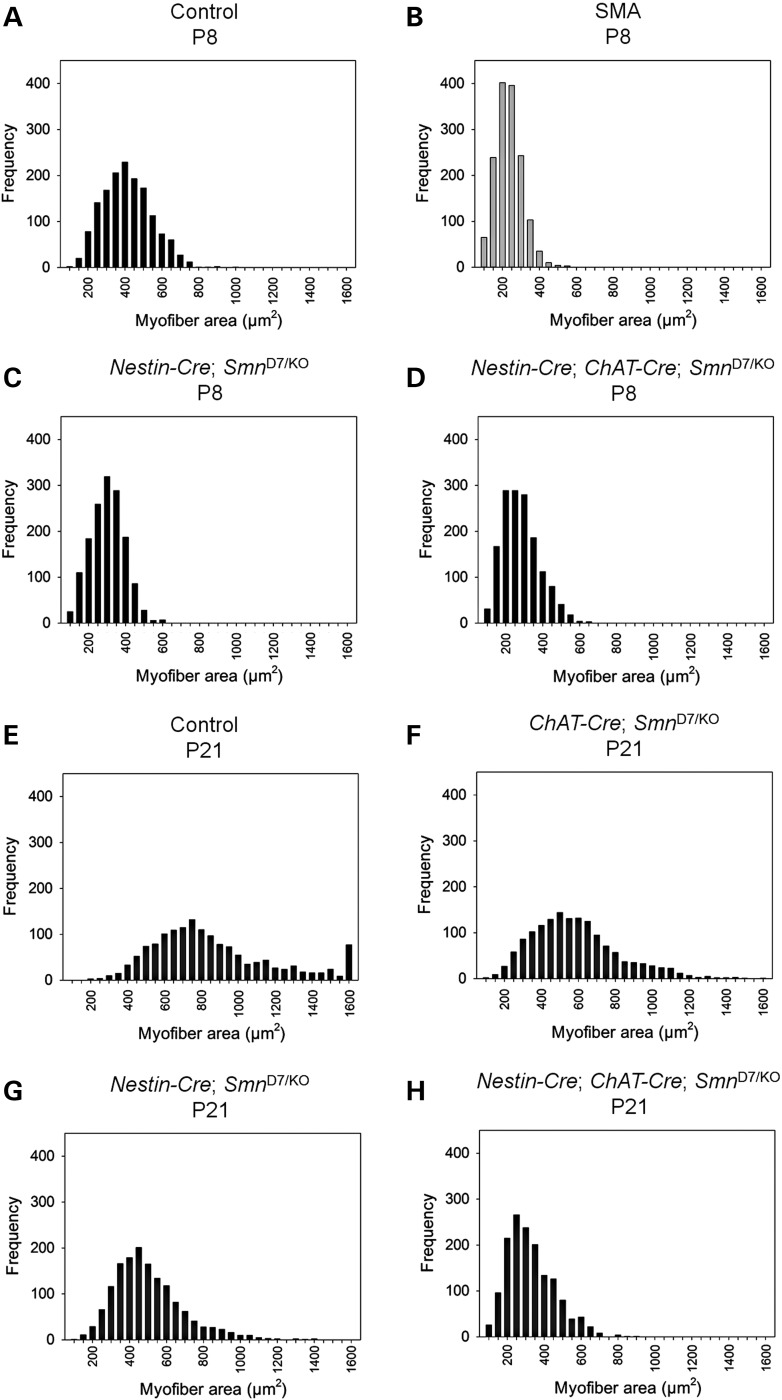
We examined the fiber size of the gastrocnemius muscle in Nestin-Cre and Nestin-Cre + ChAT-Cre deletion mice at P8 (Fig. 5A–D) and ChAT-Cre, Nestin-Cre and Nestin-Cre + ChAT-Cre deletion mice at P21 (Fig. 5E–H). We found that Nestin-Cre + ChAT-Cre Smn deletion mice possess a fiber size distribution that is very similar to the fiber sizes found in SMNΔ7 SMA mice at the same time point (Fig. 5B and D). The mean fiber size in Nestin-Cre + ChAT-Cre Smn deletion mice was 257.4 ± 2.5 µm2 compared with 211 ± 1 µm2 for SMNΔ7 SMA mice (n = 1500 fibers per group) (Fig. 5B and D) (51). At P21 the decreased fiber size is quite dramatic as Nestin-Cre + ChAT-Cre Smn deletion mice had a mean fiber size of 302.7 ± 3.2 µm2 as compared with a mean of 835.9 ± 8.8 µm2 for controls (n = 1500 fibers per group) (Fig. 5H and E). The decrease in fiber size in Nestin-Cre Smn-deletion mice is not as large (mean 477.6 ± 4.8 µm2, median 447.0 µm2) and ChAT-Cre deletion mice (mean 559.6 ± 6.1 µm2, median 530.5 µm2) were most similar to control mice at P21. There is no comparison to SMNΔ7 SMA mice at this time point as SMNΔ7 SMA mice only live 14 days. The decrease in muscle fiber size illustrates that loss of Smn in the neurons and motor neurons results in denervation and subsequent muscle atrophy.
Figure 5.
Muscle fiber size upon deletion with Nestin-Cre and ChAT-Cre. The gastrocnemius muscle from (A) control, (B) SMA, (C) Nestin-Cre Smn-deletion mice and (D) Nestin-Cre + ChAT-Cre Smn-deletion mice were measured at P8. Myofiber size in control mice at P8 (mean 387.8 ± 3.3 µm2; median 379.5 µm2) is significantly different from that of Nestin-Cre Smn-deletion mice (mean 277.5 ± 2.3 µm2; median 277.0 µm2), Nestin-Cre + ChAT-Cre Smn-deletion mice (mean 257.4 ± 2.5 µm2; median 247.0 µm2, P < 0.001). The Nestin-Cre + ChAT-Cre Smn-deletion mice however have myofiber areas similar to that of SMNΔ7 SMA mice at P8 (Mean 210.6 ± 1.9 µm2; median 205.1 µm2) (51). (E–H) The difference in myofiber area is more dramatic at P21 where (E) control mice (mean 835.9 ± 8.8 µm2; median 757.0 µm2) and (F) ChAT-Cre deletion mice (mean 559.6 ± 6.1 µm2, median 530.5 µm2) display larger fibers as compared with (G) Nestin-Cre deletion mice (mean 477.6 ± 4.8 µm2, median 447.0 µm2) and (H) Nestin-Cre + ChAT-Cre Smn-deletion mice (mean 302.7 ± 3.2 µm2; median 281.0 µm2, P < 0.001). There is no comparison to SMNΔ7 SMA mice at this time point because the SMNΔ7 SMA mice only live for 14 days. For each group, a total of 1500 fibers were measured from 3 mice (approx. 500 fibers per mouse).
Other neural Cre drivers used in this study
Additionally, we used the neural drivers SYN1-iCre, rSyn1-Cre and Gad2-Cre (Table 1) to eliminate and replace Smn (43–46). We found significantly improved survival of SMA mice when Smn was replaced with human Synapsin 1-iCre (SYN1-iCre). More than half of the mice studied (6 of 11 mice) survived for greater than one year of age with only an 18% decrease in weight (Supplementary Material, Fig. S3A and B). However, when we examined the expression pattern of the SYN1-iCre line we found nearly ubiquitous RFP staining throughout the spinal cord, muscle, heart lung, liver, pancreas, spleen, kidney and adrenal gland as well as blood vessels, connective tissue and epithelial cells (n = 3 mice) (Supplementary Material, Fig. S4). Thus SYN1-iCre is essentially a ubiquitous driver and we would expect results similar to eliminating Smn with our ubiquitous Sox2-Cre control (38).
Table 1.
Cre drivers used in this study
| Jax number | Strain name | Abbreviation | Expression | Description | References |
|---|---|---|---|---|---|
| 008454 | Tg(Sox2-Cre)1Amc/J | Sox2-Cre | Ubiquitous | e6.5, mouse Sox2 promoter | (39) |
| 003771 | B6.Cg-Tg (Nes-Cre)1Kln/J |
Nes-Cre | All neurons and glia | e10.5, rat Nestin promoter | (40) |
| 006410 | B6;129S6-ChATtm1(cre)Low1/J | ChAT-Cre | Cholinergic neurons | e10.5, endogenous mouse ChAT promoter | (41, 42) |
| 012687 | B6(129S4)-Tg(SYN1-icre/mRFP1) 9934Rdav/J |
SYN1-iCre | All neurons | Human Synapsin I promoter | (43) |
| 003966 | B6.Cg-Tg (Syn1-cre) 671Jxm/J |
rSyn1-Cre | All neurons | e12.5, rat Synapsin I promoter | (44) |
| 010802 | Gad2tm2(cre)Zjh/J | Gad2-Cre | Gad2/GAD65 (glutamic acid decarboxylase) positive neurons | e10.5, endogenous mouse Gad2 promoter | (45,46) |
When we examined the expression pattern of rat Synapsin1-Cre, (rSyn1-Cre), another neuronal specific Cre driver, with tdTomato-RFP we found less than 50% of the mice that were positive for the rSyn1-Cre transgenic insertion by genotyping had any RFP staining. It appears that some mice carrying the rSyn1-Cre transgene, confirmed with multiple sets of genotyping primers from both the rSyn1 promoter and Cre, were not expressing the driver. Additionally, when rSyn1-Cre was used to eliminate Smn (SmnD7) only four of nine mice displayed reduced survival and an SMA-like phenotype (mean 25.5 ± 12.5 days) (data not shown). The other five mice displayed no phenotype and no change in survival. Thus the rSyn1-Cre driver was not pursued. It is possible that some of the rSyn1-Cre we were testing had undergone some sort of recombination and no longer expressed Cre. Thus it is paramount to test each Cre line with a reporter gene to ensure accurate interpretation of results. Finally, we investigated the glutamic acid decarboxylase 2-Cre driver (Gad2-Cre). When Smn expression was eliminated with Gad2-Cre no phenotype was generated (n = 7). Conversely, when replacing Smn expression with Gad2-Cre the mice developed an SMA-like phenotype and died on average at 14.5 ± 5.0 days (n = 4) (data not shown) exactly like SMNΔ7 SMA mice.
Percent recombination events in LCM isolated motor neuron as determined by ddPCR
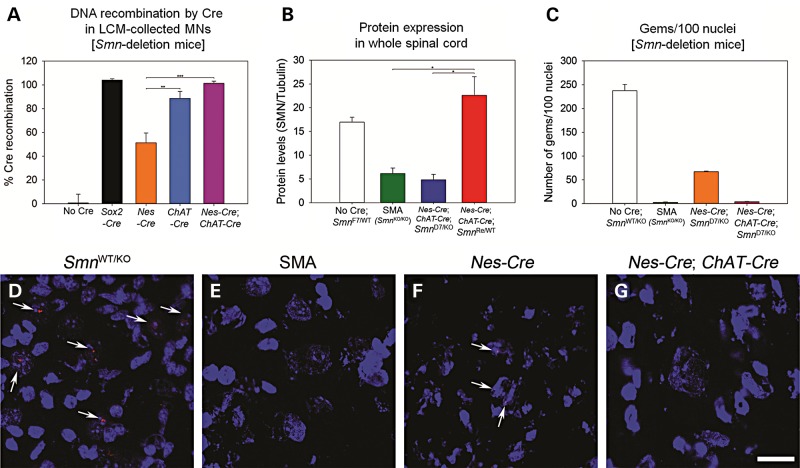
We chose to perform droplet digital PCR (ddPCR, BioRad) to determine the percentage of recombination occurring specifically in motor neurons (Fig. 6A). Only a small fraction (less than 10%) of total spinal cord extract is comprised of motor neurons. The number of recombination events that occurs for each driver line is of utmost importance for proper interpretation of results because Cre drivers often have mosaic or incomplete expression. We isolated approximately 200 motor neurons per sample with laser capture microdissection (LCM) at P10-P12 (n = 3 per genotype) and performed ddPCR to identify the recombination of the Smn-deletion allele. The number of recombination events was normalized to the total number of motor neurons per reaction as determined by Smn intron 1 primers. We found no recombination in mice that did not contain a Cre driver (0.6 ± 7.4%, n = 3) and complete recombination using the Sox2-Cre ubiquitous driver (104.0 ± 1.1%, n = 3) (38). Recombination in Nestin-Cre + ChAT-Cre Smn-deletion motor neurons was also complete (101.3 ± 1.7%, n = 3) and we determined that half of all motor neurons underwent recombination with just the Nestin-Cre driver (51.3 ± 8.1%, n = 3, P = 0.004 versus Nestin-Cre + ChAT-Cre). The ChAT-Cre driver alone resulted in 88.7 ± 5.8%, n = 3, P = 0.004 versus Nestin-Cre. The difference in recombination efficiencies between ChAT-Cre alone and Nestin-Cre + ChAT-Cre was not statistically significant (P = 0.274) illustrating that the recombination occurring in the motor neuron is dependent on the ChAT-Cre driver.
Figure 6.
Determination of Cre activity to deplete or replace Smn. (A) Percent recombination of SmnF7 in motor neurons as determined by ddPCR. Motor neurons were isolated with LCM from P10-P12 mice. There was no recombination in mice that did not contain a Cre driver (0.6 ± 7.4%, n = 3). The Sox2-Cre ubiquitous driver resulted in recombination in all motor neurons (104.0 ± 1.1%, n = 3) (38). Nestin-Cre + ChAT-Cre drivers also gave complete recombination in all motor neurons (101.3 ± 1.7%, n = 3). The ChAT-Cre driver alone resulted in 88.7 ± 5.8% (n = 3, P = 0.004 versus Nestin-Cre). On average only half of all motor neurons underwent recombination with just the Nestin-Cre driver (51.3 ± 8.1%, n = 3, P = 0.004). The difference in recombination efficiencies between ChAT-Cre alone and Nestin-Cre + ChAT-Cre was not statistically significant (P = 0.274). All values are normalized to two copies of Smn intron 1. Percent recombination values were determined by multiplex ddPCR. (B) SMN protein expression as determined by western blot analysis. The alleles used are depicted on the x axis and amount of SMN relative to tubulin on the y. Note that the amount of SMN protein found upon deletion of Smn with Nestin-Cre + ChAT-Cre is not statistically different from that of the SMA sample (P = 0.782). Conversely the replacement of Smn with Nestin-Cre + ChAT-Cre is not statistically different from that of the control sample (P = 0.322) (C) The number of SMN positive gems in motor neurons. All groups are statistically different by one-way ANOVA except for SMA versus Nestin-Cre + ChAT-Cre which are not different. Thus gem counts indicate equivalent levels of SMN protein in SMA- and Smn-deletion by Nestin-Cre + ChAT-Cre. (D) Smn+/− spinal cord section revealing many motor neurons with gems. (E) Smn−/− (SMA) sample showing lack of gems in motor neurons. (F) Nestin-Cre Smn-deletion sample shows a reduced number of gems in motor neurons. (G) Nestin-Cre + ChAT-Cre, Smn-deletion motor neurons reveal very few gems, similar to the SMA sample (n = 3 for each group).
Determination of SMN protein levels in total spinal cord and motor neurons
We analyzed the depletion and replacement of SMN in the spinal cord using the Nestin-Cre + ChAT-Cre drivers. In Nestin-Cre + ChAT-Cre mice that possessed the floxed Smn exon 7 allele, along with an Smn null allele (4.8 ± 1.1), the SMN levels were significantly reduced from no-Cre controls (17.0 ± 1.0) to SMA levels (6.2 ± 1.1) (Fig. 6B and Supplementary Material, Fig. S5B). The difference between Nestin-Cre + ChAT-Cre replacement of Smn with one reversion allele resulted in complete recovery of SMN levels (22.6 ± 3.9) back to no-Cre control levels (P < 0.01, ANOVA). In addition we examined the SMN protein levels in just motor neurons, collected by LCM, using ELISA (Supplementary Material, Fig. S5A). We found that Nestin-Cre (922.8 ± 168.9 pg SMN/MN) was not as efficient at removal of Smn exon 7 as was deletion with the Nestin-Cre + ChAT-Cre (689.7 ± 274.8 pg SMN/MN) or Sox2-Cre (469.9 ± 23.2 pg SMN/MN) drivers. Due to the large variability in this small sample size the changes did not rise to statistical significance. Finally, we measured the number of gems present in the motor neuron nuclei to assess the amount of SMN protein present in our Smn-deletion lines. We counted the number of gems present in 100 nuclei for each sample (n = 3 in each group) (Fig. 6C). We found the control with one wild-type Smn allele to have the most gems (237.4 ± 13.2 gems/100 nuclei, P < 0.001) while SMA sample had the least (2.7 ± 0.9 gems/100 nuclei, P < 0.001). The Nestin-Cre, Smn D7/KO line was again not as efficient at deletion of Smn from the motor neurons (67.1 ± 1.4 gems/100 nuclei, P < 0.001) while in the Nestin-Cre + ChAT-Cre; SmnD7/KO sample (4.0 ± 0.6 gems/100 nuclei) we identified a similar number of gems per nuclei as found in the SMA control mice. There was no statistical difference between Nestin-Cre + ChAT-Cre; SmnD7/KO and SMA gem counts. Representative spinal cord images of SMN gems are shown in Figure 6D and E. Thus protein measurements by western blot, ELISA on LCM isolated motor neuron tissue and gem counts all show that the Nestin-Cre driver is not efficient at recombination (either by deletion or replacement of Smn alleles) in the motor neuron.
Electrophysiological studies of the functional output of the motor neuron upon deletion and replacement of Smn
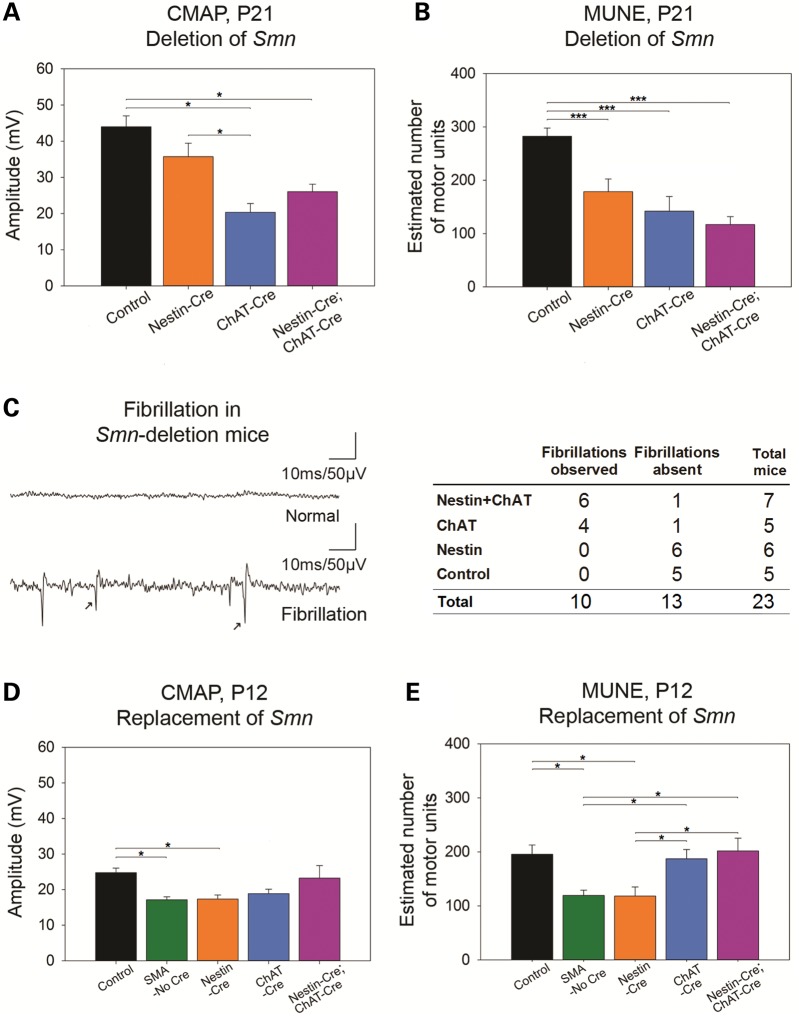
Electrophysiological studies including compound muscle action potential (CMAP), motor unit number estimation (MUNE) and electromyography (EMG) were recorded from the sciatic-innervated hindlimb muscles in mice with targeted Smn-deletion and in SMA mice with targeted SMN restoration. In mice with Smn-deletion, electrophysiological studies were performed at P21 to allow assessment of the impact of reduced SMN levels on motor unit function after completion of pruning of polyneuronal innervation at approximately 2 weeks of age (52,53). The goal was to assess loss of motor unit function in comparison with control animals with a WT copy of Smn. The findings of these studies demonstrate loss of motor unit function in the form of reduced CMAP amplitudes and reduced MUNE when SMN is reduced. Loss of motor unit function was most prominent in animals with deletion of Smn in motor neurons and glia (Nestin-Cre + ChAT-Cre) (Fig. 7A and B, Supplementary Material, Fig. S6). Furthermore electromyographic (EMG) findings of fibrillations, which results from repetitive depolarization of single muscle fibers, indicating active denervation of muscle fibers, (54) was observed in Nestin-Cre + ChAT-Cre animals. EMG analysis revealed fibrillations in hindlimb muscles of mice with deletion of Smn with Nestin-Cre + ChAT-Cre and ChAT-Cre but not Nestin-Cre, further supporting the requirement of SMN in the motor neuron (Fig. 7C). Of note, results of CMAP, MUNE and EMG in the ChAT-Cre mice at P21 have been previously published (53). Interestingly, in mice with Nestin-Cre deletion of Smn, MUNE and CMAP were slightly reduced. This is somewhat surprising as the motor neuron is the driving force behind these measures and recombination in motor neurons upon Nestin-Cre deletion was low (Fig. 6A). We suggest that a possible reason behind these observations is the role of glia, in particular astrocytes, in supporting the health and function of the motor neuron.
Figure 7.
Electrophysiological measures of motor unit function following targeted Smn-deletion and restoration. (A) Compound muscle action potential (CMAP) at P21 in mice with Smn-deletion. Compound muscle action potential (CMAP) amplitudes at postnatal day 21 (P21) were reduced in ChAT-Cre Smn-deletion (n = 9, 20.4 ± 1.9, P < 0.05) and Nestin-Cre + ChAT-Cre Smn-deletion (n = 13, 26.1 ± 2.1, P = <0.05) but not Nestin-Cre Smn-deletion mice (n = 14, 35.7 ± 3.7) compared with controls (n = 13, 44.0 ± 2.9 mV). CMAP amplitudes in ChAT-Cre Smn-deletion mice were reduced compared with Nestin-Cre Smn-deletion mice (P < 0.05). (B) Motor unit number estimation (MUNE) at P21 in mice with Smn-deletion. Motor unit number estimates (MUNE) at P21 were reduced in ChAT-Cre Smn-deletion (n = 9, 142 ± 22), Nestin-Cre + ChAT-Cre Smn-deletion (n = 13, 117 ± 15), and Nestin-Cre Smn-deletion mice (n = 14, 179 ± 24) compared with controls (n = 13, 283 ± 15) (P < 0.001). (C) Electromyography of limb muscles of mice with Smn deletion. Fibrillations were noted in both ChAT-Cre Smn-deletion (four of five mice) and Nestin-Cre + ChAT-Cre Smn-deletion mice (six of seven mice) but are absent in Nestin-Cre Smn-deletion mice (zero of six) and control mice (zero of five) (P < 0.05). (D) CMAP amplitudes at P12 in mice with Smn-restoration. CMAP amplitudes at P12 in mice with Nestin-Cre + ChAT-Cre Smn-restoration (n = 9, 23.2 ± 3.3) were similar to control mice (n = 13, 24.8 ± 1.2 mV). Conversely, compared with control mice, CMAP amplitudes were reduced in mice with Nestin-Cre-restoration of Smn (n = 8, 17.3 ± 1.1 mV) and in SMA mice with no Cre (i.e. no Smn-restoration) (n = 12, 17.1 ± 0.8 mV) (P < 0.05). CMAP amplitudes were reduced though does not reach statistical significance in SMA mice with ChAT-Cre-restoration of Smn (n = 14, 18.8 ± 1.2 mV). (E) MUNE at P12 in mice with Smn-restoration. MUNE at P12 in mice with ChAT-Cre Smn-restoration (n = 14, 187 ± 16) and Nestin-Cre + ChAT-Cre Smn-restoration (n = 9, 201 ± 22) are similar to controls (n = 13, 196 ± 16, P = 0.962), but MUNE are reduced in mice with Nestin-Cre-restoration of Smn (n = 8, 118 ± 16) and SMA mice with no Cre (i.e. no Smn-restoration) (119 ± 9) (P < 0.05). Compared with mice with Nestin-Cre-restoration of Smn, MUNE at P12 in mice with ChAT-Cre Smn-restoration (P = 0.042) and Nestin-Cre + ChAT-Cre Smn-restoration (P = 0.024) are increased. Similarly, compared with SMA mice with no Cre, MUNE in mice with ChAT-Cre Smn-restoration (P = 0.031) and Nestin-Cre + ChAT-Cre Smn-restoration (P = 0.017) are increased. (*P ≤ 0.05, ***P ≤ 0.001).
CMAP and MUNE were also performed at P12 in SMA mice with targeted Smn-restoration. The earlier time point was chosen to allow comparison to control SMA mice that only live 14 days. Upon replacement of Smn we found improved CMAP amplitudes and MUNE values with Nestin-Cre + ChAT-Cre and ChAT-Cre replacement of Smn but not with Nestin-Cre (Fig. 7D and E). These findings are consistent with electrophysiological measurements in mice following the deletion of Smn. Thus CMAP and MUNE demonstrate that elimination of Smn reduces the functional output of the motor unit while replacement of Smn restores the functional output.
Discussion
Spinal muscular atrophy (SMA) is caused by homozygous loss or mutation of SMN1, retention of SMN2 and reduction of SMN levels (3,55,56). The SMN2 gene differs from SMN1 essentially by a single nucleotide in exon 7 that results in the majority of the transcript from SMN2 lacking exon7, and the SMNΔ7 mRNA transcript produces a protein that does not oligomerize efficiently and gets rapidly degraded (6,7,57,58). SMN2 copy number varies in the population and there is an inverse correlation of SMN2 copy number with phenotypic severity (4,59). Furthermore the 859G>C variant of SMN2 has been shown to increase the incorporation of exon7 and result in milder SMA in patients (60,61). While it is clear that SMN reduction causes SMA and that the amount of SMN is critical in determining phenotypic severity, SMN levels are predicted to be decreased in all tissues. It remains unclear where high levels of SMN (above that produced by two copies of SMN2) are required, and whether this varies in different severities of SMA.
The current study is the first to use Cre recombinase to both delete and replace Smn using the same Cre driver to study the effects of reduced or restored levels of SMN in glia, motor neurons, autonomic neurons and other neurons in the motor circuit. Furthermore, we have performed a complete characterization of the expression patterns of the Cre drivers used to understand the specificity of drivers, or lack thereof, to allow full interpretation of the effects on weight, survival and motor unit function in the SMNΔ7 mouse.
Prior studies used behavioral analysis, physiological NMJ patch clamp recordings of single synapses ex vivo or morphological assessment of NMJs and motor neurons to indirectly draw conclusions about motor unit function (30,31). In contrast, the present study uses electrophysiological techniques (CMAP, MUNE and EMG) to assess motor unit number and function in vivo, similar to techniques that have shown abnormalities in clinical studies of patients with SMA (62,63). NMJ patch clamp recordings in the SMNΔ7 mouse have revealed transmission abnormalities of reduced evoked endplate current amplitudes (64). These patch clamp NMJ abnormalities are corrected with both systemic SMN restoration and with SMN restoration in the motor neuron but not with restoration in muscle (14,27,28). Though motor deficits have been noted in mice with selectively reduced levels of SMN in motor neurons, NMJ patch clamp recordings have not been performed (26). For obvious reasons these measures have not been performed in human SMA. However abnormalities on CMAP, MUNE and EMG are key features in patients with SMA (62,63). Prior to the availability of molecular diagnostic testing, EMG was an important diagnostic tool as the presence of fibrillations is a marker of denervation and a typical feature of type 1 SMA, and today it still has utility in atypical cases (53,65). CMAP and MUNE are particularly useful to quantify motor neuron or axon dysfunction, and these measures have been shown to correlate with disease severity and function in several cohorts of patients with SMA (62,63,66–68).
We have developed CMAP, MUNE and EMG methods for recording even in neonatal mice (53). We used CMAP, MUNE and EMG in the present studies due to the clinical relevance and for the key ability to analyze the functional output and connectivity of the motor neuron in vivo. We have previously shown that these measures are responsive to restoration of SMN protein via intracerebroventricular injection of antisense oligonucleotide to ISS-N1 and scAAV9-SMN in the SMNΔ7 mouse and scAAV9-SMN in a pig model of SMA (53,69). Importantly, our results reveal that decreasing and restoring the level of SMN in the motor neuron is sufficient to both cause and prevent the losses of motor unit function that are characteristic of human SMA. Our results support that SMN level in the motor neuron is central to the pathogenesis of motor neuron loss and that restoration of SMN levels in the motor neuron will be critical for therapeutic implementation in patients with SMA. While expression of high levels of SMN in just motor neurons does not rescue survival of the mouse it does rescue the function of the motor neuron. In human SMA improving the function of the motor neuron is likely to be critical in effective therapy. The mouse has other affected organs such as the heart upon low SMN levels (70–72), however additional organ involvement does not appear to be a prevalent feature of SMA in man (73).
The clinical and pathological features of human SMA are remarkably restricted to the neuromuscular system (2,53,74,75). Yet, some studies have indicated the potential importance of SMN in the periphery (76,77). In mouse models of SMA, various non-motor features have been noted, namely distal limb necrosis, endocrine abnormalities, dysautonomia, cardiac defects and gastrointestinal abnormalities (19,70,78,79). It has been suggested that the autonomic nervous system may play a role in some of these phenotypic features (19,25,80). For instance, do alterations in the vascular system and the heart (70–72) in mice relate to the autonomic nervous system? Though some similar atypical features in patients with SMA have very rarely been presented in the literature, these single case reports are restricted to most severe form of SMA, type 0 and 1a, with onset at or before birth (53,81–85). Thus whether these reports relate to the more typical type 1 SMA situation remains questionable.
In a recent study by Hua et al. (86), the importance of peripheral SMN restoration was proposed. In this study, a different model, the Taiwanese SMA mouse, was used which has a deletion of mouse Smn exon7 and either 2 copies of SMN2 (for a severe phenotype) or 4 copies of SMN2 (for a milder phenotype) (79,86). The mouse locus in this model can still produce SMN transcript lacking exon7. Splice-correcting ASO was administered via subcutaneous injection in severe Taiwanese mice dramatically extending survival similar to prior studies (21,86). Importantly in the neonatal period, ASO can cross the blood brain barrier and exert partial correction of splicing to SMN2. Therefore to limit the induction of exon 7 the authors included a blocking ASO that was administered to the CNS via intracerebral ventricular injection (86). Yet, despite delivery of the blocking ASO some increase in SMN exon7 inclusion occurred in the spinal cord. Indeed we have previously shown in the SMNΔ7 mouse model that even a small increase in leaky expression from the human skeletal actin muscle promoter HSA-SMN transgene elicited a major impact on phenotype (13). Therefore it is important to consider how the phenotypes of the SMNΔ7 and the severe Taiwanese mice (with two copies of SMN2) compare. While the survival of the SMNΔ7 and severe Taiwanese models are similar (2 weeks), the variability between the two models regarding the involvement of the motor system compared with other organ systems is significant (19,33,78,79,87). Indeed Hua et al. reported that the longissimus capitis muscle in the severe Taiwanese did not show any completely denervated NMJs (86). In contrast, the same muscle in the SMNΔ7 mouse shows partial-to-complete denervation of 80% of synapses and complete denervation in 28% of synapses (88). Given the weaker denervation phenotype in the severe Taiwanese mice a small increase of SMN in the CNS may be sufficient to overcome the CNS requirement. Furthermore, it is difficult to interpret the survival outcome of the Taiwanese mouse given the severity of dysfunction in several organ systems (19,33,78,79,87). Iascone et al. have commented that using denervation as a quantitative readout for SMNΔ7 mice and comparing it to post-mortem samples from type 1 patients on a muscle-by-muscle basis they have shown a remarkable overlap between the mouse and human neuromuscular phenotype (73). This indicates that the SMNΔ7 mouse model recapitulates the neuromuscular phenotype of patients. The SMNΔ7 SMA mouse does have defects in the heart (70–72) yet this defect does not appear to match human patient studies (73).
It is critical to consider whether key phenotypic features that are relevant to human SMA, such as motor neuron loss and denervation, are rescued when assessing where high levels of SMN are required. Our study shows that the effects of SMN level on weight and survival were decoupled from the effects on electrophysiological motor unit function. Despite significant impact on motor unit function following deletion or restoration of SMN levels in the motor neuron, the reduction or improvement of survival and weight was not dramatic unless SMN was reduced or restored in motor neurons plus neurons and glia (with ChAT-Cre + Nestin-Cre). Though both weight and survival are well-established and important preclinical endpoints and readouts of sufficiency of SMN restoration, these parameters do not appear to be directly related to the functional status of the motor unit. First, we have found that replacement of SMN in just motor neurons does not correct all phenotypic features in the SMNΔ7 mouse, but it does completely correct the electrophysiological output of the motor neuron. Conversely, upon decreasing SMN in motor neurons solely there is no impact on weight or survival, but there is a significant decrease in CMAP and MUNE. Second, expression in neurons and glia (with Nestin-Cre alone) has a considerable impact on survival but less so on motor unit function as indicated by the CMAP and MUNE read out. When SMN was reduced to SMA levels with Nestin-Cre, which is expressed in all (most) neurons and glia but not efficiently in motor neurons, no fibrillations were detected on EMG whereas with ChAT-Cre clear fibrillations are observed indicating denervation. The combination of ChAT-Cre + Nestin-Cre results in animals that either had the most severe phenotype or the most complete rescue. In fact, upon rescue some animals showed a normal life span indicating that the tissues where SMN was expressed at high levels was sufficient for normal function. In this condition (ChAT-Cre + Nestin-Cre) SMN is expressed in the autonomic nervous system. We and others have previously shown that the SMNΔ7 mice have a marked cardiac phenotype (70–72), which is partially corrected by delivery of SMN postnatally with scAAV9-SMN. This results in transduction of the autonomic neurons that innervate the heart. In the current study Nestin-Cre is likely to result in the restoration or depletion of SMN from most neurons including the autonomic neurons. The cardiac phenotype is important in the mice and certainly one possible reason for improved survival when using ChAT-Cre + Nestin-Cre for rescue of SMA mice. However, as indicated by Iascone et al. (73), the cardiac defects are not a common feature of SMA in patients and as such the mouse model does not completely mimic the human situation. Indeed they concluded that multi-organ dysfunction, including cardiac and vascular defects, is not a general feature of SMA. In addition the motor neuron circuit has been found to be defective in SMNΔ7 SMA mice (89) and this will also be corrected by the Nestin-Cre driver. Recently Rindt et al. (90) reported that expression of high levels of SMN in astrocytes benefited SMA mice. The Nestin-Cre driver is very well expressed in astrocytes (40). Thus the changes in the mouse with the ChAT-Cre + Nestin-Cre drivers can result from expression of SMN in glia and all neurons as well as motor neurons (40).
While we have shown some patchy expression of Nestin-Cre in the periphery, we feel this limited expression is unlikely to be enough to correct the function of these tissues. Instead, others have shown in human SMA-derived iPS cells differentiated into astrocytes that Glial Fibrillary Protein (GFAP) is increased and astrocyte morphology is altered (91). It is quite likely that the extent of glial involvement in the SMA phenotype is still not fully understood. However it is possible that a secreted protein from one of these tissues could also impact the phenotype.
SMN has a well-established canonical function in the assembly of RNP complexes. SMN is required for the assembly of Sm proteins onto snRNA and assembly of the Lsm10-11-Sm protein ring onto U7 snRNA. SMN has also been suggested to have a role in the assembly of other RNA complexes such as those that function in mRNA transport (92,93). Since SMN is known to function in splicing, it is quite possible that the alteration of splicing of particular genes could be affected by reduction of SMN in the motor neuron. While abnormal mRNA spliced forms have been reported it is clear that the majority of genes are spliced normally in SMA mice (94,95). Given the other organ involvement in SMA mice in addition to the replication of the neuromuscular phenotype, one wonders if the genes affected by SMN deficiency have the same sensitivity to depletion of SMN due to variance in the intron sequence and structure between species (96,97). This has been demonstrated in the case of resistin (98). Thus in the human some introns may be more sensitive to SMN deficiency than others. It is possible that in the mouse SMN must be reduced to a greater extent to uncover the motor phenotype, resulting in the dysfunction of additional genes that give rise to other organ involvement. If this is the case in humans remains to be determined. The benefit from using both ChAT-Cre and Nestin-Cre together could be due to correction of these other genes with a less pronounced effect on a gene that is sensitive to SMA deficiency. The newly developed pig model of SMA will be important in discerning intron sensitivity upon reduced SMN levels among species (69).
Materials and Methods
Mouse breeding
This study was carried out in strict accordance with the recommendations of the Institutional Animal Care and Use Committee and University Laboratory Animal Resources at The Ohio State University under protocol number 2008A-0089. Floxed Smn alleles used in this study are listed in Table 1 and described here. Mice containing the floxed Smn exon 7: SmnF7, Smn1tm1Jme/J(JAX #006138) or the conditional inversion Smn allele: SmnINV, Smn1tm3(SMN2/Smn1)Mrph/J (JAX007249) were crossed onto the SMNΔ7 SMA mouse model: (FVB.Cg-Tg(SMN2)89Ahmb Smn1tm1MsdTg(SMN2*delta7)4299Ahmb/J (JAX #005025). Cre drivers used are listed in Table 2. Cre drivers used include: Nestin-Cre, Tg(Nes-Cre)1Kln/J (JAX #003771) (40), ChAT-Cre, ChATtm1(cre)Low1/J (JAX #006410) (Lowell BB; Olson D; Yu J. 2006 direct submission to JAX); SYN1-iCre, Tg(SYN1-icre/mRFP1)9934Rdav/J (JAX #012687)(Davis R. 2009 direct submission to JAX), rSyn1-Cre, Tg(Syn1-cre)671Jxm/J (JAX #003966) (44) and Sox2-Cre, Tg(Sox2-Cre)1Amc/J (JAX #008454) (39). The HB9:GFP,Tg(Hlxb9-GFP)1Tmj/J (JAX#005029), line was used to identify motor neurons and the reporter line tdTomato,Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (JAX #007914), was used to identify recombinant tissues. All Cre drivers were also crossed onto the SMNΔ7 SMA mouse model background. To generate experimental animals the Cre drivers in the SMNΔ7 SMA background were crossed to either the SmnF7 line or the SmnINV line. After Cre activation the SmnF7 allele is referred to as SmnD7 (for deletion of exon 7) and the SmnINV line is referred to as SmnRe (for reversion of exon 7). All experimental animals contain the SmnKO allele in addition to the SmnF7 or SmnINV alleles (i.e. Cre driver; SmnD7/KO;SMN2+/+; SMNΔ7+/+ or Cre driver; SmnRe/KO; SMN2+/+; SMNΔ7+/+). To eliminate the possibility of germline recombination, known to occur in the Nestin-Cre line, only experimental offspring contained both floxed alleles and Cre drivers (99). Floxed allele and Cre driver breeder mice were maintained separately. Controls used in this study contained the Cre driver and a wild-type Smn allele (i.e. Cre driver; SmnD7/WT; SMN2+/+; SMNΔ7+/+ or Cre driver; SmnRe/WT; SMN2+/+; SMNΔ7+/+).
Genotyping
Neonatal mice were tattooed and tail snips were obtained for PCR as previously described (19). The primers used to detect Cre drivers and Smn alleles are listed in Table 3.
Phenotypic assessment of mice
Mice were weighed daily starting at P1. At 21 days after birth pups were weaned and weighed weekly. Observation of weakness and necrosis was made every day, and then every week during weighing. Mice were sacrificed according to our IACUC approved protocol before they lost 30% of their maximum body weight. An equal number of males and females were studied in each genotypic grouping. All investigators were blinded to genotype during phenotypic assessment. The number of animals required for analysis was determined as described previously (100).
Immunohistochemistry
Neonatal mice were transcardially perfused with PBS followed by 4% paraformaldehyde at P7. Tissues were cryopreserved in 30% sucrose overnight, embedded in Tissue-Tek OCT (Fisher Scientific) and flash frozen in liquid nitrogen-cooled isopentane. Cryostat section (14 µm) were blocked with 4% goat serum in PBS and stained with antibodies including chicken anti-RFP (1:2500, Millipore AB3528), rabbit anti-GFP (1:1000, Molecular Probes A11122), goat anti-ChAT (1:50, Millipore, AB144P) or mouse anti-NeuN 1:50, Millipore, MAB377) for 2 h, followed by 1 h of secondary antibody incubation with Alexa-488 (1:1000, Molecular Probes A11008 and Alexa-594 (1:1000, Molecular Probes A11042) and mounted in Flouromount-G (Southern Biotech). Confocal imaging was performed using Leica DM IRE2 with Leica TCS SL point scanning laser confocal system with photomultiplier tube detection. Image acquisition, overlays and scale bars were produced with Leica Confocal Software v2.61 and subsequential image processing was performed with Adobe Photoshop CS2.
For the whole-body sagittal section, P2 mouse pups, following perfusion with PBS and 4% paraformaldehyde, were injected with molten 2% agarose in their body cavities. The perfused pups were then subjected to a gradient of 10, 20 and 30% sucrose prior to freezing in liquid nitrogen-cooled isopentane and cryosectioning at 42 µm. Primary antibody was incubated overnight and rest of the immunostaining is as described above. The whole-body sections were imaged with a Leica MZ-16FA stereomicroscope with PhotoFluor LM-75 light source and Leica DFC-300 Fx camera. All scale bars (except whole-body) are in micrometers (µm).
Muscle fiber size analysis
Muscle fiber size was determined as previously described in references (13,51). Briefly, gastrocnemius muscle was cryosectioned at 14 µm and stained with H&E. Muscle fibers were visualized with Nikon 1600 microscope and measured with SPOT analysis software. The frequency of fiber size area was binned with Microsoft Excel 2007. Approximately 500 muscle fibers per mouse were measured for a total of 1500 muscle fibers from n = 3 mice per genotypic group. An equal number of males and females were studied in each genotypic grouping. All investigators were blinded to genotype during phenotypic assessment. The number of animals required for analysis was determined as described previously (100).
Laser capture microdissection
Lumbar spinal cord tissue was isolated at P10-P17 and flash frozen in liquid nitrogen-cooled isopentane. Spinal cords were cryosectioned (14 μm) and adhered to PEN membrane slides (Zeiss). Tissue sections were dried at room temp and then fixed in methanol for 30 s, Nissl stained in 1% cresyl violate acetate for 1 min, washed in methanol 3 times and air-dried. Sections were stored at 4°. Laser capture microdissection (LCM) was performed with the Palm Microbeam (Carl Zeiss MicroImaging) under 10× magnification. Motor neurons were identified by size and position in the ventral horn of the spinal cord. For DNA percent recombination by ddPCR: Approximately 200 motor neurons were collected per sample and processed as described previously (38). Briefly, the LCM sample was collected directly into 18 µl of cell-lysis solution (20 mm Tris–HCl pH 8, 0.02% Tween, 1 mm EDTA) in siliconized tubes (101) and frozen on dry ice. Two microliters of 2 mg/ml proteinase K solution (Invitrogen) were added to the cell-lysis solution; followed by incubation at 55°C for 2 h and inactivation at 90°C for 10 min. For ELISA: Approximately 500–600 motor neurons were collected per sample into ELISA lysis buffer (PharmOptima, Portage MI) and frozen on dry ice.
Droplet digital PCR
Each digested LCM motor neuron sample was divided in half and run in duplicate in ddPCR for an approximate 100 motor neurons per reaction (QX200, BioRad). ddPCR reactions were prepared and amplified according to the manufacturer's recommendations (BioRad). LoBind tips, plates and tubes (Eppendorf) were used to minimize DNA loss. Primers and probe used to detect the SmnF7 allele: FP 5′AAGATGACTTTGAACTCCGGGTCCT, RP 5′TGTGAGTGAACAATTCAAGCCC, probe FAM-CTGCCCATGCATCACCAAGCTTGGCAT-MGB. Primers and probe used to detect total Smn DNA are located in intron 1: FP 5′CTGTGTGACTGTGAGGGGATGTG, RP5′CCTGTGAACATCTT CATCCTGACCTAA, probe VIC-AGGCTGGCTGAAGCAAGGCAACCAGATA-MGB. The percent of motor neurons displaying recombination was normalized to the total number of Smn alleles present as detected by the Smn intron 1 primers and probe.
ELISA on LCM collected tissue
SMN protein was measured at PharmOptima (Portage, MI) using the company's proprietary electrochemiluminescence (ECL) immunoassay based on Meso Scale Discovery technology. The assay is a quantitative sandwich immunoassay, where a mouse monoclonal antibody (2B1, Liu and Dreyfuss 1996) functions as the capture antibody and a rabbit polyclonal anti-SMN antibody (Protein Tech, Cat. No. 11708-1-AP) labeled with a SULFO-TAG™ is used for detection. SMN levels are determined from a standard curve using recombinant SMN protein (Enzo Life Sciences, Cat. No. ADI-NBP-201-050) as calibrator. The dynamic range of the assay is 10 pg/ml (lower limit of quantitation) to 20 000 pg/ml (upper limit of quantitation). Assay plates were read using a Meso Scale 6000 sector imager.
Samples from LCM were diluted with lysis buffer to a total volume of 40 µl. Twenty-five microliters of the lysate was used in the SMN ECL immunoassay. SMN levels were then normalized with the number of motor neurons collected by LCM per sample.
Western blot analysis
Western blots were performed as previously described (13,19,25). Briefly, 50 µg of protein were loaded per lane. SMN was detected with mouse anti-SMN (1:1000, BD Biosciences, 610647), tubulin was detected with mouse anti-tubulin (1:10 000 Sigma, T8203). Mouse-anti HRP (1:10 000 Jackson Immuno Research) and the ECL system was used to visualize the SMN protein as described by the manufacturer (GE Healthcare Life Sciences). Blots where scanned and quantified as described (http://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/) and the area under each peak determined with ImageJ software. Quantification of tubulin was performed with LI-COR 800 secondary antibody and the LI-COR Quantitative Fluorescent Imaging Systems as previously described (102). Statistical analysis was performed with SigmaPlot.
Gem counts
SMN was detected with mouse anti-SMN (1:500, BD Biosciences, 610647) in 14 µm spinal cord cryo sections from P10 SMA, control, Nestin-Cre deletion and Nestin-Cre + ChAT-Cre deletion mice (n = 3 for each genotype). The nucleus was stained with DAPI and mounted in ProLong Gold Antifade Mountant (Life Technologies). Motor neurons were identified by their morphology and location in the spinal cord. The number of gems was counted in100 nuclei for each sample under 60× magnification using a Nikon E800 microscope.
Electrophysiological studies of sciatic CMAP and MUNE
Electrophysiological techniques were performed as previously described (53). Briefly, electromyography (EMG), compound muscle action potential amplitude (CMAP) and motor unit number estimates (MUNE) were recorded from sciatic-innervated hindlimb muscles at P21 in mice with deletion of Smn exon 7 with Nestin-Cre + ChAT-Cre drivers and compared with control mice. Similarly CMAP and MUNE were performed at P12 in mice with restoration of Smn exon 7 with Nestin-Cre + ChAT-Cre drivers and compared with SMNΔ7 SMA mice and control mice. An equal number of males and females were studied in each genotypic grouping. All investigators were blinded to genotype during phenotypic assessment. The number of animals required for analysis was determined as described previously (100).
Statistical analyses
Statistical analyses were performed as previously described for MUNE and CMAP (53). Weight curve analysis was completed with Statmod (103,104). Kaplan–Meier survival curves were performed with SigmaPlot v12 (Systat Software Inc., CA, USA) and significance was determined with the log-rank test. Morphometric measurements of muscle fiber size were performed using SPOT Advanced (v3.5.9) software (Diagnostic Instruments, Inc., MI, USA). Muscle fiber size distribution was analyzed with SigmaPlot v12.0 (Systat Software Inc., CA, USA) testing the median fiber size with Mann–Whitney Rank Sum Test and Shapiro–Wilk Normality Test (13). All error bars are represented as SEM.
Supplementary Material
Funding
Confocal images were taken at The Ohio State Neuroscience Center Core, The Ohio State University, supported by the grant P30-NS045758. This work was supported by National Institutes of Health (R01 NS038650 to A.H.M.B., R01HD060586 to A.H.M.B.) and (5K12HD001097-17 to W.D.A.); and the Marshall Heritage Foundation.
Supplementary Material
Acknowledgements
We wish to thank Kaitlyn Corlett and Xiaohui Li for genotyping assistance. We thank Dr Jill Rafael-Fortney for the use of microscope and SPOT analysis software, and Dr Christine Beattie for the use of the Leica stereomicroscope. We thank Paula Monsma, Core Manager for help with confocal imaging.
Conflict of Interest statement. None declared.
References
- 1.Roberts D.F., Chavez J., Court S.D. (1970) The genetic component in child mortality. Arch. Dis. Child, 45, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford T.O., Pardo C.A. (1996) The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis., 3, 97–110. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- 4.Burghes A.H. (1997) When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet., 61, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. (1998) SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet., 19, 63–66. [DOI] [PubMed] [Google Scholar]
- 6.Lorson C.L., Androphy E.J. (2000) An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet., 9, 259–265. [DOI] [PubMed] [Google Scholar]
- 7.Burnett B.G., Munoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. (2009) Regulation of SMN protein stability. Mol. Cell Biol., 29, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burghes A.H., Beattie C.E. (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci, 10, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. (2008) Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet., 16, 2552–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGovern V.L., Gavrilina T.O., Beattie C.E., Burghes A.H. (2008) Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet., 17, 2900–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. (2008) Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet., 17, 949–962. [DOI] [PubMed] [Google Scholar]
- 12.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W. et al. (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet., 9, 333–339. [DOI] [PubMed] [Google Scholar]
- 13.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. (2008) Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet., 17, 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H. et al. (2010) Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol., 28, 271–274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Duque S., Joussemet B., Riviere C., Marais T., Dubreil L., Douar A.M., Fyfe J., Moullier P., Colle M.A., Barkats M. (2009) Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther., 17, 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer K., Ferraiuolo L., Schmelzer L., Braun L., McGovern V., Likhite S., Michels O., Govoni A., Fitzgerald J., Morales P. et al. (2013) Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA—a dose response study in mice and nonhuman primates. Mol. Ther., 12, 2148–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. (2007) Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol., 5, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G. et al. (2011) Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med., 3, 72ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K.D., Kaspar B.K., Wilton S.D., Burghes A.H. (2012) A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet., 21, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naryshkin N.A., Weetall M., Dakka A., Narasimhan J., Zhao X., Feng Z., Ling K.K.Y., Karp G.M., Qi H., Woll M.G. et al. (2014) SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science, 345, 688–693. [DOI] [PubMed] [Google Scholar]
- 21.Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. (2011) Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature, 478, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valori C.F., Ning K., Wyles M., Mead R.J., Grierson A.J., Shaw P.J., Azzouz M. (2010) Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med., 2, 35ra42. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez E., Marais T., Chatauret N., Benkhelifa-Ziyyat S., Duque S., Ravassard P., Carcenac R., Astord S., de Moura A.P., Voit T. et al. (2011) Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet., 20, 681–693. [DOI] [PubMed] [Google Scholar]
- 24.Lutz C.M., Kariya S., Patruni S., Osborne M.A., Liu D., Henderson C.E., Li D.K., Pellizzoni L., Rojas J., Valenzuela D.M. et al. (2011) Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest., 121, 3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le T.T., McGovern V.L., Alwine I.E., Wang X., Massoni-Laporte A., Rich M.M., Burghes A.H. (2011) Temporal requirement for high SMN expression in SMA mice. Hum. Mol. Genet., 20, 3578–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park G.H., Maeno-Hikichi Y., Awano T., Landmesser L.T., Monani U.R. (2010) Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J. Neurosci., 30, 12005–12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez T.L., Kong L., Wang X., Osborne M.A., Crowder M.E., Van Meerbeke J.P., Xu X., Davis C., Wooley J., Goldhamer D.J. et al. (2012) Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J. Neurosci., 32, 8703–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paez-Colasante X., Seaberg B., Martinez T.L., Kong L., Sumner C.J., Rimer M. (2013) Improvement of neuromuscular synaptic phenotypes without enhanced survival and motor function in severe spinal muscular atrophy mice selectively rescued in motor neurons. PLoS One, 8, e75866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gogliotti R.G., Quinlan K.A., Barlow C.B., Heier C.R., Heckman C.J., Didonato C.J. (2012) Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J. Neurosci., 32, 3818–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee A.J., Awano T., Park G.H., Monani U.R. (2012) Limited phenotypic effects of selectively augmenting the SMN protein in the neurons of a mouse model of severe spinal muscular atrophy. PLoS One, 7, e46353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor A.S., Glascock J.J., Rose F.F., Lutz C., Lorson C.L. (2013) Restoration of SMN to Emx-1 expressing cortical neurons is not sufficient to provide benefit to a severe mouse model of Spinal Muscular Atrophy. Transgenic Res., 22, 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frugier T., Tiziano F.D., Cifuentes-Diaz C., Miniou P., Roblot N., Dierich A., Le Meur M., Melki J. (2000) Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet., 9, 849–858. [DOI] [PubMed] [Google Scholar]
- 33.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. (2005) SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet., 14, 845–857. [DOI] [PubMed] [Google Scholar]
- 34.Cifuentes-Diaz C., Frugier T., Tiziano F.D., Lacene E., Roblot N., Joshi V., Moreau M.H., Melki J. (2001) Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J. Cell Biol., 152, 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitte J.M., Davoult B., Roblot N., Mayer M., Joshi V., Courageot S., Tronche F., Vadrot J., Moreau M.H., Kemeny F. et al. (2004) Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol., 165, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberdoerffer P., Otipoby K.L., Maruyama M., Rajewsky K. (2003) Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res., 31, e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. (1997) Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA, 94, 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer C.C., McGovern V.L., Murray J.D., Janssen P.M.L., Burghes A.H.M.. (2015) Low levels of Survival Motor Neuron protein is sufficient for normal muscle function in the SMNΔ7 mouse model of SMA. Hum. Mol. Genet., in press. doi: 10.1093/hmg/ddv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi S., Lewis P., Pevny L., McMahon A.P. (2002) Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev., 119 Suppl 1, S97–S101. [DOI] [PubMed] [Google Scholar]
- 40.Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schutz G. (1999) Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet., 23, 99–103. [DOI] [PubMed] [Google Scholar]
- 41.Tallini Y.N., Shui B., Greene K.S., Deng K.Y., Doran R., Fisher P.J., Zipfel W., Kotlikoff M.I. (2006) BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol. Genomics, 27, 391–397. [DOI] [PubMed] [Google Scholar]
- 42.Ivanova E., Hwang G.S., Pan Z.H. (2010) Characterization of transgenic mouse lines expressing Cre recombinase in the retina. Neuroscience, 165, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel G., Greengard P., Südhof T.C. (1991) Characterization of tissue-specific transcription by the human synapsin I gene promoter. Proc. Natl. Acad. Sci. USA, 88, 3431–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., Parada L.F. (2001) Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes. Dev., 15, 859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., Kvitsiani D., Kvitsani D., Fu Y., Lu J. et al. (2011) A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron, 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katarova Z., Sekerková G., Prodan S., Mugnaini E., Szabó G. (2000) Domain-restricted expression of two glutamic acid decarboxylase genes in midgestation mouse embryos. J. Comp. Neurol., 424, 607–627. [DOI] [PubMed] [Google Scholar]
- 47.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci., 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arber S., Han B., Mendelsohn M., Smith M., Jessell T.M., Sockanathan S. (1999) Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron, 23, 659–674. [DOI] [PubMed] [Google Scholar]
- 49.Hinckley C.A., Hartley R., Wu L., Todd A., Ziskind-Conhaim L. (2005) Locomotor-like rhythms in a genetically distinct cluster of interneurons in the mammalian spinal cord. J. Neurophysiol., 93, 1439–1449. [DOI] [PubMed] [Google Scholar]
- 50.Shneider N.A., Brown M.N., Smith C.A., Pickel J., Alvarez F.J. (2009) Gamma motor neurons express distinct genetic markers at birth and require muscle spindle-derived GDNF for postnatal survival. Neural. Dev., 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer C.C., McGovern V.L., Wise D.O., Glass D.J., Burghes A.H. (2014) Deletion of atrophy enhancing genes fails to ameliorate the phenotype in a mouse model of spinal muscular atrophy. Neuromuscul. Disord., 24, 436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tapia J.C., Wylie J.D., Kasthuri N., Hayworth K.J., Schalek R., Berger D.R., Guatimosim C., Seung H.S., Lichtman J.W. (2012) Pervasive synaptic branch removal in the mammalian neuromuscular system at birth. Neuron, 74, 816–829. [DOI] [PubMed] [Google Scholar]
- 53.Arnold W.D., Porensky P.N., McGovern V.L., Iyer C.C., Duque S., Li X., Meyer K., Schmelzer L., Kaspar B.K., Kolb S.J. et al. (2014) Electrophysiological biomarkers in spinal muscular atrophy: preclinical proof of concept. Ann. Clin. Transl. Neurol., 1, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willmott A.D., White C., Dukelow S.P. (2012) Fibrillation potential onset in peripheral nerve injury. Muscle Nerve, 46, 332–340. [DOI] [PubMed] [Google Scholar]
- 55.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. (1997) Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet., 16, 265–269. [DOI] [PubMed] [Google Scholar]
- 56.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. (1997) The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet., 6, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 57.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA, 96, 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet., 8, 1177–1183. [DOI] [PubMed] [Google Scholar]
- 59.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. (1997) Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet., 60, 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prior T.W., Krainer A.R., Hua Y., Swoboda K.J., Snyder P.C., Bridgeman S.J., Burghes A.H., Kissel J.T. (2009) A positive modifier of spinal muscular atrophy in the SMN2 gene. Am. J. Hum. Genet., 85, 408–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernal S., Alias L., Barcelo M.J., Also-Rallo E., Martinez-Hernandez R., Gamez J., Guillen-Navarro E., Rosell J., Hernando I., Rodriguez-Alvarez F.J. et al. (2010) The c.859G>C variant in the SMN2 gene is associated with types II and III SMA and originates from a common ancestor. J. Med. Genet., 47, 640–642. [DOI] [PubMed] [Google Scholar]
- 62.Swoboda K.J., Prior T.W., Scott C.B., McNaught T.P., Wride M.C., Reyna S.P., Bromberg M.B. (2005) Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann. Neurol., 57, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hausmanowa-Petrusewicz I., Karwanska A. (1986) Electromyographic findings in different forms of infantile and juvenile proximal spinal muscular atrophy. Muscle Nerve, 9, 37–46. [DOI] [PubMed] [Google Scholar]
- 64.Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marce M., Griffin J.W., Rich M.M., Sumner C.J. (2009) Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci., 29, 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arnold W.D., Burghes A.H. (2013) Spinal muscular atrophy: development and implementation of potential treatments. Ann. Neurol., 74, 348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewelt A., Krosschell K.J., Scott C., Sakonju A., Kissel J.T., Crawford T.O., Acsadi G., D'Anjou G., Elsheikh B., Reyna S.P. et al. (2010) Compound muscle action potential and motor function in children with spinal muscular atrophy. Muscle Nerve, 42, 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang P.B., Gooch C.L., McDermott M.P., Darras B.T., Finkel R.S., Yang M.L., Sproule D.M., Chung W.K., Kaufmann P., de Vivo D.C. (2014) The motor neuron response to SMN1 deficiency in spinal muscular atrophy. Muscle Nerve, 49, 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaufmann P., McDermott M.P., Darras B.T., Finkel R.S., Sproule D.M., Kang P.B., Oskoui M., Constantinescu A., Gooch C.L., Foley A.R. et al. (2012) Prospective cohort study of spinal muscular atrophy types 2 and 3. Neurology, 79, 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duque S.I., Arnold W.D., Odermatt P., Li X., Porensky P.N., Schmelzer L., Meyer K., Kolb S.J., Schümperli D., Kaspar B.K. et al. (2015) A large animal model of Spinal Muscular Atrophy and correction of phenotype. Ann. Neurol., 3, 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bevan A.K., Hutchinson K.R., Foust K.D., Braun L., McGovern V.L., Schmelzer L., Ward J.G., Petruska J.C., Lucchesi P.A., Burghes A.H. et al. (2010) Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet., 19, 3895–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heier C.R., Satta R., Lutz C., DiDonato C.J. (2010) Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet., 19, 3906–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shababi M., Habibi J., Yang H.T., Vale S.M., Sewell W.A., Lorson C.L. (2010) Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum. Mol. Genet., 19, 4059–4071. [DOI] [PubMed] [Google Scholar]
- 73.Iascone D.M., Henderson C.E., Lee J.C. (2015) Spinal muscular atrophy: from tissue specificity to therapeutic strategies. F1000Prime Rep., 7, 04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harding B.N., Kariya S., Monani U.R., Chung W.K., Benton M., Yum S.W., Tennekoon G., Finkel R.S. (2015) Spectrum of neuropathophysiology in spinal muscular atrophy type I. J. Neuropathol. Exp. Neurol., 74, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monani U.R. (2005) Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron, 48, 885–896. [DOI] [PubMed] [Google Scholar]
- 76.Hamilton G., Gillingwater T.H. (2013) Spinal muscular atrophy: going beyond the motor neuron. Trends. Mol. Med., 19, 40–50. [DOI] [PubMed] [Google Scholar]
- 77.Shababi M., Lorson C.L., Rudnik-Schoneborn S.S. (2014) Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J. Anat., 224, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schreml J., Riessland M., Paterno M., Garbes L., Rosbach K., Ackermann B., Kramer J., Somers E., Parson S.H., Heller R. et al. (2013) Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585. Eur. J. Hum. Genet., 21, 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. (2000) A mouse model for spinal muscular atrophy. Nat. Genet., 24, 66–70. [DOI] [PubMed] [Google Scholar]
- 80.Gombash S., Cowley C., Fitzgerald J., Iyer C.C., Fried D., McGovern V., Williams K., Burghes A., Christofi F., Gulbransen B.D., Foust K. (2015) SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice. Hum. Mol. Genet., 13, 3847–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rudnik-Schoneborn S., Vogelgesang S., Armbrust S., Graul-Neumann L., Fusch C., Zerres K. (2010) Digital necroses and vascular thrombosis in severe spinal muscular atrophy. Muscle Nerve, 42, 144–147. [DOI] [PubMed] [Google Scholar]
- 82.Rudnik-Schoneborn S., Heller R., Berg C., Betzler C., Grimm T., Eggermann T., Eggermann K., Wirth R., Wirth B., Zerres K. (2008) Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J. Med. Genet., 45, 635–638. [DOI] [PubMed] [Google Scholar]
- 83.Distefano G., Sciacca P., Parisi M.G., Parano E., Smilari P., Marletta M., Fiumara A. (1994) Heart involvement in progressive spinal muscular atrophy. A review of the literature and case histories in childhood. Pediatr. Med. Chir., 16, 125–128. [PubMed] [Google Scholar]
- 84.Bowerman M., Murray L.M., Beauvais A., Pinheiro B., Kothary R. (2012) A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology. Neuromuscul. Disord., 22, 263–276. [DOI] [PubMed] [Google Scholar]
- 85.Arai H., Tanabe Y., Hachiya Y., Otsuka E., Kumada S., Furushima W., Kohyama J., Yamashita S., Takanashi J., Kohno Y. (2005) Finger cold-induced vasodilatation, sympathetic skin response, and R-R interval variation in patients with progressive spinal muscular atrophy. J. Child Neurol., 20, 871–875. [DOI] [PubMed] [Google Scholar]
- 86.Hua Y., Liu Y.H., Sahashi K., Rigo F., Bennett C.F., Krainer A.R. (2015) Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev., 29, 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bevan A.K., Duque S., Foust K.D., Morales P.R., Braun L., Schmelzer L., Chan C.M., McCrate M., Chicoine L.G., Coley B.D. et al. (2011) Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther., 19, 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ling K.K., Gibbs R.M., Feng Z., Ko C.P. (2012) Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum. Mol. Genet., 21, 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mentis G.Z., Blivis D., Liu W., Drobac E., Crowder M.E., Kong L., Alvarez F.J., Sumner C.J., O'Donovan M.J. (2011) Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron, 69, 453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lorson C.L., Rindt H., Shababi M. (2010) Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum. Mol. Genet., 19, R111–R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGivern J.V., Patitucci T.N., Nord J.A., Barabas M.E., Stucky C.L., Ebert A.D. (2013) Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia, 61, 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li D.K., Tisdale S., Lotti F., Pellizzoni L. (2014) SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol., 32, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hubers L., Valderrama-Carvajal H., Laframboise J., Timbers J., Sanchez G., Cote J. (2011) HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet., 20, 553–579. [DOI] [PubMed] [Google Scholar]
- 94.Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. (2009) Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet., 5, e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Z., Pinto A.M., Wan L., Wang W., Berg M.G., Oliva I., Singh L.N., Dengler C., Wei Z., Dreyfuss G. (2013) Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA, 110, 19348–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang J., Lu Z.X., Tokheim C.J., Miller S.E., Xing Y. (2015) Species-specific exon loss in human transcriptomes. Mol. Biol. Evol., 32, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie J. (2014) Differential evolution of signal-responsive RNA elements and upstream factors that control alternative splicing. Cell Mol. Life Sci., 71, 4347–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghosh S., Singh A.K., Aruna B., Mukhopadhyay S., Ehtesham N.Z. (2003) The genomic organization of mouse resistin reveals major differences from the human resistin: functional implications. Gene, 305, 27–34. [DOI] [PubMed] [Google Scholar]
- 99.Zhang J., Dublin P., Griemsmann S., Klein A., Brehm R., Bedner P., Fleischmann B.K., Steinhauser C., Theis M. (2013) Germ-line recombination activity of the widely used hGFAP-Cre and nestin-Cre transgenes. PLoS One, 8, e82818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Butchbach M.E., Edwards J.D., Burghes A.H. (2007) Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol. Dis., 27, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sambrook J., Fritsch E.F., Maniatis T. (1987) Molecular Cloning. A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. E.1–E.2. [Google Scholar]
- 102.Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S.E., Nagy A. (2005) Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res., 33, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elso C.M., Roberts L.J., Smyth G.K., Thomson R.J., Baldwin T.M., Foote S.J., Handman E. (2004) Leishmaniasis host response loci (lmr1–3) modify disease severity through a Th1/Th2-independent pathway. Genes. Immun., 5, 93–100. [DOI] [PubMed] [Google Scholar]
- 104.Baldwin T., Sakthianandeswaren A., Curtis J.M., Kumar B., Smyth G.K., Foote S.J., Handman E. (2007) Wound healing response is a major contributor to the severity of cutaneous leishmaniasis in the ear model of infection. Parasite. Immunol., 29, 501–513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.