Abstract
Interpretation of biological mechanisms underlying genetic risk associations for prostate cancer is complicated by the relatively large number of risk variants (n = 100) and the thousands of surrogate SNPs in linkage disequilibrium. Here, we combined three distinct approaches: multiethnic fine-mapping, putative functional annotation (based upon epigenetic data and genome-encoded features), and expression quantitative trait loci (eQTL) analyses, in an attempt to reduce this complexity. We examined 67 risk regions using genotyping and imputation-based fine-mapping in populations of European (cases/controls: 8600/6946), African (cases/controls: 5327/5136), Japanese (cases/controls: 2563/4391) and Latino (cases/controls: 1034/1046) ancestry. Markers at 55 regions passed a region-specific significance threshold (P-value cutoff range: 3.9 × 10−4–5.6 × 10−3) and in 30 regions we identified markers that were more significantly associated with risk than the previously reported variants in the multiethnic sample. Novel secondary signals (P < 5.0 × 10−6) were also detected in two regions (rs13062436/3q21 and rs17181170/3p12). Among 666 variants in the 55 regions with P-values within one order of magnitude of the most-associated marker, 193 variants (29%) in 48 regions overlapped with epigenetic or other putative functional marks. In 11 of the 55 regions, cis-eQTLs were detected with nearby genes. For 12 of the 55 regions (22%), the most significant region-specific, prostate-cancer associated variant represented the strongest candidate functional variant based on our annotations; the number of regions increased to 20 (36%) and 27 (49%) when examining the 2 and 3 most significantly associated variants in each region, respectively. These results have prioritized subsets of candidate variants for downstream functional evaluation.
Introduction
Prostate cancer is the most common non-skin cancer and the second leading cause of cancer death among men in the USA. The risk of prostate cancer varies across racial/ethnic populations, with the incident rate in African Americans being 1.6 times that in European Americans, and 2.6 times that in Asian Americans (1). Genome-wide association studies (GWAS) and large-scale collaborative replication efforts have identified 100 prostate cancer risk variants (2–15) (referred to as index variants), mainly in populations of European or Asian ancestry. Whether the associations with these risk variants generalize and define the biologically relevant variation in other populations are important questions. In prior studies, examining previously identified risk variants in men of African ancestry (16,17), we have noted directionally consistent associations at the majority of risk loci (83%) which suggests that the underlying functional variant is common and shared across populations. Fine-mapping in these regions in men of African ancestry revealed markers that have greater statistical significance and larger effect sizes [odds ratios (ORs)] for 27 (out of 82) index variants in this population (17). Due to the varying linkage disequilibrium (LD) patterns and allele frequencies observed across racial/ethnic groups, studies in diverse populations, and most notably African-American populations, have been suggested to increase power for fine-mapping by reducing the number of proxies that are correlated with the underlying functional allele (18–21).
Since the vast majority of index variants (and their proxies) are located in regions outside of protein-coding exons, identifying biologically functional candidate variants and the genes they influence are substantial challenges in human genetics. It is now clear that GWAS trait-associated variants are enriched among regulatory elements (22–25). Recently, we and others have developed approaches to identify candidate functional variants by intersecting genetic information with epigenetic marks that characterize regulatory elements (26–30). Identifying the target gene of a regulatory element also poses a challenge since regulatory elements can act over great distances. Expression quantitative trait loci (eQTL) analysis has emerged as a powerful method to nominate candidate genes (31–33). Such approaches have led to the identification of putative functional variants and candidate genes for a number of prostate cancer risk regions, including 8q24 (34–36), 10q11/MSMB (37), 6q22/RFX6 (38) and 8p21/NKX3.1 (39).
In the present study, we combined multiethnic fine-mapping results with detailed tissue-specific functional annotation and eQTL data for prostate cancer. Specifically, we conducted genotyping and imputation-based fine-mapping of 67 regions (see Materials and Methods) in a large multiethnic sample comprised of 17 524 prostate cancer cases and 17 519 controls from populations of European (8600 cases and 6946 controls), African (5327 cases and 5136 controls), Japanese (2563 cases and 4391 controls) and Latino (1034 cases and 1046 controls) ancestry to further refine the complexity of prostate cancer-associated variants as well as elucidate novel risk variants (i.e. secondary signals) for this malignancy. We used epigenetic and gene expression information to functionally annotate the most-associated variants in an attempt to identify a subset of variants in each region to be prioritized for functional testing.
Results
Statistical fine-mapping
The 67 regions contained 69 index risk variants; 3p11-p12 and 4q22 each harbored 2 index SNPs (see below). In the analysis of 17 524 prostate cancer cases and 17 519 controls (Supplementary Material, Tables S1–S3; Supplementary Material, File S1), a high degree of directional consistency of the per-allele ORs was noted with the index signals in these populations, consistent with what we previously observed in many of these same samples/populations (16,17,40). Of the 69 risk alleles, 68 were available (frequency ≥0.01) in populations of European ancestry and all 68 alleles (100%) were positively associated with risk, with 50 (74%) nominally statistically significant (P < 0.05); whereas these proportions (positive OR versus nominally significant) were 90% (62/69) and 33% (23/69) in the African, 84% (54/64) and 41% (26/64) in the Japanese and 81% (55/68) and 25% (17/68) in the Latino ancestry populations, respectively. We observed significant effect heterogeneity across populations for six index SNPs (Phet < 9.1 × 10−4 and I2 > 80.0%; see Materials and Methods), two of which (rs2660753/3p12 and rs9600079/13q22) were directionally inconsistent, while the other four (rs12653946/5p15, rs1512268/8p21, rs7501939/17q12 and rs1859962/17q24) were directionally consistent but had large differences in estimated effect sizes across populations (Supplementary Material, Table S4).
Using a region-specific threshold of statistical significance (see Materials and Methods), we found 55 of 67 (82%) regions contained signals that were significantly associated with prostate cancer risk (Supplementary Material, Table S4). Among these 55 regions, the index SNP remained the most significantly associated marker at 10 regions, while a correlated variant was marginally more significantly associated (r2 ≥ 0.2 and <1 order of magnitude change in the P-value compared with the index SNP) at 15 regions (Supplementary Material, Table S4). The effect sizes (ORs) of the index SNP and the most-associated correlated variant in these 15 regions were similar in magnitude in both the multiethnic sample and the racial/ethnic population in which the discovery GWAS was conducted (referred to as the discovery GWAS population), with no statistically significant heterogeneity noted.
In 30 regions, combined data from multiple populations revealed variants that were more significantly associated with risk than the index variant, which we defined as a >1 order of magnitude change in the P-value (Table 1; Supplementary Material, Fig. S1). A complete list of these variants can be found in Supplementary Material, Table S4. The most significantly associated markers at three regions (rs13017478/2p21, rs58235267/2p15 and rs76925190/3q26) were weakly correlated with the index variant in each of these respective regions (r2 range, 0.15–0.18), but were still able to capture the index signals by conditional analysis (see Materials and Methods; Supplementary Material, Table S5). In these 30 regions, the ORs of the most-associated markers demonstrated marked directional consistency for 27 (90%) regions, compared with only 18 of the 32 (56%) index variants (Table 1). However, each of the 55 regions had a set of risk-associated SNPs from the meta-analysis with similar effect sizes and corresponding P-values. While this set of markers is statistically indistinguishable they define a relatively small subset that most likely contain the underlying functional variant in each region.
Table 1.
The index variants and most significantly associated markers in 30 known prostate cancer susceptibility regions.
| Index variant | Region, ethnicitya | Multiethnic |
European |
African |
Japanese |
Latino |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Most-associated marker | r2 with index in EUR/AFR/ASNb | Allelesc | ORd | Pe | Phetf | RAFg | ORd | Pe | RAFg | ORd | Pe | RAFg | ORd | Pe | RAFg | ORd | Pe |
| rs11902236 | 2p25.1, EUR | T/C | 1.01 | 0.72 | 0.055 | 0.27 | 1.06 | 0.036 | 0.62 | 0.98 | 0.56 | 0.11 | 0.92 | 0.13 | 0.24 | 0.94 | 0.4 |
| rs7575106 | 0.20/0.08/— | G/A | 1.11 | 0.0011 | 0.033 | 0.07 | 1.16 | 0.0013 | 0.09 | 1.11 | 0.038 | 0 | – | – | 0.06 | 0.80 | 0.099 |
| rs13385191 | 2p24.1, ASN | G/A | 1.08 | 0.0003 | 0.043 | 0.24 | 1.03 | 0.24 | 0.06 | 1.00 | 0.99 | 0.56 | 1.16 | 6.1 × 10−5 | 0.28 | 1.14 | 0.053 |
| rs9306894 | 0.45/0.27/0.82 | G/A | 1.10 | 5.8 × 10−8 | 0.21 | 0.37 | 1.08 | 0.0011 | 0.12 | 1.05 | 0.28 | 0.57 | 1.17 | 1.1 × 10−5 | 0.32 | 1.12 | 0.1 |
| rs1465618 | 2p21, EUR | T/C | 1.10 | 2.3 × 10−6 | 0.013 | 0.22 | 1.08 | 0.0097 | 0.11 | 1.02 | 0.62 | 0.68 | 1.22 | 3.6 × 10−7 | 0.41 | 1.04 | 0.6 |
| rs13017478 | 0.17/0.02/0.73 | C/T | 1.10 | 7.4 × 10−8 | 0.013 | 0.70 | 1.09 | 0.00091 | 0.80 | 1.07 | 0.081 | 0.71 | 1.23 | 4.4 × 10−7 | 0.79 | 0.96 | 0.65 |
| rs721048 | 2p15, EUR | A/G | 1.07 | 0.0091 | 0.21 | 0.18 | 1.05 | 0.13 | 0.04 | 1.12 | 0.15 | 0.05 | 1.01 | 0.94 | 0.17 | 1.24 | 0.0088 |
| rs58235267 | 0.15/0.03/0.00 | G/C | 1.13 | 3.9 × 10−12 | 0.73 | 0.51 | 1.12 | 1.7 × 10−5 | 0.44 | 1.16 | 1.5 × 10−6 | 0.73 | 1.11 | 0.013 | 0.54 | 1.09 | 0.2 |
| rs10187424 | 2p11.2, EUR | T/C | 1.07 | 3.6 × 10−5 | 0.24 | 0.58 | 1.04 | 0.12 | 0.36 | 1.07 | 0.016 | 0.63 | 1.10 | 0.0079 | 0.66 | 1.18 | 0.015 |
| rs1561198 | 0.82/0.40/0.99 | C/T | 1.09 | 3.6 × 10−7 | 0.69 | 0.54 | 1.08 | 0.0015 | 0.32 | 1.07 | 0.02 | 0.63 | 1.10 | 0.011 | 0.63 | 1.17 | 0.02 |
| rs2660753h | 3p12.1, EUR | T/C | 1.07 | 0.00057 | 5.1 × 10−7 | 0.10 | 1.14 | 0.00047 | 0.50 | 0.95 | 0.061 | 0.27 | 1.20 | 4.7 × 10−6 | 0.21 | 1.23 | 0.0071 |
| rs76668454 | 0.37/0.02/0.28 | C/T | 1.29 | 4.6 × 10−19 | 0.19 | 0.07 | 1.27 | 1.2 × 10−7 | 0.05 | 1.15 | 0.041 | 0.16 | 1.34 | 1.3 × 10−9 | 0.13 | 1.46 | 8.5 × 10−5 |
| rs2055109h | 3p11.2, ASN | C/T | 1.08 | 0.00024 | 0.024 | 0.23 | 1.05 | 0.088 | 0.12 | 1.06 | 0.21 | 0.11 | 1.27 | 2.5 × 10−5 | 0.17 | 1.07 | 0.43 |
| rs76668454 | 0.06/0.00/0.30 | C/T | 1.29 | 4.6 × 10−19 | 0.19 | 0.07 | 1.27 | 1.2 × 10−7 | 0.05 | 1.15 | 0.041 | 0.16 | 1.34 | 1.3 × 10−9 | 0.13 | 1.46 | 8.5 × 10−5 |
| rs7611694 | 3q13.2, EUR | A/C | 1.05 | 0.0028 | 0.068 | 0.58 | 1.08 | 0.0017 | 0.65 | 0.99 | 0.63 | 0.20 | 1.11 | 0.024 | 0.67 | 1.06 | 0.37 |
| rs12629813 | 0.80/0.04/0.71 | C/T | 1.08 | 1.3 × 10−5 | 0.97 | 0.57 | 1.08 | 0.0029 | 0.57 | 1.07 | 0.027 | 0.23 | 1.09 | 0.066 | 0.64 | 1.10 | 0.15 |
| rs10934853 | 3q21.3, EUR | A/C | 1.06 | 0.00039 | 0.037 | 0.28 | 1.12 | 1.1 × 10−5 | 0.71 | 1.04 | 0.24 | 0.50 | 1.01 | 0.7 | 0.40 | 0.97 | 0.67 |
| rs4857837 | 0.90/0.18/0.11 | A/G | 1.13 | 2.1 × 10−11 | 0.15 | 0.28 | 1.10 | 0.00053 | 0.30 | 1.16 | 1.2 × 10−6 | 0.15 | 1.21 | 0.00015 | 0.29 | 1.04 | 0.56 |
| rs10936632 | 3q26.2, EUR | A/C | 1.08 | 2.7 × 10−5 | 0.22 | 0.51 | 1.11 | 1.0 × 10−5 | 0.26 | 1.05 | 0.16 | 0.34 | 1.04 | 0.46 | 0.37 | 1.00 | 0.95 |
| rs76925190 | 0.18/0.04/0.10 | A/C | 1.22 | 1.4 × 10−13 | 0.15 | 0.81 | 1.28 | 1.8 × 10−12 | 0.96 | 1.06 | 0.47 | 0.79 | 1.15 | 0.015 | 0.79 | 1.19 | 0.048 |
| rs1894292 | 4q13.3, EUR | G/A | 1.05 | 0.002 | 0.3 | 0.52 | 1.07 | 0.006 | 0.68 | 1.03 | 0.42 | 0.66 | 1.08 | 0.032 | 0.64 | 0.96 | 0.55 |
| rs4694176 | 0.53/0.04/0.55 | C/A | 1.07 | 3.9 × 10−5 | 0.7 | 0.58 | 1.08 | 0.0017 | 0.79 | 1.10 | 0.0089 | 0.65 | 1.04 | 0.27 | 0.50 | 1.04 | 0.54 |
| rs12500426i | 4q22.3, EUR | A/C | 1.03 | 0.052 | 0.12 | 0.46 | 1.05 | 0.043 | 0.40 | 0.98 | 0.5 | 0.44 | 1.04 | 0.3 | 0.55 | 1.14 | 0.043 |
| rs60063444 | 0.65/0.13/0.83 | T/C | 1.08 | 5.9 × 10−5 | 0.79 | 0.40 | 1.07 | 0.0042 | 0.11 | 1.10 | 0.048 | 0.39 | 1.05 | 0.17 | 0.51 | 1.12 | 0.075 |
| rs17021918i | 4q22.3, EUR | C/T | 1.05 | 0.003 | 0.95 | 0.65 | 1.05 | 0.038 | 0.78 | 1.06 | 0.093 | 0.63 | 1.04 | 0.34 | 0.72 | 1.08 | 0.28 |
| rs60063444 | 0.30/0.00/0.42 | T/C | 1.08 | 5.9 × 10−5 | 0.79 | 0.40 | 1.07 | 0.0042 | 0.11 | 1.10 | 0.048 | 0.39 | 1.05 | 0.17 | 0.51 | 1.12 | 0.075 |
| rs2242652 | 5p15.33, EUR | G/A | 1.12 | 5.9 × 10−6 | 0.43 | 0.80 | 1.13 | 0.001 | 0.85 | 1.07 | 0.12 | 0.81 | 1.21 | 0.0035 | 0.85 | 1.15 | 0.22 |
| rs7726159 | 0.43/0.59/0.27 | C/A | 1.11 | 1.7 × 10−7 | 0.92 | 0.67 | 1.12 | 0.00013 | 0.81 | 1.11 | 0.0053 | 0.65 | 1.10 | 0.025 | 0.71 | 1.06 | 0.49 |
| rs12653946 | 5p15.33, EUR | T/C | 1.13 | 6.8 × 10−14 | 4.6 × 10−6 | 0.42 | 1.10 | 8.2 × 10−5 | 0.42 | 1.07 | 0.018 | 0.46 | 1.33 | 2.8 × 10−15 | 0.49 | 1.02 | 0.76 |
| rs4975758 | 0.82/0.49/0.96 | G/C | 1.15 | 2.2 × 10−16 | 2.7 × 10−5 | 0.47 | 1.11 | 5.0 × 10−5 | 0.33 | 1.11 | 0.0012 | 0.46 | 1.34 | 1.3 × 10−15 | 0.50 | 1.04 | 0.55 |
| rs1933488 | 6q25.2, EUR | A/G | 1.03 | 0.082 | 0.28 | 0.58 | 1.06 | 0.02 | 0.56 | 0.99 | 0.81 | 0.19 | 1.05 | 0.26 | 0.58 | 0.97 | 0.67 |
| rs13215045 | 0.70/0.32/0.32 | C/T | 1.06 | 0.0003 | 0.71 | 0.69 | 1.09 | 0.0016 | 0.56 | 1.05 | 0.081 | 0.38 | 1.03 | 0.37 | 0.72 | 1.06 | 0.45 |
| rs6465657 | 7q21.3, EUR | C/T | 1.06 | 0.0045 | 0.15 | 0.46 | 1.09 | 0.00051 | 0.89 | 1.01 | 0.92 | 0.90 | 1.04 | 0.48 | 0.69 | 0.94 | 0.36 |
| rs138101303 | 0.22/0.03/0.11 | A/G | 1.11 | 7.3 × 10−5 | 0.12 | 0.71 | 1.17 | 8.2 × 10−6 | 0.82 | 1.06 | 0.2 | 0.86 | 1.01 | 0.87 | 0.80 | 1.00 | 0.99 |
| rs1512268 | 8p21.2, EUR | T/C | 1.17 | 2.5 × 10−21 | 0.00012 | 0.43 | 1.10 | 6.8 × 10−5 | 0.65 | 1.15 | 2.3 × 10−6 | 0.38 | 1.34 | 1.3 × 10−15 | 0.47 | 1.20 | 0.0037 |
| rs1160267 | 0.99/0.64/0.99 | G/A | 1.18 | 4.5 × 10−23 | 0.00017 | 0.43 | 1.10 | 4.3 × 10−5 | 0.72 | 1.19 | 8.2 × 10−8 | 0.38 | 1.34 | 1.2 × 10−15 | 0.47 | 1.20 | 0.0041 |
| rs817826 | 9q31.2, ASN | C/T | 1.04 | 0.11 | 0.36 | 0.14 | 1.02 | 0.58 | 0.30 | 1.07 | 0.027 | 0.06 | 0.97 | 0.64 | 0.11 | 0.94 | 0.51 |
| rs1746824 | 0.02/0.38/0.25 | C/T | 1.06 | 0.00095 | 0.032 | 0.28 | 1.01 | 0.6 | 0.48 | 1.10 | 0.0012 | 0.29 | 1.03 | 0.44 | 0.31 | 1.22 | 0.0039 |
| rs2252004 | 10q26.12, ASN | C/A | 1.08 | 0.00022 | 0.021 | 0.90 | 1.08 | 0.047 | 0.49 | 1.03 | 0.39 | 0.79 | 1.21 | 2.0 × 10−5 | 0.70 | 1.05 | 0.48 |
| rs77929344 | —/0.00/0.48 | T/C | 1.31 | 9.3 × 10−7 | 1 | 0 | – | – | <0.01 | – | – | 0.87 | 1.31 | 9.3 × 10−7 | 0 | – | – |
| rs1938781 | 11q12.1, ASN | G/A | 1.08 | 1.2 × 10−5 | 0.023 | 0.21 | 1.03 | 0.4 | 0.33 | 1.07 | 0.02 | 0.31 | 1.19 | 9.6 × 10−6 | 0.24 | 1.13 | 0.091 |
| rs12223473 | 0.02/0.16/0.50 | G/T | 1.18 | 4.6 × 10−7 | 0.66 | <0.01 | – | – | 0.08 | 1.22 | 0.00012 | 0.19 | 1.15 | 0.0022 | 0.07 | 1.20 | 0.16 |
| rs10875943 | 12q13.12, EUR | C/T | 1.06 | 0.0004 | 0.33 | 0.30 | 1.08 | 0.0049 | 0.63 | 1.02 | 0.45 | 0.81 | 1.13 | 0.0093 | 0.32 | 1.06 | 0.39 |
| rs11168963 | 0.92/0.58/0.71 | G/A | 1.08 | 2.4 × 10−5 | 0.91 | 0.29 | 1.07 | 0.0086 | 0.57 | 1.07 | 0.021 | 0.76 | 1.11 | 0.021 | 0.31 | 1.07 | 0.36 |
| rs902774 | 12q13.13, EUR | A/G | 1.12 | 1.7 × 10−5 | 0.0032 | 0.16 | 1.20 | 3.9 × 10−8 | 0.08 | 0.97 | 0.55 | 0.06 | 1.21 | 0.19 | 0.14 | 1.00 | 0.97 |
| rs55958994 | 0.82/0.01/— | T/C | 1.21 | 2.8 × 10−13 | 0.34 | 0.13 | 1.25 | 2.1 × 10−10 | 0.14 | 1.17 | 0.00013 | 0 | – | – | 0.07 | 1.09 | 0.5 |
| rs1270884 | 12q24.21, EUR | A/G | 1.08 | 2.3 × 10−5 | 0.21 | 0.48 | 1.11 | 2.9 × 10−5 | 0.20 | 1.03 | 0.47 | 0.20 | 1.03 | 0.49 | 0.38 | 1.14 | 0.046 |
| rs10774740 | 0.41/0.21/0.59 | G/T | 1.10 | 2.2 × 10−8 | 0.12 | 0.61 | 1.14 | 1.1 × 10−7 | 0.32 | 1.06 | 0.062 | 0.31 | 1.04 | 0.27 | 0.53 | 1.14 | 0.039 |
| rs9600079 | 13q22.1, ASN | T/G | 1.03 | 0.035 | 7.8 × 10−5 | 0.44 | 1.02 | 0.41 | 0.53 | 0.97 | 0.22 | 0.39 | 1.19 | 1.1 × 10−6 | 0.40 | 1.03 | 0.65 |
| rs7327286 | 0.19/0.01/0.83 | G/A | 1.13 | 6.1 × 10−10 | 0.19 | 0.18 | 1.09 | 0.0047 | 0.17 | 1.11 | 0.0071 | 0.40 | 1.20 | 3.3 × 10−7 | 0.38 | 1.09 | 0.19 |
| rs8008270 | 14q22.1, EUR | C/T | 1.11 | 3.0 × 10−6 | 0.029 | 0.81 | 1.17 | 2.4 × 10−7 | 0.72 | 1.04 | 0.17 | <0.01 | – | – | 0.89 | 1.06 | 0.56 |
| rs62003517 | 0.98/0.01/1.00 | C/G | 1.15 | 2.0 × 10−7 | 0.33 | 0.81 | 1.18 | 2.2 × 10−7 | 0.95 | 1.05 | 0.44 | <0.01 | – | – | 0.90 | 1.15 | 0.18 |
| rs11649743 | 17q12 A, EUR | G/A | 1.16 | 3.9 × 10−12 | 0.2 | 0.80 | 1.15 | 3.2 × 10−6 | 0.93 | 1.07 | 0.22 | 0.72 | 1.20 | 8.5 × 10−6 | 0.83 | 1.30 | 0.0018 |
| rs11658433 | 0.87/0.38/0.67 | A/C | 1.19 | 5.6 × 10−14 | 0.55 | 0.79 | 1.16 | 1.9 × 10−6 | 0.94 | 1.19 | 0.014 | 0.72 | 1.23 | 3.6 × 10−6 | 0.82 | 1.30 | 0.0038 |
| rs7501939 | 17q12 B, EUR | C/T | 1.16 | 2.5 × 10−19 | 1.9 × 10−6 | 0.61 | 1.20 | 1.5 × 10−12 | 0.48 | 1.05 | 0.062 | 0.70 | 1.34 | 6.6 × 10−14 | 0.68 | 1.04 | 0.52 |
| rs11263763 | 0.71/0.18/0.91 | A/G | 1.20 | 3.9 × 10−26 | 0.00031 | 0.54 | 1.22 | 2.9 × 10−15 | 0.62 | 1.10 | 0.0028 | 0.70 | 1.35 | 4.6 × 10−14 | 0.61 | 1.11 | 0.12 |
| rs11650494 | 17q21.32, EUR | A/G | 1.10 | 0.00049 | 0.86 | 0.08 | 1.09 | 0.047 | 0.23 | 1.10 | 0.0035 | <0.01 | – | – | 0.06 | 1.02 | 0.86 |
| rs111834333 | 0.87/0.18/— | T/C | 1.15 | 6.0 × 10−6 | 0.043 | 0.08 | 1.08 | 0.078 | 0.12 | 1.24 | 1.1 × 10−6 | 0 | – | – | 0.06 | 0.97 | 0.84 |
| rs1859962 | 17q24.3, EUR | G/T | 1.10 | 7.0 × 10−9 | 9.8 × 10−6 | 0.49 | 1.20 | 1.1 × 10−13 | 0.29 | 1.00 | 0.91 | 0.27 | 1.01 | 0.79 | 0.61 | 1.14 | 0.041 |
| rs6501436 | 0.96/0.08/0.94 | G/A | 1.14 | 1.5 × 10−14 | 0.005 | 0.50 | 1.20 | 1.0 × 10−13 | 0.22 | 1.13 | 0.00047 | 0.28 | 1.01 | 0.74 | 0.61 | 1.15 | 0.039 |
| rs8102476 | 19q13.2, EUR | C/T | 1.07 | 4.5 × 10−5 | 0.017 | 0.54 | 1.11 | 1.3 × 10−5 | 0.76 | 1.09 | 0.015 | 0.37 | 0.97 | 0.34 | 0.50 | 1.07 | 0.28 |
| rs11083450 | 0.77/0.58/0.51 | T/C | 1.09 | 7.7 × 10−7 | 0.2 | 0.49 | 1.13 | 1.0 × 10−6 | 0.67 | 1.06 | 0.04 | 0.31 | 1.03 | 0.38 | 0.42 | 1.03 | 0.63 |
| rs9623117 | 22q13.1, EUR | C/T | 1.07 | 0.00079 | 0.13 | 0.21 | 1.09 | 0.0023 | 0.77 | 1.04 | 0.24 | 0.03 | 0.92 | 0.41 | 0.17 | 1.21 | 0.019 |
| rs6001833 | 0.38/0.26/0.18 | A/G | 1.09 | 2.9 × 10−5 | 0.86 | 0.25 | 1.08 | 0.0027 | 0.77 | 1.09 | 0.012 | <0.01 | – | – | 0.18 | 1.14 | 0.11 |
aThe racial/ethnic population in which the index variant was initially identified.
bPairwise correlation with the index variant in European (EUR), African (AFR) and Asian (ASN) populations from 1000 Genomes Project. Missing correlation as indicated by ‘—’ was due to at least one of the SNPs being monomorphic in the given population.
cRisk allele/reference allele. Risk allele is the allele associated with increased risk in the multiethnic sample.
dPer-allele odds ratio of the risk allele.
eP-value from the 1 d.f. Wald test of trend.
fP-value from 3 d.f. χ2-test of heterogeneity across four populations.
gRAF, risk allele frequency in controls.
h,iTwo index SNPs in the same region.
Interestingly, two index SNPs located 357 kb apart and previously reported as independent signals (rs2660753/3p12 (41) and rs2055109/3p11 (11)), could be explained by the most-associated marker in the region after fine-mapping (rs76668454). Both index SNPs are modestly correlated with rs76668454 in the discovery GWAS populations (r2 ≥ 0.30). This scenario was also observed at 4q22 with the two index SNPs (rs12500426 and rs17021918), located 48 kb apart, captured by marker rs60063444 (Supplementary Material, Table S6).
When evaluating the most-associated variants instead of the index SNPs, associations at three regions (rs76668454/3p12, rs7327286/13q22 and rs6501436/17q24) were no longer significantly heterogeneous across populations. However, three other regions (rs4975758/5p15, rs1160267/8p21 and rs11263763/17q12) remained significantly heterogeneous, likely due to the larger estimated effect sizes observed in the Japanese population (Table 1).
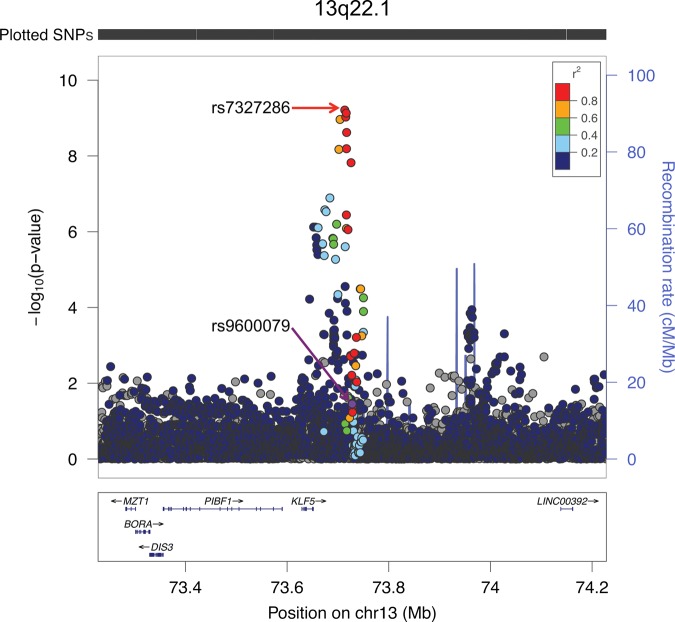
An example of a region illustrating the improvement in the association signal through multiethnic fine-mapping is shown in Figure 1. At 13q22, the index SNP rs9600079, originally identified in this Japanese sample (42) (Table 1), was not significantly associated with prostate cancer risk in the other racial/ethnic populations (European: OR = 1.02, P = 0.41; African: OR = 0.97, P = 0.22; Latino: OR = 1.03, P = 0.65). In testing all common variants that are correlated with rs9600079 in Asians (r2 ≥ 0.2), the most-associated variant in the multiethnic meta-analysis was rs7327286 (overall: P = 6.1 × 10−10), which is located 15 kb upstream from the index SNP (Fig. 1). This variant is highly correlated with the index SNP in Asians (r2 = 0.83), but is minimally correlated in Europeans (r2 = 0.19) and Africans (r2 = 0.01). It was more statistically significant and had a larger effect than the index SNP in each population and overall, statistical evidence for heterogeneity no longer remained (Phet = 0.19 versus Phet of index = 7.8 × 10−5).
Figure 1.
A Regional Association Plot of the Prostate Cancer Risk Region at Chromosome 13q22.1. The −log10 P-values are from the multiethnic meta-analysis. The index SNP (rs9600079), originally discovered in this Japanese sample (42), is designated by a purple circle. The r2 shown is estimated in Asians from 1000 Genomes Project (ASN 1KGP) in relation to rs9600079. Gray circles are SNPs not in ASN 1KGP (r2 cannot be estimated). The top red circle represents the marker (rs7327286) that is most strongly associated with risk in this region. The plot was generated using LocusZoom (http://csg.sph.umich.edu/locuszoom/).
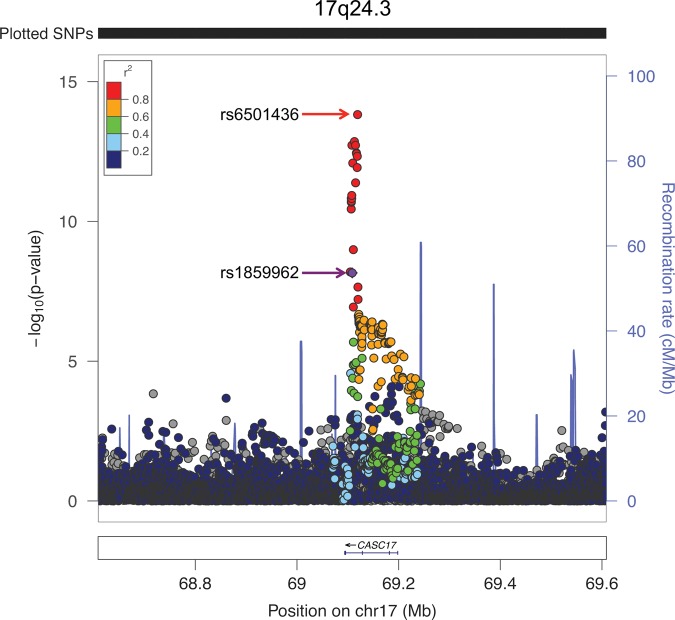
At 17q24 (Fig. 2), the index SNP rs1859962 was originally reported in a European GWAS (5). The association with the index SNP was significantly heterogeneous across racial/ethnic populations (Phet = 9.8 × 10−6; Table 1), with the largest effect and the most significant association observed in Europeans (OR = 1.20, P = 1.1 × 10−13). When examining all correlated (r2 ≥ 0.2) variants in Europeans, rs6501436, a SNP located 10 kb downstream from the index SNP, was the most-associated marker in the multiethnic analysis (P = 1.5 × 10−14). This marker is strongly correlated with the index SNP in both European (r2 = 0.96) and Asian ancestry populations (r2 = 0.94), but is minimally correlated (r2 = 0.08) in Africans. Moreover, in men of African ancestry, this SNP was more significantly associated with risk than the index SNP (OR = 1.13, P = 4.7 × 10−4 versus OR = 1.00, P = 0.91). The effect heterogeneity of rs6501436 was no longer significant across populations (Phet = 0.005 versus Phet of index = 9.8 × 10−6).
Figure 2.
A Regional Association Plot of the Prostate Cancer Risk Region at Chromosome 17q24.3. The −log10 P-values are from multiethnic meta-analysis. The index SNP (rs1859962), originally discovered in a European GWAS (5), is designated by a purple circle. The r2 shown is estimated in Europeans from 1000 Genomes Project (EUR 1KGP) in relation to rs1859962. Gray circles are SNPs not in EUR 1KGP (r2 cannot be estimated). The top red circle represents the marker (rs6501436) that is most strongly associated with risk in this region. The plot was generated using LocusZoom (http://csg.sph.umich.edu/locuszoom/).
Investigating associations in multiple populations also aided in deciphering potentially ethnic-specific risk variants. As an example, at 10q26, the index variant rs2252004, initially identified in this Japanese sample where the signal is the strongest (OR = 1.21, P = 2.0 × 10−5), is common in all populations (RAF range, 0.49–0.90) and is only weakly associated with risk in Europeans (OR = 1.08, P = 0.04; Table 1). In examining all variants correlated with rs2252004 (r2 > 0.2, ASN 1KGP), the most-associated marker, rs77929344, was only found in Japanese (RAF = 0.87; OR = 1.31, P = 9.3 × 10−7). Markers correlated with rs2252004 were only modestly associated with prostate cancer risk in the other populations (P-values >0.003; region-specific threshold for significance P = 0.001; Supplementary Material, Table S4), suggesting that this may be a Japanese-specific risk signal.
We also identified evidence of potential secondary signals in two regions through conditional analyses (at P < 5.0 × 10−6; see Materials and Methods; Supplementary Material, Table S7). At 3q21, rs13062436, located 179 kb from the index SNP (rs10934853), was significantly associated with prostate cancer risk when conditioning on the index signal (OR = 1.14, P = 5.0 × 10−8; Supplementary Material, Fig. S2). Similarly at 3p12, rs17181170 was significantly associated with risk in conditional analyses (OR = 1.10, P = 5.9 × 10−8; Supplementary Material, Fig. S3). As expected, both of these novel risk variants are uncorrelated with the index SNPs or the most-associated markers for the index signals in each population (r2 ≤ 0.06).
Functional annotation
Multiethnic fine-mapping in each region defined sets of alleles based on statistical significance, with many having similar effect sizes (Supplementary Material, Table S4). To further prioritize which of the most-associated variants have putative functionality, we mapped them relative to epigenetic marks and transcription factor binding data from publically available sources (see Materials and Methods). Here we limited the annotation to the 55 regions that were significantly associated with prostate cancer risk in the multiethnic analysis (as described above) and the 666 variants in these regions that had P-values that were within 1 order of magnitude of the most-associated marker (referred to as ‘top-order’ variants). Since this deterministic approach relies heavily on P-value rankings, we compared this approach with the ranking distribution obtained by re-sampling the effects of all candidate SNPs in each region for each population from a multivariate distribution (see Materials and Methods). At 49 (out of 55, 89%) regions, the set of top-order variants contained the top-ranked SNP when resampling (Supplementary Material, Table S8). Moreover, 84% on average (100% median) of the top-order SNPs were within the 95% joint posterior probabilities from resampling. For 28 regions, the entire set of top-order SNPs was included within the 95% joint posterior probabilities.
Of the 666 variants, 193 (29%) overlapped with functional marks and could be assigned into one of four predicted functional categories: missense variants, enhancers, promoter or promoter-proximal enhancers and untranslated regions of coding exons (UTR) (Supplementary Material, Tables S9 and S10). Of the 193 SNPs, 2 were non-synonymous substitutions (rs2292884/H347R in MLPH and rs699664/R325Q in GGCX), 152 (79%) were present in enhancers, 29 (15%) were in promoters, and 10 (5%) were located within untranslated regions. The most statistically associated variants in 12 of the 55 (22%) regions represented the best functional candidates in the region, whereas this number increased to 20 (36%) and 27 (49%) when examining the 2 or 3 most significantly associated markers in each region, respectively (Supplementary Material, Table S9). To determine whether the top-order variants are enriched for enhancer annotations, we examined 721 371 SNPs (MAF > 1%) and insertion/deletion variants in the 1KGP database within windows of 1 Mb (centered on the index SNP) at the 55 regions and found that 102 735 (14%) overlapped with the features that we used to identify and annotate enhancers. Compared with this figure, our 666 top-order variants contained 152 enhancer overlaps (23%), constituting a 1.6-fold enrichment over background (P = 1.1 × 10−9).
We also performed a cis-eQTL analysis with the top-order variants (n = 666) in 145 prostate tumor samples to further assess whether the risk signals defined by the top SNPs may also be associated with an eQTL. The cis-eQTL associations in this sample based on the index variants or correlated markers (r2 > 0.5) in the CEU 1KGP population have been published previously (35). In 11 regions, we found suggestive evidence of cis-eQTL associations (P < 9.1 × 10−4; see Materials and Methods) between top-order SNPs and the expression of one or more nearby genes (Supplementary Material, Table S9) with the most significant associations observed at 5p15 and IRX4 (P = 2.4 × 10−15) (43), 6q21 and SESN1 (P = 6.3 × 10−8), 6q25 and RGS17 (P = 2.5 × 10−6), 11q22 and MMP7 (P = 2.3 × 10−6), and at 17p13 with VPS53 (P = 1.3 × 10−6) and FAM57A (P = 6.4 × 10−6). Five cis-eQTL associations (SLC6A19, C10orf32, CTBP2, MMP7 and FAM57A) were not identified previously when focusing on the index variant or those correlated with the index SNP in the European ancestry population (35). To determine whether the top-order variants were enriched for cis-eQTL associations, we examined all common variants (MAF ≥ 1%) in the 55 regions where fine-mapping was conducted. Of 334 357 variants in these regions, 4822 (1.4%) were significant cis-eQTLs. In comparison, 87 out of the 666 top-order variants in the same regions were significant cis-eQTLs (13%), constituting a 9-fold enrichment over background (P = 5.0 × 10−55).
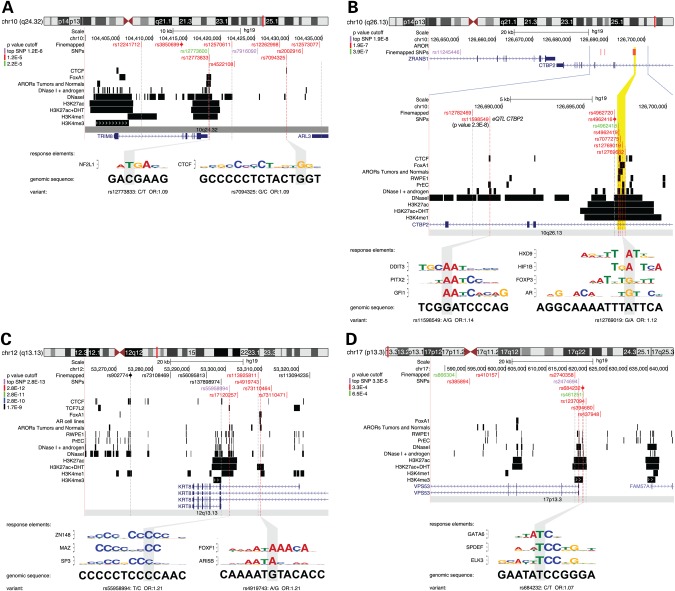
Four of the regions were unique in that the statistical association, functional annotation and eQTL evidence converged on a small number of variants (Fig. 3). These regions are discussed below; detailed annotation information for all regions is provided in the Supplementary Material, Figure S4 and Supplementary Material, File S1.
Figure 3.
Genome Browser Views of Four Candidate Risk Regions. SNP locations are shown relative to epigenetic features in LNCaP (and other) cell lines. Intervals for significant peak regions are denoted with black rectangles for each dataset indicated at left. SNPs are colored by order of P-value magnitude relative to the top SNP (lilac) in the region: first order red, second order green, third order blue, all others black. Red dotted lines guide the exact position of the best functional candidates relative to epigenetic features. Gray lines guide the position of other SNPs. For the most notable SNPs in enhancer or promoter regions an alignment of the surrounding DNA sequence to a response element match is shown, with gray boxes indicating position of the SNP. (A) chromosome 10q24.32 region. (B) chromosome 10q2.13 region. (C) chromosome 12q13.13 region. (D) chromosome 17p13.3 region.
At 10q24, the most promising functional candidate is variant rs12773833 (7 annotations, P = 1.8 × 10−6), one of the most-associated variants in the region. It overlaps with ChIPseq peaks for FOXA1, the Androgen Receptor in tumor and normal prostate tissue samples, DNaseI, H3K27Ac and H3K4me1 and is an eQTL (P = 6.5 × 10−4) with the AS3MT gene ∼200 kb telomeric (Fig. 3A; Supplementary Material, Table S9). A second candidate, rs7094325 (P = 1.7 × 10−6), is an eQTL for AS3MT (P = 5.1 × 10−4) and C10orf32 (P = 7.7 × 10−4; Supplementary Material, Table S9), overlaps a CTCF peak in LNCaP (Fig. 3A) and disrupts a potential CTCF response element (Supplementary Material, Table S10).
At 10q26, the top-order SNPs include rs11598549 (P = 2.3 × 10−8), which is situated in a DNaseI site (Fig. 3B) and is an eQTL with CtBP2 (P = 7.9 × 10−4; Supplementary Material, Table S9). Other top-order SNPs, rs4962419, rs7077275, rs12769019 and rs12769682 (P < 2 × 10−7), also overlap the DNaseI hypersensitive and transcription-factor bound region of this enhancer, with the low-risk A allele of rs12769019 predicted to disrupt an Androgen Receptor response element (Fig. 3B; Supplementary Material, Table S10). A recent report confirmed that the Androgen Receptor binds to this region and that the risk allele of rs12769019 mediates increased androgen responsiveness of the enhancer in a reporter assay (44).
At 12q13, the most significantly associated SNP rs55958994 (P = 2.8 × 10−13; Supplementary Material, Table S9) overlaps 6 annotations (Fig. 3C) and is situated within a DNaseI hypersensitive site in an H3K27Ac and H3K4me1 marked active promoter-proximal enhancer of KRT8. In a previous fine-mapping study in this African sample we reported rs55958994 as the most significant association in the region (17). The multiethnic results presented here with a larger sample and multiple populations further support rs55958994 as the most-associated marker and best functional candidate in the region.
At 17p13, the index SNP rs684232 (five annotations, P = 5.3 × 10−5) and the most-associated SNP rs2474694 (four annotations, P = 3.3 × 10−5) are in different DNaseI hypersensitive sites within the same H3K27Ac and H3K4me3 marked promoter of the VPS53 gene (Fig. 3D). Both SNPs are also eQTLs with VPS53 (rs684232 P = 4.6 × 10−6; rs2474694 P = 4.6 × 10−4) and FAM57A (rs684232 P = 2.3 × 10−5; rs2474694 P = 2.3 × 10−5; Supplementary Material, Table S9). Of the two, rs684232 is predicted to disrupt a GATA6 binding site, whereas rs2474694 is located within the 5′ UTR of VPS53.
Discussion
In this multiethnic fine-mapping study of prostate cancer risk loci, 55 regions (out of 67 examined) passed region-specific significance thresholds, and in 25 regions, the index SNP and correlated variants were most effective in denoting the risk association. In 30 regions, we identified markers that are more statistically associated with risk than the index SNP in the multiethnic sample. In applying information on functional annotation from prostate cancer cell lines we were able to further define variants in many regions that may be considered high-priority functional candidates for experimental testing.
GWAS have revealed 100 genetic risk variants for prostate cancer, more than any other common cancer. Although most of the associations were originally identified in populations of European ancestry, the vast majority of these risk variants are also associated with risk in other populations, pointing to common shared functional variants (16,17,40). Some loci do not replicate despite adequate statistical power. Given the directional consistency noted across populations, this observation suggests that the index variant is not the biologically relevant allele or is not a valid proxy for the true causal variant in all populations. Fine-mapping using trans-ethnic populations has been shown both in theory (45–48) and in practice (18–21) for a number of traits to have better performance than using a single population of relatively homogeneous ancestry. Based on the expectation that most functional alleles are common and shared, the success of this approach requires having good coverage on the genetic variation across populations. Here we used GWAS genotyping plus imputation based on a high-density reference panel from 1KGP, which is currently the most comprehensive and efficient approach for testing common alleles at risk regions.
The primary goal of our study was to reduce the complexity in each region defined by an index signal and to identify a subset of variants that most likely contain functional variation affecting prostate cancer risk in all populations. However, this does not rule out the possibility of additional risk variants in these regions as demonstrated at 3q21 and 3p12, or the possibility of allelic heterogeneity in a region in these populations. Additionally, multiple variants may jointly contribute to the disease risk in a non-additive fashion such as through haplotypes or gene–gene interactions, which requires further investigation.
In selecting regions for fine-mapping, we applied a conservative region-specific significance threshold to reduce the potential for identifying and reporting false-positive associations. Several methods for prioritizing the best candidate markers in a region have been proposed and utilized in trans-ethnic fine-mapping studies (16,18–21,47). In general, these methods attempt to identify a single variant or so-called ‘best marker’ in a region among a group of variants mutually correlated. For example, MANTRA (47) leverages heterogeneity across multiple ethnic groups to improve power and provide a credible set of variants. When applied to our data, the credible set often contained hundreds of SNPs with a ranking similar to the P-value ranking (data not shown). Instead, we opted to leverage the differential LD structure and investigate SNPs with consistent estimates across populations ranked by P-value, in order to provide a narrower list of top-order variants as promising candidates for functional annotation. To account for the uncertainty from estimation on the resulting ranking of P-values, we used an empirical resampling approach to estimate the probability that each SNP is the most-associated marker within each region. For the vast majority (89%) of the regions that we examined, the top-ranked SNP from resampling is included within our defined set of top-order variants. Although requiring validation, this suggests that the underlying functional allele, which is expected to have the largest effect size and smallest P-value, has a good probability of being included within the subset of alleles that we have defined in each region (shown in Supplementary Material, Table S4).
Compared with past fine-mapping studies for prostate cancer, our study is empowered by improved imputation coverage, increased sample size and different LD structure from multiethnic populations. Previous fine-mapping efforts limited to single populations include region-specific studies of 5p15 (TERT) (49), 8p21 (NKX3.1) (39), 10q11 (MSMB) (50), 11q13 (51–53), 17q12 (HNF1B) (54,55) and 19q13 (KLK3) (56,57) in European or Asian ancestry populations as well as an initial characterization of the known regions in part of this African ancestry sample (16,17). In a concurrent fine-mapping study, which is the largest fine-mapping study to date in men of European ancestry, Al Olama et al. (79) examined 64 regions in 25 779 prostate cancer cases and 26 218 controls, with the majority of samples genotyped with the iCOGS array (14). Leveraging individual-level data, they used stepwise regression to suggest more significant markers at 47 regions. In total, they indicate 1623 variants as candidates via statistical fine-mapping (21.6 SNPs per region on average; median of 13). Of these variants, they identify 403 with additional functional annotation with an average of 5.4 per region (median = 3). In comparison, our multiethnic study suggests a total of 666 SNPs in 55 regions (average of 12.1 SNPs per region and a median of 6). With additional functional annotation our study identifies an average of 3.5 markers (median = 2). Comparing final conclusions for the 46 overlapping regions across the two studies, there are 29 (63.0%) regions with at least one overlapping functional candidate marker. Moreover, 11 of the 46 regions (23.9%) have a single overlapping candidate—a strong candidate marker for future more detailed functional investigation. For these overlapping regions, Al Olama et al. reported novel secondary signals (P < 1.0 × 10−5) with 13 variants in these regions. In our study, nine of these variants were nominally associated with prostate cancer risk (P < 0.05), with two markers correlated (r2 range, 0.47–0.93, EUR 1KGP) with the secondary signals we identified at 3q21 and 3p12. We believe that these contrasting findings highlight the value of using both homogeneous as well as multiethnic samples in fine-mapping, with the latter focused on prioritizing risk variants that generalize across populations and thus, are most likely to be or tag the biologically functional variant.
In order to assess the variants for biological relevance we determined their locations relative to chromatin biofeatures and other primary sequence features in cell types that reflect the epithelial cell types of origin for this cancer. These include PrEC and RWPE1, which are immortalized, non-transforming prostate epithelial cell lines, and LNCaP, a prostate cancer cell line that was originally isolated from lymph node metastases. Previously, we examined GWAS risk variants and proxies within biofeatures in these cell types and identified 727 putative functional SNPs in LD (r2 > 0.5) with the index SNP in 77 risk regions (58). A large proportion (667, or 88%) of these SNPs fell within 217 distinct enhancer regions. While this subset of variants likely captures many true biologically functional variants, associations with prostate cancer risk were not directly assessed. Here we show that through multiethnic association mapping we are able to eliminate many of the proxies and reduce the number of candidate functional variants to a much smaller subset within one or two enhancers in many regions. In particular, we were able to narrow down our list of candidates from 666 top-order SNPs to 193 within chromatin features of biological interest in the cell type of origin, of which 152 (79%) are located in 82 distinct enhancer regions, with an average of 1.5 enhancers per region over 55 regions. Contrast this with the previous study where we found 217 putative risk enhancers over 77 regions, equivalent to 2.8 enhancers per risk region. We also evaluated eQTLs to further highlight potential functional alleles and target genes that may be altered by inherited variation. Through combining different sources of evidence we have attempted to reduce the number of variants and hypotheses for researchers to test in follow-up studies designed to reveal the biological mechanisms underlying genetic risk.
We showed that relative to random selections of SNPs from 1KGP, our top-order set was highly enriched for enhancer and promoter regulatory elements, consistent with previous findings that the majority of GWAS signals for many cancers overlap enhancer regions (58,59). Moreover, our finding of enrichment strongly suggests that there is a real bias toward perturbation of regulatory sequences in risk for prostate cancer. It also suggests, because of the types of disruptions, that risk is mediated biologically at the level of point-disruptions of enhancer–promoter interactions mediated by transcription factor binding to the chromosome at the location of the SNP. More studies are needed to assess the gene regulation for each risk enhancer, identify potential gene targets, and determine which targets mediate phenotypes consistent with increased risk for cancer using cell-based assays. Combining new technologies, including CRISPR-cas9 RNA-mediated genome editing for knockout and allele replacement experiments and 4C chromatin interaction assays to identify physical interactions between risk enhancers and their target regions, are expected to yield insight into the mechanism of GWAS identified risk at many of the regions.
In summary, we have characterized 55 prostate cancer risk regions through statistical multiethnic fine-mapping, functional annotation and cis-eQTL analyses and have revealed variants in ∼50% of these regions that may be functional and in turn should be prioritized in future experimental testing to understand biological mechanisms at prostate cancer risk regions.
Materials and Methods
Study populations and genotyping
We combined data from studies with existing high-density SNP genotyping in prostate cancer GWAS in the following populations: European ancestry [8600 cases and 6946 controls from the Cancer of the Prostate in Sweden (CAPS) (60), Breast and Prostate Cancer Cohort Consortium (BPC3) (10) and PEGASUS]; African ancestry [5327 cases and 5136 controls from the African Ancestry Prostate Cancer GWAS Consortium (AAPC) (8) and the Ghana Prostate Study (61)]; Japanese ancestry [2563 cases and 4391 controls from GWAS in Japanese in the Multiethnic Cohort (MEC) (40), and in Biobank Japan (11,42)]; and Latino ancestry [a GWAS of 1034 cases and 1046 controls from the MEC (40)]. Details of each study are provided in the Supplementary Material, File S1 and Supplementary Material, Table S1. Genotyping of each study was performed using Illumina or Affymetrix GWAS arrays and quality control procedures of each GWAS have been described previously and are provided in Supplementary Material, Table S2. Imputation was performed in each study using a cosmopolitan reference panel from the 1000 Genomes Project (1KGP; March, 2012). Across each region, genotyped SNPs, imputed SNPs and insertion/deletion variants ≥1% frequency were examined for association with prostate cancer risk. SNPs with an imputation r2 [‘info score’ (62)] < 0.3 were not tested for association. The vast majority of SNPs reported in this paper (581/666: 87%) had an r2 ≥ 0.8 in all studies. This study was approved by the Institutional Review Board at the University of Southern California.
Statistical analysis
Here we focus on 69 of the 100 known risk variants (referred to as index SNPs) in 67 regions; exclusions include 23 regions/variants where multiethnic fine-mapping in this sample has already been reported (15), 8q24 (2,4,42,63,64) and 11q13 which harbor multiple independent risk variants (51–53) and will be reported separately, and 19q13 (KLK3, a gene that encodes PSA) which is a risk region for low-grade prostate cancer (56,57,65). Regions that contain two index SNPs are 3p11-p12 (rs2055109/3p11 and rs2660753/3p12) and 4q22 (rs12500426 and rs17021918). Within ±500 kb of each index SNP, association testing with prostate cancer risk was conducted within each study using unconditional logistic regression, adjusted for global ancestry in an additive model. We summarized the ethnic-specific and overall effect using a fixed-effect inverse-variance-weighted meta-analysis. For each SNP, we report a per-allele OR, 95% confidence interval (CI), and a P-value obtained from a 1-degree of freedom Wald test.
In our multiethnic fine-mapping, we focused initially on SNPs that are correlated (r2 ≥ 0.2) with the index SNP in the racial/ethnic population in which the original discovery was made, and are more statistically significant (>1 order of magnitude change in P-value in the multiethnic sample). The r2 threshold of 0.2 was lowered when demonstrated through conditional analyses that a more weakly correlated variant could account for the association signal defined by the index variant. To determine statistical significance within each region, we applied a region-specific threshold to correct for multiple independent tests conducted for SNPs correlated (r2 ≥ 0.2) with the index SNP in the original GWAS population. For each region, an empirically determined threshold accounting for the number of correlated SNPs was estimated as 0.05 divided by the number of tags that can capture all of the common tested SNPs [minor allele frequency (MAF) ≥ 0.05] at r2 > 0.8 in AFR 1KGP. These region-specific significance cutoffs range from 3.9 × 10−4 to 5.6 × 10−3 and are conservative given that African ancestry populations often require more ‘tags’ and represent only approximately one-third of our study sample, and thus, will reduce the number of false-positive signals. Of the 67 regions examined, the significance of the index SNP or those correlated with the index SNP (at r2 ≥ 0.2) surpassed region-specific thresholds for 55 regions (Supplementary Material, Table S4). In these regions we evaluated effect heterogeneity across racial/ethnic populations using Cochran's Q test and I2 statistics (66). Statistically significant heterogeneous effects were defined as those with Phet < 9.1 × 10−4 (0.05/55 regions) and I2 > 80.0%. For 15 regions in which the most-associated SNP, or a SNP with nominal P-value < 10−5, was weakly correlated with the index variant (r2 < 0.2), we performed conditional analysis with both SNPs in the same model to determine if a secondary signal could be identified. A secondary signal was defined as significantly associated with prostate cancer risk in the overall meta-analysis at P < 5.0 × 10−6, with no impact on the effect or degree of statistical significance of the index SNP or most significantly associated marker in the region (16).
For each statistically significant region, we defined a ‘top-set’ of SNPs as those with P-values within one order of magnitude change from the most-associated marker. This definition was used for ease of implementation and interpretation. However, several new statistical approaches for fine-mapping have been presented and rely on the conversion of a marginal P-value to a posterior probability (67), Bayes Factors or scaled Bayes Factor (68) for re-ranking the SNPs and providing probabilistic interpretations for the final set of SNPs selected (i.e. credible sets). The main advantage of these approaches is that they incorporate the corresponding SNP variance, weighted relative to a prior variance, to re-rank the SNPs. The re-ranking of the SNPs is dependent upon the specification of the prior and Wakefield (69) discusses several options including a prior with effect size-MAF dependence and a prior equivalent to the ranking via P-values. Importantly, in Wakefield (70) it is noted that these priors implicitly assume that the SNPs are independent, thus they may not be appropriate for fine-mapping and may lead to false conclusions when relying on final probabilistic interpretations (i.e. 95% credible sets). While more sophisticated hierarchical modeling approaches may be necessary, such as those in Conti and Witte (71), these lack the ease of implementation. Moreover, the major strength of our overall approach is the leveraging of differential linkage disequilibrium across multiple ethnic groups to narrow the set of SNPs. In this situation, it is unclear how sensitive final SNP rankings are to various pre-specified priors. To account for the potential uncertainty in the ranking of P-values due to effect estimation within each ethnic group, we performed a resampling approach similar in spirit to the methods described in Zaitlen et al. (46). That is, for all candidate SNPs (r2 ≥ 0.2 with index in the original GWAS population), we resampled the ethnic-specific effect estimates from a multivariate normal distribution with a mean given by the estimated ethnic-specific marginal maximum likelihood estimates (MLE) and structured covariance matrix with the diagonal elements equal to the estimated variance for each marginal effect and the off-diagonal elements equal to the approximated covariance for two marginal effects , where ρij is the estimated pairwise correlation between SNPi and SNPj from 1KGP for the corresponding racial/ethnic population. For each region, resampled ethnic-specific effect estimates were then meta-analyzed in a fixed-effect model, weighted by the estimated variance. This approach provides posterior probabilities (under a prior with a point mass at the MLE) for the joint ranking of all candidate SNPs within a region and avoids assumptions of independence.
Functional annotation
For each region we mapped the most-associated SNPs (Supplementary Material, Tables S9 and S10) to putative functional domains using bedtools software (bedtools.readthedocs.org/en/latest/#) and in-house python scripts to assemble a matrix of positive overlaps. We used a number of publicly available prostate epithelia and PrCa ENCODE datasets of chromatin features to identify putative enhancer/regulatory regions in each risk region (58,72). These datasets included LNCaP and RWPEI DnaseI HS sites (GSE32970) ENCODE; PrEC DNaseI HS sites (GSE29692) ENCODE; LNCaP CTCF ChIP-seq peaks (GSE33213) ENCODE; LNCaP H3K27ac and TCF7L2 (GSE51621) (58), H3K4me3 and H3K4me1 histone modification ChIP-seq peaks (GSE27823) (73); FoxA1 ChIP-seq peaks (GSE28264) (74); Androgen Receptor (AR) ChIP-seq peaks (75) and AR binding sites (GSE28219) (76); NKX3-1 ChIP-seq peaks (GSE28264) (74). We also included AR ChIP-seq data on 7 normal and 13 tumor prostate tissue (M. Pomerantz et al., submitted for publication).
We subsequently classified the putative functionality of each SNP according to the mapped features. These fall into four categories, promoter, enhancer, coding disruptions and untranslated exonic regions. For coding exons we used dbSNP (77) to assess the nature of disruptions in protein-coding sequence. For 5′ or 3′ UTR SNPs we reported overlap with miRcode highcons predicted target sequences (78). To assess potential disruptions of transcription factor response elements, we performed motif analysis as previously described (58), reporting motifs with a >85% match and 70% difference between the reference and effect alleles in the position frequency matrix describing the motif. To calculate enrichment of SNPs in enhancers we used the hypergeometric distribution as implemented in base packages of R. The hypergeometric distribution measures the probability of k successes in n draws without replacement from a finite population given that the entire makeup of the population is known.
Cis-eQTL analysis
For the most-associated variants in each region we examined the associations with expression of nearby genes in 145 prostate tumor samples from the TCGA database (February 2013). If a variant was not represented in the TCGA data, the genotypes were imputed using IMPUTE2 (62). A cis-eQTL analysis was performed for these variants and any transcript within a 1 Mb interval (500 kb on either side). Gene expression values were adjusted for somatic copy number and CpG methylation as previously described (32). Each risk variant was corrected for the number of transcripts in the interval. Significant associations were defined as a nominal P-value of < 9.1 × 10−4, which is a Bonferroni correction for the number of regions examined (n = 55).
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) GAME-ON U19 initiative for prostate cancer (ELLIPSE, CA148537). The BPC3 was supported by NIH/NCI cooperative agreements U01-CA98233 to D.J.H., U01-CA98710 to S.M.G., U01-CA98216 to E.R., and U01-CA98758 to B.E.H., and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics). ATBC, PEGASUS/PLCO and the Ghana Prostate Study were supported in part by the Intramural Research Program of the NIH and NCI. Additionally, this research was supported by U.S. Public Health Service contracts N01-CN-45165, N01-RC-45035, N01-RC-37004 and HHSN261201000006C from the NCI, Department of Health and Human Services. CAPS was supported by the Cancer Risk Prediction Center (CRisP; www.crispcenter.org), a Linneus Centre (Contract ID 70867902) financed by the Swedish Research Council (grant no. K2010-70X-20430-04-3), the Swedish Cancer Foundation (grant no. 09-0677), the Hedlund Foundation, the Söderberg Foundation, the Enqvist Foundation, ALF funds from the Stockholm County Council. Stiftelsen Johanna Hagstrand och Sigfrid Linnér's Minne, Karlsson's Fund for urological and surgical research. The AAPC studies were supported as follows: The MEC and the genotyping in AAPC were supported by NIH grants CA63464, CA54281, CA1326792, CA148085 and HG004726. Functional annotation was supported by grant CA136924. Genotyping of the PLCO samples was funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, NCI, NIH. LAAPC was funded by grant 99-00524V-10258 from the Cancer Research Fund, under Interagency Agreement #97-12013 (University of California contract # 98-00924V) with the Department of Health Services Cancer Research Program. Cancer incidence data for the MEC and LAAPC studies have been collected by the Los Angeles Cancer Surveillance Program of the University of Southern California with Federal funds from the NCI, NIH, Department of Health and Human Services, under Contract No. N01-PC-35139, and the California Department of Health Services as part of the state-wide cancer reporting program mandated by California Health and Safety Code Section 103885, and grant number 1U58DP000807-3 from the Centers for Disease Control and Prevention. KCPCS was supported by NIH grants CA056678, CA082664 and CA092579, with additional support from the Fred Hutchinson Cancer Research Center and the Intramural Program of the National Human Genome Research Institute. MDA was support by grants, CA68578, ES007784, DAMD W81XWH-07-1-0645, and CA140388. GECAP was supported by NIH grant ES011126. CaP Genes was supported by CA88164 and CA127298. IPCG was support by DOD grant W81XWH-07-1-0122. DCPC was supported by NIH grant S06GM08016 and DOD grants DAMD W81XWH-07-1-0203, DAMD W81XWH-06-1-0066 and DOD W81XWH-10-1-0532. CPS-II is supported by the American Cancer Society. SELECT is funded by Public Health Service grants CA37429 and 5UM1CA182883 from the National Cancer Institute. SCCS is funded by NIH grant CA092447. SCCS sample preparation was conducted at the Epidemiology Biospecimen Core Lab that is supported in part by the Vanderbilt Ingram Cancer Center (CA68485). Data on SCCS cancer cases used in this publication were provided by the Alabama Statewide Cancer Registry; Kentucky Cancer Registry; Tennessee Department of Health, Office of Cancer Surveillance; Florida Cancer Data System; North Carolina Central Cancer Registry, North Carolina Division of Public Health; Georgia Comprehensive Cancer Registry; Louisiana Tumor Registry; Mississippi Cancer Registry; South Carolina Central Cancer Registry; Virginia Department of Health, Virginia Cancer Registry; Arkansas Department of Health, Cancer Registry. The Arkansas Central Cancer Registry is fully funded by a grant from National Program of Cancer Registries, Centers for Disease Control and Prevention (CDC). Data on SCCS cancer cases from Mississippi were collected by the Mississippi Cancer Registry which participates in the National Program of Cancer Registries (NPCR) of the Centers for Disease Control and Prevention (CDC). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the CDC or the Mississippi Cancer Registry. JAPC and LAPC were supported by NIH grants CA63464, CA54281 and CA098758, the NIH/National Human Genome Research Institute by U01 HG004726-01 and the Genome Coordinating Center was supported by U01 HG004446. BBJ was supported by the Ministry of Education, Culture, Sports, Sciences and Technology of the Japanese government, and this study was supported in part by the Princess Takamatsu Cancer Research Fund and Takeda Science Foundation Award (to H.N.). G.A. Coetzee was supported by grant R01 CA136924 and M.L. Freedman was supported by grant R01 GM107427. We should like to thank all of the men who took part in these studies.
Supplementary Material
References
- 1.Brawley O.W. (2012) Prostate cancer epidemiology in the United States. World J. Urol., 30, 195–200. [DOI] [PubMed] [Google Scholar]
- 2.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J. et al. (2007) Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet., 39, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J., Sulem P., Rafnar T., Bergthorsson J.T., Manolescu A., Gudbjartsson D., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Blondal T. et al. (2008) Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet., 40, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Olama A.A., Kote-Jarai Z., Giles G.G., Guy M., Morrison J., Severi G., Leongamornlert D.A., Tymrakiewicz M., Jhavar S., Saunders E. et al. (2009) Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet., 41, 1058–1060. [DOI] [PubMed] [Google Scholar]
- 5.Eeles R.A., Kote-Jarai Z., Al Olama A.A., Giles G.G., Guy M., Severi G., Muir K., Hopper J.L., Henderson B.E., Haiman C.A. et al. (2009) Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet., 41, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudmundsson J., Sulem P., Gudbjartsson D.F., Blondal T., Gylfason A., Agnarsson B.A., Benediktsdottir K.R., Magnusdottir D.N., Orlygsdottir G., Jakobsdottir M. et al. (2009) Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet., 41, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Li G., Wiley K.E., Kim S.T., Zhu Y., Zhang Z., Hsu F.C. et al. (2009) Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res., 69, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haiman C.A., Chen G.K., Blot W.J., Strom S.S., Berndt S.I., Kittles R.A., Rybicki B.A., Isaacs W.B., Ingles S.A., Stanford J.L. et al. (2011) Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat. Genet., 43, 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kote-Jarai Z., Olama A.A., Giles G.G., Severi G., Schleutker J., Weischer M., Campa D., Riboli E., Key T., Gronberg H. et al. (2011) Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat. Genet., 43, 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumacher F.R., Berndt S.I., Siddiq A., Jacobs K.B., Wang Z., Lindstrom S., Stevens V.L., Chen C., Mondul A.M., Travis R.C. et al. (2011) Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet., 20, 3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akamatsu S., Takata R., Haiman C.A., Takahashi A., Inoue T., Kubo M., Furihata M., Kamatani N., Inazawa J., Chen G.K. et al. (2012) Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat. Genet., 44, 426–429, S421. [DOI] [PubMed] [Google Scholar]
- 12.Al Olama A.A., Kote-Jarai Z., Schumacher F.R., Wiklund F., Berndt S.I., Benlloch S., Giles G.G., Severi G., Neal D.E., Hamdy F.C. et al. (2012) A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J., Mo Z., Ye D., Wang M., Liu F., Jin G., Xu C., Wang X., Shao Q., Chen Z. et al. (2012) Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat. Genet., 44, 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eeles R.A., Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S. et al. (2013) Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet., 45, 385–391, 391e381-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al Olama A.A., Kote-Jarai Z., Berndt S.I., Conti D.V., Schumacher F., Han Y., Benlloch S., Hazelett D.J., Wang Z., Saunders E. et al. (2014) A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haiman C.A., Chen G.K., Blot W.J., Strom S.S., Berndt S.I., Kittles R.A., Rybicki B.A., Isaacs W.B., Ingles S.A., Stanford J.L. et al. (2011) Characterizing genetic risk at known prostate cancer susceptibility loci in African Americans. PLoS Genet., 7, e1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y., Signorello L.B., Strom S.S., Kittles R.A., Rybicki B.A., Stanford J.L., Goodman P.J., Berndt S.I., Carpten J., Casey G. et al. (2014) Generalizability of established prostate cancer risk variants in men of African ancestry. Int. J. Cancer, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C.T., Buchkovich M.L., Winkler T.W., Heid I.M., Borecki I.B., Fox C.S., Mohlke K.L., North K.E. et al. , African Ancestry Anthropometry Genetics, C., Consortium, G., (2014) Multi-ethnic fine-mapping of 14 central adiposity loci. Hum. Mol. Genet., 23, 4738–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franceschini N., van Rooij F.J., Prins B.P., Feitosa M.F., Karakas M., Eckfeldt J.H., Folsom A.R., Kopp J., Vaez A., Andrews J.S. et al. (2012) Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am. J. Hum. Genet., 91, 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Waite L.L., Jackson A.U., Sheu W.H., Buyske S., Absher D., Arnett D.K., Boerwinkle E., Bonnycastle L.L., Carty C.L. et al. (2013) Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet., 9, e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Replication, D.I.G., Meta-analysis, C., Asian Genetic Epidemiology Network Type 2 Diabetes, C., South Asian Type 2 Diabetes, C., Mexican American Type 2 Diabetes, C., Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples, C. Mahajan A., Go M.J., Zhang W., Below J.E. et al. (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet., 46, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickrell J.K. (2014) Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet., 94, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker S.C., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Program N.C.S. et al. (2013) Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA, 110, 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet., 6, e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coetzee S.G., Rhie S.K., Berman B.P., Coetzee G.A., Noushmehr H. (2012) FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res., 40, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grisanzio C., Werner L., Takeda D., Awoyemi B.C., Pomerantz M.M., Yamada H., Sooriakumaran P., Robinson B.D., Leung R., Schinzel A.C. et al. (2012) Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc. Natl. Acad. Sci. USA, 109, 11252–11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coetzee G.A., Jia L., Frenkel B., Henderson B.E., Tanay A., Haiman C.A., Freedman M.L. (2010) A systematic approach to understand the functional consequences of non-protein coding risk regions. Cell Cycle, 9, 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. (2009) Mapping complex disease traits with global gene expression. Nat. Rev. Genet., 10, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q., Seo J.H., Stranger B., McKenna A., Pe'er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. (2013) Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell, 152, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westra H.J., Franke L. (2014) From genome to function by studying eQTLs. Biochimica et Biophys. Acta, 1842, 1896–1902. [DOI] [PubMed] [Google Scholar]
- 34.Jia L., Landan G., Pomerantz M., Jaschek R., Herman P., Reich D., Yan C., Khalid O., Kantoff P., Oh W. et al. (2009) Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet., 5, e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q., Stram A., Chen C., Kar S., Gayther S., Pharoah P., Haiman C., Stranger B., Kraft P., Freedman M.L. (2014) Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum. Mol. Genet., 23, 5294–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmadiyeh N., Pomerantz M.M., Grisanzio C., Herman P., Jia L., Almendro V., He H.H., Brown M., Liu X.S., Davis M. et al. (2010) 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. USA, 107, 9742–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomerantz M.M., Shrestha Y., Flavin R.J., Regan M.M., Penney K.L., Mucci L.A., Stampfer M.J., Hunter D.J., Chanock S.J., Schafer E.J. et al. (2010) Analysis of the 10q11 cancer risk locus implicates MSMB and NCOA4 in human prostate tumorigenesis. PLoS Genet., 6, e1001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Q., Whitington T., Gao P., Lindberg J.F., Yang Y., Sun J., Vaisanen M.R., Szulkin R., Annala M., Yan J. et al. (2014) A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet., 46, 126–135. [DOI] [PubMed] [Google Scholar]
- 39.Akamatsu S., Takata R., Ashikawa K., Hosono N., Kamatani N., Fujioka T., Ogawa O., Kubo M., Nakamura Y., Nakagawa H. (2010) A functional variant in NKX3.1 associated with prostate cancer susceptibility down-regulates NKX3.1 expression. Hum. Mol. Genet., 19, 4265–4272. [DOI] [PubMed] [Google Scholar]
- 40.Cheng I., Chen G.K., Nakagawa H., He J., Wan P., Laurie C.C., Shen J., Sheng X., Pooler L.C., Crenshaw A.T. et al. (2012) Evaluating genetic risk for prostate cancer among Japanese and Latinos. Cancer Epidemiol. Biomarkers Prev., 21, 2048–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kote-Jarai Z., Easton D.F., Stanford J.L., Ostrander E.A., Schleutker J., Ingles S.A., Schaid D., Thibodeau S., Dork T., Neal D. et al. (2008) Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol. Biomarkers Prev., 17, 2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takata R., Akamatsu S., Kubo M., Takahashi A., Hosono N., Kawaguchi T., Tsunoda T., Inazawa J., Kamatani N., Ogawa O. et al. (2010) Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet., 42, 751–754. [DOI] [PubMed] [Google Scholar]
- 43.Xu X., Hussain W.M., Vijai J., Offit K., Rubin M.A., Demichelis F., Klein R.J. (2014) Variants at IRX4 as prostate cancer expression quantitative trait loci. Eur. J. Hum. Genet., 22, 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takayama K.I., Suzuki T., Fujimura T., Urano T., Takahashi S., Homma Y., Inoue S. (2014) CtBP2 Modulates the Androgen Receptor to Promote Prostate Cancer Progression. Cancer Res., in press. [DOI] [PubMed] [Google Scholar]
- 45.Teo Y.Y., Ong R.T., Sim X., Tai E.S., Chia K.S. (2010) Identifying candidate causal variants via trans-population fine-mapping. Genet. Epidemiol., 34, 653–664. [DOI] [PubMed] [Google Scholar]
- 46.Zaitlen N., Pasaniuc B., Gur T., Ziv E., Halperin E. (2010) Leveraging genetic variability across populations for the identification of causal variants. Am. J. Hum. Genet., 86, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris A.P. (2011) Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol., 35, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ong R.T., Wang X., Liu X., Teo Y.Y. (2012) Efficiency of trans-ethnic genome-wide meta-analysis and fine-mapping. Eur. J. Hum. Genet., 20, 1300–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kote-Jarai Z., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Dadaev T., Jugurnauth-Little S., Ross-Adams H., Al Olama A.A., Benlloch S., Halim S. et al. (2013) Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum. Mol. Genet., 22, 2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lou H., Yeager M., Li H., Bosquet J.G., Hayes R.B., Orr N., Yu K., Hutchinson A., Jacobs K.B., Kraft P. et al. (2009) Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc. Natl. Acad. Sci. USA, 106, 7933–7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng S.L., Stevens V.L., Wiklund F., Isaacs S.D., Sun J., Smith S., Pruett K., Wiley K.E., Kim S.T., Zhu Y. et al. (2009) Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol. Biomarkers Prev., 18, 1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung C.C., Ciampa J., Yeager M., Jacobs K.B., Berndt S.I., Hayes R.B., Gonzalez-Bosquet J., Kraft P., Wacholder S., Orr N. et al. (2011) Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Hum. Mol. Genet., 20, 2869–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung C.C., Boland J., Yeager M., Jacobs K.B., Zhang X., Deng Z., Matthews C., Berndt S.I., Chanock S.J. (2012) Comprehensive resequence analysis of a 123-kb region of chromosome 11q13 associated with prostate cancer. Prostate, 72, 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berndt S.I., Sampson J., Yeager M., Jacobs K.B., Wang Z., Hutchinson A., Chung C., Orr N., Wacholder S., Chatterjee N. et al. (2011) Large-scale fine mapping of the HNF1B locus and prostate cancer risk. Hum. Mol. Genet., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Purcell L.D., Gao Z., Hsu F.C., Kim S.T., Liu W., Zhu Y. et al. (2008) Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet., 40, 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kote-Jarai Z., Amin Al Olama A., Leongamornlert D., Tymrakiewicz M., Saunders E., Guy M., Giles G.G., Severi G., Southey M., Hopper J.L. et al. (2011) Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum. Genet., 129, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parikh H., Wang Z., Pettigrew K.A., Jia J., Daugherty S., Yeager M., Jacobs K.B., Hutchinson A., Burdett L., Cullen M. et al. (2011) Fine mapping the KLK3 locus on chromosome 19q13.33 associated with prostate cancer susceptibility and PSA levels. Hum. Genet., 129, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hazelett D.J., Rhie S.K., Gaddis M., Yan C., Lakeland D.L., Coetzee S.G., Ellipse/GAME-ON consortium, Practical consortium, Henderson, B.E., Noushmehr, H. et al. (2014) Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet., 10, e1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhie S.K., Hazelett D.J., Coetzee S.G., Yan C., Noushmehr H., Coetzee G.A. (2014) Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genomics, 15, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duggan D., Zheng S.L., Knowlton M., Benitez D., Dimitrov L., Wiklund F., Robbins C., Isaacs S.D., Cheng Y., Li G. et al. (2007) Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J. Natl. Cancer Inst., 99, 1836–1844. [DOI] [PubMed] [Google Scholar]
- 61.Cook M.B., Wang Z., Yeboah E.D., Tettey Y., Biritwum R.B., Adjei A.A., Tay E., Truelove A., Niwa S., Chung C.C. et al. (2014) A genome-wide association study of prostate cancer in West African men. Hum. Genet., 133, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J. et al. (2006) A common variant associated with prostate cancer in European and African populations. Nat. Genet., 38, 652–658. [DOI] [PubMed] [Google Scholar]
- 64.Yeager M., Chatterjee N., Ciampa J., Jacobs K.B., Gonzalez-Bosquet J., Hayes R.B., Kraft P., Wacholder S., Orr N., Berndt S. et al. (2009) Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet., 41, 1055–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J. et al. (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet., 40, 316–321. [DOI] [PubMed] [Google Scholar]
- 66.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. (2003) Measuring inconsistency in meta-analyses. Bmj, 327, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onengut-Gumuscu S., Chen W.M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E. et al. (2015) Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet., 47, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The Wellcome Trust Case Control Consortium, Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S. et al. (2012) Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet., 44, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakefield J. (2009) Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol., 33, 79–86. [DOI] [PubMed] [Google Scholar]
- 70.Wakefield J. (2007) A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet., 81, 208–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conti D.V., Witte J.S. (2003) Hierarchical modeling of linkage disequilibrium: genetic structure and spatial relations. Am. J. Hum. Genet., 72, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. (2012) The accessible chromatin landscape of the human genome. Nature, 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A. et al. (2011) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature, 474, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan P.Y., Chang C.W., Chng K.R., Wansa K.D., Sung W.K., Cheung E. (2012) Integration of regulatory networks by NKX3–1 promotes androgen-dependent prostate cancer survival. Mol. Cell. Biol., 32, 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andreu-Vieyra C., Lai J., Berman B.P., Frenkel B., Jia L., Jones P.A., Coetzee G.A. (2011) Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol. Cell. Biol., 31, 4648–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma N.L., Massie C.E., Ramos-Montoya A., Zecchini V., Scott H.E., Lamb A.D., MacArthur S., Stark R., Warren A.Y., Mills I.G. et al. (2013) The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell, 23, 35–47. [DOI] [PubMed] [Google Scholar]
- 77.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeggari A., Marks D.S., Larsson E. (2012) miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics, 28, 2062–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al Olama A.A., Dadaev T., Hazelett D.J., Li Q., Leongamornlert D., Saunders E.J., Stephens S., Cieza-Borrella C., Whitmore I., Garcia S.B. et al. Multiple novel prostate cancer susceptibility signals identified by fine-mapping of known risk loci among Europeans. Hum. Mol. Genet., 24, 5589–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.