Abstract
Aims/hypothesis
Zfp69 was previously identified by positional cloning as a candidate gene for obesity-associated diabetes. C57BL/6J and New Zealand obese (NZO) mice carry a loss-of-function mutation due to the integration of a retrotransposon. On the NZO background, the Zfp69 locus caused severe hyperglycaemia and loss of beta cells. To provide direct evidence for a causal role of Zfp69, we investigated the effects of its overexpression on both a lean [B6-Tg(Zfp69)] and an obese [NZO/B6-Tg(Zfp69)] background.
Methods
Zfp69 transgenic mice were generated by integrating the cDNA into the ROSA locus of the C57BL/6 genome and characterised.
Results
B6-Tg(Zfp69) mice were normoglycaemic, developed hyperinsulinaemia, and exhibited increased expression of G6pc and Pck1 and slightly reduced phospho-Akt levels in the liver. During OGTTs, glucose clearance was normal but insulin levels were significantly higher in the B6-Tg(Zfp69) than in control mice. The liver fat content and plasma triacylglycerol levels were significantly increased in B6-Tg(Zfp69) and NZO/B6-Tg(Zfp69) mice on a high-fat diet compared with controls. Liver transcriptome analysis of B6-Tg(Zfp69) mice revealed a downregulation of genes involved in glucose and lipid metabolism. Specifically, expression of Nampt, Lpin2, Map2k6, Gys2, Bnip3, Fitm2, Slc2a2, Ppargc1α and Insr was significantly decreased in the liver of B6-Tg(Zfp69) mice compared with wild-type animals. However, overexpression of Zfp69 did not induce overt diabetes with hyperglycaemia and beta cell loss.
Conclusions/interpretation
Zfp69 mediates hyperlipidaemia, liver fat accumulation and mild insulin resistance. However, it does not induce type 2 diabetes, suggesting that the diabetogenic effect of the Zfp69 locus requires synergy with other as yet unidentified genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-015-3703-8) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Diabetes, Hepatosteatosis, Insulin resistance, Lipid metabolism, Zfp69
Introduction
Multiple studies have demonstrated the shared contribution of both genetic and environmental factors in the development of type 2 diabetes mellitus [1, 2]. Genome-wide association and linkage studies have markedly improved our understanding of the genetic basis of type 2 diabetes in humans [3–5] and rodents [6–10], respectively. In mouse studies, diabetogenic quantitative trait loci (QTL) have been mapped by intercross of inbred strains with diabetes-related phenotypes, leading to the identification of several candidate genes for diabetes susceptibility or resistance [6, 8–10].
In an intercross of NON (non-obese non-diabetic) and NZO (New Zealand obese) mice, Leiter et al mapped the NON-derived diabetogenic locus Nidd1 to chromosome 4 [8]. This locus contributed substantially to the development of hyperglycaemia and hypoinsulinemia [8]. A diabetogenic locus partially overlapping with Nidd1 (Nidd/SJL) was identified in a backcross population of Swiss Jim Lambert (SJL) and NZO mice. The SJL-derived QTL caused severe hyperglycaemia and hypoinsulinaemia [9]. Moreover, when combined with the obesity QTL, Nob1, the diabetogenic effect from Nidd/SJL was greatly enhanced by a high-fat diet (HFD), which strongly suggests that Nidd/SJL contains a gene for obesity-associated diabetes [9].
By sequencing and gene expression profiling of the critical region of Nidd/SJL, we identified the zinc finger domain transcription factor Zfp69 as the most likely candidate gene within the QTL. Mouse strains such as SJL and NON carry the diabetogenic allele of Zfp69, which generates a normal full length mRNA comprising a Krüppel-associated box (KRAB) and a Znf-C2H2 domain [11]. By contrast, carriers of the retrotransposon IAPLTR1a in intron 3 of Zfp69 (NZO, C57BL/6J) produce a truncated mRNA and are less diabetes prone (NZO) or fully protected (C57BL6/J) [11].
In order to provide additional evidence for a causal role of Zfp69 and to investigate the mechanism of its diabetogenic potency, we generated a transgenic mouse line overexpressing the gene on the B6 and NZO × B6 background, and studied glucose homeostasis and fat distribution. Zfp69 induced the accumulation of liver fat and a mild insulin resistance, confirming the role of Zfp69 as a diabetogenic gene.
Methods
Generation of a transgenic mouse line overexpressing Zfp69
Zfp69 cDNA tagged with a C-terminal Myc epitope was fused to the ubiquitin C promoter. For integration into the ROSA locus, the construct was flanked by fragments corresponding with the sequence of this locus. A Zfp69 transgenic mouse line was generated with C57BL/6J mice as background strain (Ozgene, Perth, Western Australia, Australia). To generate obese NZO/B6 F1 hybrid mice, B6-Tg(Zfp69) hemizygote male mice were mated with NZO/HIBomDife female mice (R. Kluge, German Institute of Human Nutrition, Nuthetal, Germany).
The animals were housed in a controlled environment (20 ± 2°C, 12 h/12 h light/dark cycle), fed a standard diet (SD; V153x R/M-H, Ssniff, Soest, Germany) or a HFD (45% energy from fat, D12451, Research Diets, New Brunswick, NJ, USA). All animal experiments were approved by the ethics committee of the State Office of Environment, Health and Consumer Protection (State of Brandenburg, Germany).
Study design
Male mice [B6-wild-type (WT) and B6-Tg(Zfp69); NZO/B6-WT and NZO/B6-Tg(Zfp69)] were fed SD or HFD. Body weight and blood glucose levels were measured weekly from 4 to 16 weeks of age, and then every other week until 24 weeks of age. Body composition was analysed at 8 and 16 weeks of age by computed tomography (CT). OGTT was performed at week 18. Animals were killed in a postprandial state at 24 weeks of age and plasma insulin and pancreatic insulin content were estimated.
Quantitative real-time PCR
Total RNA was extracted and cDNA synthesis was performed as described previously [12] for quantitative real-time PCR (qPCR) via the LightCycler 480 system and FastStart Universal probe Master mix (Roche, Mannheim, Germany). Primers are listed in Electronic Supplementary Material (ESM) Table 1.
Nuclear extract preparation and western blotting
Liver samples (8 weeks) were homogenised and nuclear extracts were isolated by a kit according to manufacturer’s instructions (Thermo Scientific NE-PER, Bonn, Germany). Nuclear extracts were analysed by western blot with a primary antibody against ZFP69 [11]. For the detection of phospho-Akt (pAKT) and total-Akt (tAKT), liver lysates were analysed by western blot with antibodies against pAKT, tAKT, both raised in rabbit, at 1:1,000 dilution (Merck Millipore, Darmstadt, Germany) and β-actin raised in mouse at 1:5,000 dilution (Sigma, Munich, Germany).
Plasma analysis
Blood glucose was measured with a Glucometer Elite (Bayer, Leverkusen, Germany). Insulin was measured with ELISAs from DRG Diagnostics (Marburg, Germany), and triacylglycerols were measured with Triglyceride Reagent from Sigma. Measurements were performed in a blinded manner.
Pancreatic insulin content and isolation of islets of Langerhans
Detection of total pancreatic insulin, isolation of pancreatic islets and detection of glucose-stimulated insulin secretion were determined as previously described [13].
Quantification of beta cell area
Pancreases were fixed in 4% paraformaldehyde for 24 h. Evenly spaced 12 μm sections were stained for insulin (DAKO, Hamburg, Germany) to determine the beta cell area. Secondary Cy3-conjugated antibodies (Life Technologies, Darmstadt, Germany) and DAPI (Sigma) for cell-nuclei were used. The cross-sectional and insulin-positive areas were quantified using Fiji/ImageJ (Fiji, Dresden, Germany). Relative insulin-positive area was determined by quantification of cross-sectional insulin-positive area divided by cross-sectional area of the whole pancreas and presented as 100% of weight.
OGTT
B6 mice were fasted for 6 h prior to an oral or intraperitoneal application of glucose (20% solution, 2 g/kg body weight), while NZO/B6 were fasted for 16 h to reach basal glucose levels before the application. Glucose and insulin concentrations were detected at indicated time points.
Insulin tolerance test
For the insulin tolerance test (ITT), B6-WT and B6-Tg(Zfp69) mice (6 h fasted) were intraperitoneally injected with insulin (1 U for SD; 1.25 U for HFD) and the blood glucose levels were estimated from the tail-tips.
Immunohistochemistry
Paraffin sections of the liver of B6-WT and B6-Tg(Zfp69) mice were prepared as described earlier [14]. Sections were incubated with anti-Plin2 antibody (Progen Biotechnik, Heidelberg, Germany) in combination with fluorescence-conjugated Alexa488-antibody (Life Technologies) and analysed with a Leica TCS SP2 Laser Scan inverted microscope (Leica, Wetzlar, Germany).
CT
CT was performed using LaTheta LCT-200 (Hitachi-Aloka, Tokyo, Japan). Subcutaneous fat and visceral fat of B6-WT and B6-Tg(Zfp69) at 8 and 16 weeks of age on SD were determined as previously described [15].
Liver fat
Liver samples were ground in liquid nitrogen and dissolved in HB buffer (10 mM NaH2PO4, 1 mM EDTA, 1% polyoxyethylene-10-tridecyl-ether, pH 7.4). The triacylglycerol concentration was measured with the Randox TR210 kit (Randox, Wülfrath, Germany) according to the manufacturer’s instructions.
Microarray
Liver RNA was purified from four animals each (8 weeks) from B6-WT and B6-Tg(Zfp69) on SD and used for microarray analysis (Agilent Whole Genome Mouse 4 × 44K arrays, Source Bioscience, Berlin, Germany).
Results
C57BL/6J transgenic mice overexpressing Zfp69
In order to investigate the impact of Zfp69 on glucose homeostasis and fat distribution, its myc-tagged cDNA fused to the ubiquitin C promoter was integrated into the ROSA locus of B6 genome (ESM Fig. 1a). At 8 weeks of age, Zfp69 expression levels were examined in various tissues of the transgenic mouse line. As anticipated, Zfp69 was markedly overexpressed in all tissues of the transgenic mice (ESM Fig. 1b), whereas mRNA levels were below the detection level (Ct value <35 by qPCR) in B6-WT. This increase in Zfp69 mRNA levels gave rise to protein levels in liver nuclei that were twofold higher in Zfp69 transgenic mice than in SJL (ESM Fig. 1c).
The Zfp69 transgenic mice developed normally and did not show any alteration in body weight increment as compared with B6-WT mice (ESM Fig. 2).
Mild insulin resistance in B6-Tg(Zfp69) mice
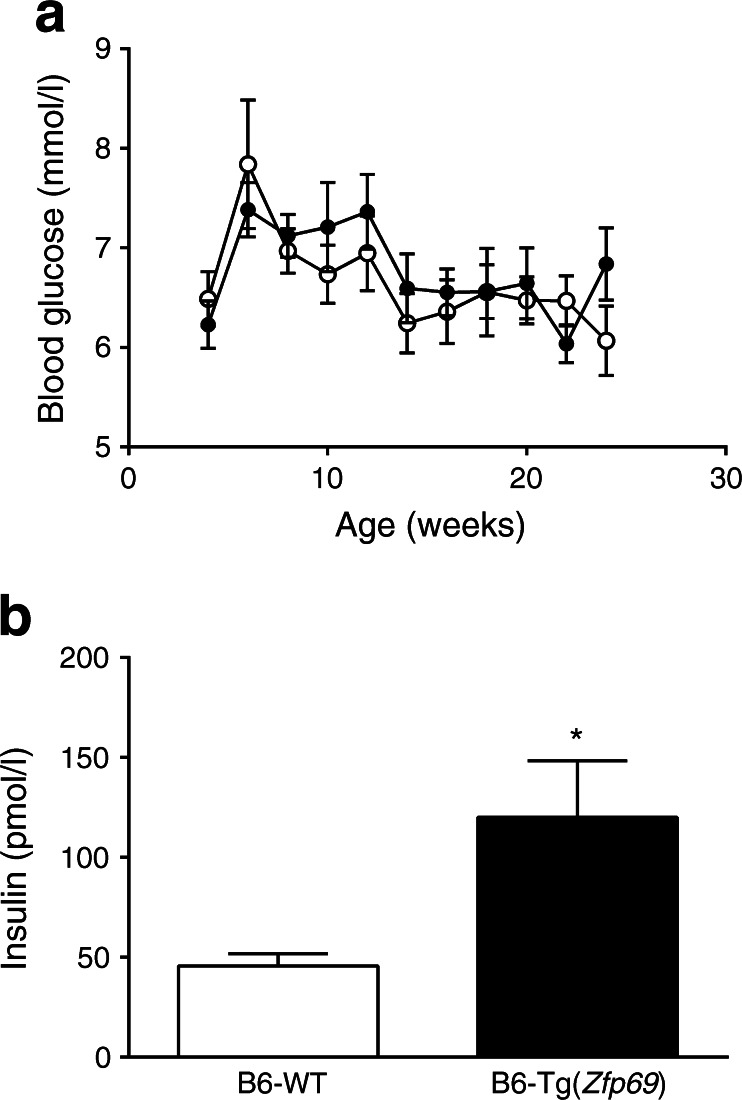
To examine the effect of Zfp69 on glucose metabolism, blood glucose and insulin levels were compared between control and transgenic mice. Zfp69 overexpression did not result in increased blood glucose levels in the fed status at any time point (Fig. 1a).
Fig. 1.
Increased plasma insulin levels in B6-Tg(Zfp69) mice. (a) Samples for measurement of randomly fed blood glucose levels were collected in the morning from mice on SD. White circles, B6-WT; black circles, B6-Tg(Zfp69). (b) Plasma insulin levels were measured in the postprandial state at the age of 24 weeks. Data are presented as mean ± SE of 7–11 mice. *p < 0.05 by Student’s t test
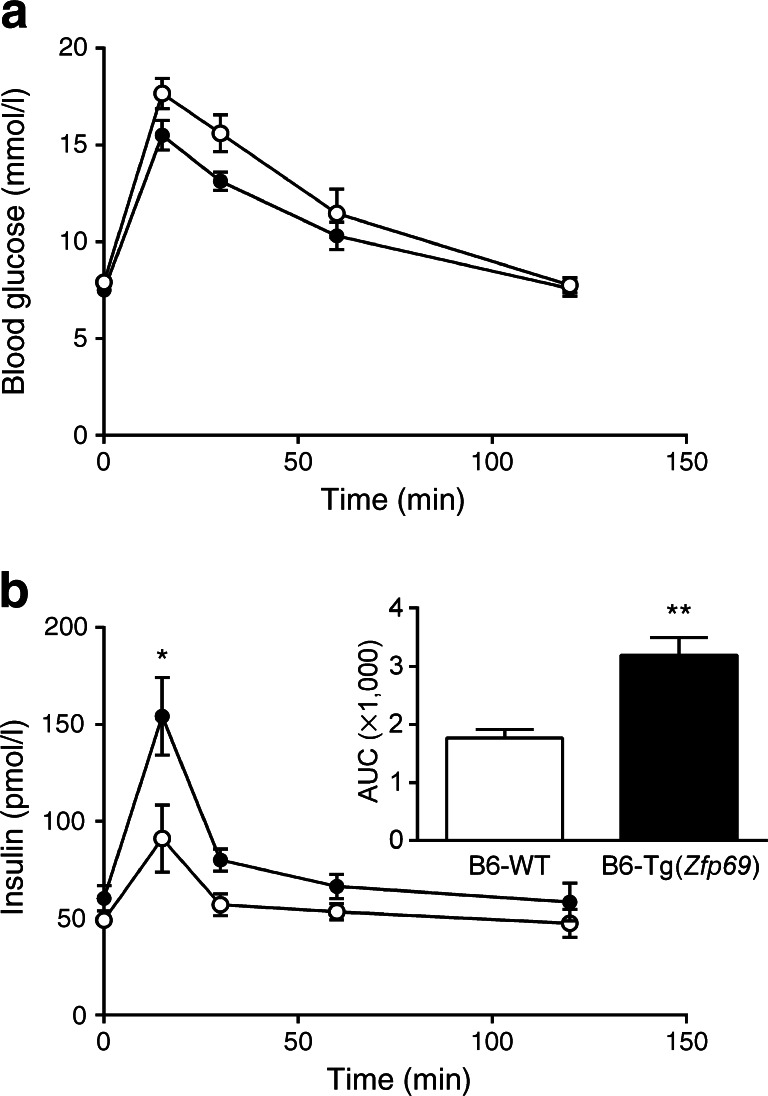
However, plasma insulin levels at 24 weeks of age were significantly higher in B6-Tg(Zfp69) than in WT mice (Fig. 1b), whereas total pancreatic insulin content did not differ between the groups (ESM Fig. 3a). Beta cell area in the pancreas did not show any difference between the genotypes (ESM Fig. 3b). Furthermore, glucose and glucose plus palmitate stimulation of isolated islets, as well as potassium chloride treatment, increased the secretion of insulin from both B6-WT and B6-Tg(Zfp69) islets (ESM Fig. 3c) without significant differences between genotypes. Glucose clearance during an OGTT at week 18 was not different between B6-WT and B6-Tg(Zfp69) mice (Fig. 2a), but the insulin concentration was significantly increased in B6-Tg(Zfp69) mice (Fig. 2b), suggesting that Zfp69 overexpression causes mild insulin resistance. Additionally, in intraperitoneal GTT (IP-GTT) blood glucose levels were not different between B6-WT and B6-Tg(Zfp69) mice (ESM Fig. 4a). However, insulin levels during IP-GTT showed only a tendency to increase (ESM Fig. 4b).
Fig. 2.
Increased insulin levels during OGTT in B6-Tg(Zfp69) mice. Mice kept on SD until 18 weeks of age were fasted for 6 h before oral glucose gavage (2 g/kg body weight). (a) Blood glucose and (b) corresponding insulin levels were measured at indicated time points. Area under the curve (AUC) values of insulin are depicted in the small panel in (b) (pmol/l × min). White circles, B6-WT; black circles, B6-Tg(Zfp69). Data are presented as mean ± SE of 10–11 animals. *p < 0.05, **p < 0.01 by Student’s t test
Consistent with the assumption of a hepatic insulin resistance, the expression of proteins involved in glucose homeostasis, such as glucose-6-phosphatase (encoded by G6pc) and phosphoenolpyruvate carboxykinase1 (encoded by Pck1), was significantly increased in livers of B6-Tg(Zfp69) mice at week 24 (ESM Fig. 5).
Effect of Zfp69 on glucose metabolism in obese mouse models
Since the diabetogenic effect of the Zfp69 locus required obesity, we challenged control and B6-Tg(Zfp69) mice with a HFD, which markedly increased body weight. In addition, NZO mice were crossed with B6-Tg(Zfp69) mice to produce obese NZO/B6 F1 progeny overexpressing Zfp69. Like B6-Tg(Zfp69) mice, NZO/B6-Tg(Zfp69) mice overexpressed Zfp69 in fat tissues, muscle and liver (ESM Fig. 6a). Blood glucose in the fed status did not differ between NZO/B6-WT and NZO/B6-Tg(Zfp69) mice (ESM Fig. 6b). As expected, the body weight of NZO/B6 F1 mice was twofold higher than that of B6 mice, with no differences between genotypes (ESM Fig. 6c), suggesting that Zfp69 overexpression does not alter growth or fat accumulation.
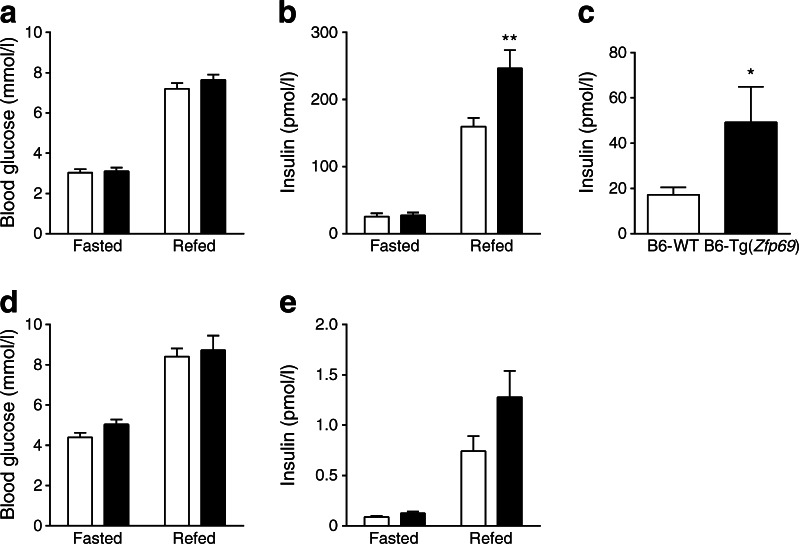
In order to investigate whether or not Zfp69 expression causes any alteration in glucose homeostasis, blood glucose levels and the corresponding insulin levels were measured after an overnight fast and 2 h after refeeding. Blood glucose levels did not differ between the genotypes in either condition (Fig. 3a), but insulin levels were significantly higher in the postprandial state of 8-week-old B6-Tg(Zfp69) mice on HFD (Fig. 3b). Interestingly, in week 18, B6-Tg(Zfp69) mice showed significantly higher fasted insulin concentrations than B6-WT mice (Fig. 3c), but with no changes in the corresponding blood glucose levels (data not shown). Similarly, NZO/B6-Tg(Zfp69) mice exhibited a tendency towards higher insulin levels after refeeding (Fig. 3e), whereas the blood glucose levels were not different between the genotypes (Fig. 3d).
Fig. 3.
Increased postprandial insulin levels in obese mouse models expressing Zfp69. (a) Blood glucose and (b) corresponding insulin levels of 8-week-old B6-WT and B6-Tg(Zfp69) mice were measured after an overnight fast and 2 h after refeeding. (c) Insulin levels in 16 h fasted status of B6-WT and B6-Tg(Zfp69) mice fed on an HFD at 18 weeks of age. (d) Blood glucose and (e) corresponding insulin levels measured in 11-week-old NZO/B6-WT and NZO/B6-Tg(Zfp69) mice after an overnight fast and 2 h refeeding. Mice were kept on HFD until the experiment. White bars, WT mice; black bars, transgenic mice. Data are presented as mean ± SE of 9–11 animals. *p < 0.05, **p < 0.01 by two-way ANOVA with Bonferroni’s multiple comparisons test
In OGTT of B6-Tg(Zfp69) mice on HFD (week 22) blood glucose levels were not different (ESM Fig. 7a), whereas corresponding insulin levels in Zfp69 transgenic mice tended to be increased (ESM Fig. 7b). ITTs revealed a tendency towards increased blood glucose levels in B6-Tg(Zfp69) mice on a HFD (ESM Fig. 8b) compared with controls, indicating impaired insulin sensitivity.
Zfp69 overexpression increases liver fat and plasma triacylglycerol levels and decreases pAKT
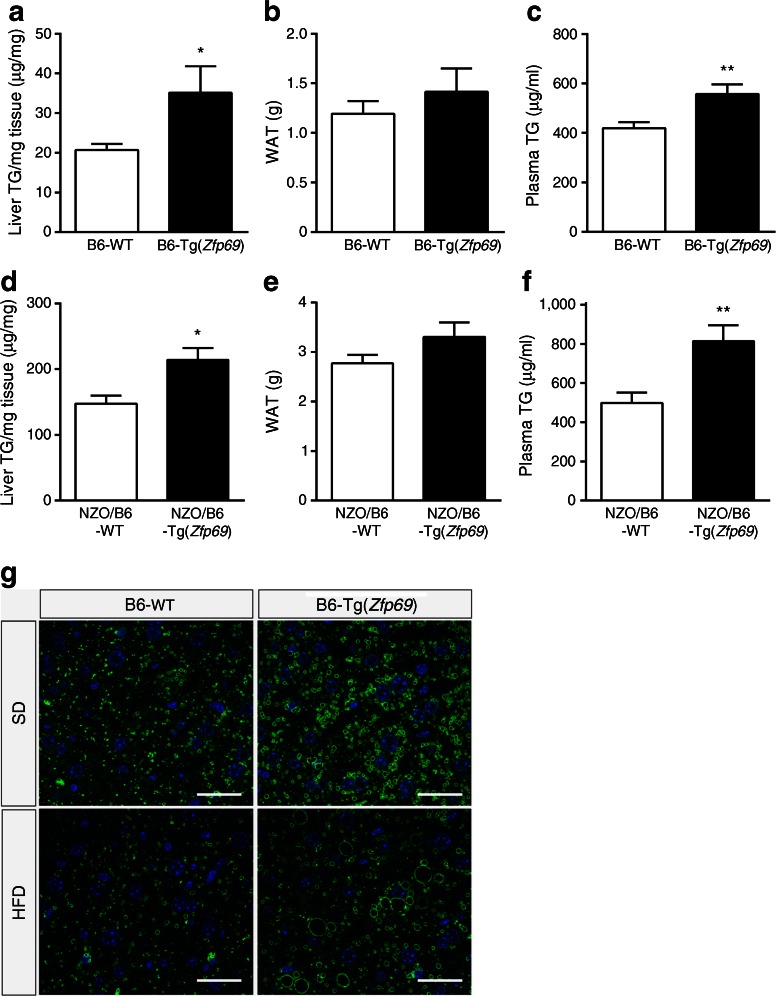
Lean mice receiving a HFD as well as obese mice often develop hepatosteatosis which participates in the development of insulin resistance [16]. To test whether or not Zfp69 overexpression enhances hepatic fat storage, we measured the liver triacylglycerol concentrations and detected significantly elevated levels, in both B6-Tg(Zfp69) and NZO/B6-Tg(Zfp69) mice on HFD (Fig. 4a and d); on SD this effect did not reach statistical significance (ESM Fig. 9). Lipid droplets, visualised by Plin2 staining were greater in numbers and larger in size in livers of B6-Tg(Zfp69) mice on both SD and HFD compared with WT mice (Fig. 4g). Hepatosteatosis was not due to hyperphagia because food intake did not differ between control and B6-Tg(Zfp69) mice on HFD (ESM Fig. 10).
Fig. 4.
Increase in liver fat and plasma triacylglycerol (TG) levels in obese mice expressing Zfp69. B6-WT and transgenic mice fed with HFD were killed at week 24 at postprandial status. (a) Liver triacylglycerol levels and (b) gonadal fat mass (white adipose tissue [WAT]) was determined. (c) Plasma triacylglycerol levels were measured at a postprandial state in B6 background mice at 12 weeks of age. NZO/B6 mice (20 weeks old) fed on an HFD were killed at postprandial status. (d) Liver triacylglycerol levels and (e) gonadal fat mass were determined. (f) Plasma triacylglycerol levels of NZO/B6 mice were estimated at postprandial status at week 23. Data are presented as mean ± SE of 7–9 animals. *p < 0.05, **p < 0.01 by Student’s t test. (g) Plin2 staining of the liver sections of B6-WT and B6-Tg(Zfp69). Livers were taken after 6 h fasting from mice on SD (20 weeks) and HFD (22 weeks). Scale bar, 30 μm
The mass of the white gonadal fat pad was not affected by Zfp69 overexpression in these mice (Fig. 4b and e). However, plasma triacylglycerol levels were significantly higher in Zfp69 transgenic mice on B6 and NZO/B6 background (Fig. 4c and f) compared with controls, as observed in the Zfp69 overexpressing congenic mice in a previous study [11]. Furthermore, we evaluated the fat distribution of B6-WT and B6-Tg(Zfp69) mice at 8 and 16 weeks of age using CT but did not detect any difference (data not shown).
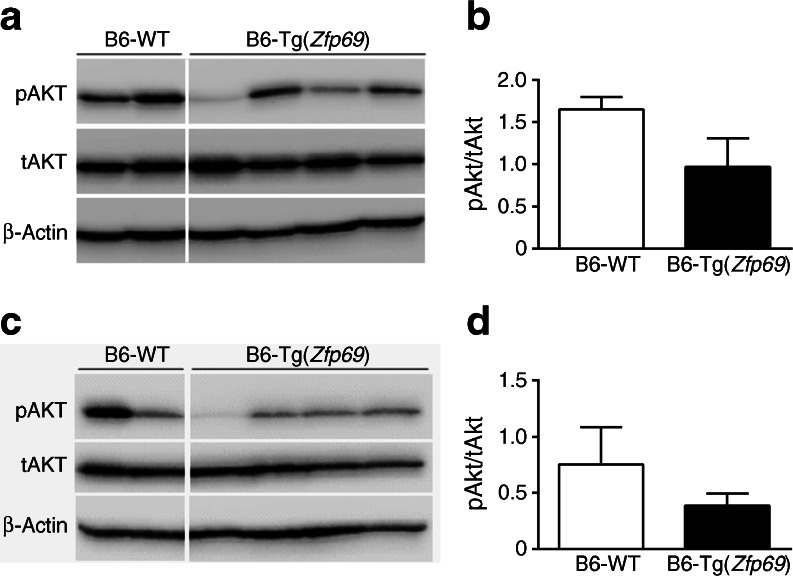
In order to examine whether or not Zfp69 overexpression affects hepatic insulin sensitivity, we investigated levels of pAKT in livers of B6 mice 20 min after intraperitoneal insulin application. We detected a tendency towards decreased pAKT levels in B6-Tg(Zfp69) on SD and HFD compared with B6-WT livers (Fig. 5).
Fig. 5.
Slightly reduced hepatic pAKT levels in B6-Tg(Zfp69) mice. B6 mice on SD (20 weeks) were killed 20 min after an intraperitoneal injection of insulin (1 U). Western blots of pAKT and tAKT in the liver of B6-WT and B6-Tg(Zfp69) mice on (a) SD and (c) HFD. Quantification of pAKT/tAKT ratio of (b) SD and (d) HFD fed mice. β-actin was determined as a loading control. Data are presented as mean ± SE of two to four animals
Zfp69 suppresses the expression of genes involved in glucose and lipid metabolism
In order to further investigate the effects of Zfp69 overexpression on the molecular regulation of glucose and lipid metabolism in the liver, we compared the liver transcriptome of 8-week-old B6-WT and B6-Tg(Zfp69) mice by a microarray analysis. Overall, 76 genes met the predefined criteria for greater than 1.5-fold differential expression (signal intensity on microarray >90, p < 0.05; Student’s t test of four analyses per group). In livers from B6-Tg(Zfp69) mice, 20 genes were upregulated and 56 genes were downregulated (Tables 1 and 2).
Table 1.
Upregulated genes in the liver of B6-Tg(Zfp69) mice
| Gene bank ID | Gene symbol | Description | Ratio |
|---|---|---|---|
| NM_007919 | Cela2a | Chymotrypsin-like elastase family, member 2A | 6.55 |
| NM_025350 | Cpa1 | Carboxypeptidase A1 | 6.00 |
| NM_025583 | Ctrb1 | Chymotrypsinogen B1 | 5.55 |
| NM_001033875 | Ctrc | Chymotrypsin C (caldecrin) | 5.52 |
| NM_025469 | Clps | Colipase, pancreatic | 5.23 |
| NM_019738 | Nupr1 | Nuclear protein 1 | 3.99 |
| NM_007446 | Amy1 | Amylase 1, salivary | 2.46 |
| NM_024440 | Derl3 | Der1-like domain family, member 3 | 2.21 |
| NM_177388 | Slc41a2 | Solute carrier family 41, member 2 | 2.13 |
| NM_013769 | Tjp3 | Tight junction protein 3 | 1.79 |
| NM_175138 | Dnaic1 | Dynein, axonemal, intermediate chain 1 | 1.78 |
| NM_138302 | Ecgf1 (also known as Tymp) | Endothelial cell growth factor 1 (platelet-derived) | 1.66 |
| NM_146116 | Tubb2c (also know as Tubb4b) | Tubulin, beta 2c | 1.65 |
| NM_009777 | C1qb | Complement component 1, q subcomponent, beta polypeptide | 1.57 |
| NM_013612 | Slc11a1 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 1.57 |
| NM_007572 | C1qa | Complement component 1, q subcomponent, alpha polypeptide | 1.55 |
| NM_017372 | Lyzs (also known as Lyz2) | Lysozyme | 1.52 |
| NM_153074 | Lrrc25 | Leucine rich repeat containing 25 | 1.51 |
| NM_001037859 | Csf1r | Colony stimulating factor 1 receptor | 1.51 |
| NM_013632 | Pnp | Purine-nucleoside phosphorylase | 1.51 |
Livers were collected from mice on SD at 8 weeks of age
Genes with a greater than 1.5-fold increased expression are listed
Table 2.
Downregulated genes in the liver of B6-Tg(Zfp69) mice
| Gene bank ID | Gene symbol | Description | Ratio |
|---|---|---|---|
| NM_201256 | Eif4ebp3 | Eukaryotic translation initiation factor 4E binding protein 3 | 0.42 |
| NM_032002 | Nrg4 | Neuregulin 4 | 0.45 |
| NM_177368 | Tmtc2 | Transmembrane and tetratricopeptide repeat containing 2 | 0.50 |
| NM_008898 | Por | P450 (cytochrome) oxidoreductase | 0.52 |
| NM_013631 | Pklr | Pyruvate kinase liver and red blood cell | 0.55a |
| NM_207665 | Olfr144 (also known as Olfr1537) | Olfactory receptor 144 | 0.55 |
| NM_021524 | Nampt | Nicotinamide phosphoribosyltransferase | 0.56b |
| NM_007812 | Cyp2a5 | Cytochrome P450, family 2, subfamily a, polypeptide 5 | 0.56 |
| NM_011943 | Map2k6 | Mitogen-activated protein kinase 6 | 0.57c |
| NM_145572 | Gys2 | Glycogen synthase 2 | 0.59d |
| NM_009760 | Bnip3 | BCL2/adenovirus E1B interacting protein 1, NIP3 | 0.60e |
| NM_011391 | Slc16a7 | Solute carrier family 16 (monocarboxylic acid transporters), member 7 | 0.61 |
| NM_172668 | Lrp4 | Low density lipoprotein receptor-related protein 4 | 0.63 |
| NM_010361 | Gstt2 | Glutathione S-transferase, theta 2 | 0.64 |
| NM_173397 | Fitm2 | Fat storage-inducing transmembrane protein 2 | 0.65f |
| NM_009647 | Ak3l1 (also known as Ak4) | Adenylate kinase 3 alpha-like 1 | 0.65 |
| NM_007618 | Serpina6 | Serine (or cysteine) peptidase inhibitor, clade A, member 6 | 0.65 |
| NM_007520 | Bach1 | BTB and CNC homology 1 | 0.66 |
| NM_172838 | Slc16a12 | Solute carrier family 16 (monocarboxylic acid transporters), member 12 | 0.66 |
| NM_031884 | Abcg5 | ATP-binding cassette, sub-family G (WHITE), member 5 | 0.67 |
| NM_008772 | P2ry1 | Purinergic receptor P2Y, G-protein coupled 1 | 0.68 |
| NM_172563 | Hlf | Hepatic leukemia factor | 0.68 |
| NM_001081131 | Dhtkd1 | Dehydrogenase E1 and transketolase domain containing 1 | 0.68 |
| NM_022882 | Lpin2 | Lipin 2 | 0.68g |
| NM_009030 | Rbbp4 | Retinoblastoma binding protein 4 | 0.69 |
| NM_026003 | Smarca2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 0.69 |
| NM_024198 | Gpx7 | Glutathione peroxidase 7 | 0.69 |
| NM_008813 | Enpp1 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 0.70 |
| NM_145076 | Trim24 | Tripartite motif protein 24 | 0.71 |
| NM_024289 | Osbpl5 | Oxysterol binding protein-like 5 | 0.71 |
| NM_031197 | Slc2a2 | Solute carrier family 2 (facilitated glucose transporter), member 2 | 0.71h |
| NM_008188 | Thumpd3 | THUMP domain containing 3 | 0.71 |
| NM_001081260 | Tnks1bp1 | Tankyrase 1 binding protein 1 | 0.71 |
| NM_020567 | Gmnn | Geminin | 0.72 |
| NM_028790 | Acot12 | Acyl-CoA thioesterase 12 | 0.72 |
| NM_008904 | Ppargc1a | Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha | 0.72i |
| NM_022722 | Dpys | Dihydropyrimidinase | 0.72 |
| NM_183262 | Stk35 | Serine/threonine kinase 35 | 0.72 |
| NM_146078 | Ubr2 | Ubiquitin protein ligase E3 component n-recognin 2 | 0.73 |
| NM_011200 | Ptp4a1 | Protein tyrosine phosphatase 4a1 | 0.73 |
| NM_198300 | Cpeb3 | Cytoplasmic polyadenylation element binding protein 3 | 0.73 |
| NM_009981 | Pcyt1a | Phosphate cytidylyltransferase 1, choline, alpha isoform | 0.73 |
| NM_009738 | Bche | Butyrylcholinesterase | 0.74 |
| NM_178378 | Iqcg | IQ motif containing G | 0.74 |
| NM_011864 | Papss2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | 0.74 |
| NM_177327 | Wwp1 | WW domain containing E3 ubiquitin protein ligase 1 | 0.74 |
| NM_172907 | Olfml1 | Olfactomedin-like 1 | 0.74 |
| NM_145823 | Pitpnc1 | Phosphatidylinositol transfer protein, cytoplasmic 1 | 0.74 |
| NM_010568 | Insr | Insulin receptor | 0.74j |
| NM_013505 | Dsc2 | Desmocollin 2 | 0.74 |
| NM_001013391 | Cpsf6 | Cleavage and polyadenylation specific factor 6 | 0.74 |
| NM_011387 | Slc10a1 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | 0.74 |
| NM_007996 | Fdx1 | Ferredoxin 1 | 0.74 |
| NM_177321 | Mia2 | Melanoma inhibitory activity 2 | 0.74 |
| NM_022996 | Ndfip1 | Nedd4 family interacting protein 1 | 0.75 |
| NM_153599 | Cdk8 | Cyclin-dependent kinase 8 | 0.75 |
Livers were collected from mice on SD at 8 weeks of age
Genes with reduced expression by >25% are listed
a–jThe ten genes selected by a literature survey and a MetaCore-based pathway enrichment analysis as being related to diabetes and lipid metabolism
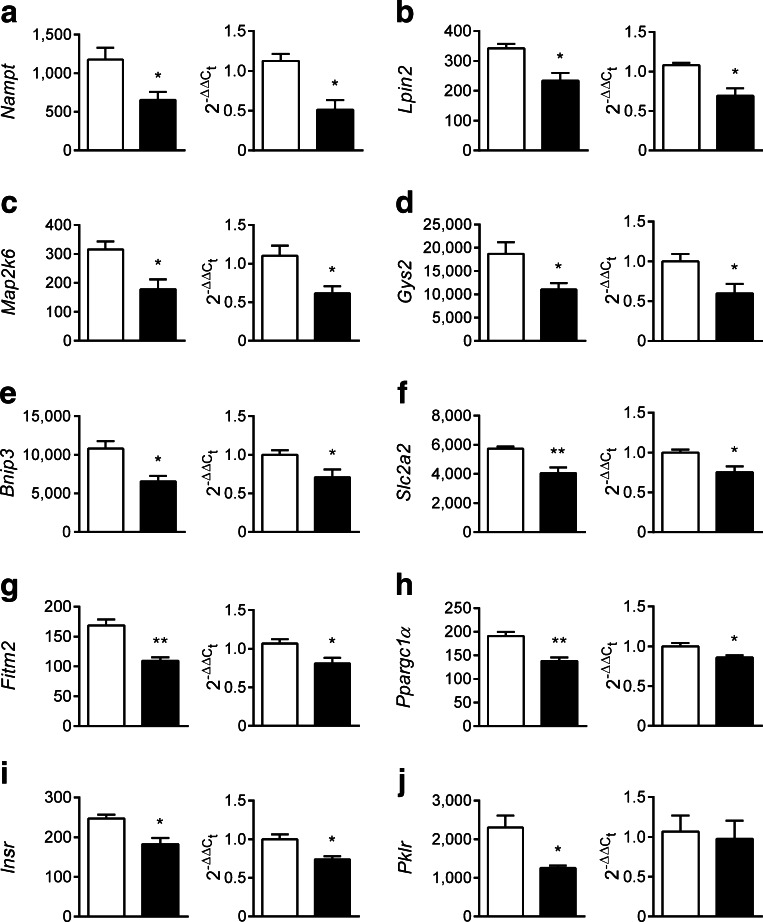
Since ZFP69 is an inhibitory regulator of gene expression by analogy, we further analysed the genes that were suppressed in livers of B6-Tg(Zfp69) mice. According to a literature survey and a MetaCore-based pathway enrichment analysis, ten genes with a reduced expression by >25% were related to diabetes and lipid metabolism (Table 2). The Zfp69-dependent suppression of nine of these genes, Nampt, Lpin2, Map2k6, Gys2, Bnip3, Fitm2, Slc2a2, Ppargc1α and Insr was validated by qPCR (Fig. 6).
Fig. 6.
Suppression of hepatic genes involved in glucose and lipid metabolism in response to Zfp69 overexpression. Genes involved in glucose and lipid metabolism were selected by MetaCore-based pathway enrichment analysis among genes exhibiting >25% decreased expression in the liver of B6-Tg(Zfp69) mice at 8 weeks of age on SD. (a) Nampt (b) Lpin2 (c) Map2k6 (d) Gys2 (e) Bnip3 (f) Slc2a2 (g) Fitm2 (h) Ppargc1α (i) Insr and (j) Pklr. Each gene is presented by the signal intensity on microarray (left graph) and the expression validation by qPCR (right graph). White bars, B6-WT; black bars, B6-Tg(Zfp69). Data are presented as mean ± SE of three to four animals. *p < 0.05, **p < 0.01 by Student’s t test
Discussion
Zfp69 has been previously suggested to be a causal gene in the diabetes loci Nidd1 and Nidd/SJL [11]. In this study, we present direct evidence in support of this conclusion, and demonstrate that Zfp69 increases liver fat content and plasma triacylglycerol concentrations and causes moderate insulin resistance.
B6-Tg(Zfp69) mice displayed elevated insulin levels in OGTTs at weeks 18 and 24 compared with B6-WT animals. Interestingly, IP-GTT did not significantly increase insulin levels. As IP-GTT by-passes incretin stimulation, in contrast to OGTT, it can be speculated that Zfp69 overexpression increases incretin release and thereby insulin secretion. Along these lines, we did not detect an elevated glucose-stimulated insulin secretion in isolated islets of transgenic mice.
Elevated liver triacylglycerol concentrations, slightly reduced hepatic pAKT levels and higher glucose levels during ITT of HFD fed mice indicate that Zpf69 mediates a moderate insulin resistance. Accordingly, hepatic expression of G6pc and Pck1 was significantly increased, as shown by others in the insulin-resistant state [17, 18]. The higher liver fat and plasma triacylglycerol levels could result either from hepatic insulin resistance or from Zfp69 overexpression in adipose tissue. Similarly, in a previous study, we observed hepatosteatosis and hyperlipidaemia in recombinant congenic mice carrying the Zfp69 locus of SJL mice and hypothesised that this is due to reduced lipid storage capacity of adipocytes [11]. However, as Zfp69 transgenic mice did not exhibit smaller fat pads, elevated plasma triacylglycerol concentrations appear not to be a consequence of elevated lipolysis.
Overall, the phenotype of the transgenic mice is much weaker than that of recombinant congenic mice carrying the Zfp69 locus of SJL mice [11]. Contrary to our expectations, we did not observe hyperglycaemia or beta cell failure in B6-Tg(Zfp69) or in NZO/B6-Tg(Zfp69) mice. A possible explanation for the mild phenotype could be that Zfp69 needs other diabetogenic gene variants in order to produce hyperglycaemia and beta cell failure. We have previously shown that the introgression of Nidd/SJL encompassing the intact Zfp69 into the NZO background induced hypoinsulinaemia due to beta cell failure, resulting in hyperglycaemia [9]. However, when the Nidd/SJL locus was introgressed into B6-ob mice, it caused an altered fat distribution (hepatosteatosis and a reduced white gonadal fat depot) as well as mild hyperglycaemia, but not beta cell failure [11], consistent with the assumption that B6 mice carry diabetes suppressing genes [6]. Moreover, previous studies showed that Nidd/SJL interacts with NZO genes (e.g. on chromosomes 1 and 15) that enhance the diabetogenic effect of Nidd/SJL [9, 11].
In addition, the possibility that the Nidd/SJL locus harbours additional diabetogenic genes that are in linkage disequilibrium with Zfp69 cannot be discounted. Indeed, a linkage study of an F2 intercross between B6 and DBA/2 mouse lines with a deficiency in the leptin receptor (db/db) identified an interval of chromosome 4 that was associated with the traits blood glucose and plasma triacylglycerols [19], confirming that the region is responsible for obesity-induced diabetes. Furthermore, from their genome-wide analysis on an (B6xC3H/HeJ)F2 intercross with a deficiency in apolipoprotein E, Logsdon et al concluded that Zfp69 variants are associated with body weight, blood glucose and cholesterol levels [20].
We compared the liver transcriptome of B6-WT and B6-Tg(Zfp69) mice at an early time point (week 8) before insulin resistance occurs in order to exclude secondary effects of insulin resistance and to examine the direct effect of Zfp69 overexpression. Microarray analysis revealed that Zfp69 overexpression decreased the expression of several genes involved in glucose and lipid metabolism. Interestingly, none of the upregulated genes in the microarray was related to glucose or lipid metabolism.
Nampt plays a key role in NAD synthesis, which is needed for the enzymatic activity of sirtuin1 (SIRT1), an important regulator of glucose and lipid metabolism [21] and thereby regulates hepatic triacylglycerol homeostasis [22]. Decreased expression of Nampt could participate in the development of hepatic steatosis and insulin resistance by reducing SIRT1 activity. Hepatic deletion of SIRT1 impaired peroxisome proliferator-activated receptor α function, decreased fatty acid beta-oxidation and caused hepatic steatosis [23].
Lpin2 is one of three members of the lipin family that act as phosphatidate phosphatases. These enzymes are required for glycerolipid biosynthesis and also act as transcriptional co-activators that regulate expression of lipid metabolising genes. Interestingly, polymorphisms in the LPIN1 and LPIN2 genes are associated with traits of metabolic disease, including insulin sensitivity, diabetes and increased blood pressure, as well as the response to thiazolidinediones [24]. Furthermore, Lpin1 expression levels in adipose tissue and liver are positively correlated with insulin sensitivity [25]. Thus, a reduced Lpin2 expression in the liver of B6-Tg(Zfp69) mice might affect lipid metabolism leading to insulin resistance.
Gys2 catalyses the rate-limiting step in the synthesis of glycogen. The liver-specific deletion of Gys2 resulted not only in a marked reduction of glycogen storage but also in impaired glucose tolerance [26]. Mitogen-activated protein kinase (MAPK) kinase 6 (MAP2K6) is a dual-specificity protein kinase that activates the stress-activated protein p38 MAPK [27]. In the livers of obese mice, p38 MAPK activity was markedly reduced compared with that of lean mice [28]. The liver-specific overexpression of constitutively active MAP2K6 (MKK6Glu) in obese ob/ob mice markedly reduced plasma insulin concentrations and improved glucose tolerance [28], indicating that reduced Map2k6 expression as detected in B6-Tg(Zfp69) mice might impair glucose homeostasis.
Reduced Bnip3 in mice overexpressing Zfp69 can be also linked to elevated hepatic fat storage because deletion of Bnip3 in mice resulted in increased lipid synthesis in the liver [29]. Bnip3 localises to the outer mitochondrial membrane and plays an important role in mitophagy and mitochondrial dynamics and is thereby vital in the adaptive response to changes in energy balance arising from deficiencies in oxygen or glucose availability [30].
Fitm2 encodes the fat storage-inducing transmembrane protein2, an evolutionarily conserved protein that is directly involved in fat storage. It is located in the endoplasmic reticulum and involved in partition of triacylglycerol into lipid droplets [31]. In a three-stage association study performed with an east Asian population FITM2 was shown to associate with type 2 diabetes [32].
Ppargc1α is a transcriptional coactivator that is a central inducer of mitochondrial biogenesis [33] and a regulator of gluconeogenesis [34, 35]. As increased hepatosteatosis was shown to correlate with reduced Ppargc1α expression [36], it can be speculated that lower Ppargc1α levels in the liver of B6-Tg(Zfp69) mice participate in ectopic fat storage and subsequently cause insulin resistance.
The molecular mechanism by which Zfp69 regulates the expression of target genes remains to be elucidated and further studies are required to provide direct functional evidence that Zfp69 modulates these candidates directly. By sequence homology, Zfp69 is a member of KRAB domain zinc finger proteins, which are assumed to suppress expression of target genes [37]. Our microarray results reflect this to some extent, with considerably more genes downregulated in the liver than upregulated.
In conclusion, our analysis of the phenotype of transgenic mice overexpressing Zfp69 indicates that Zfp69 plays a role lipid metabolism, liver fat accumulation and presumably in incretin release. Thus, the data are consistent with the conclusion that Zfp69 is a causal gene of the diabetes locus Nidd/SJL. However, the data also suggest that Zfp69 is not the only factor mediating the diabetogenic effect of this locus.
Electronic supplementary material
(PDF 138 kb)
(PDF 97.1 kb)
(PDF 244 kb)
(PDF 127 kb)
(PDF 120 kb)
(PDF 281 kb)
(PDF 122 kb)
(PDF 90.1 kb)
(PDF 58.5 kb)
(PDF 103 kb)
(PDF 127 kb)
Acknowledgements
We thank the following colleagues at the German Institute of Human Nutrition, Potsdam-Rebruecke, Germany, M. Niehaus, A. Teichmann and M. Rath for technical assistance, S. Sartig for animal care, A. Kamitz, N. Hallahan, and W. Jonas for critical readings.
Funding
This study was supported by the German Federal Ministry of Education and Research (NGFNplus: 01GS0821, NEUROTARGET: 01GI0847; DZD: 01GI0922 and 01GI0925) and the state of Brandenburg, Germany.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
BC acquired and analysed the data, and wrote and edited the manuscript. MS, NS and DJ acquired and analysed data and revised the manuscript. SS and HGJ designed and directed the study and edited the manuscript. AS designed and directed the study, analysed the data and wrote and edited the manuscript. AS is the guarantor of this work. All authors approved the final version of the manuscript.
Abbreviations
- CT
Computed tomography
- HFD
High-fat diet
- IP-GTT
Intraperitoneal GTT
- ITT
Insulin tolerance test
- KRAB
Krüppel-associated box
- MAPK
Mitogen-activated protein kinase
- MAP2K6
MAPK kinase 6
- NON
Non-obese non-diabetic
- NZO
New Zealand obese
- qPCR
Quantitative real-time PCR
- QTL
Quantitative trait locus
- SD
Standard diet
- SIRT1
Sirtuin1
- SJL
Swiss Jim Lambert
- WT
Wild-type
References
- 1.Scott RA, Fall T, Pasko D, et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independently of obesity. Diabetes. 2014;63:4378–4387. doi: 10.2337/db14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgio E, Lopomo A, Migliore L. Obesity and diabetes: from genetics to epigenetics. Mol Biol Rep. 2015;42:799–818. doi: 10.1007/s11033-014-3751-z. [DOI] [PubMed] [Google Scholar]
- 3.Dimas AS, Lagou V, Barker A, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2013;6:2158–2171. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara K, Fujita H, Johnson TA, et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet. 2014;23:239–246. doi: 10.1093/hmg/ddt399. [DOI] [PubMed] [Google Scholar]
- 5.Kluth O, Matzke D, Schulze G, Schwenk RW, Joost HG, Schurmann A. Differential transcriptome analysis of diabetes resistant and sensitive mouse islets reveals significant overlap with human diabetes susceptibility genes. Diabetes. 2014;63:4230–4238. doi: 10.2337/db14-0425. [DOI] [PubMed] [Google Scholar]
- 6.Clee SM, Nadler ST, Attie AD. Genetic and genomic studies of the BTBR ob/ob mouse model of type 2 diabetes. Am J Ther. 2005;12:491–498. doi: 10.1097/01.mjt.0000178781.89789.25. [DOI] [PubMed] [Google Scholar]
- 7.Clee SM, Yandell BS, Schueler KM, et al. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet. 2006;38:688–693. doi: 10.1038/ng1796. [DOI] [PubMed] [Google Scholar]
- 8.Leiter EH, Reifsnyder PC, Flurkey K, Partke HJ, Junger E, Herberg L. NIDDM genes in mice: deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes. 1998;47:1287–1295. doi: 10.2337/diab.47.8.1287. [DOI] [PubMed] [Google Scholar]
- 9.Plum L, Giesen K, Kluge R, et al. Characterisation of the mouse diabetes susceptibility locus Nidd/SJL: islet cell destruction, interaction with the obesity QTL Nob1, and effect of dietary fat. Diabetologia. 2002;45:823–830. doi: 10.1007/s00125-002-0796-7. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt C, Gonzaludo NP, Strunk S, et al. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol Genomics. 2008;34:42–53. doi: 10.1152/physiolgenomics.00267.2007. [DOI] [PubMed] [Google Scholar]
- 11.Scherneck S, Nestler M, Vogel H, et al. Positional cloning of zinc finger domain transcription factor Zfp69, a candidate gene for obesity-associated diabetes contributed by mouse locus Nidd/SJL. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchmann J, Meyer C, Neschen S, et al. Ablation of the cholesterol transporter adenosine triphosphate-binding cassette transporter G1 reduces adipose cell size and protects against diet-induced obesity. Endocrinology. 2007;148:1561–1573. doi: 10.1210/en.2006-1244. [DOI] [PubMed] [Google Scholar]
- 13.Schulz N, Himmelbauer H, Rath M, et al. Role of medium- and short-chain L-3-hydroxyacyl-CoA dehydrogenase in the regulation of body weight and thermogenesis. Endocrinology. 2011;152:4641–4651. doi: 10.1210/en.2011-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaschke A, Chung B, Hesse D, et al. The GTPase ARFRP1 controls the lipidation of chylomicrons in the Golgi of the intestinal epithelium. Hum Mol Genet. 2012;21:3128–3142. doi: 10.1093/hmg/dds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lubura M, Hesse D, Neumann N, Scherneck S, Wiedmer P, Schurmann A. Non-invasive quantification of white and brown adipose tissues and liver fat content by computed tomography in mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Meijer VE, Le HD, Meisel JA, Sharma AK, Popov Y, Puder M. Tumor necrosis factor alpha-converting enzyme inhibition reverses hepatic steatosis and improves insulin sensitivity markers and surgical outcome in mice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamino T, Orimo M, Shimizu I, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009;15:1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Lee J, Reno CM, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis RC, van Nas A, Castellani LW, et al. Systems genetics of susceptibility to obesity-induced diabetes in mice. Physiol Genomics. 2012;44:1–13. doi: 10.1152/physiolgenomics.00003.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logsdon BA, Hoffman GE, Mezey JG. Mouse obesity network reconstruction with a variational Bayes algorithm to employ aggressive false positive control. BMC Bioinf. 2012;13:53. doi: 10.1186/1471-2105-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai S, Yoshino J. The importance of NAMPT/NAD/SIRT1 in the systemic regulation of metabolism and ageing. Diabetes Obes Metab. 2013;15(Suppl 3):26–33. doi: 10.1111/dom.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J Biol Chem. 2011;286:14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bou Khalil M, Blais A, Figeys D, Yao Z. Lipin - the bridge between hepatic glycerolipid biosynthesis and lipoprotein metabolism. Biochim Biophys Acta. 2010;1801:1249–1259. doi: 10.1016/j.bbalip.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Reue K. The lipin family: mutations and metabolism. Curr Opin Lipidol. 2009;20:165–170. doi: 10.1097/MOL.0b013e32832adee5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irimia JM, Meyer CM, Peper CL, et al. Impaired glucose tolerance and predisposition to the fasted state in liver glycogen synthase knock-out mice. J Biol Chem. 2010;285:12851–12861. doi: 10.1074/jbc.M110.106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allred DR, Staehelin LA. Implications of cytochrome b6/f location for thylakoidal electron transport. J Bioenerg Biomembr. 1986;18:419–436. doi: 10.1007/BF00743013. [DOI] [PubMed] [Google Scholar]
- 28.Han D, Moon S, Kim H, et al. Detection of differential proteomes associated with the development of type 2 diabetes in the Zucker rat model using the iTRAQ technique. J Proteome Res. 2011;10:564–577. doi: 10.1021/pr100759a. [DOI] [PubMed] [Google Scholar]
- 29.Glick D, Zhang W, Beaton M, et al. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol. 2012;32:2570–2584. doi: 10.1128/MCB.00167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rikka S, Quinsay MN, Thomas RL, et al. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross DA, Zhan C, Silver DL. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc Natl Acad Sci U S A. 2011;108:19581–19586. doi: 10.1073/pnas.1110817108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 34.Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z, Boss O. Targeting PGC-1 alpha to control energy homeostasis. Expert Opin Ther Targets. 2007;11:1329–1338. doi: 10.1517/14728222.11.10.1329. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Kamat A, Pergola P, Swamy A, Tio F, Cusi K. Metabolic factors in the development of hepatic steatosis and altered mitochondrial gene expression in vivo. Metab Clin Exp. 2011;60:1090–1099. doi: 10.1016/j.metabol.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 138 kb)
(PDF 97.1 kb)
(PDF 244 kb)
(PDF 127 kb)
(PDF 120 kb)
(PDF 281 kb)
(PDF 122 kb)
(PDF 90.1 kb)
(PDF 58.5 kb)
(PDF 103 kb)
(PDF 127 kb)