Abstract
Burkholderia pseudomallei is a soil dwelling Gram-negative bacteria predominates in Southeast Asia zone and the tropical part of Australia. Genetic diversity has been explored among various populations and environments worldwide. To date, little data is available on MLST profiling of clinical B. pseudomallei isolates in peninsular Malaysia. In this brief report, thirteen culture positive B. pseudomallei cases collected from a single population of Terengganu state in the Western Peninsular Malaysia and were confirmed by In-house TTS1-PCR. Isolates were subjected for multi-locus sequence typing (MLST) to explore their genotypic diversity and to investigate for possible clonal clustering of a certain sequence type. Patient’s clinical information was examined to investigate for clinical correlation among the different genotypes. In spite of small sample set, MLST results indicated predictive results; considerable genotypic diversity, predominance and novelty among B. pseudomallei collected over a single geographically-located population in Malaysia. Massive genotypic heterogeneity was observed; 8 different sequence types with predominance of sequence type 54 and discovery of two novel sequence types. However, no clear pathogenomic or organ tropism clonal relationships were predicted.
Keywords: Burkholderia pseudomallei, melioidosis, multi-locus sequence typing, Malaysia
Introduction
Burkholderia pseudomallei is a soil saprophyte Gram-negative bacteria and is the causative agent of melioidosis. It was first isolated in patients with fatal pneumonia. The infection (melioidosis) is usually acquired by inoculation and inhalation the bacteria causing a wide spectrum clinical disease, particularly in those with predisposing factors such as diabetes mellitus. There is marked heterogeneity in clinical presentation and disease severity among melioidosis patients. The most severe manifestations of melioidosis are pneumonia and severe sepsis [1]. Melioidosis is known to predominate exclusively in Southeast Asia and Northern Australia, but still can be recognized in other countries worldwide [2]. Regional variations in melioidosis was reported; with prostatic abscesses and encephalomyelitis being common in Australia, and parotid abscesses and hepatosplenic suppuration being described in Thailand [3,4]. The reason behind this diversity is still unclear whether it was due to host, bacterial, or environmental factors [1].
The study of epidemiology using molecular methods gave more details regarding bacterial diversity and distribution and improved the knowledge about disease pathogenesis [5]. In Malaysia, many molecular epidemiological studies were performed on environmental, animal and clinical isolates of B. pseudomallei using pulsed-field gel electrophoresis (PFGE) [6,7], random amplification of polymorphic DNA (RAPD) [8], ribotyping [9,10] and multi-locus sequence typing (MLST) [1]. To date, most genotyping studies described the genetic relatedness among the studied strains without correlating the genotypes with clinical characteristics in melioidosis patients. This short report provided the first MSLT-based molecular epidemiology study on clinical B. pseudomallei strains in Peninsular Malaysia.
Methodology
Collection and preparation of bacterial strains
Archived isolates of B. pseudomallei were previously cultivated from different kinds of clinical samples obtained from thirteen melioidosis patients residing in Terengganu state who were admitted to Hospital Universiti Sains Malaysia (HUSM) at Kelantan state. B. pseudomallei isolates were identified by typical biochemical reactions using API20NE® (bioMérieux, France) before being stocked under -80°C. Only a single isolate from each patient was obtained to preserve the assumption of independence of observations. Samples collection and B. pseudomallei isolation, complete identification and archiving were done as part of routine diagnostics following the standard protocol at the Medical Microbiology & Parasitology Laboratory, HUSM. B. pseudomallei was identified by the growth of silver-whitish colonies on blood agar, Gram negative staining, typical biochemical reaction on triple sugar iron, oxidase positive, motility, growth at 42°C and colistin-resistance. Commercially available identification system, API20NE® (bioMérieux, France). Once needed, the bacteria were reactivated by overnight aerobic cultivation on Tryptone soya agar (Oxoid Ltd., Basingstoke, UK).
DNA preparation
Dense bacterial suspensions were prepared in Tryptone soya broth and subjected for genomic DNA extraction using DNeasy tissue kit (Qiagen Inc., Hilden, Germany) according to manufacturer instructions, with minor modification of the last step in which the final incubation in the elution step was prolonged to 30 minutes to increase DNA yield.
In-house PCR and multi-locus sequence typing (MLST)
Further identification confirmation was done by PCR assay designed, optimized and validated in our laboratory settings that targeted 316bp in sctQ putative gene which is located adjacent to orf1-sctQ gap region of type three secretion system 1 (TTS 1) (Zueter, A., unpublished protocol).
MLST protocol developed previously [11] was applied in this report, in which PCR primers for amplifying seven house-keeping genes were used. For every gene, the PCR mixture contained 5 µl of extracted DNA in 45 µl of a PCR mastermix containing 5x MyTaq Red reaction buffer (Bioline Ltd UK), which comprises 5 mM dNTPs, 15 mM MgCl2, stabilizers and enhancers, 400 nM of each forward and reverse primer, 5 U/μl MyTaq DNA polymerase (Bioline Ltd UK), and PCR-grade water. Amplification was performed in SuperCycler Trinity (Kyratec, Australia) PCR using standard settings. The thermal profile included initial denaturation at 95°C for 4 min, followed by 30 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 60 s. The samples were then maintained at 72°C for a further 10 min, cooled to 4°C, and stored at -20°C. No-template (PCR-grade water) and positive B. pseudomallei controls were included in each run to rule out amplification failure or possible contamination. Using 1.5% agarose gel, PCR products electrophoresis was performed and the separated amplicons were visualized by ultraviolet light transilluminator and computerized image analyzing system (G-Box, Syngene, USA). The resulted amplicons were purified using PCR purification kit (GE Healthcare, UK) and sequenced by a commercially available sequencing service (1st BASE, Singapore).
Data analysis
Sequence quality for raw forward and reverse sequences for every gene was checked using Sequence ScannerTM software version 2.0 (Life technology, UK). Raw forward and reverse sequences for every gene were aligned with MLST reference allele sequence obtained from MLST database website (http://bpseudomallei.mlst.net/) using ClustalW multiple alignment provided by BioEdit V7.0 software (Ibis Therapeutics, USA). The resulting sequences were trimmed and edited. The final sequence was given a digit, so that every isolate was represented by a code of 7 digits called allelic profile, which was assigned as sequence type (ST) and correspond to the allelic numbers at the seven loci in the order: ace-gltB-gmhD-lepA-lipA-narK-ndh. New allelic profiles were confirmed by repeated PCR and sequencing for loci that differed from an existing allele profile that already cataloged in the B. pseudomallei MLST database. Confirmed novel sequence types were assigned new allelic profile numbers and were submitted, as well as all existing sequence types encountered in this report, along with isolate information to the Burkholderia MLST website curator. The achieved sequence types were deposited in MLST database and analyzed by eBURST Java application and Molecular evolutionary genetic analysis (MEGA) version 6 software that was combined with patient’s clinical history. Classification of human cases was done according on previous literatures [12,13].
Ethical approval by the Universiti Sains Malaysia Research Ethics Committee (Human) was obtained prior to the commencement of this study (USM/PPP/JEPeM [235.4 (2.5)]).
Results
All B. pseudomallei isolates were successfully confirmed with sctQ-PCR assay. Thirteen isolates were typed by MLST. Eight different sequence types were identified, of which ST 54 was the most frequent sequence type with predomination up to 50% of the population analyzed. In addition, Two novel sequence types were reported in this study and assigned with new codes ST1322 (MLST-ID3662) and ST1325 (MLST-ID3665) at MLST database and. All sequence types identified in this study, except novel STs, were previously reported in Southeast Asia countries and all allelic profiles were referenced in MLST database (Figure 1).
Figure 1.
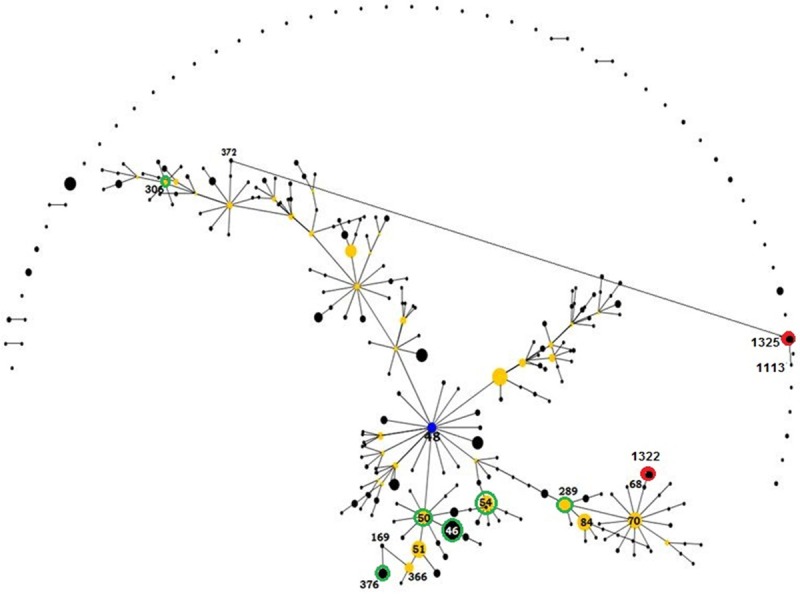
An eBURST analysis showing the genetic relationship among sequence types (STs) obtained from this study and their relatedness to the historical STs from Thailand, Singapore and Malaysia. Black dots refer to STs, blue dot refer to major ancestor, yellow dots refer to subgroup ancestors, and green and red hollows refer to existing and novel STs, respectively, encountered in this study.
The majority of our isolates corresponded to a single large clonal complex (CC48) that represented a predicted ancestor for a major group containing 288 STs, from which many subgroups were emerged with founders identified in our study including ST 50, ST 54, ST 289, and ST 306. Our STs were close to each other, and having several locus variants of regional sequence types, except for ST 1325 which was newly identified in this study and noticed to be singleton. Novel sequence types ST 1322 and ST 1325 were genetically unrelated to each other. However, ST 1322 showed genetic relatedness of single locus variation (SLV) with ST 68 and double locus variations (DLV) with 8 sequence types, among them three were subgroup founders; ST 84, ST 289 and ST 70. Sequence type 1325 is a singleton showed the least genetic relatedness; having only two double locus variants. Type two diabetes mellitus and male gender were the most frequent risk factors for melioidosis, which was mainly bacteremic. No clear pathogenomic or organ tropism clonal relationships were predicted (Figure 2).
Figure 2.
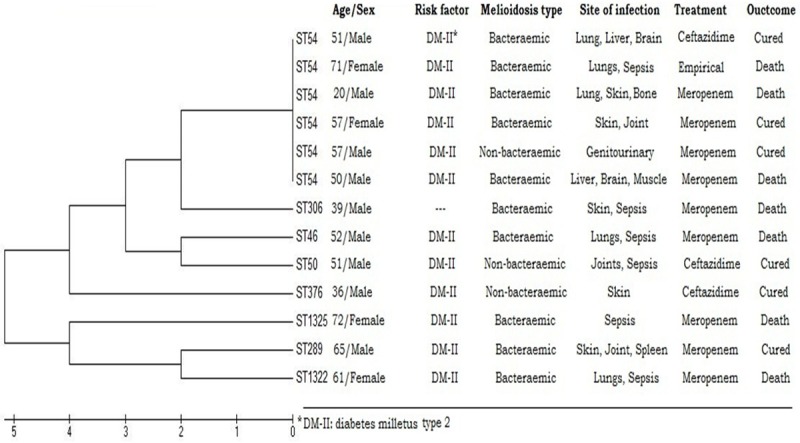
Evolutionary relationships of STs inferred using the UPGMA method with 103 replicates bootstrapping showed no significant clonal organ tropism.
Discussion
Melioidosis has emerged as an important cause of morbidity, mortality, and fatal community-acquired bacteraemic pneumonia in Northern Australia and Southeast Asia. The history of melioidosis in Malaysia began with description of infection in animal as far back as 1913. Human cases were subsequently published in 1932 [14]. The clinical, serological and phenotypic epidemiology of melioidosis in Malaysia has been studied extensively in various Malaysian states [15-20].
A variety of molecular tools have been used to infer epidemiology and genetic relatedness between isolates of B. pseudomallei, particularly in Northern Australia and Thailand [21,22]. In Malaysia, molecular studies on B. pseudomallei were also enrolled into melioidosis projects in the past 20 years. In one study, ribotyping analysis and pulsed-field gel electrophoresis (PFGE) was applied on 49 clinical isolates of B. pseudomallei archived from melioidosis patients over 18 years, in which five ribotype patterns were identified, with predomination of 2 ribotypes over 90% of the isolates, PFGE on the other hand had shown further discriminative ability among a single ribotype; therefore it was postulated that PFGE enabled greater discrimination between isolates of the same ribotype and among isolates from certain clinical source within a ribotype [9]. Further studies had explored the wide diversity of B. pseudomallei distribution in various Malaysian states [6,23].
Multi-locus sequence genotyping (MLST) showed perfect discriminative capabilities comparable to PFGE. Godoy et al [11] has successfully established an MLST scheme for B. pseudomallei and demonstrated its utility for epidemiological study of melioidosis, in which the genetic relationships between closely related B. pseudomallei, B. thailandensis and B. mallei has been resolved.
To date, little data is available on MLST profiling of clinical B. pseudomallei isolates in peninsular Malaysia. Few published studies profiled B. pseudomallei sequence types isolated from various clinical, animal and environmental samples from the island part of Malaysia (Sabah and Sarawak states) [24].
In the present report, MLST was done on clinical B. pseudomallei isolates collected from single population community of Northern Terengganu state, which is located on the Eastern coast of peninsular Malaysia on the borders of Kelantan state. MLST showed massive heterogeneity of sequence types in spite of limited number of isolates; 8 out of 13 different sequence types with two novel STs. This result agreed with previous literatures that suggested wide diversity of B. pseudomallei over a single geographical area of a given endemic country. In addition, ST 54 showed predomination and was recovered from 6 patients with different clinical pictures, predicting possible evidence for genotype colonization in population residing in the same geographical area. Genotype colonization was reported previously in Malaysia and other countries using various molecular typing methods [25,26].
The global distribution of our sequence types was displayed in global MLST maps (http://bpseudomallei.mlst.net/earth/maps/). Almost all STs, except ST 1325, corresponded to a single large clonal complex (CC48) and to major subgroup founders: ST 50, ST 70 and ST 289. All sequence types identified in this study, except ST 1322 and ST1325, were previously reported in Southeast Asia countries, including: Thailand, Malaysia, Indonesia, Laos, Bangladesh and Vietnam. Therefore, and in accordance with the geographical location of Terengganu state, probable dissemination of these genotypes via environmental factors and population travelling was suggested. In addition, it was noted that major subgroup founders were identified in Terengganu from which other sequence types, some of them are exclusive for Malaysia, were emerged; this indicates the historical colonization of B. pseudomallei in this area. It was not surprising that ST 54 predominated in almost half of patients, as well as the presence of sequence types of Thai origin, due to their endemicity in neighboring states from which transmission might have occurred. In addition, ST 1322 was shown to be a single locus variant of Thai ST 68, and ST 1325, a double locus variant of Thai ST 1113 and ST 372 that were identified in environmental samples, suggested point mutation of existing neighbor sequence types in the same region.
Melioidosis was not restricted to certain age group, in this study it was reported in adults and elderly. Pediatric melioidosis was uncommon in endemic areas [27]; thus far no cases of pediatric melioidosis were reported. Out of 13 patients, only 4 were females and 9 males. Male gender was a known risk factor due to increased exposure to infection reservoir. Lungs were the most common primary organ of infection that disseminated to multi-organs in some patients, whereas soft tissue was the second most primary site of infection. Bone, liver brain and spleen involvement was also seen among multi-organ melioidosis, suggesting various nonspecific organ involvement which was supported previously [19]. Diabetes mellitus type 2 was noted to be the major risk factor seen in most patients, since it has been reported as the main predisposing factor for bacteraemic melioidosis [14]. To a lesser extent, steroid treatment, chronic kidney disease, and defective immunity as a result of Down syndrome also predisposed to melioidosis. However, when those clinical and demographical data were correlated with their corresponding genotypes, no significant correlations were found, as has been noted in previous literature [1,28]. However, significance of such correlation in this report cannot be appropriately concluded due to the small sample size.
Conclusion
In spite of small sample set, MLST results indicated considerable genotypic diversity, predominance and novelty among B. pseudomallei collected over a single geographically-located population in Malaysia. Given a larger sample size, as well as collection from many other states, more illustrated genetic variability with clinical correlations will be anticipated. Diabetes mellitus remained the main risk factor for melioidosis, thus a higher index of clinical suspicion of melioidosis among medical doctors is very important in endemic areas when dealing with febrile diabetic patients.
Acknowledgements
We would like also to thank Azlan Abdullah and Nurleem Mursheed for their help in the procurement of the isolates. This project was funded by Malaysian Ministry of Education Exploratory Research Grant Scheme (ERGS) grant, no. 203/PPSP/6730024 awarded to Azian Harun.
Disclosure of conflict of interest
None.
References
- 1.Cheng AC, Godoy D, Mayo M, Gal D, Spratt BG, Currie BJ. Isolates of Burkholderia pseudomallei from Northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J Clin Microbiol. 2004;42:5477–5483. doi: 10.1128/JCM.42.12.5477-5483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puthucheary SD. Melioidosis in Malaysia. Med J Malaysia. 2009;64:266–274. [PubMed] [Google Scholar]
- 3.White NJ. Melioidosis. Lancet. 2003;361:1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 4.Currie BJ, Fisher DA, Howard DM, Burrow JN. Neurological melioidosis. Acta Trop. 2000;74:145–151. doi: 10.1016/s0001-706x(99)00064-9. [DOI] [PubMed] [Google Scholar]
- 5.Currie BJ, Jacups SP, Cheng AC, Fisher DA, Anstey NM, Huffam SE, Krause VL. Melioidosis epidemiology and risk factors from a prospective whole-population study in northern Australia. Trop Med Int Health. 2004;9:1167–1174. doi: 10.1111/j.1365-3156.2004.01328.x. [DOI] [PubMed] [Google Scholar]
- 6.Azura MN, Norazah A, Kamel AG, Zorin SA. DNA fingerprinting of septicemic and localized Burkholderia pseudomallei isolates from Malaysian patients. Southeast Asian J Trop Med Public Health. 2011;42:114–121. [PubMed] [Google Scholar]
- 7.Chua KH, See KH, Thong KL, Puthucheary SD. DNA fingerprinting of human isolates of Burkholderia pseudomallei from different geographical regions of Malaysia. Trop Biomed. 2010;27:517–524. [PubMed] [Google Scholar]
- 8.Radu S, Lihan S, Idris A, Ling OW, Al-Haddawi MH, Rusul G. Genotypic and phenotypic relationship in Burkholderia pseudomallei indicates colonization with closely related isolates. Southeast Asian J Trop Med Public Health. 1999;30:760–763. [PubMed] [Google Scholar]
- 9.Vadivelu J, Puthucheary SD, Mifsud A, Drasar BS, Dance DA, Pitt TI. Ribotyping and DNA macrorestriction analysis of isolates of Burkholderia pseudomallei from cases of melioidosis in Malaysia. Trans R Soc Trop Med Hyg. 1997;91:358–360. doi: 10.1016/s0035-9203(97)90107-3. [DOI] [PubMed] [Google Scholar]
- 10.Vadivelu J, Puthucheary SD, Drasar BS, Dance DA, Pitt TL. Stability of strain genotypes of Burkholderia pseudomallei from patients with single and recurrent episodes of melioidosis. Trop Med Int Health. 1998;3:518–521. doi: 10.1046/j.1365-3156.1998.00262.x. [DOI] [PubMed] [Google Scholar]
- 11.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Currie BJ, Fisher DA, Howard DM, Burrow JN, Lo D, Selva-Nayagam S, Anstey NM, Huffam SE, Snelling PL, Marks PJ, Stephens DP, Lum GD, Jacups SP, Krause VL. Endemic melioidosis in tropical northern Australia: A 10-year prospective study and review of the literature. Clin Infect Dis. 2000;31:981–986. doi: 10.1086/318116. [DOI] [PubMed] [Google Scholar]
- 13.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein R, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 14.Currie BJ. Melioidosis: Evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med. 2015;36:111–125. doi: 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 15.Strauss JM, Alexander AD, Rapmund G, Gan E, Dorsey AE. Melioidosis in Malaysia. 3. Antibodies to Pseudomonas pseudomallei in the human population. Am J Trop Med Hyg. 1969;18:703–707. [PubMed] [Google Scholar]
- 16.Puthucheary SD, Parasakthi N, Lee MK. Septicaemic melioidosis: A review of 50 cases from Malaysia. Trans R Soc Trop Med Hyg. 1992;86:683–685. doi: 10.1016/0035-9203(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 17.Vadivelu J, Puthucheary SD, Gendeh GS, Parasakthi N. Serodiagnosis of melioidosis in Malaysia. Singapore Med J. 1995;36:299–302. [PubMed] [Google Scholar]
- 18.How SH, Ng KH, Jamalludin AR, Shah A, Rathor Y. Melioidosis in Pahang, Malaysia. Med J Malaysia. 2005;60:606–613. [PubMed] [Google Scholar]
- 19.Deris ZZ, Hasan H, Siti Suraiya MN. Clinical characteristics and outcomes of bacteraemic melioidosis in a teaching hospital in a northeastern state of Malaysia: A five-year review. J Infect Dev Ctries. 2010;4:430–435. doi: 10.3855/jidc.491. [DOI] [PubMed] [Google Scholar]
- 20.Hassan MR, Pani SP, Peng NP, Voralu K, Vijayalakshmi N, Mehanderkar R, Aziz NA, Michael E. Incidence, risk factors and clinical epidemiology of melioidosis: a complex socioecological emerging infectious disease in the Alor Setar region of Kedah, Malaysia. BMC Infect Dis. 2010;10:302. doi: 10.1186/1471-2334-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng AC, Ward L, Godoy D, Norton R, Mayo M, Gal D, Spratt BG, Currie BJ. Genetic diversity of Burkholderia pseudomallei isolates in Australia. J Clin Microbiol. 2008;46:249–254. doi: 10.1128/JCM.01725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chantratita N, Wuthiekanun V, Thanwisai A, Limmathurotsakul D, Vesaratchavest M, Feil EJ, Amornchai P, Tumapa S, Day NP, Peacock SJ. Genetic diversity and microevolution of Burkholderia pseudomallei in the environment. PLoS Negl Trop Dis. 2008;2:e182. doi: 10.1371/journal.pntd.0000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radua S, Ling OW, Srimontree S, Lulitanond A, Hin WF, Yuherman , Lihan S, Rusul G, Mutalib AR. Characterization of Burkholderia pseudomallei isolated in Thailand and Malaysia. Diagn Microbiol Infect Dis. 2000;38:141–145. doi: 10.1016/s0732-8893(00)00189-9. [DOI] [PubMed] [Google Scholar]
- 24.Podin Y, Sarovich DS, Price EP, Kaestli M, Mayo M, Hii K, Ngian H, Wong S, Wong I, Wong J, Mohan A, Ooi M, Fam T, Tuanyok A, Keim P, Giffard PM, Currie BJ. Burkholderia pseudomallei isolates from Sarawak, Malaysian Borneo, are predominantly susceptible to aminoglycosides and macrolides. Antimicrob Agents Chemother. 2014;58:162–166. doi: 10.1128/AAC.01842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haase A, Melder A, Smith-Vaughan H, Kemp D, Currie B. RAPD analysis of isolates of Burkholderia pseudomallei from patients with recurrent melioidosis. Epidemiol Infect. 1995;115:115–121. doi: 10.1017/s0950268800058179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vesaratchavest M, Tumapa S, Day NP, Wuthiekanun V, Chierakul W, Holden MT, White NJ, Currie BJ, Spratt BG, Feil EJ, Peacock SJ. Nonrandom distribution of Burkholderia pseudomallei clones in relation to geographical location and virulence. J Clin Microbiol. 2006;44:2553–2557. doi: 10.1128/JCM.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sam IC, Puthucheary SD. Melioidosis in children from Kuala Lumpur, Malaysia. Ann Trop Paediatr. 2006;26:219–224. doi: 10.1179/146532806X120318. [DOI] [PubMed] [Google Scholar]
- 28.Cheng AC, Day NP, Mayo MJ, Gal D, Currie BJ. Burkholderia pseudomallei strain type, based on pulsed-field gel electrophoresis, does not determine disease presentation in melioidosis. Microbes Infect. 2005;7:104–109. doi: 10.1016/j.micinf.2004.08.020. [DOI] [PubMed] [Google Scholar]


