Abstract
AIM: To examine the protein expression alterations in liver injury/repair network regulation as a response to gut-derived lipopolysaccharide (LPS) treatment, in order to anticipate the possible signal molecules or biomarkers in signaling LPS-related liver injury.
METHODS: Male BALB/c mice were treated with intra-peritoneal (i.p.) LPS (4 mg/kg) and sacrificed at 0, 6, 24 and 30 h to obtain livers. The livers were stained with hematoxylin and eosin for histopathologic analyses. Total liver protein was separated by two-dimensional gel electrophoresis (2-DE). The peptide mass of liver injury or repair related proteins were drawn up and the protein database was searched to identify the proteins.
RESULTS: Observations were as follows: (1) TRAIL-R2 was down regulated in livers of LPS-treated mice. TNFAIP1 was significantly up regulated at 6 h, then down-regulated at 24, 30 h with silent expression during senescent stage. (2) The amount of metaxin 2 and mitochondria import inner membrane translocase subunit TIM8a (TIMM8A) was increased upon treatment with LPS. (3) P34 cdc2 kinase was significantly up-regulated 30 h after LPS administration with silent expression during senescent, 6, 24 h treated stage. (4) The amount of proteasome activator 28 alpha subunit (PA28), magnesium dependent protein phosphatase (MDPP) and lysophospholipase 2 was decreased 6 h after LPS treatment but recovered or up-regulated 24 and 30 h after LPS treatment.
CONCLUSION: LPS-treated mouse liver displaying a time-dependent liver injury can result in expression change of some liver injury or repair related proteins.
INTRODUCTION
Human intestinal tract accumulates Gram-negative bacteria that supply lipopolysaccharide (LPS). Gut-derived LPS reaches the first target organ liver through portal venous flow and results in hepatic clearance or tissue damage. Recent studies have shown that peritoneal macrophages in cirrhotic patients may secret angiogenic products when stimulated by LPS and accumulate in liver injury[1-3]. In vivo experiments also indicate LPS may cause liver injury in normal subjects and exaggerate liver injury in alcoholic liver disease[4-7]. That is, chronic liver diseases suffering from subsequent LPS attack would result in much severer clinical findings. The precise mechanisms underlying LPS-treated liver injury remain unclear. Several studies of liver protein expression exposed to some hepatotoxic agents have revealed certain changes which illustrated the pathogenesis of liver injury in some extent[2,4,8,9]. However, most reported researches focused only on some partial signaling or effecter molecules on a limited view[4,5,7]. Protein expression changes of liver exposed to LPS remains to be established.
The goal of the present study was to dynamically examine protein expression alterations in liver tissues from mice exposed to LPS administration, in order to anticipate of the possible signal molecules or biomarkers in signaling LPS-related liver injury.
MATERIALS AND METHODS
Reagents
Ultra pure reagents for polyacrylamide gel preparation were obtained from Bio-Rad. Eighteen cm immobilized pH gradient (IPG) strips (pH3-10 L), IPG buffer and dry strip cover fluid were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). 3-[(3-cholamidopropyl) dimethylammonio]-propanesulfonate (CHAPS), glycine, ammonium persulphate (APS), TEMED, trifluoroacetic acid were obtained from Amresco (Solon, OH, USA). Dithiothreitol (DTT), PMSF, Iodoacetamide, urea, thiourea, α-cyano-4-hydroxycinnaic acid (CCA), TPCK-Trypsin, E. coli LPS 0111:B4 were purchased from Sigma Chemical (St Louis, MO, USA). Acetonitrile was HPLC grade. Ten μL ZipTipTMC18 tip were purchased from Millipore Company (USA).
Animal treatment protocol
Male BALB/c mice (supplied by Laboratory Research Animal Center, 2nd Xiangya Hospital, Central South University), 6 wk of age and weighing 19-21 g, were used. The animals were housed individually in mouse gang cages in cleaning-grade controlled room. The Hunan Association accredited the house facility for Accreditation of Laboratory Animal Care, and all animal handling procedures were in conform to the Guide for the Care and Use of Laboratory Animals. Four groups of five mice each received intra-peritoneal (i.p) RPMI-1640 medium treatment (control) and LPS (diluted in RPMI-1640 medium) 4 mg/kg for 6, 24, 30 h, respectively. Mice were killed by decapitation, liver samples were removed and flash-frozen in liquid nitrogen and kept at -70 °C until proteomics analysis. For histopathologic research, liver slices from the right lateral lobes were immersion-fixed in 40 g/L neutral buffered formaldehyde, routinely processed to 5-μm thick paraffin sections, stained with hematoxylin and eosin (HE) and examined by light microscopy.
Sample preparation[10]
Liver samples were homogenized in eight volumes of 8 mol/L urea, 2 mol/L thiourea, 40 g/L CHAPS, 20 g/L Triton X-100, 5 g/L DTT, 1 mmol/L PMSF, 1 mmol/L EDTA, 40 mmol/L Tris. Homogenates were centrifuged at 42000 g at 20 °C for 1 h (LE-80 K ultracentrifuge, Type 90 rotor, Beckman). Supernatant was removed, divided into several aliquots and stored at -70 °C until to be used. Protein concentration of each sample was measured by Bradford method[11].
Two-dimensional gel electrophoresis[11,12]
The first-dimensional isoelectrical focus (IEF) was performed on precast 18 cm IPG strips at 20 °C with a maximum current setting of 50 μA/strip using an Amersham Pharmacia IPGphor IEF unit. The strips were rehydrated at 30 V for 16 h in 350 μL samples containing 8 mol/L urea, 20 g/L CHAPS, 20 mmol/L DTT, 5 g/L IPG buffer. Eight hundred μg proteins was applied to each IPG strip. After rehydration, IEF run was carried out using the following conditions: 500 V, 500 Vh; 1000 V, 1000 Vh; 8000 V, 40000 Vh. Then, the strips were subjected to a two-step equilibration. The first was with an equilibration buffer consisting of 6 mol/L urea, 300 g/L glycerol, 20 g/L SDS, 50 mmol/L Tris (pH8.8) and 10 g/L DTT. The second equilibration buffer was the same, only with DTT substituted by 25 g/L iodoacetamide. After the strips were transferred onto the second-dimensional 0.75 mm thick 125 g/L sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, the strips were sealed in place with 7.5 g/L agarose. Electrophoresis was carried out at a constant current of 25 μA/gel at 12 °C until bromophenol blue reached the edge of gels.
Silver staining
Gels were fixed in 400 mL/L ethanol, 100 mL/L acetic acid in water for 30 min, and then sensitized with 2 g/L sodium thiosulfate for 30 min. After the gels were rinsed twice with water for 5 min each, they were incubated in 2.5 g/L silver nitrate for 20 min. After the silver nitrate was discarded and rinsed with three changes of distilled water for 5 min each, the gels were visualized in 25 g/L sodium carbonate in 0.004% formalin until the desired intensity was attained. Then the gels were incubated with 14.6 g/L EDTA disodium dihydrate for 10 min to stop the development. The staining procedure was performed by three rinses with water for 5 min each.
Quantitative gel pattern analysis
Stained 2-D gels were scanned on a UMAX powerLook III scanner (Amersham Biosciences). Image analysis and Group Wise statistical comparisons were performed using the Image Master 2D v3.01 software (Amersham Pharmacia). The protein spot intensity was arbitrarily calculated by integrating the optical density over the spot area to search for treatment-related protein change. The amount of protein was marked by volumes (A × area) and expressed as mean ± SD. Statistical difference was analyzed by analysis of Variance (ANOVA). The cutoff of quantitative protein changes between control and treated groups was made at P < 0.05.
Protein digestion and protein identification by MALDI-TOF-MS[10,12]
Destaining Protein spots of interest were excised from the 2-D gels and washed 3 times in distilled water, destained with 15 mmol/L potassium ferricyanide in 50 mmol/L sodium thiosulfate, dehydrated in acetonitrile and finally dried in a centrifugal vaporizer.
Reduction and alkylation Gel pieces were incubated in 100 mmol/L NH4HCO3 by adding 10 mmol/L DTT at 57 °C for 1 h. After they were cooled, 100 mmol/L NH4HCO3 with 55 mmol/L iodoacetamide was added and incubated at room temperature in dark for 30 min. Supernatant was removed and samples were washed twice in 100 mmol/L NH4HCO3 and dehydrated in 1000 g/L acetonitrile, finally dried in a centrifugal vaporizer.
In-gel digestion Gel pieces were rehydrated in a digestion buffer containing 0.1 mg/mL TPK-trypsin for 30 min. After excess buffer was discarded, the gel pieces were covered with digestion buffer and proteins were in-gel digested for 24 h at 37 °C.
Deionization Digested peptides were pipetted out and into to make them attached in a 10 μL ZipTipTMC18 tip, washed twice with 1mL/L trifluoroacetic acid.
MALDI-TOF-MS for protein identification The matrix solution was saturated in 500 g/L acetonitrile, 1 g/L TFA in water. Peptides were eluted with 5 μL MALDI matrix, 2.0 μL mixed solution was loaded to the sample plate target and allowed to air-evaporate. MALDI experiments were performed on a Perspective Biosystem Voyager-DE STR time-of-flight mass spectrometer (Applied Biosystems Voyager system 4307, USA) equipped with delayed ion extraction. Data were acquired in the delayed ion extraction mode using a 20 KV accelerating voltage and a 100 ns extraction delay time. Dual microchannel plate detection was utilized in the reflector mode with the ion signal recorded using a transient digitizer. The performance of the mass spectrometer produced sufficient mass resolution to produce isotopic multiplet for each ion species below a mass-to-charge (m/z) of 3000. All MALDI mass spectra were internally calibrated using masses from two trypsin autolysis products (monoisotopic masses 2163.05 and 2273.15). The software package Protein Prospector (prospector. ucsf.edu) was used to identify protein spots. The mouse nonredundant (NR) database Genpept.5.1.2002 was used in searches. The search parameters used were as follows: Cysteines modification mode as carbamidomethylation, maximum allowed peptide mass error 50 ppm, more than four peptide mass hits required for protein match, up to one enzymatic missed cleavage. A restriction was placed on species of origin, pI (experimental pI +/- 1.00 pH unit) and protein mass (experimental molecular mass +/- 50%).
RESULTS
General health and histopathology
All subjects were inspected for general health before, during and after LPS exposure. Throughout the study, no animal experienced mortality that was determined to be related to the exposure. Nine of 10 animal that were treated by exposure to LPS for 24, 30 h experienced visible signs of laziness and retardation. Light microscopic examination of HE sections from LPS-treated mice revealed that a large number of inflammatory cells infiltrated portal areas with additional focal hepatocellular necrosis 24, 30 h after i.p. LPS (4 mg/kg)[13].
2-DE and relative abundance change
The resolution in 2-DE silver-stained gels resulted in approximately 1000 spots. The volumes (A × area) and coordinates of each spot (pI and MW) were determined to select the fine right proteins among candidates. There were significant alterations in 40 protein spots following LPS treatment when compared with each other, P < 0.01. These 40 proteins were identified by MALDI-TOF-MS (data not shown). Figure 1 (A-D) shows the location of 8 injury/repair related proteins in illustrated partial 2-DE maps of control and LPS-treated mice, respectively. Table 1 summarizes the relative amount of these considered proteins. The amount of protein 1 was 9800 in the control and down-regulated to 1130 in 6-h group, absent in 24-, 30-h group. Protein 5 was significantly up-regulated 6 h after LPS treatment, then down-regulated 24 h and 30 h after LPS treatment with silent expression in the control. Proteins 2 and 13 were increased upon treatment with LPS. Protein 9 was significantly up-regulated 30 h after LPS treatment with silent expression in control, 6, 24 h treated stage. Proteins 6, 447 and 524 were decreased in amount 6 h after LPS treatment, recovered and up-regulated 24 and 30 h after LPS treatment.
Figure 1.
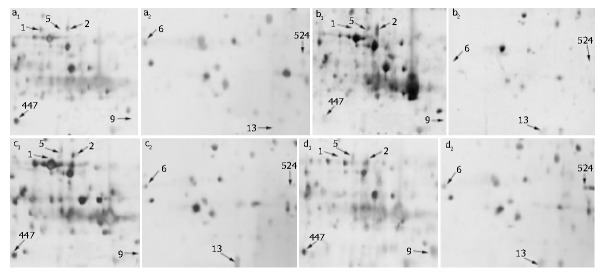
Partial 2-D images of mouse liver proteins from (a1, a2) a control animal and animals treated with 4 mg/kg i.p LPS (b1, b2) 6 h, (c1, c 2) 24 h, (d1, d2) 30 h. The spots representing proteins whose levels changed following LPS treatment are shown. a1, b1, c1, d1 represent the relative same partial map after different LPS-treated onset. a2, b2, c2, d2 represent another relative same partial map.
Table 1.
Relative amount of 8 injury/repair related liver proteins in response to LPS treatment (mean ± SD)
| Liver protein | Control |
LPS |
||
| 6 h | 24 h | 30 h | ||
| 1 | 9800 ± 32 | 1130 ± 41 | absent | absent |
| 2 | 9952 ± 21 | 11020 ± 26 | 34060 ± 19 | 29800 ± 24 |
| 5 | absent | 38040 ± 18 | 19500 ± 42 | 20540 ± 34 |
| 6 | 12420 ± 24 | absent | 15500 ± 32 | 17200 ± 30 |
| 9 | absent | absent | 1200 ± 35 | 41500 ± 29 |
| 13 | absent | 4500 ± 42 | 8900 ± 44 | 10500 ± 32 |
| 447 | 43200 ± 21 | 3200 ± 35 | absent | 54400 ± 32 |
| 524 | absent | absent | absent | 9500 ± 23 |
Identity of injury/repair related proteins
The proteins were cut from 2-DE gels and subjected to tryptic digestion. The 8 proteins analyzed by peptide mass fingerprinting yielded searchable masses and resulted in the identification (Table 2). Figure 2 indicates the peptide mass fingerprint of MDPP (spot447) as a representative.
Table 2.
Results from search of Genpept.5.1.2002 database for Musculus protein sequences with tryptic peptide masses
| Protein NO. | MOWSE score | Masses matched (%) | Coverage (%) | Protein Mr | Protein PI | Accession NO. | Protein name |
| 1 | 36.2 | 4 | 27 | 42677 | 6.5 | Q9QZM4 | TRAIL receptor 2 |
| 2 | 56.3 | 5 | 45 | 35646 | 6.0 | P47802 | Metaxin 2 |
| 5 | 29.5 | 4 | 32 | 36134 | 8.0 | O70479 | TNF-α induced protein 1 |
| 6 | 47.9 | 4 | 26 | 28673 | 5.7 | P97371 | Proteasome activator 28-alpha subunit (PA28) |
| 9 | 76.1 | 4 | 20 | 34191 | 7.8 | P11440 | P34 cdc2 kinase |
| 13 | 25.3 | 4 | 26 | 11043 | 5.1 | Q9WVA2 | Mitochondria import inner membrane translocase subunit TIM8 A (TIMM8A) |
| 447 | 23.2 | 4 | 22 | 42795 | 5.0 | 452526 | Magnesium dependent protein phosphatase (MDPP) |
| 524 | 37.3 | 4 | 25 | 24794 | 6.7 | P47713 | Lysophospholipase 2 |
Figure 2.
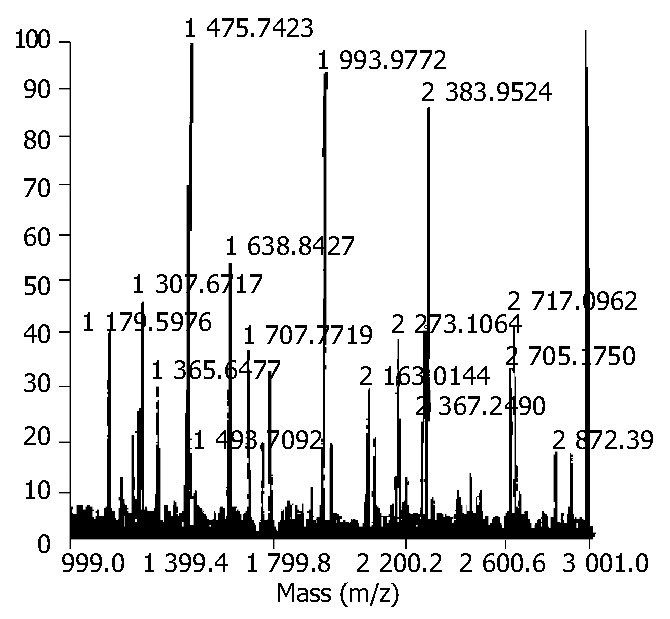
Peptide mass fingerprint of MDPP (spot447). The spot was in-gel digested with trypsin. After desalted, the peptide mixture was analyzed by MALDI-TOF-MS. All MALDI mass spectra were internally calibrated using masses from two trypsin autolysis products (monoisotopic masses 2163.05 and 2273.15).
DISCUSSION
It has been proved that gut-derived LPS provokes liver inflammation with focal hepatocyte injury and increases the sensitivity of hepatotoxicity to carbon tetrachloride, concanavalin A, D-galactosamine and ethanol induced liver damage[4-6,14]. The role of LPS signaling pathway in the initiation and early propagation of immune response has been deeply demonstrated[8,9,15]. In this study, liver injury occurred apparently 24 h after LPS administration. It indicated that LPS could promote hepatotoxicity by indirect mechanisms. Some immediate signaling or effecter molecules may play a pivotal role in this delayed injurious response.
In our research, TNF-related apoptosis-inducing ligand receptor 2 (TRAIL-R2), TNF-α inducible endothelial protein 1 (TNFAIP1) and mitochondria apoptosis signaling related proteins metaxin 2, mitochondria import inner membrane translocase subunit TIM8a (TIMM8A) were demonstrated to be involved in the initiation of liver injury of LPS-treated mice and consistent with the histological change of liver tissue.
A recent research showed that LPS administration could result in elevated circulating TNF-α and up-regulation of gene transcription for TNF-α as early as 2 h after administration[16]. Then, TNF-α related apoptosis pathway played a control role in mediating the tissue injury. TRAIL was recently reported to promote apoptosis of carcinogenic cells by binding to the transmembrane receptors. The resulting death-inducing signaling complex consisting of TRAIL, TRAIL-R2 and caspase-8, initiated the subsequent cascade of Caspases mediating apoptosis[17,18]. The down-regulation of TRAIL-R2 in livers of LPS-treated mice could result in rather a protective mode of normal hepatocytes. The induction of TNFAIP1 has been shown to be apoptosis-specific, suggesting that TNF-α related apoptosis is triggered in liver cells of LPS-treated mice at earlier stage[19].
Metaxin 2 is bound to the cytosolic face of mitochondria outer membrane by its interaction with membrane-bound metaxin 1, and this complex may play a role in protein import into mammalian mitochondria. The requirement of metaxin in TNF-induced cell death has been confirmed by in vitro[20,21]. It has been found that TIMM8A exists in the mitochondria intermembrane space which mediates the import and insertion of inner membrane proteins[22-24]. These two mitochondria proteins are required for normal physiological balance. Treatment-mediated changes of these proteins were likely to associated with mitochondria apoptosis. How mitochondria related pro- or anti-apoptotic proteins are involved in liver injury needs to be further demonstrated.
Tissue repair associated proteins, p34 cdc2 kinase and proteasome related proteins, showed some controversial expression in response to LPS-treated liver injury. p34 cdc2 kinase belongs to the Ser/Thr family of protein kinases. The sequential activation and inactivation of p34 cdc2 kinase of eukaryotic cells are required for it to entry into mitosis and to exit from it. In addition, a sustained activation of it during mitotic arrest has been associated with anti-microtubule agents-induced apoptosis[25,26]. In this study, we found significant histopathological changes in mice livers 24 h after LPS treatment. p34 cdc2 kinase of mice livers was significantly up-regulated r 30 h after LPS administration with silent expression during senescent, 6-, 24-h treated stage. It indicated that the enhanced expression of p34 cdc2 kinase was subsequently present to severe hepatic damage. It is likely that up-regulation of p34 cdc2 kinase in LPS-treated mice was more reflective of mitogenic response rather than antimitogenic activities. That is, cell division and tissue repair occurred as events opposing to tissue injury and could limit the extent of liver injury. It seemed possible that p34 cdc2 kinase could act by promoting mitosis and tissue repair, thus contributing to tissue repair response to LPS-treated liver injury.
The biological implications of amount in abundance of proteosome related proteins in mice liver response, including proteasome activator 28 alpha subunit (PA28), magnesium dependent protein phosphatase (MDPP) and lysophospholipase 2, which decreased in amount 6 h after LPS treatment but recovered or up-regulated 24 and 30 h after LPS treatment, were even less clear. Previous study demonstrated that PA28 could bind to and activate the proteasome by the alpha subunit[27]. MDPP is a new member of the halo acid dehalogenase family which has been found to be competent to catalyzing dephosphorylation of tyrosine-phosphorylated proteins[28]. Lysophospholipase 2 was implicated in the initiation of inflammatory response by mediating cell division and differentiation[1,29].
It is concluded that proteosome related proteins play a major role in the initiation of tissue repair program opposing to tissue damage. While the function, pathway interactions and ultimate biological outcomes of these changes in protein expression remains to be explained by further research.
In summary, the current investigation used 2-DE to characterize the proteome of LPS-treated mice liver that displayed a time-dependent liver injury in some liver injury or repair related proteins. In this study, 8 liver injury or repair related proteins were identified whose expression levels were altered at different time points after LPS treatment. TNF related apoptosis induced by LPS treatment could provoke an initiation response in tissue repair.
Footnotes
Supported by the Science and Development Foundation of Hunan Province No.99SSY2002-22
Edited by Wang XL and Xu FM
References
- 1.Pérez-Ruiz M, Ros J, Morales-Ruiz M, Navasa M, Colmenero J, Ruiz-del-Arbol L, Cejudo P, Clária J, Rivera F, Arroyo V, et al. Vascular endothelial growth factor production in peritoneal macrophages of cirrhotic patients: regulation by cytokines and bacterial lipopolysaccharide. Hepatology. 1999;29:1057–1063. doi: 10.1002/hep.510290416. [DOI] [PubMed] [Google Scholar]
- 2.Hanck C, Manigold T, Böcker U, Kurimoto M, Kölbel CB, Singer MV, Rossol S. Gene expression of interleukin 18 in unstimulated peripheral blood mononuclear cells of patients with alcoholic cirrhosis. Gut. 2001;49:106–111. doi: 10.1136/gut.49.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathurin P, Deng QG, Keshavarzian A, Choudhary S, Holmes EW, Tsukamoto H. Exacerbation of alcoholic liver injury by enteral endotoxin in rats. Hepatology. 2000;32:1008–1017. doi: 10.1053/jhep.2000.19621. [DOI] [PubMed] [Google Scholar]
- 4.Uesugi T, Froh M, Arteel GE, Bradford BU, Thurman RG. Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology. 2001;34:101–108. doi: 10.1053/jhep.2001.25350. [DOI] [PubMed] [Google Scholar]
- 5.Josephs MD, Bahjat FR, Fukuzuka K, Ksontini R, Solorzano CC, Edwards CK, Tannahill CL, MacKay SL, Copeland EM, Moldawer LL. Lipopolysaccharide and D-galactosamine-induced hepatic injury is mediated by TNF-alpha and not by Fas ligand. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1196–R1201. doi: 10.1152/ajpregu.2000.278.5.R1196. [DOI] [PubMed] [Google Scholar]
- 6.Han DW. Intestinal endotoxemia as a pathogenetic mechanism in liver failure. World J Gastroenterol. 2002;8:961–965. doi: 10.3748/wjg.v8.i6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seki E, Tsutsui H, Nakano H, Tsuji N, Hoshino K, Adachi O, Adachi K, Futatsugi S, Kuida K, Takeuchi O, et al. Lipopolysaccharide-induced IL-18 secretion from murine Kupffer cells independently of myeloid differentiation factor 88 that is critically involved in induction of production of IL-12 and IL-1beta. J Immunol. 2001;166:2651–2657. doi: 10.4049/jimmunol.166.4.2651. [DOI] [PubMed] [Google Scholar]
- 8.Kinser S, Copple BL, Roth RA, Ganey PE. Enhancement of allyl alcohol hepatotoxicity by endotoxin requires extrahepatic factors. Toxicol Sci. 2002;69:470–481. doi: 10.1093/toxsci/69.2.470. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Cai SR, Gao C, Sladek FM, Ponder KP. Lipopolysaccharide results in a marked decrease in hepatocyte nuclear factor 4 alpha in rat liver. Hepatology. 2001;34:979–989. doi: 10.1053/jhep.2001.28885. [DOI] [PubMed] [Google Scholar]
- 10.Steiner S, Gatlin CL, Lennon JJ, McGrath AM, Aponte AM, Makusky AJ, Rohrs MC, Anderson NL. Proteomics to display lovastatin-induced protein and pathway regulation in rat liver. Electrophoresis. 2000;21:2129–2137. doi: 10.1002/1522-2683(20000601)21:11<2129::AID-ELPS2129>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 11.Kristensen DB, Kawada N, Imamura K, Miyamoto Y, Tateno C, Seki S, Kuroki T, Yoshizato K. Proteome analysis of rat hepatic stellate cells. Hepatology. 2000;32:268–277. doi: 10.1053/jhep.2000.9322. [DOI] [PubMed] [Google Scholar]
- 12.Fountoulakis M, Berndt P, Boelsterli UA, Crameri F, Winter M, Albertini S, Suter L. Two-dimensional database of mouse liver proteins: changes in hepatic protein levels following treatment with acetaminophen or its nontoxic regioisomer 3-acetamidophenol. Electrophoresis. 2000;21:2148–2161. doi: 10.1002/1522-2683(20000601)21:11<2148::AID-ELPS2148>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Liu XW, You Y, Lu FG. [TLR4 mRNA expression and liver injury in LPS-induced mouse] Hunan Yike Daxue Xuebao. 2003;28:217–220. [PubMed] [Google Scholar]
- 14.Enomoto N, Takei Y, Hirose M, Ikejima K, Miwa H, Kitamura T, Sato N. Thalidomide prevents alcoholic liver injury in rats through suppression of Kupffer cell sensitization and TNF-alpha production. Gastroenterology. 2002;123:291–300. doi: 10.1053/gast.2002.34161. [DOI] [PubMed] [Google Scholar]
- 15.Muta T, Takeshige K. Essential roles of CD14 and lipopolysaccharide-binding protein for activation of toll-like receptor (TLR)2 as well as TLR4 Reconstitution of TLR2- and TLR4-activation by distinguishable ligands in LPS preparations. Eur J Biochem. 2001;268:4580–4589. doi: 10.1046/j.1432-1327.2001.02385.x. [DOI] [PubMed] [Google Scholar]
- 16.Barton CC, Barton EX, Ganey PE, Kunkel SL, Roth RA. Bacterial lipopolysaccharide enhances aflatoxin B1 hepatotoxicity in rats by a mechanism that depends on tumor necrosis factor alpha. Hepatology. 2001;33:66–73. doi: 10.1053/jhep.2001.20643. [DOI] [PubMed] [Google Scholar]
- 17.Wu GS, Burns TF, Zhan Y, Alnemri ES, El-Deiry WS. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res. 1999;59:2770–2775. [PubMed] [Google Scholar]
- 18.Srivastava RK. TRAIL/Apo-2L: mechanisms and clinical applications in cancer. Neoplasia. 2001;3:535–546. doi: 10.1038/sj.neo.7900203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swift S, Blackburn C, Morahan G, Ashworth A. Structure and chromosomal mapping of the TNF-alpha inducible endothelial protein 1 (Edp1) gene in the mouse. Biochim Biophys Acta. 1998;1442:394–398. doi: 10.1016/s0167-4781(98)00180-8. [DOI] [PubMed] [Google Scholar]
- 20.Abdul KM, Terada K, Yano M, Ryan MT, Streimann I, Hoogenraad NJ, Mori M. Functional analysis of human metaxin in mitochondrial protein import in cultured cells and its relationship with the Tom complex. Biochem Biophys Res Commun. 2000;276:1028–1034. doi: 10.1006/bbrc.2000.3589. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Ono K, Kim SO, Kravchenko V, Lin SC, Han J. Metaxin is required for tumor necrosis factor-induced cell death. EMBO Rep. 2001;2:628–633. doi: 10.1093/embo-reports/kve135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roesch K, Curran SP, Tranebjaerg L, Koehler CM. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum Mol Genet. 2002;11:477–486. doi: 10.1093/hmg/11.5.477. [DOI] [PubMed] [Google Scholar]
- 23.Ye K, Zhou J, Landen JW, Bradbury EM, Joshi HC. Sustained activation of p34(cdc2) is required for noscapine-induced apoptosis. J Biol Chem. 2001;276:46697–46700. doi: 10.1074/jbc.C100550200. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann S, Rothbauer U, Mühlenbein N, Neupert W, Gerbitz KD, Brunner M, Bauer MF. The C66W mutation in the deafness dystonia peptide 1 (DDP1) affects the formation of functional DDP1.TIM13 complexes in the mitochondrial intermembrane space. J Biol Chem. 2002;277:23287–23293. doi: 10.1074/jbc.M201154200. [DOI] [PubMed] [Google Scholar]
- 25.Selengut JD. MDP-1 is a new and distinct member of the haloacid dehalogenase family of aspartate-dependent phosphohydrolases. Biochemistry. 2001;40:12704–12711. doi: 10.1021/bi011405e. [DOI] [PubMed] [Google Scholar]
- 26.Schoemaker MH, Ros JE, Homan M, Trautwein C, Liston P, Poelstra K, van Goor H, Jansen PL, Moshage H. Cytokine regulation of pro- and anti-apoptotic genes in rat hepatocytes: NF-kappaB-regulated inhibitor of apoptosis protein 2 (cIAP2) prevents apoptosis. J Hepatol. 2002;36:742–750. doi: 10.1016/s0168-8278(02)00063-6. [DOI] [PubMed] [Google Scholar]
- 27.Song X, von Kampen J, Slaughter CA, DeMartino GN. Relative functions of the alpha and beta subunits of the proteasome activator, PA28. J Biol Chem. 1997;272:27994–28000. doi: 10.1074/jbc.272.44.27994. [DOI] [PubMed] [Google Scholar]
- 28.Wang A, Johnson CA, Jones Y, Ellisman MH, Dennis EA. Subcellular localization and PKC-dependent regulation of the human lysophospholipase A/acyl-protein thioesterase in WISH cells. Biochim Biophys Acta. 2000;1484:207–214. doi: 10.1016/s1388-1981(00)00020-2. [DOI] [PubMed] [Google Scholar]
- 29.Khan S, van den Broek M, Schwarz K, de Giuli R, Diener PA, Groettrup M. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]


