Abstract
AIM: To investigate the association between cytochrome P450 2C19 (CYP2C19) gene polymorphism and cancer susceptibility by genotyping of CYP2C19 poor metabolizers (PMs) in cancer patients.
METHODS: One hundred and thirty-five cases of esophagus cancer, 148 cases of stomach cancer, 212 cases of lung cancer, 112 cases of bladder cancer and 372 controls were genotyped by allele specific amplification-polymerase chain reaction (ASA-PCR) for CYP2C19 PMs. The frequencies of PMs in cancer groups and control group were compared.
RESULTS: The frequencies of PMs of CYP2C19 were 34.1% (46/135) in the group of esophagus cancer patients, 31.8% (47/148) in the stomach cancer patients, 34.4% (73/212) in the group of lung cancer patients, only 4.5% (5/112) in the bladder cancer patients and 14.0% (52/372) in control group. There were statistical differences between the cancer groups and control group (esophagus cancer, χ2 = 25.65, P < 0.005, OR = 3.18, 95%CI = 2.005-5.042; stomach cancer, χ2 = 21.70, P < 0.005, OR = 2.86, 95%CI = 1.820-4.501; lung cancer, χ2 = 33.58, P < 0.005, OR = 3.23, 95%CI = 1.503-6.906; bladder cancer, χ2 = 7.50, P < 0.01, OR = 0.288, 95%CI = 0.112-0.740).
CONCLUSION: CYP2C19 PMs have a high incidence of esophagus cancer, stomach cancer and lung cancer, conversely they have a low incidence of bladder cancer. It suggests that CYP2C19 may participate in the activation of procarcinogen of esophagus cancer, stomach cancer and lung cancer, but may involve in the detoxification of carcinogens of bladder cancer.
INTRODUCTION
Individuals vary widely in their susceptibility to carcinogens. One attractive genetic mechanism to account for this variability is the activity of polymorphically expressed cytochrome P450 enzymes that activate procarcinogens or conversely detoxify carcinogens. Cytochrome P450 2C19 (CYP2C19) is a clinically important metabolic enzyme responsible for the metabolism of a number of therapeutic drugs, such as S-mephenytoin, omeprazole, diazepam, proguanil, propranolol and certain antidepressants[1]. Recently, there were several papers concerning the CYP2C19 polymorphism and cancer susceptibility. Wadelius et al[2] found no association between CYP2C19 polymorphism and prostate cancer. Roddam et al[3] reported an increased risk of CYP2C19 poor metabolizers (PMs) to develop adult acute leukaemia and Sachse et al[4] found CYP2C19*2 had an decreased risk of colorectal cancer. Normally, Oriental people had a higher incidence of CYP2C19 poor metabolizers, which was usually about 13%-16%, but in Caucasian people it was only 1%-3% as well. So, for the purpose of investigating association between CYP2C19 polymorphism and cancer susceptibility, it is more easy to draw a conclusion in Oriental population. The aim of this study was to evaluate the relationship between CYP2C19 polymorphism and susceptibility to different kind of cancer by means of CYP2C19 genotyping among Chinese subjects.
MATERIALS AND METHODS
Reagents
Taq DNA polymerase, dNTPs, PCR buffer and 25 mmol/L MgCl2 were purchased from Promega USA, Primers (Sangon, Shanghai), Igepal CA-630 (Sigma, USA), DNA Ladder (Huamei, Shanghai), Agarose (Pharmacia, Sweden), SmaI and HamHI were obtained from MBI, USA. Other chemicals were of analytical grade.
Equipments
PCR machine (Hybaid, USA), electrophoresis apparatus (Pharmacia, Sweden), gel imaging system (Stratagene, USA), high speed centrifuge (Hitachi, Japan) and electro-balance (Mettle, France) were used.
Subjects
Cancer patients were from No 1 and No 2 hospitals affiliated to Zhejiang University. Healthy controls were recruited randomly around the same area. This study was approved by the Medical Institutional Review Board of the University and all subjects were provided informed consent prior to their participation. All the subjects were Chinese Han Nationality.
Methods
DNA extraction and detection of CYP2C19*2 and CYP2C19*3 A 5 mL blood sample was collected from each subject and DNA was extracted from blood for CYP2C19 genotyping according to Lahiri et al[5]. For the detection of CYP2C19*2, paired PCR reactions were set, one containing CYP2C19*2 primers and the other containing wild-type primers (Table 1). Each PCR reaction (50 μL) containing 10 mmol/L Tris-HCl, pH8.3, 50 mmol/L KCl, 2.0 mmol/L MgCl2, 0.2 mmol/L of each dNTPs, 0.2 μmol/L of each primers and 50-1000 ng of DNA template, 1 unit of Taq DNA polymerase was amplified through 35 cycles, each cycle consisting of denaturation at 94 °C for 1 min, primer annealing at 61 °C for 1 min, and extension at 72 °C for 1.5 min. Finally, a further extension was carried out at 72 °C for 10 min. The amplicon was analyzed on 20 g/L agarose gel electrophoresis. For detection of CYP2C19*3, paired PCR reactions were set, one containing CYP2C19*3 primers and the other containing wild-type primers (Table 2). Each PCR reaction (50 μL) containing 10 mmol/L Tris-HCl pH8.3, 50 mmol/L KCl, 2.0 mmol/L MgCl2, 0.2 mmol/L of each dNTPs, 0.2 μmol/L of each primers and 50-1000 ng of DNA template, 1 unit of Taq DNA polymerase was amplified through 35 cycles, each cycle consisting of denaturation at 94 °C for 1 min, primer annealing at 58 °C for 1 min and extension at 72 °C for 30 s. Finally, a further extension was performed at 72 °C for 10 min. The amplicon was analyzed on 20 g/L agarose gel electrophoresis.
Table 1.
Primers designed for detection of CYP2C19*2
| Genotype | Sequences | |
| Wild-type | Forward | 5’-AATTACAACCAGAGAGCTTGGC-3’ |
| Reverse | 5’-GTAATTTGTTATGGGTTCCC-3’ | |
| Mutant | Forward | 5’-AATTACAACCAGAGAGCTTGGC-3’ |
| Reverse | 5’-GTAATTTGTTATGGGTTCCT-3’ | |
Table 2.
Primers designed for detection of CYP2C19*3
| Genotype | Sequences | |
| Wild-type | Forward | 5’-TATTATTATCTGTTAACTAATATGA-3’ |
| Reverse | 5’-AACTTGGCCTTACCTGGATC-3’ | |
| Mutant | Forward | 5’-TATTATTATCTGTTAACTAATATGA-3’ |
| Reverse | 5’-AACTTGGCCTTACCTGGATT-3’ | |
The genotypes were judged by shown up of goal fragments in the paired PCR amplifications. For CYP2C19*2 allele, the goal fragment (139 bp band) could be seen only in the lane of CYP2C19*2 PCR reaction means homozygous CYP2C19*2 allele (*2/*2), only in the lane of wild-type reaction means homozygous wild-type (*1/*1), and in both reaction means heterozygous CYP2C19*2 (*1/*2). For CYP2C19*3 allele, the goal fragment (253 bp band) could be seen only in the lane of CYP2C19*3 PCR reaction means homozygous CYP2C19*3 allele (*3/*3), only in the lane of wild-type reaction means homozygous wild-type (*1/*1), and in both reaction means heterozygous CYP2C19*3 (*1/*3). In the consideration of both alleles, the other genotype could be *2/*3.
Quality control checks of PCR procedures indicated DNA samples genotyped in a double-blinded fashion yielded the same alleles as found during previous genotyping of DNA. The genotypes were checked for reliability by comparing with PCR-RFLP procedure[6,7].
RESULTS
Genotyping of CYP2C19
Four pairs of primers were designed according to CYP2C19 wild-type sequence and the mutations in CYP2C19*2 and CYP2C19*3 by the principle of allele-specific amplification. PCR amplification conditions were tested to find the optimum annealing temperature and MgCl2 concentration. The method was proved to be quick, accurate and less contamination (Figure 1, Figure 2).
Figure 1.
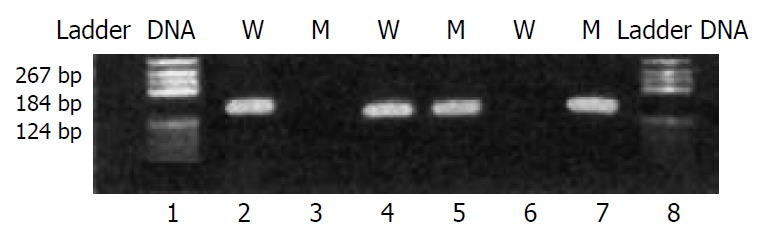
Typical electrophoresis pattern in detection of CYP2C19*2. Goal fragment was 139 bp. Lanes 1 and 8: DNA ladder; Lanes 2, 4 and 6: wild-type primers; Lanes 3, 5 and 7: mutant primers; Sample 1: homozygous wild-type (lanes 2 and 3); Sample 2: heterozygote (lanes 4 and 5); Sample 3: homozygous mutant (lanes 6 and 7).
Figure 2.
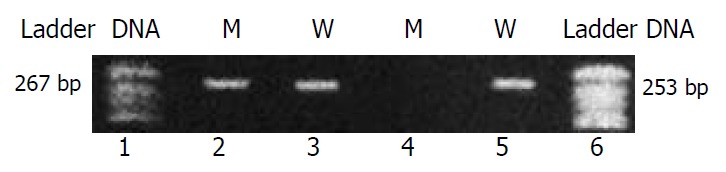
Typical electrophoresis pattern in detection of CYP2C19*3. Goal fragment was 253 bp. Lanes 1 and 6: DNA ladder; Lanes 2 and 4: mutant primers; Lanes 3 and 5: wild-type primers. Sample 1: heterozygote (lanes 2 and 3); Sample 2: homozygous wild-type (lanes 4 and 5); No homozygous mutant found in this study.
Frequency study of poor metabolizers in the cancer groups and the control group
The frequencies of poor metabolizers of CYP2C19 (genotypes of *2/*2 and *2/*3, no *3/*3 was found in this study) were of 34.1% (46/135) in the group of esophagus cancer patients, 31.8% (47/148) in the stomach cancer patients, 34.4% (73/212) in the group of lung cancer patients, only 4.5% (5/112) in the bladder cancer patients and 14.0% (52/372) in control group. There were statistical differences between cancer groups and control group (esophagus cancer, χ2 = 25.65, P < 0.005, OR = 3.18, 95%CI = 2.005-5.042; stomach cancer, χ2 = 21.70, P < 0.005, OR = 2.86, 95%CI = 1.820-4.501; lung cancer, χ2 = 33.58, P < 0.005, OR = 3.23, 95%CI = 1.503-6.906; bladder cancer, χ2 = 7.50, P < 0.01, OR = 0.288, 95%CI = 0.112-0.740). It was obviously that CYP2C19 PMs had a high incidence of esophagus cancer, stomach cancer and lung cancer, conversely CYP2C19 PMs had a low incidence of bladder cancer (Table 3).
Table 3.
Frequencies of EMs and PMs in different cancer groups and control group
| Group | Total |
EMs (*1/*1, *1/*2 , *1/*3) |
PMs (*2/*2,*2/*3) |
||
| cases | Cases | % | Cases | % | |
| Esophagus cancer | 135 | 89 | 65.9 | 46 | 34.1 |
| Stomach cancer | 148 | 101 | 68.2 | 47 | 31.8 |
| Lung cancer | 212 | 139 | 65.6 | 73 | 34.4 |
| Bladder cancer | 112 | 107 | 95.5 | 5 | 4.5 |
| Control | 372 | 320 | 86.0 | 52 | 14.0 |
Esophagus cancer, χ2 = 25.65, P < 0.005, OR = 3.18, 95%CI = 2.005-5.042; Stomach cancer, χ2 = 21.70, P < 0.005, OR = 2.86, 95%CI = 1.820-4.501; Lung cancer, χ2 = 33.58, P < 0.005, OR = 3.23, 95%CI = 1.503-6.906; Bladder cancer, χ2 = 7.50, P < 0.01, OR = 0.288, 95%CI = 0.112-0.740.
DISCUSSION
With the completion of the Human Genome Project, great opportunities exist to investigate the effects of genetic variation of the cancer susceptibility to environment exposure. It is known to all cigarette smoke can cause lung cancer, but the fact is not every smoker suffering from lung cancer. What is the mechanism of which smoker is more susceptible to lung cancer, which smoker may not catch the disease. One of the answers may lie on the gene polymorphisms of drug metabolising enzymes. This rule may also apply to esophagus cancer, stomach cancer and bladder cancer.
Cytochrome P450s are the main drug metabolizing enzymes in human body, and are always found to participate in the metabolism of carcinogens or procarcinogens. Some are involved in the activation of procarcinogens, some may take part in the inactivation of carcinogens. That depends on what kind of carcinogens and what kind of cancers, and what kind of mechanism of carcinogenesis.
CYP2C19-one of the most important cytochrome P450s, is known as a key enzyme in the in vivo metabolism of a number of related hydantoins and barbiturates, as well as in the metabolism of structurally unrelated drugs such as omeprazole, lansoprazole, progunil, mephenytoin and citalopram[1]. Individuals can be divided into two groups, poor metabolizers (PMs) and extensive metabolizers (EMs), depending on the hydroxylation ability of S-mephenytoin. There are two main enzyme deficient alleles called CYP2C19*2 (CYP2C19m1) and CYP2C19*3 (CYP2C19m2). CYP2C19*2 is a single base pair G681→A mutation in exon 5 of CYP2C19 and accounts for 75% and 85% of Oriental and Caucasian mutant alleles, respectively[6]. CYP2C19*3 is a single base pair G636→A mutation in exon 4 of CYP2C19 which results in a premature stop codon[7]. It accounts for 10%-25% of Oriental mutant alleles and is rare in Caucasians. An individual who inherits two mutant CYP2C19 alleles, whatever same kind (*2/*2, *3/*3) or different kind (*2/*3), has a reduced capacity to metabolize CYP2C19 substrates and is a PM. Individuals who are homozygous (*1/*1) or heterozygous (*1/*2, *1/*3) for wild-type CYP2C19*1 have efficient enzyme to metabolize CYP2C19 substrates and are EMs. Although there are several other reports about rare enzyme defect alleles, it is recognized that the purpose of prediction of CYP2C19 phenotype can be achieved by genotyping CYP2C19 only with CYP2C19*2 and CYP2C19*3 in Chinese population[8].
In this study, 135 esophagus cancer patients, 148 stomach cancer patients, 212 lung cancer patients, 112 bladder cancer patients and 372 controls were genotyped for CYP2C19. Among them, 34.1% of esophagus cancer patients, 31.8% of stomach cancer patients and 34.4% of lung cancer patients, but only 4.5% of bladder cancer patients and 14.0% of healthy controls were genotyped as CYP2C19 PMs. Statistical analysis of the frequencies of PMs in cancer groups and control group showed significant differences (Table 3). It means CYP2C19 PMs are more susceptible to esophagus cancer, stomach cancer and lung cancer, but it is unsusceptible to Bladder cancer.
Several studies on CYP2C19 polymorphism and its association with carcinogenesis have shown self-contradiction results[2-4]. However, our data indicated that CYP2C19 polymorphism was associated with esophagus cancer, stomach cancer, lung cancer and bladder cancer. Furthermore, we found that CYP2C19 PMs had increased risk of esophagus cancer, stomach cancer and lung cancer, and a decreased risk of bladder cancer. However, Klose et al[9] reported that CYP2C19 was only expressed in liver and duodenum. How it functioned so differently in different organs remained a mystery. But from the organs listed above, it is deducible that CYP2C19 PMs are more susceptible to the cancers of upper or systemic organs, such as esophagus, stomach, lung and blood. And they are unsusceptible to the cancers of lower organs, like bladder and colorectal cancers. Prostate cancer was another example which showed no relationship between CYP2C19 polymorphism and carcinogenesis. It implies that different type of cancers may have different oncological mechanisms.
The genetic background and living environment of an individual are most important factors for carcinogensis[10]. The relationship between genetic polymorphism of CYP genes[11,12], ABO blood groups[13] and well water pollution are widely recognized. But more efforts needed to elucidate the correlation of different types of cancer to so many different factors, the development of biochip technology may speed it up[14].
Footnotes
Supported by Research funding from Health Bureau of Zhejiang Province (G20030697) and Research Fund from Hangzhou Tobacco Factory
Edited by Kumar M Proofread by Xu FM
References
- 1.Xie HG, Kim RB, Wood AJ, Stein CM. Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol. 2001;41:815–850. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- 2.Wadelius M, Autrup JL, Stubbins MJ, Andersson SO, Johansson JE, Wadelius C, Wolf CR, Autrup H, Rane A. Polymorphisms in NAT2, CYP2D6, CYP2C19 and GSTP1 and their association with prostate cancer. Pharmacogenetics. 1999;9:333–340. doi: 10.1097/00008571-199906000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Roddam PL, Rollinson S, Kane E, Roman E, Moorman A, Cartwright R, Morgan GJ. Poor metabolizers at the cytochrome P450 2D6 and 2C19 loci are at increased risk of developing adult acute leukaemia. Pharmacogenetics. 2000;10:605–615. doi: 10.1097/00008571-200010000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Sachse C, Smith G, Wilkie MJ, Barrett JH, Waxman R, Sullivan F, Forman D, Bishop DT, Wolf CR. A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis. 2002;23:1839–1849. doi: 10.1093/carcin/23.11.1839. [DOI] [PubMed] [Google Scholar]
- 5.Lahiri DK, Nurnberger JI. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 7.de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 8.Shu Y, Zhou HH. Individual and ethnic differences in CYP2C19 activity in Chinese populations. Acta Pharmacol Sin. 2000;21:193–199. [PubMed] [Google Scholar]
- 9.Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol. 1999;13:289–295. doi: 10.1002/(sici)1099-0461(1999)13:6<289::aid-jbt1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimura K, Hanaoka T, Ohnami S, Ohnami S, Kohno T, Liu Y, Yoshida T, Sakamoto H, Tsugane S. Allele frequencies of single nucleotide polymorphisms (SNPs) in 40 candidate genes for gene-environment studies on cancer: data from population-based Japanese random samples. J Hum Genet. 2003;48:654–658. doi: 10.1007/s10038-003-0096-1. [DOI] [PubMed] [Google Scholar]
- 11.Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K. Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev. 2000;9:3–28. [PubMed] [Google Scholar]
- 12.Ribeiro Pinto LF, Teixeira Rossini AM, Albano RM, Felzenszwalb I, de Moura Gallo CV, Nunes RA, Andreollo NA. Mechanisms of esophageal cancer development in Brazilians. Mutat Res. 2003;544:365–373. doi: 10.1016/j.mrrev.2003.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Su M, Lu SM, Tian DP, Zhao H, Li XY, Li DR, Zheng ZC. Relationship between ABO blood groups and carcinoma of esophagus and cardia in Chaoshan inhabitants of China. World J Gastroenterol. 2001;7:657–661. doi: 10.3748/wjg.v7.i5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landi S, Gemignani F, Gioia-Patricola L, Chabrier A, Canzian F. Evaluation of a microarray for genotyping polymorphisms related to xenobiotic metabolism and DNA repair. Biotechniques. 2003;35:816–820, 822, 824-7. doi: 10.2144/03354mt03. [DOI] [PubMed] [Google Scholar]


