Abstract
AIM: To identify the new alternative splicing variants of human CYP2D6 in human extratumoral liver tissue with RT-PCR and sequencing.
METHODS: Full length of human CYP2D6 cDNAs was amplificated by reverse transcription-polymerase chain reaction (RT-PCR) from a human extratumoral liver tissue and cloned into pGEM-T vector. The cDNA was sequenced. Exons from 1 to 4 of human CYP2D6 cDNAs were also amplificated by RT-PCR from extratumoral liver tissues of 17 human hepatocellular carcinomas. Some RT-PCR products were sequenced. Exons 1 to 4 of CYP2D6 gene were amplified by PCR from extratumoral liver tissue DNA. Two PCR products from extratumoral liver tissues expressing skipped mRNA were partially sequenced.
RESULTS: One of the CYP2D6 cDNAs had 470 nucleotides from 79 to 548 (3’ portion of exons 1 to 5’portion of exon 4), and was skipped. Exons 1 to 4 of CYP2D6 cDNA were assayed with RT-PCR in 17 extratumoral liver tissues. Both wild type and skipped mRNAs were expressed in 4 samples, only wild type mRNA was expressed in 5 samples, and only skipped mRNA was expressed in 8 samples. Two more variants were identified by sequencing the RT-PCR products of exons 1 to 4 of CYP2D6 cDNA. The second variant skipped 411 nucleotides from 175 to 585. This variant was identified in 4 different liver tissues by sequencing the RT-PCR products. We sequenced partially 2 of the PCR products amplified of CYP2D6 exon 1 to exon 4 from extratumoral liver tissue genomic DNA that only expressed skipped mRNA by RT-PCR. No point mutations around exon 1, intron 1, and exon 4, and no deletion in CYP2D6 gene were detected. The third variant was the skipped exon 3 , and 153 bp was lost.
CONCLUSION: Three new alternative splicing variants of CYP2D6 mRNA have been identified. They may not be caused by gene mutation and may lose CYP2D6 activity and act as a down-regulator of CYP2D6.
INTRODUCTION
Over 90% medications are metabolized by cytochrome P450 (CYP) family of liver isoenzymes[1]. The most important enzymes are CYP1A2, 3A4, 2C9/19, 2D6 and 2E1. Although CYP2D6 accounts for < 2% of the total CYP liver enzyme, nevertheless it mediates metabolism in 25% of drugs in clinical use such as antipsychotics, antidepressants, beta-blockers, antiarrhythmic agents and opiates[2]. CYP2D6 exhibits an extensive polymorphism[3]. Over 40 CYP2D6 allelic variants have been reported[4].
Pre-mRNA splicing involves precise removal of introns from pre-mRNA, such that exons are spliced together to form mature RNAs with intact translation reading frames. Splicing requires exon recognition, followed by accurate cleavage and rejoining, which are determined by the invariant GU and AG intronic dinucleotides at the 5’ (donor) and 3’ (acceptor) exon-intron junctions, respectively[5]. Human genes typically contain multiple introns, and in many cases the exons can be joined in more than one way to generate multiple mRNAs, encoding distinct protein isoforms. This process named alternative splicing is a major mechanism for modulating the expression of cellular and viral genes and enables a single gene to increase its coding capacity, allowing synthesis of several structurally and functionally distinct protein isoforms[6-9].
CYP2D subfamily comprises CYP2D6 gene and psudogenes, i.e. CYP2D7P and CYP2D8BP. Six mRNA splice variants of CYP2D have been identified in human liver[10], breast[11], lung[12], and brain tissues[13]. Variants a and b retain intron 5 and 6, respectively; variant b’ has missed the 3’ 91 bp portion of exon 6; variant c has missed exon 6; variant d retains 57 bp portion of intron 6; variant e retains the 3’ portion of intron 6 and has missed the 61 bp fragment at the 3’ end of exon 6. Forms b’ and c are variants of CYP2D6, and forms d, c, b, b’ are variants of CYP2D7P[10-13]. All of these CYP2D splice variant mRNAs disrupt open reading frames. If they are translated, they would not have CYP2D6 function.
Three new pre-mRNA alternative splicing variants of human CYP2D6 in human extratumoral liver tissue were identified by RT-PCR and sequencing.
MATERIALS AND METHODS
Materials
Moloney murine leukemia virus (M-MuLV) reverse transcriptase was supplied by MBI Fermentas AB, Lithuania. Random hexamer primers, T4 DNA ligase and pGEM-T vector system were supplied by Promega Corp. PCR primers, DNA sequence primers, dNTPs and Taq DNA polymerase were synthesized or supplied by Shanghai Sangon Biotechnology Co. DNA sequencing kit was purchased from Perkin-Elmer Corp. Diethyl pyrocarbonate (DEPC) was from Sigma Chemical Co. DNA gel extraction kit was from Hangzhou V-gene Biotechnology Ltd. Other chemical reagents used were all of analytical purity from commercial sources. Extratumoral liver tissue samples were collected from patients undergoing hepatocellular carcinoma resection at affiliated hospitals of Zhejiang University School of Medicine and stored at -70 °C.
Cloning and sequencing of human CYP2D6 cDNA from a human extratumoral liver tissue
Total RNA was extracted from a human extratumoral liver tissue from a Han nationality Chinese with AGPC method[14]. RT-PCR amplifications were described before[15,16]. Two specific 28 mer oligonucleotide PCR primers were designed according to cDNA sequence of CYP2D6 reported by Gonzalez et al[10] (GenBank accession no. NM_000106). The sequence of sense primer (CYP2D6 F) corresponds to base position of -39 to -12, i.e. 5’-AGGTGTGTCTCGAGGAGCCCATTTGGTA-3’, with a restriction site of XhoI (underlined), and the antisense one (CYP2D6 R), corresponds to base position from 1503 to 1530, i.e. 5’-TGGCTAGGGATCCGGCTGGGGACTAGGT-3’, with a restriction site of BamH I (underlined). The anticipated PCR products were 1.569 kb in length. PCR was performed at 94 °C for 5 min, then 35 cycles, each at 94 °C for 60 s, at 62 °C for 60 s, at 72 °C for 2 min, and a final extension at 72 °C for 10 min. An aliquot (10 μL) from PCR was subjected to electrophoresis in a 10 g/L agarose gel. The PCR products were ligated with pGEM-T vector, and transformed into E. coli DH5α. The CYP2D6 cDNA cloned in pGEM-T was sequenced by dideoxy chain-termination method marked with BigDye with primers of T7 and SP6 promoters and a specific primer of 5-ACCTCATGAATCACGGCAGT-3 (nt 1069 to 1088) on Perkin-Elmer-ABI Prism 310 automated DNA sequencer.
RT-PCR and sequencing analysis of liver transcripts of CYP2D6 exons 1 to 4
Transcripts of CYP2D6 exons 1 to 4 were assayed with RT-PCR using 17 extratumoral liver tissues from Han nationality Chinese, with CYP2D6 F and CYP2D6 4R primer: 5’-GCAGAAAGCCCGACTCCTCCTTCA-3’ (nt 638 to 661). Simultaneously the beta-actin (GenBank accession no. NM_001101) cDNA fragment was amplified in the same Eppendorf tube as an internal control[17]. The sequences of sense and antisense primers used for amplification of beta-actin cDNA fragment were 5’- TCCCTGGAGAAGAGCTACGA-3’ (nt 776 to 795) and 5’-CAAGAAAGGGTGTAACGCAAC-3’ (nt 1217 to 1237) respectively. The anticipated PCR products of CYP2D6 exons 1 to 4 were 700 bp in length, and those of beta-actin were 462 bp. PCR was performed at 94 °C for 3 min, then 35 cycles, each at 94 °C for 30 s, at 62 °C for 30 s, at 72 °C for 45 s, and a final extension at 72 °C for 7 min. An aliquot (10 μL) from PCR was subjected to electrophoresis in a 17 g/L agarose gel. Several RT-PCR products of CYP2D6 exons 1 to exon 4 were shorter than the anticipated full length of 700 bp. The shorter RT-PCR products were separated by agarose gel electrophoresis and extracted using DNA gel extraction kit according to the manufacturer’s instructions, and then sequenced by dideoxy chain-termination method marked with BigDye with primer of CYP2D6 F on Perkin-Elmer-ABI Prism 310 automated DNA sequencer.
Sequencing identification of exon 1 to exon 4 of CYP2D6 gene
Genomic DNA of human extratumoral liver tissues expressed full length of CYP2D6 exons 1 to 4 only (3 samples), or only exons skipped (2 samples) or both of full length and skipped ones (3 samples) were extracted according to the methods reported by Gross-Bellard et al[18]. The segment of CYP2D6 gene from exons 1 to 4 was amplified with CYP2D6 F and CYP2D6 4R primers. The anticipated PCR products were 2043 bp (from 1581 to 3623) in length. Two PCR products from extratumoral liver tissues that only expressed skipped mRNA were partially sequenced by RT-PCR.
RESULTS
Identification of a new alternative splicing variant from a cDNA clone
During cloning of CYP2D6 cDNA, a 1.1 kb RT-PCR product was obtained, which was much shorter than anticipated 1.57 kb (Figure 1).
Figure 1.
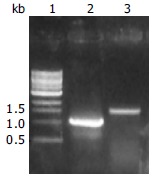
Electrophoresis of RT-PCR products of CYP2D6 cDNA. Lane 1: 1 kb DNA marker, 2: Amplification of 1.1 kb in size, 3: Amplification of full length of 1.57 kb.
The cDNA obtained was then inserted into pGEM-T vector and sequenced. Compared with human wild type CYP2D6 cDNA sequence reported by Kimura et al[19] (GenBank accession no.M33388), there were 470 nucleotides from 84 to 553, i.e., the 3’ end portion 97 bp of exon 1 (180 bp), exon2, exon 3, and the 5’ end portion 48 bp of exon 4 (161 bp) were skipped in the cDNA shorter in length (Figures 2, Figures 3). Substitution, insertion and deletion occurred in 13 base pairs, i.e. 620A→G, 712G→T, 1196T→G, 1401T→G, 1405C→G, 1408A→G, 1410T→C, 1432C→T, 1433A→C, 1435G→C, 1400 insG, 1442T→C, 1443T→A, 1449C del, 1457G→C. This cDNA had a premature stop codon at codon 96 and was named variant f.
Figure 2.
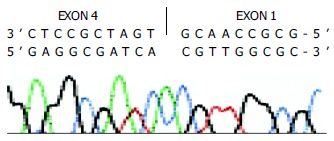
Partial sequences of the cloned human CYP2D6 cDNA. The upper sequence represents the sense strand and the underside sequence represents the sequenced anti-sense strand.
Figure 3.

Comparison of sequences of CYP2D6 cDNA, CYP2D7 BP and 3 new CYP2D6 alternative splicing variants, f, g, h. The different bases compared with wild type CYP2D6 are indicated in boldface and “_”, and the border of exons is indicated by ‘!’. The skipped parts are indicated by ‘-’.
RT-PCR analysis of transcripts of CYP2D6 exons 1 to 4 in 17 liver extratumoral tissues and HepG2 cells, two more new alternative splicing variants were identified by sequencing
In 17 liver tissues, 4 samples expressed both wild type and skipped mRNA, 5 samples expressed only wild-type mRNA, and 8 samples expressed only skipped mRNA. Representative electrophoresis of RT-PCR products of exons 1 to 4 of CYP2D6 from human extratumoral liver tissues and HepG2 cells with beta-actin as internal control are shown in Figure 4.
Figure 4.
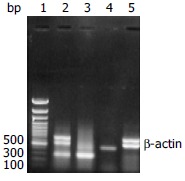
Representative electrophoresis of RT-PCR products of exons 1 to 4 of CYP2D6 from human liver tissues and HepG2 cells with beta-actin as internal control (464 bp). Lane 1: 100 bp marker, lane 2: A sample having both full length of 700 bp and shorter 300 bp, lane 3: A sample having only shorter ones (300 bp), lane 4: HepG2 cells having no CYP2D6 expressed, Lane 5: A sample having only full length 700 bp of exons 1 to 4 of CYP2D6.
The sequencing data of RT-PCR products demonstrated that the second variant lost 411 nucleotides (137 amino acid residues) from 177 to 587, i.e., the 3’ end portion 4 bp of exon 1 (180 bp), exon 2, exon 3, and the 5’ end portion 83 bp of exon 4 (161 bp) and gained a base substitution 100 C→T, resulting in one amino acid exchange P34S, as compared with the human wild type CYP2D6 cDNA sequence reported by Kimura et al[19] (GenBank accession no.M33388) (Figure 5A, Figure 2). This variant was identified in 4 different extratumoral liver tissues by sequencing RT-PCR products and was named variant g.
Figure 5.
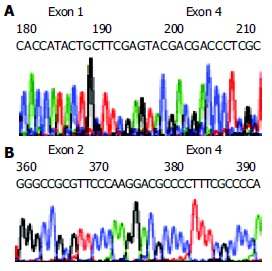
Partial sequences of two alternative splicing variants of CYP2D6. A: Part of skipped exon 1, exon 2, exon 3, and part of exon 4. B: Skipped exon 3.
The third variant had skipped exon 3, losing 153 bp from nucleotides 353 to 505 or 51 amino acid residues from 117 to 167, and had 2 base substitutions 100 C→T and 336 T→C, resulting in one changed amino acid P34S and a samesense mutation 112F, respectively. This variant was named variant h (Figure 5B, Figure 2).
No mutation events around splicing sites of exon 1 and exon 4 nor gene deletion from exon 1 to exon 4 of CYP2D6 gene
The segment of CYP2D6 gene at exons 1 to 4 was amplified with CYP2D6 F and CYP2D6 4R primers by PCR. All amplified products from 8 samples gave anticipated 2.04 kb in length but no 0.3 kb PCR products as predicted from the possibility that the shorter cDNA was originated from deleted mutations in CYP2D6 gene rather than due to alternative splicing (Figure 6). Two PCR products from extratumoral liver tissues expressed only skipped exon were purified and partially sequenced. One showed 3 mutations, i.e., 1719 (nt 100) C→T in exon 1, resulting in P34S, 3280 (nt 408) G→C in exon 3, and samesense mutations 136 V, 3056 G→C in intron 2. Another one showed 3 mutations, i.e., 1719 (nt 100) C→T in exon 1, resulting in P34S, 1958 T→C in intron 1, and 3056 G→C in intron 2.
Figure 6.
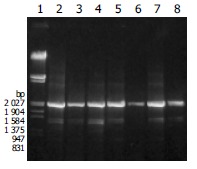
Electrophoresis of PCR products from genomic DNA amplified using CYP2D6 F and CYP2D6 4R primers. Lane 1: λ/EcoR I and Hind III marker, lanes 2-4: PCR products from livers expressing full and short length RT-PCR products, Lanes 5, 6: PCR products from livers expressing full length RT-PCR products, Lanes 7, 8: PCR products from livers expressing only short length RT-PCR products.
DISCUSSION
The mechanism of exon missed in cDNA may include gene deletion, alternative splicing, template sequence recombination present in PCR amplification[20-22], and retroviral recombination[23-25]. One or several exons missed entirely could ascribe to alternative splicing. The first mechanism of formation of recombinant molecules during PCR is mediated by premature termination of chain synthesis. Therefore, a short extension time or increased dissociation of polymerases from the template may promote recombination. The optimum values to minimize recombinant molecule formation could prolong the extension time and could be achieved using 30 or 34 amplification cycles respectively. The second mechanism is template switching, it implicates that a fragment elongating on one template, can continue this elongation on another homologous template. The high template concentrations could facilitate template switching and de novo synthesize recombinant molecules. We performed PCR with lower template concentrations, 1 min/kb extension, and 32 to 35 amplification cycles to minimize the PCR recombination. During synthesis of the first DNA strand, reverse transcriptase could switch templates from one to other copy of RNA, a phenomenon known as copy-choice. The term ‘template switching’ implies that the nascent DNA strand is transferred from one RNA (the ‘donor’) to the other (the ‘acceptor’). This process depends on a ribonuclease (RNase H) activity, carried by reverse transcriptase, which could degrade the RNA template once it has been copied[26]. The RNA recombination is a very rare event. In this study, MMLV without RNase H activity was used and skipped mRNA was found in 12 out of 17 liver tissues by RT-PCR assay. Variant g was identified in 4 different extratumoral liver tissues by sequencing RT-PCR products. These facts indicated that the exon missed variant g did not come from PCR recombination or retroviral recombination. PCR amplification and sequencing identification of exon 1 to exon 4 of CYP2D6 gene from genomic DNA indicated that the exon missed variant g came from neither gene mutation around splicing sites of exon 1 and exon 4 nor gene deletion.
Burset et al[27] analyzed canonical and non-canonical splice sites in mammalian genomes, and found that 99.24% contained canonical dinucleotides GT and AG for donor and acceptor sites, respectively, 0.69% contained GC-AG, 0.05% contained AT-AC and only 0.02% contained other types of non-canonical splice sites. Some non-canonical splice sites seemed to be involved in immunoglobulin gene expression and the others in alternative splicing events.
CYP2D6 gene has 9 exons and 8 introns. All alternative splicing CYP2D reported are located in exon 6 to exon 7. No alternative splicing in exon 1 to exon 4 has been reported. We identified 3 new alternative splicing CYP2D6 cDNA in exon 1 to exon 4 in human extratumoral liver tisssues. The skipped mRNA was not an unusually phenomenon, for 12 samples expressed skipped mRNA in 17 human liver tissue, whereas 9 samples expressed wild-type mRNA in this study.
We cloned a cDNA variant of human CYP2D6, the variant f with some exons skipped. The five nt 5’-CAACG-3’, located at the boundary of partially skipped exons 1 and 4 (Figure 6). It looked as the 3’ end portion of skipped exon 1 or the 5’ end portion of exon 4. If we looked it as the 3’ end portion of the skipped exon 1, then the splicing donor and acceptor sites would be CT-CG, which were not found as splice site pairs[27]. If we looked it as the 5’ end portion of exon 4, then there would be 470 nucleotides from 79 to 548, i.e., the 3’ end portion 102 bp of exon 1 (180 bp), exon 2, exon 3, and the 5 end portion 43 bp of exon 4 (161 bp) were skipped. The splicing donor and acceptor sites would be the non-canonical splice sites:CA-AG[27].
CYP2D gene exists in a number of structurally polymorphic haplotypes and also comprises several variants of CYP2D7P and CYP2D8P pseudogenes. CYP2D6 is highly homologous to CYP2D7 and CYP2D8 pseudogenes. The characteristic base substitution, insertion and deletion of the variant f from 1401 to 1457 in exon 9 indicated that this exon was derived from CYP2D7BP[28] (GenBank accession no.X58468). So this cDNA clone may be transcripted from CYP2D6*36 (Trivial name: CYP2D6Ch2 or CYP2D6*10C) gene, which has gene conversion to CYP2D7 in exon 9[29]. Johansson et al[29] using allele-specific polymerase chain reaction analysis of genomic DNA from 90 Chinese individuals revealed that CYP2D6*10 (CYP2D6Ch1) allele was the most common one, CYP2D6*36 was not a common allele[30] and so we got an alternative splicing variant from this allele only once.
Another alternative splicing CYP2D6 cDNA, variant g, has 7 bp 5’-GCTTCGA-3’, which locates at the boundary of partially skipped exons 1 and 4 (Figure 6). It can be regarded as the 3’ end portion of skipped exon 1 or the 5’ end portion of exon 4. The splicing donor and acceptor sites would be CC-GA or GC-CC which have not been found as splice site pairs[27]. If we regarded it as the skipped nucleotides from 175 to 585, then 411 nucleotides (137 amino acid residues), i.e., the 3’ end portion 6 bp of exon 1 (180 bp), exon 2, exon 3, and the 5’ end portion 81 bp of exon 4 (161 bp) were skipped. The splicing donor and acceptor sites would be the non-canonical splice sites: GA-TC[27]. As this variant has been identified in 4 different liver tissues by RT-PCR products sequencing, it could not be the RT-PCR artificial ones.
The third variant, variant h, having skipped the entire exon 3, would use the most common non-canonical GC-AG splice sites (Figure 6). It has an amino acid change of P34S which is a common polymorphism occurring in CYP2D6*4, 10, 14, 36, 37. This variant might come from CYP2D6*10, the most common allele in Chinese.
The cDNA sequence of CYP2D6 is differed from that of CYP2D7P at nucleotides 629 to 639, CYP2D6[19] was AGGAGGGACTG whereas CYP2D7P[28] was AGGGAGGGATCG (Figure 6). All sequencing data of the alternative splicing cDNA and PCR products originated from related genomic DNA showed the characteristics of CYP2D6, so they were all derived from CYP2D6 but not from CYP2D7P.
Variant f has premature stop codon at codon 96, which would cause loss of its enzyme activity. According to the homology modeling study of human CYP2 family enzyme reported by Lewis, the substrate could recognize site (SRS) 1 of human CYP2D6 located between amino acid residues 101 and 123 and the SRS2 located between amino acid residues 204 to 215. Variant g which lost 137 amino acid residues from 58 to 194, would lose SRS1, and might not have CYP2D6 activity. Variant h having skipped the entire exon 3, would lose 51 amino acid residues from 117 to 167, where part of SRS1 located. It might not have CYP2D6 activity.
All 3 CYP2D6 splice variants might not have CYP2D6 function. But the possibility of the expression of proteins with novel function(s) could not be excluded. The splice variants might have a role in the posttranscriptional down-regulation of the expression of CYP2D6. Formation of variant mRNAs at the expense of full-length mRNAs would ultimately result in a diminished expression of CYP2D6 protein.
Three new CYP2D6 pre-mRNA alternative splicing variants in human extratumoral liver tissues have been identified. Futher work is needed to clarify if they are commonly existed in liver tissue or only in extratumoral liver tissue, and their relation with hepatocellular carcinoma as well.
Footnotes
Supported by the National Key Basic Research and Development Program of China, No. 2002CB512901, National Natural Science Foundation of China, No.39770868 and Natural Science Foundation of Zhejiang Province, No.397490
Edited by Ren SY and Wang XL Proofread by Xu FM
References
- 1.Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- 2.Bertilsson L, Dahl ML, Dalén P, Al-Shurbaji A. Molecular genetics of CYP2D6: clinical relevance with focus on psychotropic drugs. Br J Clin Pharmacol. 2002;53:111–122. doi: 10.1046/j.0306-5251.2001.01548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cascorbi I. Pharmacogenetics of cytochrome p4502D6: genetic background and clinical implication. Eur J Clin Invest. 2003;33 Suppl 2:17–22. doi: 10.1046/j.1365-2362.33.s2.3.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 5.Nissim-Rafinia M, Kerem B. Splicing regulation as a potential genetic modifier. Trends Genet. 2002;18:123–127. doi: 10.1016/s0168-9525(01)02619-1. [DOI] [PubMed] [Google Scholar]
- 6.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 7.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 8.Kriventseva EV, Koch I, Apweiler R, Vingron M, Bork P, Gelfand MS, Sunyaev S. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- 9.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, Gelboin HV, Hardwick JP, Meyer UA. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988;331:442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- 11.Huang Z, Fasco MJ, Kaminsky LS. Alternative splicing of CYP2D mRNA in human breast tissue. Arch Biochem Biophys. 1997;343:101–108. doi: 10.1006/abbi.1997.0153. [DOI] [PubMed] [Google Scholar]
- 12.Huang Z, Fasco MJ, Spivack S, Kaminsky LS. Comparisons of CYP2D messenger RNA splice variant profiles in human lung tumors and normal tissues. Cancer Res. 1997;57:2589–2592. [PubMed] [Google Scholar]
- 13.Woo SI, Hansen LA, Yu X, Mallory M, Masliah E. Alternative splicing patterns of CYP2D genes in human brain and neurodegenerative disorders. Neurology. 1999;53:1570–1572. doi: 10.1212/wnl.53.7.1570. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Dong H, Cai Z, Yu Y. Stable expression of human cytochrome CYP2B6 and CYP1A1 in Chinese hamster CHL cells: their use in micronucleus assays. Chin Med Sci J. 1997;12:148–155. [PubMed] [Google Scholar]
- 16.Qian Y, Yu Y, Cheng X, Luo J, Xie H, Shen B. Molecular events after antisense inhibition of hMSH2 in a HeLa cell line. Mutat Res. 1998;418:61–71. doi: 10.1016/s1383-5718(98)00108-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhuge J, Luo Y, Yu YN. Heterologous expression of human cytochrome P450 2E1 in HepG2 cell line. World J Gastroenterol. 2003;9:2732–2736. doi: 10.3748/wjg.v9.i12.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross-Bellard M, Oudet P, Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973;36:32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- 19.Kimura S, Umeno M, Skoda RC, Meyer UA, Gonzalez FJ. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989;45:889–904. [PMC free article] [PubMed] [Google Scholar]
- 20.Judo MS, Wedel AB, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaphiropoulos PG. Non-homologous recombination mediated by Thermus aquaticus DNA polymerase I. Evidence supporting a copy choice mechanism. Nucleic Acids Res. 1998;26:2843–2848. doi: 10.1093/nar/26.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shammas FV, Heikkilä R, Osland A. Fluorescence-based method for measuring and determining the mechanisms of recombination in quantitative PCR. Clin Chim Acta. 2001;304:19–28. doi: 10.1016/s0009-8981(00)00374-0. [DOI] [PubMed] [Google Scholar]
- 23.Negroni M, Buc H. Mechanisms of retroviral recombination. Annu Rev Genet. 2001;35:275–302. doi: 10.1146/annurev.genet.35.102401.090551. [DOI] [PubMed] [Google Scholar]
- 24.Negroni M, Buc H. Retroviral recombination: what drives the switch. Nat Rev Mol Cell Biol. 2001;2:151–155. doi: 10.1038/35052098. [DOI] [PubMed] [Google Scholar]
- 25.Chetverin AB. The puzzle of RNA recombination. FEBS Lett. 1999;460:1–5. doi: 10.1016/S0014-5793(99)01282-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu S, Li W, Cao ZJ. Does MMLV-RT lacking RNase H activity have the capability of switching templates during reverse transcription. FEBS Lett. 2002;520:185. [Google Scholar]
- 27.Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heim MH, Meyer UA. Evolution of a highly polymorphic human cytochrome P450 gene cluster: CYP2D6. Genomics. 1992;14:49–58. doi: 10.1016/s0888-7543(05)80282-4. [DOI] [PubMed] [Google Scholar]
- 29.Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjöqvist F, Ingelman-Sundberg M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994;46:452–459. [PubMed] [Google Scholar]
- 30.Chida M, Ariyoshi N, Yokoi T, Nemoto N, Inaba M, Kinoshita M, Kamataki T. New allelic arrangement CYP2D6*36 x 2 found in a Japanese poor metabolizer of debrisoquine. Pharmacogenetics. 2002;12:659–662. doi: 10.1097/00008571-200211000-00011. [DOI] [PubMed] [Google Scholar]


