Abstract
AIM: To characterize the protein files in blood from same patients with esophageal squamous cell carcinoma (ESCC) before and after operation at the high-incidence area for ESCC in Henan Province, China.
METHODS: Two-dimensional electrophoresis, silver staining and ImageMaster 2-DE analysis software were applied to the determination of protein files in the blood obtained from normal controls and ESCC patients before and after operation.
RESULTS: A total of 655, 662 and 677 protein spots were identified, respectively, from the normal controls and ESCC patients before and after operation. No significant difference in the number of protein spots was observed between the normal group and ESCC patients. A total of seven protein spots were identified with a dramatic difference among the samples before and after operation. Six protein spots were up-regulated and one protein spot was down-regulated in the group after operation compared with those in normal and before operation. Three protein spots were further characterized by matrix-assisted laser desorption/ionization time of flying mass spectrometry (MALDI-TOF-MS). The proteins from these three spots were identified as serum amyloid A (SAA), amyloid related serum protein and haptoglobin.
CONCLUSION: serum amyloid A, amyloid related serum protein and haptoglobin may be related with ESCC and/or surgery. The significance of these proteins needs to be further characterized. The present study provides informative data for the establishment of serum protein profiles related with ESCC.
INTRODUCTION
Esophageal squamous cell carcinoma (ESCC) is one of the six most common malignant diseases in the world with a remarkable geographical distribution. The ratio between the high- and low-incidence areas could be as high as 500:1. The prognosis of ESCC is very poor, the five-year survival rate is only about 10% for the patients at late or advanced stage. China is a country with the highest incidence and mortality rate of ESCC in the world. There are about 300000 new ESCC patients identified all over the world each year, half of them occur in China. Linzhou City (formerly Linxian County) and its neighbouring counties in Henan Province have been well-recognized as the highest incidence area in the world, the average incidence ratio of males and females is 161 and 103 per 100000, respectively[1]. ESCC remains the leading cause of cancer related-deaths in these areas. Unclear molecular mechanism, and lacking of sensitive and specific biomarkers for early diagnosis may be the reasons for the unchanged ESCC incidence pattern[2].
Many studies have been focused on gene level for early diagnosis of ESCC. However, because alterations in DNA and RNA may or may not induce similar protein changes, genetic changes could not reflect the stage and progression of the disease directly and objectively[3]. Proteomics based on two-dimensional electrophoresis and mass spectrometry is a new method for identification of cancer-specific protein markers[4]. In this study, we analyzed the serum protein changes in ESCC patients before and after operation and compared them with normal controls by proteomic methods to find the specific ESCC-related proteins.
MATERIALS AND METHODS
Blood samples
Blood samples were collected from ESCC patients in Yaocun Esophageal Cancer Hospital of Linzhou, Henan Province. Of the ESCC patients, there were 4 males and 2 females with an average age of 63 years (range, 52-74 years). All the blood samples were collected two times from each patient, just before and one week after the operation. The control blood samples (10 mL/subject) were collected in our laboratory from the normal people with matched ages as the patients. There were not any abnormalities identified by physical and biochemical examinations in volunteers of the control group.
Reagents
Electrophoresis reagents, including 400 g/L acrylamide solution, N, N-methylenebisacryl amide, N, N, N’, N’-tetramethylethylen -ediamine, urea, tris-base, glycine, glycerol, 3-[(3-cholannidopropyl) -dimethylammonio]-1-propanesulfonate (CHAPS), sodium dodecyl sulfate (SDS), dithiothreitol (DTT), ammonium persulfate, bromophenol blue, immobiline drystrips, immobilized pHgradient buffer and silver nitrate were from Amersham Pharmacia Biotechnology Inc. (Uppsala, Sweden). Iodoacetamide was from Acros (New Jersey, USA), sequence grade trypsin was from Washington Biochemical Corporation and trifluoroacetic acid (TFA) was from Fluka (Switzerland). All other reagents were of analytical grade.
Serum concentration detection
Serum samples were thawed and diluted by dH2O as 1:5 (2 μL serum was added to 8 μL dH2O), 1:10 (2 μL diluent was added to 18 μL dH2O), 1:200 (4 μL diluent was added to 796 μL dH2O) respectively, up to 1:10000 dilution and then 800 μL 1:10000 diluent was added to 200 μL protein assay reagent and absorbance was measured at 595 nm, finally the concentration of protein in serum was calculated.
First dimensional electrophoresis (Isoelectric focusing, IEF)
Precast IPG strips (pH3-10 linear, 18 cm, Amersham Pharmacia Biotechnology Inc.) was used in the first dimension. A total amount of 250 μg proteins was diluted to a total volume of 350 μL with the buffer (8 mol/L urea, 20 g/L CHAPS, 5 g/L IPG buffer 3-10, 20 mol/L DTT and a trace of bromophenol blue). After being loaded on IPG strips, IEF was carried out according to the following protocol: rehydration for 6 h at 0 V, 10 h at 30 V, 1 h at 500 V, 1 h at 1000 V and 7 h at 8000 V. The current was limited to 50 μA per gel.
Second dimensional electrophoresis (SDS-polyacrylamide gel electrophoresis, SDS-PAGE)
After IEF separation, the strips were immediately equilibrated for 2 × 15 min with equilibration solution (50 mmol/L Tris-HCl, pH6.8, 6 mol/L urea, 300 g/L glycerol and 20 g/L SDS). Then 20 mmol/L DTT was included in the first equilibration solution, and 20 g/L iodoacetamide was added in the second equilibration step to alkylate thiols. Thirteen percent SDS-PAGE gels were handled to become 1 mm thick. The strips were held in place with 5 g/L agarose dissolved in SDS/Tris running buffer and electrophoresis was carried out at constant power (2.5 W/gel for 40 min and 15 W/gel for 6 h) and temperature (20 °C) using Ettan Dalt II system (Amersham Pharmacia Biotechnology Inc.).
Silver staining
Gels were stained with silver nitrate according to the instructions of the silverstaining kit[5] (Amersham Pharmacia Biotechnology Inc.).
Gel scanning and image analysis
Protein profiles were obtained in normal controls and ESCC patients before and after operation through correcting the background to detect, match and quantify the spots by ImageMasterTM two-dimensional Elite analysis software (Amersham Biosciences).
In-gel protein digestion
Individual protein spots were excised from the gel by Ettan spot picker (Amersham Pharmacia Biotechnology Inc.), destained with the solution (15 mmol/L potassium ferricyanide, 50 mmol/L sodium thiosulfate) and washed till opaque and colorless with 25 mmol/L ammonium bicarbonate/500 g/L acetonitrile. After being dried with vacuum concentrator (SpeedVac Plus, USA), the gel was rehydrated with 3-10 μL of trypsin solution (10 ng/μL) at 4 °C for 30 min, followed by incubation at 37 °C overnight. Tryptic peptides were eluted and dried on SpeedVac vacuum concentrator.
Protein identification by MALDI mass spectrometry
The protein spots were analyzed by matrix-assisted laser desorption/ionization time of flying mass spectrometry (MALDI-TOF-MS) and SWISS-PROT database[6].
RESULTS
A total of 655, 662 and 677 protein spots were identified, respectively, from the normal control and ESCC patients before and after operation. No significant difference in number of protein spots was observed between normal group and ESCC patients. A total of seven protein spots were identified with a dramatic difference among the samples before and after operation. Six protein spots were up-regulated and one protein spot was down-regulated in the group after operation compared with those in normal and before operation. Of these six protein spots, three spots were further characterized by MALDI-TOF-MS. The proteins from these three spots were identified as serum amyloid A (SAA), amyloid related serum protein and haptoglobin (Figures 1, Figures 2).
Figure 1.
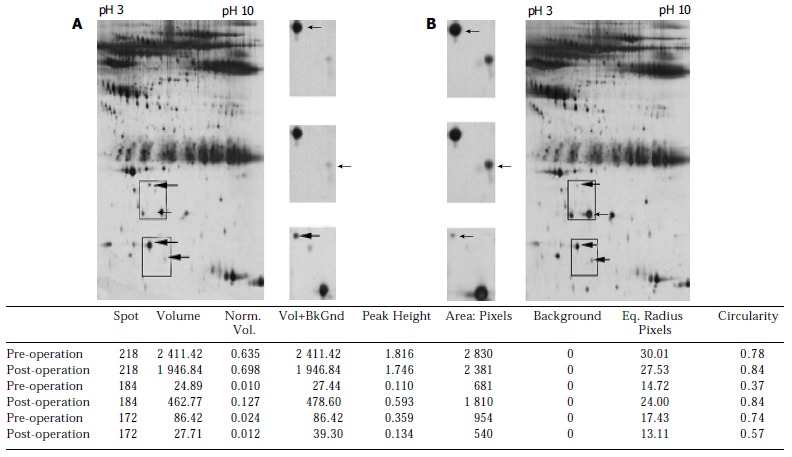
Representative of two-dimensional electrophoresis profiles and three-matched protein spots (arrows) analysis of the sera from esophageal squamous cell carcinoma patients before (A) and after (B) operation with different acidity from pH3 to pH10. Norm: normalization, Vol: volume, BkGnd: background, Eq: equality.
Figure 2.
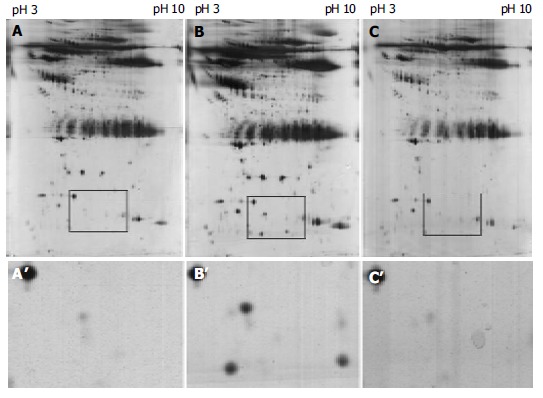
Representative of two-dimensional electrophoresis patterns of sera protein from the same esophageal squamous cell carcinoma patient before (A) and after (B) operation and the normal subject (C) with different acidity from pH3 to pH10. A’, B’ and C’ are the magnification of A, B and C, respectively, for the sera protein with marked difference.
DISCUSSION
In the present study, we found that serum amyloid A (SAA) and its isoform amyloid related serum protein, and haptoglobin were significantly increased after operation in ESCC patients compared with those in normal and pre-operation by mass spectrum. It indicated that these proteins might be related with ESCC and/or surgery. Although the literatures have reported that these acute-phase proteins are associated with tumors, such as colon cancer, the changes of these proteins may be more concerned with stress response, for example, the trauma by operation. The significance for these proteins needs to be further characterized. Although two-dimensional electrophoresis coupled with mass spectrometry is a powerful tool for screening and identification of cancer-specific protein markers, it costs time and money. The present study provided informative data for the establishment of serum protein profiles related with ESCC.
SAA is an acute-phase protein existing as various isoforms in a molecular mass range of 11-14. It is derived mainly from two genes, SAA1 and SAA2[7], mapped at chromosome 11p15.4-15.1[8]. In normal individuals, SAA is produced by hepatocytes in the liver. After its production, it is secreted into serum and rapidly binds to high-density lipoproteins, 90% of the protein particles are bound to high-density lipoprotein. A review of the literature showed that only a low level of SAA could be found in the sera of healthy individuals[9], but during the acute-phase response, its level in the blood could elevate 1000-fold after various injuries, including trauma, infection, inflammation and neoplasia[10,11].
Recently, many studies have shown that high SAA protein level is present in the serum of patients with disseminated cancer, reflecting the extent of malignant diseases, and inversely correlated with patient survival[12]. The mechanism is not clear. The mode of involvement of SAA in metastatic processes has not been elucidated. Several proposed functions for SAA proteins are compatible with the mechanism of tumor cell invasion and metastasis. These include inhibition of malignant cell attachment to extracellular matrix (ECM) proteins[13,14], induction of the expression of enzymes degrading the ECM[15] and induction of adhesion, migration, and tissue infiltration of cells[16-18]. Moreover, SAA contains functional arginine-glycine-aspartic acid (RGD) -like and tyrosine-isoleucine-glycine-serine-arginine (YIGSR) -like adhesion motifs[19], and the peptides containing these motifs could inhibit tumor cell invasion, metastasis, and angiogenesis[20,21]. Cumulatively, these findings, together with our observation of SAA alterations in blood after operation, imply that SAA may play a role in one or more steps of tumor progression or regression.
Glojnaric et al[22] indicated that although the presence of colorectal carcinoma caused an increase in serum levels of all the acute phase reactants studied, SAA protein showed the most powerful reaction in pre-operative disease stage, with the mean value of 330 mg/L as compared to the normal values of < 1.2 mg/L obtained in 30 healthy adults. SAA protein concentration increased to 487 mg/L after surgery and declined during the post-operative clinical course until the sixth chemotherapy cycle, but never returned to the normal range. In the later chemotherapy cycles, the mean SAA protein increased to 163 mg/L, probably as a result of the disease relapse. According to the statistical relations among exact confidence intervals for proportions, SAA protein showed the best specificity for colorectal carcinoma of all the acute phase proteins studied (83%-100%) and also a sensitivity of 100%. They concluded that SAA protein seems to be a reliable parameter, which could be recommended for clinical routine as a non-specific tumor marker for colorectal carcinoma.
Haptoglobin is an acute phase protein capable of binding haemoglobin and may play a role in modulating many aspects of the acute phase response[23]. The complex of haptoglobin with haemoglobin is metabolized in the heptic reticuloendothelial system[24]. Biosynthesis of haptoglobin occurs not only in the liver, but also in adipose tissue and in lung, providing antioxidant and antimicrobial activity[25]. Changes in the measured concentrations of haptoglobin in serum may help to assess the disease status of patients with inflammations, infections, malignancy, etc[26]. Dynamic detection of haptoglobin in leukemia patient plasmas showed that haptoglobin level increased obviously in leukemia patients than that in normal persons and dropped gradually with the improvement of diseases.
In conclusion, serum amyloid A, amyloid related serum protein and haptoglobin may be related with ESCC. The significance of these proteins needs to be further characterized. The present study provides informative data for the establishment of serum protein profiles related with ESCC.
Footnotes
Supported by National Science Fund for Outstanding Young Scholars of China, No.30025016; State Basic Research Development Program of China, No.G1998051206; Foundation of Henan Education Committee, No.1999125 and the US NIH Grant, No.CA65871
Edited by Kumar M and Wang XL Proofread by Xu FM
References
- 1.Wang LD, Zheng S. The mechanism of esophageal and gastric cardia carcinogenesis from the subjects at high-incidence area for esophageal cancer in henan. Zhengzhou Daxue Xuebao. 2002;37:717–729. [Google Scholar]
- 2.Wang LD, Zhou Q, Feng CW, Liu B, Qi YJ, Zhang YR, Gao SS, Fan ZM, Zhou Y, Yang CS, et al. Intervention and follow-up on human esophageal precancerous lesions in Henan, northern China, a high-incidence area for esophageal cancer. Gan To Kagaku Ryoho. 2002;29 Suppl 1:159–172. [PubMed] [Google Scholar]
- 3.Hanash SM, Bobek MP, Rickman DS, Williams T, Rouillard JM, Kuick R, Puravs E. Integrating cancer genomics and proteomics in the post-genome era. Proteomics. 2002;2:69–75. [PubMed] [Google Scholar]
- 4.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Simpson RJ. Proteins and Proteomics. A Laboratory Manual. 1st. New York Cold Spring Harbor Laboratory Press 2003: 75-76 [Google Scholar]
- 6.Bini L, Magi B, Marzocchi B, Arcuri F, Tripodi S, Cintorino M, Sanchez JC, Frutiger S, Hughes G, Pallini V, et al. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 1997;18:2832–2841. doi: 10.1002/elps.1150181519. [DOI] [PubMed] [Google Scholar]
- 7.Yamada T. Serum amyloid A (SAA): a concise review of biology, assay methods and clinical usefulness. Clin Chem Lab Med. 1999;37:381–388. doi: 10.1515/CCLM.1999.063. [DOI] [PubMed] [Google Scholar]
- 8.Watson G, See CG, Woo P. Use of somatic cell hybrids and fluorescence in situ hybridization to localize the functional serum amyloid A (SAA) genes to chromosome 11p15.4-p15.1 and the entire SAA superfamily to chromosome 11p15. Genomics. 1994;23:694–696. doi: 10.1006/geno.1994.1559. [DOI] [PubMed] [Google Scholar]
- 9.d'Eril GM, Anesi A, Maggiore M, Leoni V. Biological variation of serum amyloid A in healthy subjects. Clin Chem. 2001;47:1498–1499. [PubMed] [Google Scholar]
- 10.Cho WC, Yip TT, Yip C, Yip V, Thulasiraman V, Ngan RK, Yip TT, Lau WH, Au JS, Law SC, et al. Identification of serum amyloid a protein as a potentially useful biomarker to monitor relapse of nasopharyngeal cancer by serum proteomic profiling. Clin Cancer Res. 2004;10:43–52. doi: 10.1158/1078-0432.ccr-0413-3. [DOI] [PubMed] [Google Scholar]
- 11.Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol. 2000;7:64–69. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M, Tomita Y, Imai T, Saito T, Katagiri A, Ohara-Mikami Y, Matsudo T, Takahashi K. Significance of serum amyloid A on the prognosis in patients with renal cell carcinoma. Cancer. 2001;92:2072–2075. doi: 10.1002/1097-0142(20011015)92:8<2072::aid-cncr1547>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 13.Jensen LE, Whitehead AS. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem J. 1998;334(Pt 3):489–503. doi: 10.1042/bj3340489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershkoviz R, Preciado-Patt L, Lider O, Fridkin M, Dastych J, Metcalfe DD, Mekori YA. Extracellular matrix-anchored serum amyloid A preferentially induces mast cell adhesion. Am J Physiol. 1997;273:C179–C187. doi: 10.1152/ajpcell.1997.273.1.C179. [DOI] [PubMed] [Google Scholar]
- 15.Preciado-Patt L, Levartowsky D, Prass M, Hershkoviz R, Lider O, Fridkin M. Inhibition of cell adhesion to glycoproteins of the extracellular matrix by peptides corresponding to serum amyloid A. Toward understanding the physiological role of an enigmatic protein. Eur J Biochem. 1994;223:35–42. doi: 10.1111/j.1432-1033.1994.tb18963.x. [DOI] [PubMed] [Google Scholar]
- 16.Migita K, Kawabe Y, Tominaga M, Origuchi T, Aoyagi T, Eguchi K. Serum amyloid A protein induces production of matrix metalloproteinases by human synovial fibroblasts. Lab Invest. 1998;78:535–539. [PubMed] [Google Scholar]
- 17.Urieli-Shoval S, Shubinsky G, Linke RP, Fridkin M, Tabi I, Matzner Y. Adhesion of human platelets to serum amyloid A. Blood. 2002;99:1224–1229. doi: 10.1182/blood.v99.4.1224. [DOI] [PubMed] [Google Scholar]
- 18.Badolato R, Wang JM, Murphy WJ, Lloyd AR, Michiel DF, Bausserman LL, Kelvin DJ, Oppenheim JJ. Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J Exp Med. 1994;180:203–209. doi: 10.1084/jem.180.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto Y, Nomizu M, Yamada Y, Ito Y, Tanaka K, Sugioka Y. Inhibition of angiogenesis, tumour growth and experimental metastasis of human fibrosarcoma cells HT1080 by a multimeric form of the laminin sequence Tyr-Ile-Gly-Ser-Arg (YIGSR) Br J Cancer. 1996;73:589–595. doi: 10.1038/bjc.1996.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 21.Thorn CF, Lu ZY, Whitehead AS. Regulation of the human acute phase serum amyloid A genes by tumour necrosis factor-alpha, interleukin-6 and glucocorticoids in hepatic and epithelial cell lines. Scand J Immunol. 2004;59:152–158. doi: 10.1111/j.0300-9475.2004.01369.x. [DOI] [PubMed] [Google Scholar]
- 22.Glojnarić I, Casl MT, Simić D, Lukac J. Serum amyloid A protein (SAA) in colorectal carcinoma. Clin Chem Lab Med. 2001;39:129–133. doi: 10.1515/CCLM.2001.022. [DOI] [PubMed] [Google Scholar]
- 23.Wassell J. Haptoglobin: function and polymorphism. Clin Lab. 2000;46:547–552. [PubMed] [Google Scholar]
- 24.Kurash JK, Shen CN, Tosh D. Induction and regulation of acute phase proteins in transdifferentiated hepatocytes. Exp Cell Res. 2004;292:342–358. doi: 10.1016/j.yexcr.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Bernard D, Christophe A, Delanghe J, Langlois M, De Buyzere M, Comhaire F. The effect of supplementation with an antioxidant preparation on LDL-oxidation is determined by haptoglobin polymorphism. Redox Rep. 2003;8:41–46. doi: 10.1179/135100003125001233. [DOI] [PubMed] [Google Scholar]
- 26.Dobryszycka W. Biological functions of haptoglobin--new pieces to an old puzzle. Eur J Clin Chem Clin Biochem. 1997;35:647–654. [PubMed] [Google Scholar]


