Abstract
POD-1/TCF21 may play a crucial role in adrenal and gonadal homeostasis and represses Sf-1/SF-1 expression in adrenocortical tumor cells. SF-1 and LRH-1 are members of the Fzt-F1 subfamily of nuclear receptors. LRH-1 is involved in several biological processes, and both LRH-1 and its repressor SHP are involved in many types of cancer. In order to assess whether POD-1 can regulate LRH-1 via the same mechanism that regulates SF-1, we analyzed the endogenous mRNA levels of POD-1, SHP, and LRH-1 in hepatocarcinoma and adrenocortical tumor cells using qRT-PCR. Hereafter, these tumor cells were transiently transfected with pCMVMycPod-1, and the effect of POD-1 overexpression on E-box elements in the LRH-1 and SHP promoter region were analyzed by ChIP assay. Also, Cyclin E1 protein expression was analyzed to detect cell cycle progression. We found that POD-1 overexpression significantly decreased SHP/SHP mRNA and protein levels through POD-1 binding to the E-box sequence in the SHP promoter. Decreased SHP expression affected LRH-1 regulation and increased Cyclin E1. These findings show that POD-1/TCF21 regulates SF-1 and LRH-1 by distinct mechanisms, contributing to the understanding of POD-1 involvement and its mechanisms of action in adrenal and liver tumorigenesis, which could lead to the discovery of relevant biomarkers.
1. Introduction
POD-1 (Capsulin, Epicardin, and Tcf21) is a basic helix-loop-helix (bHLH) transcriptional regulatory protein expressed in mesenchyme cells at sites of epithelial-mesenchyme interactions in the developing urogenital, cardiovascular, respiratory, and gastrointestinal systems [1–4]. In the adrenal gland, POD-1 is expressed exclusively in the capsule region of the adrenal cortex, as shown in mice expressing a lacZ gene reporter under control of the regulatory region of POD-1 [5]. Recently, a progenitor population in the adrenal gland, characterized by the expression of WT1, GATA4, GLI1, and TCF21, was identified by cell lineage tracing analyses and used to generate steroidogenic cells in vivo [6]. In the testes of fetal mice, POD-1 repressed Steroidogenic Factor-1 (Sf-1/Nr5a1), a transcription factor required for gonadal development and sex determination, and regulated adrenal and gonadal steroidogenesis in adult mice [7, 8]. In human adrenocortical tumor cells, we found that POD-1 binds to the SF-1 E-box promoter sequence and inhibits SF-1 expression, as well as the expression of StAR protein, which is controlled by SF-1 [9].
The orphan nuclear receptor Liver Receptor Homologue-1 (LRH-1/NR5A2) also belongs to the Fzt-F1 subfamily of nuclear receptors [10]. LRH-1 is involved in development, steroidogenesis, and proliferation of liver, pancreas, and intestine cells [11, 12]. LRH-1 is critical for liver development and regulates cholesterol and bile acid homeostasis in the adult liver [11, 12]. The human adrenal gland also expresses LRH-1, although LRH-1 expression has been described to be species-specific [13, 14]. LRH-1 is active constitutively and its transcriptional activity is repressed by orphan receptors like the Small Heterodimer Partner (SHP/NR0B2) and the dosage-sensitive sex reversal-adrenal hypoplasia congenital gene on the X chromosome (DAX-1) [15, 16]. Like DAX-1, an important nuclear receptor involved in adrenal development, SHP is an unusual orphan nuclear receptor because it lacks a DNA-binding domain [17]. SHP interacts with the AF-2 transactivation domain of LRH-1 and competes with the binding of coactivators [18]. In primary hepatocarcinoma cells and nude mice, SHP/Shp inhibited tumor growth and induced apoptosis [19]. SHP also inhibits estrogen action at multiple levels, and its induction of expression or activity in the breasts is relatively specific for inhibiting estrogen signaling in breast cancer [20]. LRH-1 is also involved in tumorigenesis in pancreatic and intestinal cancer [21, 22]. Its suppression arrested the cell cycle, mediated by the downregulation of Cyclin E1, in human hepatocellular carcinoma cells [23]. Despite our previous work demonstrating that POD-1 overexpression binds and inhibits SF-1 expression, its mechanism of action in the other members of the Fzt-F1 nuclear receptor family, like LRH-1, is unknown. To fill this gap and explore the effect of POD-1 in cell cycle regulation in tumor cells, we investigated whether POD-1/TCF21 regulates LRH-1 and SHP in adrenocortical and hepatocarcinoma cell lines. In this study, we show that POD-1/TCF21 reduces SHP expression in hepatocarcinoma cells, resulting in an increased LRH-1 and Cyclin E1 expression via binding in the E-box element of SHP, through a different mechanism of action and effect other than that described for SF-1.
2. Materials and Methods
2.1. Cell Cultures
The NCI-H295R human adrenocortical tumor cell line [24] was cultured in RPMI (Gibco, USA), supplemented with 2% fetal bovine serum (Gibco, USA) and 1% ITS (Gibco, USA). The HepG2 human hepatocarcinoma tumor cell line [25] and the ACC-T36 human adrenocortical tumor cell culture [9] were cultured in DMEM (Gibco, USA) and supplemented with 10% fetal bovine serum (Gibco, USA). The Y-1 mouse adrenocortical tumor cell line [26] was grown in DMEM supplemented with 7.5% horse serum (Gibco, USA) and 2.5% fetal bovine serum (Gibco, USA). All cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2.
2.2. Cell Culture Transfection
The H295R and HepG2 cell lines were transiently transfected with pCMVMycPod-1, which was kindly provided by Dr. Masataka Nakamura (Tokyo Medical University, Japan), as described in Funato and coworkers [27]. To extract RNA, we plated 1.5 × 105 cells into 6-well tissue culture plates (Becton Dickinson Labware, Franklin Lakes, NJ, USA) and transfected them with 2 μg of plasmid DNA combined with 2 μL of X-tremeGENE HP-DNA transfection reagent (Roche Diagnostics GmbH, Mannheim, Germany). For the protein assay, we plated 5 × 105 cells into a 60 mm tissue culture dish (Becton Dickinson Labware, Franklin Lakes, NJ, USA) and transfected them with 5 μg of plasmid DNA and 5 μL of X-tremeGENE HP-DNA transfection reagent per 1 × 105 cells (Roche Diagnostics GmbH, Mannheim, Germany). For the ChIP assay, we plated 6.6 × 105 cells into a 100 mm tissue culture dish and transfected them with 10 μg of plasmid DNA and 10 μL of X-tremeGENE HP-DNA transfection reagent (Roche Diagnostics GmbH, Mannheim, Germany).
2.3. Total RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
After 48 h of transfection, we extracted total RNA using Trizol reagent (Invitrogen, USA) and treated it with TURBO DNA-free (Ambion). The cDNA was generated from 1 μg of treated total RNA using Oligo dT, RNaseOUT, and M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The primers used for qRT-PCR were hPOD-1 forward 5′-ACCCTCTTCCTCGCTTTCTC-3′ and reverse 5′-AACCCGTCACATTCCAACAT-3′; hLRH-1 forward 5′-TGCCTTGCCTCCTACAGACT-3′ and reverse 5′-AGGCTCATCTGGCTCACACT-3′; mLrh-1 forward 5′-ACCTGTGAGCCCTGAAGCTA-3′ and reverse 5′-AGAGGGTTACTGCCCGTTTT-3′; hSHP-1 forward 5′-CACTGGGTGCTGTGTGAAGT-3′ and reverse 5′-CCAATGATAGGGCGAAAGAA-3′. RT-qPCR was performed on a RotorGene6000 Corbett (Qiagen, USA) sequence detector using Platinum SYBR qPCR SuperMix-UDG (Invitrogen, USA). A cycle threshold (Ct) value in log range of amplification was selected for each sample in triplicate and was normalized to β-actin expression levels. Reactions were carried out in triplicate. A pool of commercially available RNA isolated from normal adrenal gland of 62 male/female Caucasians, aged 15–61, was used as a normal control (Clontech, Palo Alto, CA). Data were analyzed using the 2−ΔΔCt method [28].
2.4. Immunoblotting
The cells were lysed 72 h after transfection in RIPA buffer, as well as in protease and phosphatase inhibitors (Sigma Aldrich Gmbh, Steinheim, Germany). Bradford assay determined the total protein concentration. Total protein from the lysates (30 μg) was resolved by 12% SDS-PAGE and, after electrophoresis, gels were blotted onto nitrocellulose membranes. Nonspecific binding sites were blocked for 2 h with 0.1% bovine serum albumin or 5% nonfat dried milk in TBST (TRIS-buffered saline solution containing 1% Tween 20). All washes and antibody incubations were performed using TBST. The following primary antibodies were used: anti-Cyclin E1 (Sta Cruz sc-481) 1 : 500 in blocking buffer (0.1% bovine serum albumin in TBST), anti-SHP (Perseus Proteomics-PP-N7519-00) 1 : 1000 in blocking buffer (5% nonfat dried milk in TBST), and anti-actinin 1 : 1000 in TRIS-buffered saline containing 1% Tween 20. Proteins were visualized by ECL detection with secondary HRP-conjugated anti-rabbit (Amersham Hybond ECL, Freiburg, Germany) or anti-mouse antibodies (Jackson Immuno Research, Pennsylvania, USA). Immunoblot results were quantified by densitometer using the GeneSnap and GeneTools software (SynGene-Synoptic Ltd., Cambridge, UK). Ponceau staining of membranes was used to monitor protein transfer and loading.
2.5. Chromatin Immunoprecipitation (ChIP)
HepG2 cells transfected with pCMVMycPod-1 were fixed with 1% formaldehyde for 10 min. ChIP assays were performed using the ChIP-IT Express kit (Active Motif, Rixensart, Belgium) following the manufacturer's instructions. Chromatin was fragmented by sonication with eight 10-s pulses at 25 μm amplitude in a VCX130PB ultrasonic processor (Sonics & Materials, CT, USA). Most resulting chromatin fragments ranged from 200 bp to 600 bp. Sheared chromatin was incubated with 3 μg anti-MYC (Clontech) or with 1 μg IgG as negative control (ChIP-IT Control Kit-Human, Active Motif).
2.6. ChIP-PCR
The 5.0-kb upstream sequences of human Liver Receptor Homolog-1 (NR5A2, transcript ENST00000367362, Ensembl release 64, GRCh37), human Small Heterodimer Partner (NR0B2-001 ENST00000254227, Ensembl release 64, GRCh37), and androgen receptor (AR, transcript ENST00000374690, Ensembl release 64, GRCh37) were analyzed for putative E-box binding sites using MatInspector [29]. Based on this analysis and on the location of predicted E-box sites in humans [9], the following primers were designed using Primer3 [30]: AR E-box, forward 5′-CTCTGATTCTTGGGGCTGAG-3′ and reverse 5′CATGACCAAGCCAGCAGATA 3′ (113 bp amplicon); LRH-1 E-box-53, forward 5′-TCATTTCTTTGCCATTATCTGG-3′ and reverse 5′-TGGAAACTTTTGATAGGCTTTGA-3′ (120 bp amplicon); LRH-1 E-box-1300, forward 5′-CCCATACACACAACCTGCAT-3′ and reverse 5′-TGCTGGAATTATAGGCGTGA-3′ (100 bp amplicon); SHP E-box-117, forward 5′-ACCGGCCACTTCATTGACT-3′ and reverse 5′-CCAACAACCTTGACTCCAGAA-3′ (146 bp amplicon); SHP E-box-3702, forward 5′- CAGGTATGCACCACCATGTC-3′ and reverse 5′-ATCTCAGCACTTTGGGAAGG-3′, (131 bp amplicon).
As a negative control for POD-1 binding, primers amplifying sequences in intron 1-2 of AR (forward 5′-TTGTCAAAGTCTTTTCCAGTTAATTT-3′ and reverse 5′-TTAACCCTACCAAGTAAATTTGTTC-3′, 114 bp amplicon), in intron 2-3 of LRH-1 (forward 5′-CCCACTGGAAGGTGATCCTA-3′ and reverse 5′-CCCCTTTGTCTTTCCCCTTA-3′, 97 bp amplicon), and in intron 1-2 of SHP (forward 5′-GGGAGGACAGGAAAGGAGTC-3′ and reverse 5′-CCTGGGGAACTCTCATCTCA-3′, 104 bp amplicon) were used. Anti-MYC-IP, IgG-IP, and 0.1% input DNA samples were used as templates for PCR amplification. PCR reactions were performed using 5 U/μL Platinum Taq DNA Polymerase (Invitrogen) and 5 μM primers for 35 cycles of amplification with an annealing temperature of 60°C.
2.7. Statistical Analyses
Data are presented as mean ± standard deviation (SD) values of three replicate experiments. Data were analyzed using the Kruskal-Wallis test (nonparametric one-way ANOVA) or with paired t-tests, when indicated. Results were considered statistically significant when p < 0.05.
3. Results
3.1. Distinct Endogenous POD-1, SHP, and LRH-1 mRNA Levels Found in Tumor Cells
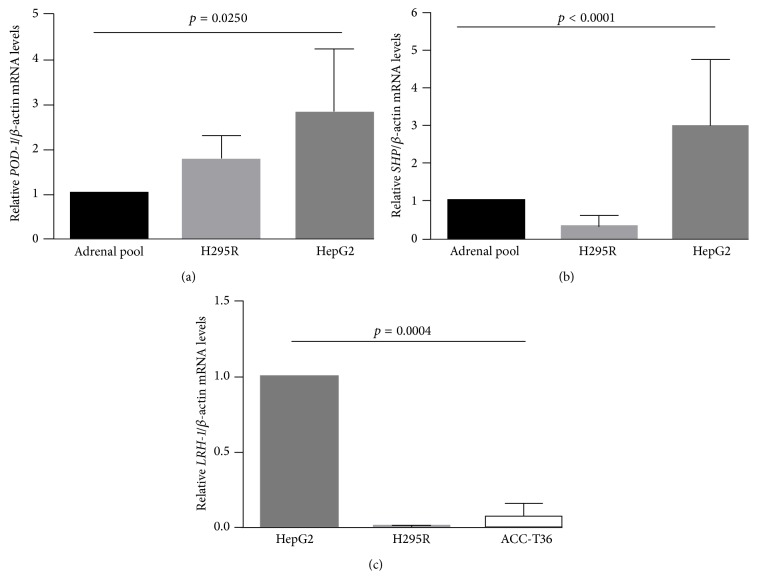
We used qRT-PCR to estimate the endogenous mRNA levels of POD-1, SHP, and LRH-1 in HepG2 and H295R tumor cell lines and in a human tumor adrenocortical cell culture, ACC-T36 cells, using qRT-PCR (Figure 1). POD-1 was significantly higher in H295R and HepG2 cells (by 1.73 ± 0.27- and 2.8 ± 0.79-fold, resp.) than in the normal adrenal pool (p = 0.025; Figure 1(a)). Endogenous SHP in both cell lines differed significantly (p < 0.0001) from the normal adrenal pool, and SHP was higher in HepG2 than H295R, respectively, by 2.99 ± 1.8- and 0.3 ± 0.2-fold (Figure 1(b)). In contrast, LRH-1 was barely detectable in H295R cells (0.006 ± 0.002-fold, p = 0.0004) and ACC-T36 cells (0.07 ± 0.03-fold, p = 0.0004) when compared to HepG2 cells (Figure 1(c)).
Figure 1.
Quantitative reverse transcription PCR (qRT-PCR) analysis of the relative POD-1/β-actin (a), SHP/β-actin (b), and LRH-1/β-actin mRNA levels (c) in the human adrenocortical tumor cell line (H295R and ACC-T36 cells) and in the human hepatocarcinoma tumor cell line (HepG2 cells). Differences were tested with a Kruskal-Wallis one-way ANOVA. Values represent means ± standard deviations from 3 experiments.
3.2. POD-1 Overexpression Reduces SHP Expression in HepG2 and H295R Cells
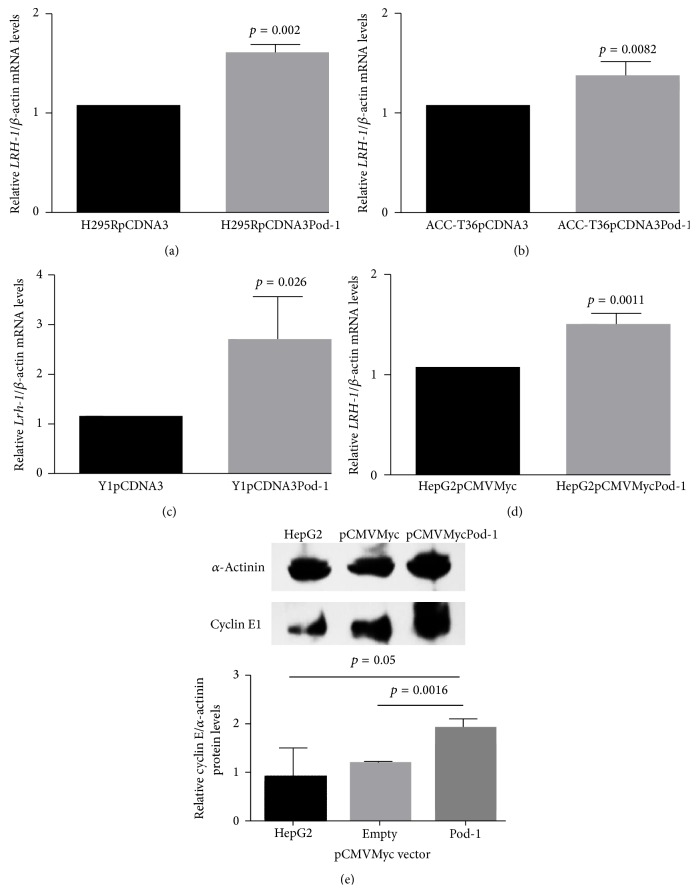
The transient transfection of pCMVMycPod-1 in HepG2 cells increased POD-1 mRNA levels (143.469 ± 16072-fold, p = 0.0009) compared to controls transfected with the empty vector (pCMVMyc) (Figure 2(a)). The POD-1 mRNA in H295R and ACC-T36 cells transfected with pCDNA3Pod or pCMVMycPod-1 showed an increase, as previously shown in França et al. [9]. Then, we determined the effect of overexpression of POD-1 on SHP/SHP mRNA and protein levels in H295R and HepG2 cells transiently transfected with the expression vector pCMVMycPod-1 (Figure 2(d)). SHP mRNA levels of H295R and HepG2 cell lines were reduced by 3.7 ± 0.08-fold (p = 0.0013; Figure 2(b)) and 2.3 ± 0.04 (p = 0.0002; Figure 2(c)), respectively, compared to cells transfected with the empty vector. SHP protein levels, showing two putative variants with approximately 28 kDa, were also significantly lower in HepG2 cells (0.29 ± 0.04 fold) than in controls (p = 0.017; Figure 2(d)).
Figure 2.
Quantitative reverse transcription RT-PCR (qRT-PCR) and immunoblotting analysis of relative SHP/SHP expression. Relative POD-1/β-Actin mRNA levels in HepG2 cells transiently transfected with the empty vector pCMVMyc or with pCMVMycPod-1 (a); SHP/β-Actin mRNA levels in H295R cells transiently transfected with the empty vector pCMVMyc or with pCMVMycPod-1 (b); SHP/β-Actin mRNA levels and SHP/α-actinin protein levels in HepG2 cells (c and d, resp.). Total RNA samples and protein samples were prepared 48 h and 72 h after transfection, respectively. Differences were tested with paired samples t-tests. Values represent means ± standard deviations from 3 experiments.
3.3. Validation of Chromatin Enrichment and Characterization of SHP-E-box Binding of POD-1
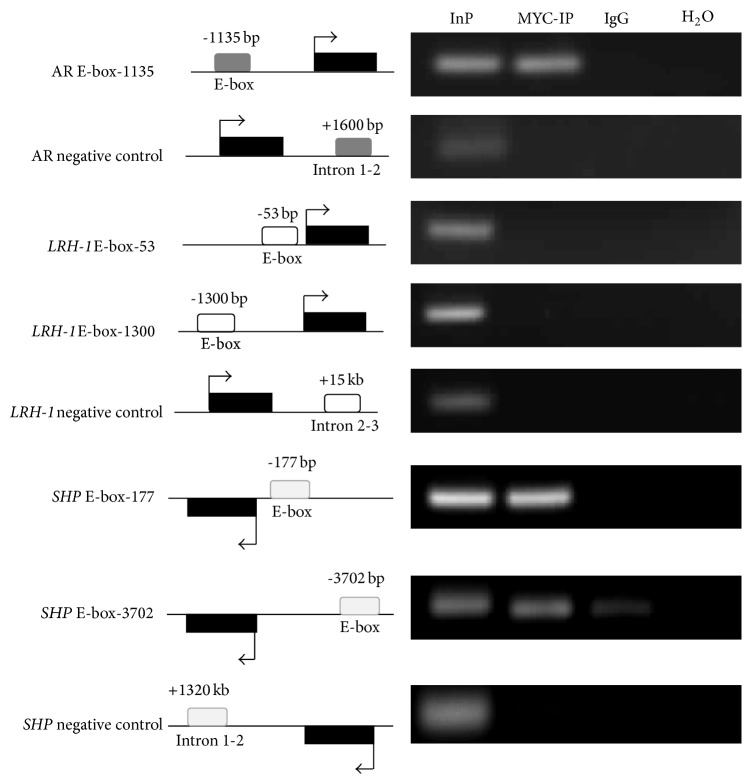
To determine whether the repressive effect of POD-1 overexpression on SHP expression was mediated through POD-1 binding to E-box elements in the SHP promoter region, chromatin immunoprecipitation (ChIP) assays were performed in HepG2 cells transfected with pCMVMycPod-1 using an anti-Myc antibody (Figure 3). To validate this approach, ChIP-PCR assays were performed using primers amplifying an E-box sequence in the promoter region of AR (E-box sequences are compared and shown in Table 1); it has been previously shown in mice and humans that POD-1 binds to an E-box element in the promoter of Ar/AR to repress transcription [9, 31]. As shown in Figure 3, immunoprecipitated chromatin was enriched for this region, whereas intron 1-2 of AR, used as a local negative control and not expected to bear E-box elements, failed to amplify. The 5.0-kb upstream sequence of human SHP and LRH-1 was analyzed for putative E-box elements using MatInspector [29], and two SHP elements were identified, located 177 and 3702 base pairs upstream of the transcription start site, respectively. Two LRH-1 elements were also identified, located 53 and 1300 bases pairs upstream of the transcription start site. ChIP-PCR assays using specifically designed primers showed that Myc immunoprecipitated DNA from HepG2-pCMVMycPod-1 cells amplified both “E-box-177” and “E-box-3702” sequences. This confirms that chromatin was enriched by ChIP, while the negative control sequence located in the intron 1-2 (1320 Kb) of SHP failed to amplify (Figure 3). Nevertheless, as shown in Figure 3, MYC-IP DNA from HepG2 transfected cells did not amplify “E-box-53” and “E-box-1300” LRH-1 sequences, as well as the ChIP negative control sequence (intron 2-3). Altogether, these results show POD-1 binding to E-box elements in the human SHP promoter site.
Figure 3.
Chromatin enrichment was confirmed by PCR amplification of the E-box region of the SHP-1 promoter from anti-MYC immunoprecipitated HepG2pCMVMycPod-1 DNA. The positions of the amplicons in relation to the Transcriptional Start Sites (TSSs) (represented by arrows) are shown. Black and open bars represent the exon and a different E-box sequence, respectively. Androgen receptor (AR), input (Inp) 0.1% DNA, Anti-MYC-IP HepG2pCMVMycPod-1 (IP), and anti-IgG (IgG).
Table 1.
Nucleotide sequence and E-box elements identified in silico using MatInspector.
| Identified E-box | Species | Sequence |
|---|---|---|
| AR E-box -1135 | Human | 5′-atgccaCGAGgcc-3′ |
| Mouse | 5′-gt gtcaggaattc-3′ | |
|
| ||
| LRH-1 E-box -53 | Human | 3′-tcatcaCATGact-5′ |
| Mouse | 3′-tggtcacatgacc-5′ | |
|
| ||
| LRH-1 E-box -1300 | Human | 5′-tggccaGGTGcgg-3′ |
| Mouse | 5′-ttgccttcagtgc-3′ | |
|
| ||
| SHP E-box -177 | Human | 5′-gtgccaCGTGggg-3′ |
| Mouse | 5′-aggccacgtggagc-3′ | |
|
| ||
| SHP E-box -3702 | Human | 3′-ggaTCACttgagg-5′ |
| Mouse | 3′-agatctctcagct-3′ | |
E-box elements were named based on distance to transcriptional start, as shown in Figure 3. Core human sequence elements, as identified by MatInspector (see Section 2), are shown in capitals.
Nucleotides conserved, in human and mouse, are shown underlined in the mouse sequence; sequence alignment was done using Clustalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), under default settings.
3.4. The Downregulation of SHP Induced by POD-1 Increased LRH-1 mRNA and Cyclin E1 Protein Levels in HepG2 Cells
To evaluate the effect of POD-1-mediated inhibition of SHP/SHP in adrenocortical carcinoma and hepatocarcinoma cells transfected with POD-1, LRH-1/Lrh-1 mRNA and Cyclin E1 protein levels were analyzed by qRT-PCR and immunoblotting (Figure 4). LRH-1 mRNA levels were significantly increased in human and mice adrenocortical tumor cells relative to controls (H295R cell line: 1.61 ± 0.04-fold, p = 0.002, Figure 4(a); ACC-T36 cells: 1.38 ± 0.07-fold, p = 0.0082, Figure 4(b); and Y1 cell line: 2.71 ± 0.4-fold, p = 0.026, Figure 4(c)). LRH-1 mRNA levels were also increased in HepG2 cells transfected with pCMVMycPod-1, compared to transfected controls (1.51 ± 0.06-fold, p = 0.0011, Figure 4(d)). POD-1 overexpression also significantly increased Cyclin E1 protein levels in the HepG2 cell line relative to controls transfected with pCMVMyc (1.94 ± 0.09 fold, p = 0.0016). Taken together, these results could be associated with a decrease of SHP-mediated protein expression.
Figure 4.
Quantitative reverse transcription RT-PCR (qRT-PCR) of Lrh-1/LRH-1 mRNA levels and immunoblotting of Cyclin E1 protein levels. LRH-1/β-actin mRNA levels in H295R cells (a), ACC-T36 cells (b), Y1 cells (c), and HepG2 cells (d) transiently transfected with the empty vector (pCDNA3 or pCMVMyc) or with pCDNA3Pod-1 or pCMVMycPod-1; immunoblotting analysis of the relative Cyclin E1/α-Actinin protein levels in HepG2 cells (e) transiently transfected with the empty vector pCMVMyc or with pCMVMycPod-1. Total RNA samples and protein samples were prepared 48 h and 72 h after transfection, respectively. Differences were tested with paired samples t-tests. Values represent means ± standard deviations of 3 experiments.
4. Discussion
Here we described that SHP is regulated by POD-1/TCF21 through binding E-box sequences to the SHP promoter in adrenocortical and hepatocarcinoma tumor cells. SHP repression affected LRH-1 regulation and cell cycle balance, which was associated with increased Cyclin E1 levels.
Previous studies show that POD-1 may play a crucial role in adrenal and gonadal homeostasis, and it has also been described to play a role in different organs [1, 3, 8]. Furthermore, TCF21 is downregulated in adrenocortical carcinoma (ACC), melanoma, lung, and head and neck squamous cell carcinomas [9, 32–34]. Consistent with these findings, our results showed that POD-1, SHP, and LRH-1 are expressed in adrenocortical and hepatocarcinoma tumor cells.
POD-1 has been shown to repress Sf-1/SF-1 expression in mouse and human adrenocortical cells [8, 9], in contrast with what was observed here with LRH-1, which is upregulated by inhibition of SHP. SF-1 and LRH-1 have structural similarities; however, they also have important functional differences [10]. LRH-1 and SHP are involved in many types of cancer such as liver, pancreatic, gut, and breast [17]. SHP appears to suppress tumorigenesis in liver cancer, inhibiting tumor growth and increasing sensitivity to apoptotic stimuli, due to repression of LRH-1-dependent Cyclin D1 transcription [35, 36]. In this study, we showed that POD-1 is able to modulate SHP expression in human adrenocortical and hepatocarcinoma cell lines, which provides new insights into the role of POD-1 in both types of cancer. Using a ChIP assay, we showed that POD-1 binds directly to the E-box SHP promoter, inhibiting its activity, but does not bind to the LRH-1 promoter E-box sequence.
In intestinal crypt cells, LRH-1 controls cell proliferation by inducing the expression of G1 Cyclins (D1 and E1) and is correlated with the rate of intestinal cell renewal. The increase of proliferation in these cells occurs after induction of Cyclins D1 and E1, enhanced by the interaction with β-catenin [21]. In vivo and in vitro approaches have shown that LRH-1 overexpression promoted pancreatic cancer cell growth, proliferation, and angiogenesis by regulating Cyclins E1 and D1 [37]. Furthermore, LRH-1 has been shown to contribute to colon tumorigenesis in vivo through its effects on the cell cycle and inflammation [38]. LRH-1 is also involved in the progression and development of pancreatic adenocarcinoma cells [22]. Consistent with these findings, we found that SHP downregulation increased LRH-1 expression, which binds to the Cyclin E1 promoter that may lead to the G1 to S phase transition of the cell cycle. These results are in contrast with our finding that POD-1 inhibits SF-1 in tumor adrenocortical cells [9], suggesting that POD-1 has different effects on the control of the cell cycle in tumor cells expressing LRH-1.
5. Conclusion
We showed that POD-1 overexpression promotes LRH-1 increase through downregulation of SHP, which may promote cell cycle progression via Cyclin E. POD-1/TCF21 has a potential role as a regulator of SF-1 and LRH-1 transcription factors via distinct mechanisms promoting different effects. These findings contribute to the understanding of how the cellular tumorigenic process is controlled in adrenocortical and liver tumors and suggests new therapeutic targets.
Acknowledgment
MMF received a scholarship (2010/0071-9) from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Foundation for the Support of Research of the State of São Paulo); CFPL received funding from FAPESP (2011/07656-3 and 2012/21839-6), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, the National Council for Scientific and Technological Development), and Pró-Reitoria de Pesquisa da Universidade de São Paulo (University of São Paulo Dean's Office for Research Projects).
Conflict of Interests
The authors declare that there is no conflict of interests that could be detrimental to the impartiality of the research reported.
References
- 1.Lu J., Richardson J. A., Olson E. N. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mechanisms of Development. 1998;73(1):23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 2.Quaggin S. E., Vanden Heuvel G. B., Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mechanisms of Development. 1998;71(1-2):37–48. doi: 10.1016/S0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- 3.Robb L., Mifsud L., Hartley L., et al. Epicardin: a novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Developmental Dynamics. 1998;213(1):105–113. doi: 10.1002/(sici)1097-0177(199809)213:1lt;105::aid-aja10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Quaggin S. E. Transcriptional regulation of podocyte specification and differentiation. Microscopy Research and Technique. 2002;57(4):208–211. doi: 10.1002/jemt.10076. [DOI] [PubMed] [Google Scholar]
- 5.Kim A. C., Barlaskar F. M., Heaton J. H., et al. In search of adrenocortical stem and progenitor cells. Endocrine Reviews. 2009;30(3):241–243. doi: 10.1210/er.2008-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandiera R., Vidal V. P. I., Motamedi F. J., et al. WT1 maintains adrenal-gonadal primordium identity and marks a population of AGP-like progenitors within the adrenal gland. Developmental Cell. 2013;27(1):5–18. doi: 10.1016/j.devcel.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura M., Kanno Y., Chuma S., Saito T., Nakatsuji N. Pod-1/Capsulin shows a sex- and stage-dependent expression pattern in the mouse gonad development and represses expression of Ad4BP/SF-1 . Mechanisms of Development. 2001;102(1-2):135–144. doi: 10.1016/s0925-4773(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 8.Cui S., Ross A., Stallings N., Parker K. L., Capel B., Quaggin S. E. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development. 2004;131(16):4095–4105. doi: 10.1242/dev.01266. [DOI] [PubMed] [Google Scholar]
- 9.França M. M., Ferraz-de-Souza B., Santos M. G., et al. POD-1 binding to the E-box sequence inhibits SF-1 and StAR expression in human adrenocortical tumor cells. Molecular and Cellular Endocrinology. 2013;371(1-2):140–147. doi: 10.1016/j.mce.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fayard E., Auwerx J., Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends in Cell Biology. 2004;14(5):250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Galarneau L., Drouin R., Bélanger L. Assignment of the fetoprotein transcription factor gene (FTF) to human chromosome band 1q32.11 by in situ hybridization. Cytogenetics and Cell Genetics. 1998;82(3-4):269–270. doi: 10.1159/000015116. [DOI] [PubMed] [Google Scholar]
- 12.Li M., Xie Y.-H., Kong Y.-Y., Wu X., Zhu L., Wang Y. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. Journal of Biological Chemistry. 1998;273(44):29022–29031. doi: 10.1074/jbc.273.44.29022. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z.-N., Bassett M., Rainey W. E. Liver receptor homologue-1 is expressed in the adrenal and can regulate transcription of 11 beta-hydroxylase. Journal of Molecular Endocrinology. 2001;27(2):255–258. doi: 10.1677/jme.0.0270255. [DOI] [PubMed] [Google Scholar]
- 14.Schoonjans K., Annicotte J.-S., Huby T., et al. Liver receptor homolog-1 controls the expression of the scavenger receptor class B type I. The EMBO Reports. 2002;3(12):1181–1187. doi: 10.1093/embo-reports/kvf238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin B., Jones S. A., Price R. R., et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Molecular Cell. 2000;6(3):517–526. doi: 10.1016/S1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 16.Sablin E. P., Woods A., Krylova I. N., Hwang P., Ingraham H. A., Fletterick R. J. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlund A., Treuter E. Ligand-independent actions of the orphan receptors/corepressors DAX-1 and SHP in metabolism, reproduction and disease. Journal of Steroid Biochemistry and Molecular Biology. 2012;130(3–5):169–179. doi: 10.1016/j.jsbmb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y.-K., Moore D. D. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. The Journal of Biological Chemistry. 2002;277(4):2463–2467. doi: 10.1074/jbc.m105161200. [DOI] [PubMed] [Google Scholar]
- 19.He N., Park K., Zhang Y., Huang J., Lu S., Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134(3):793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Kovacic A., Speed C. J., Simpson E. R., Clyne C. D. Inhibition of aromatase transcription via promoter II by short heterodimer partner in human preadipocytes. Molecular Endocrinology. 2004;18(1):252–259. doi: 10.1210/me.2003-0211. [DOI] [PubMed] [Google Scholar]
- 21.Botrugno O. A., Fayard E., Annicotte J.-S., et al. Synergy between LRH-1 and β-catenin Induces G1 cyclin-mediated cell proliferation. Molecular Cell. 2004;15(4):499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Benod C., Vinogradova M. V., Jouravel N., Kim G. E., Fletterick R. J., Sablin E. P. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):16927–16931. doi: 10.1073/pnas.1112047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S., Lan F., Huang L., et al. Suppression of hLRH-1 mediated by a DNA vector-based RNA interference results in cell cycle arrest and induction of apoptosis in hepatocellular carcinoma cell BEL-7402. Biochemical and Biophysical Research Communications. 2005;333(3):917–924. doi: 10.1016/j.bbrc.2005.05.186. [DOI] [PubMed] [Google Scholar]
- 24.Gazdar A. F., Oie H. K., Shackleton C. H., et al. Establishment and characterization of a human adrenocortical carcinoma cell line that expresses multiple pathways of steroid biosynthesis. Cancer Research. 1990;50(17):5488–5496. [PubMed] [Google Scholar]
- 25.Knowles B. B., Howe C. C., Aden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209(4455):497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 26.Yasumura Y. V., Buonassisi V., Sato G. Clonal analysis of differentiated function in animal cell cultures. I. Possible correlated maintenance of differentiated function and the diploid karyotype. Cancer Research. 1966;26(3):529–535. [PubMed] [Google Scholar]
- 27.Funato N., Ohyama K., Kuroda T., Nakamura M. Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription. The Journal of Biological Chemistry. 2003;278(9):7486–7493. doi: 10.1074/jbc.m212248200. [DOI] [PubMed] [Google Scholar]
- 28.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Cartharius K., Frech K., Grote K., et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 30.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. A. M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research. 2007;35(2):W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong C. Y., Gong E., Kim K., et al. Modulation of the expression and transactivation of androgen receptor by the basic helix-loop-helix transcription factor pod-1 through recruitment of histone deacetylase 1. Molecular Endocrinology. 2005;19(9):2245–2257. doi: 10.1210/me.2004-0400. [DOI] [PubMed] [Google Scholar]
- 32.Arab K., Smith L. T., Gast A., et al. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32(10):1467–1473. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giordano T. J., Kuick R., Else T., et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clinical Cancer Research. 2009;15(2):668–676. doi: 10.1158/1078-0432.ccr-08-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith L. T., Lin M., Brena R. M., et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):982–987. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Xu P., Park K., Choi Y., Moore D. D., Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48(1):289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Soto J., Park K., et al. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Molecular and Cellular Biology. 2010;30(6):1341–1356. doi: 10.1128/mcb.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Q., Aihara A., Chung W., et al. LRH1 as a driving factor in pancreatic cancer growth. Cancer Letters. 2014;345(1):85–90. doi: 10.1016/j.canlet.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoonjans K., Dubuquoy L., Mebis J., et al. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(6):2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]