Abstract
This study analyzes the occurrence and distribution of phylogenetic groups of 391 strains of Escherichia coli isolated from poultry, cattle, and water buffalo. The frequency of the phylogroups was A = 19%, B1 = 57%, B2 = 2.3%, C = 4.6%, D = 2.8%, E = 11%, and F = 3.3%. Phylogroups A (P < 0.001) and F (P = 0.018) were associated with E. coli strains isolated from poultry, phylogroups B1 (P < 0.001) and E (P = 0.002) were associated with E. coli isolated from cattle, and phylogroups B2 (P = 0.003) and D (P = 0.017) were associated with E. coli isolated from water buffalo. This report demonstrated that some phylogroups are associated with the host analyzed and the results provide knowledge of the phylogenetic composition of E. coli from domestic animals.
1. Introduction
Escherichia coli is a gram-negative, fermentative, rod-shaped bacterium that is the major facultative anaerobic bacterium in the intestinal tract of most animal species. E. coli cause enteric and extraintestinal diseases in animals [1]. Avian pathogenic E. coli (APEC) are associated mainly with extraintestinal infections and with cellulitis [2]. EHEC (Enterohemorrhagic Escherichia coli), especially O157:H7, cause hemorrhagic colitis and hemolytic uremic syndrome in humans. Contaminated foods of animal origin are the main form of transmission of EHEC to humans. Besides raw milk, beef and broiler carcasses are an important source of EHEC [3].
Differentiation of pathogenic strains from normal microbiota is based on the production of virulence factors and on the identification of mechanisms by which they cause disease, which allow their classification into pathotypes [1]. Combinations of genes can be used to cluster E. coli strains into phylogenetic groups. Multilocus sequence typing (MLST) data improves the understanding of E. coli phylogenetic structure and allowed strains to be classified in one of the seven phylogroups A, B1, B2, C, D, E, and F [4]. MLST is the best technique for typing E. coli but the sequence type (ST) provided in the analysis does not directly allow classification into phylogroups. Thus, it is necessary to determine the correspondence between ST and phylogroups, with the latter performed by means of the “Clermont method” [4, 5].
The understanding of E. coli structure showed that the strains belonging to the different phylogroups are not dispersed randomly and are associated with the source of isolation [4]. Phylogenetic studies are important to improve the understanding of E. coli population and the relationship of strains and their hosts and disease [6]. Therefore, this study aims to analyze the occurrence and distribution of phylogenetic groups of Escherichia coli isolated from different domestic animals.
2. Materials and Methods
Seventy-one fecal specimens were collected from water buffalo calves up to 90 days old from five farms located in Minas Gerais State, Brazil. All farms bred only Mediterranean and/or Murrah buffalos for milk production. Also, one hundred seventy-one fecal samples were collected from crossbred Holstein-Gyr calves up to 90 days old born in a dairy herd located in the city of Martinho Campos, Minas Gerais State, Brazil. One gram of feces of each animal was diluted in 3.0 mL of PBS pH 7.2. From that suspension, 1.0 mL was inoculated into 9.0 mL of buffered peptone water and incubated for 18–24 h at 37°C. After incubation, an aliquot of the preenriched cultures was plated onto MacConkey agar plate and incubated at 37°C for 18–24 h. Three E. coli colonies were identified biochemically and collected from each agar plate [7].
One hundred forty-nine poultry carcasses were collected from one slaughterhouse under Federal Sanitary Inspection localized in Tocantins State, Brazil, which slaughter broiler chickens from São Paulo, Tocantins, Goiás, and the Federal District. Swabs of the air bags and trachea carcasses (respiratory tract), liver, and heart were collected. The swabs of each organ were placed in a sterile test tube containing 0.9 mL of 0.85% saline and refrigerated until processing. The swabs were processed individually [7] for isolation and identification of E. coli. Briefly, swabs were streaked onto MacConkey Agar and incubated at 37°C for 24 hours. Then, after checking the growth of colonies on MacConkey Agar, up to three lactose positive colonies were characterized biochemically [7].
Only one E. coli isolated from each host sample was used for further phylogenetic characterization and statistical analysis. The E. coli strains were tested for chuA, yjaA, TSPE4.C2, arpA, and trpA genes by PCR and this characterized phylogenetic groups A, B1, B2, C, D, E, and F [4]. Amplified DNA was resolved on a 2% agarose gel, stained with 0.5 μg/mL of ethidium bromide and photographed under UV light.
The Shannon and Simpson diversity indexes were calculated [8]. All data analyses were carried out using the Stata/SE 12.0 software. The association between the host and phylogroups was studied using the chi-square test. The results were each expressed as P value. The result was considered to be significant at P ≤ 0.05. Correspondence analysis (CA) was used to compare the categories of host and phylogroup using the Stata/SE 12.0. In the CA analysis, the relationship between the categories is represented in a two-dimensional graph.
3. Results
A total of 391 E. coli strains were analyzed in the study. These were assigned to one of the seven phylogenetic groups (Table 1). The frequency of phylogroups was the following: A = 19%, B1 = 57%, B2 = 2.3%, C = 4.6%, D = 2.8%, E = 11%, and F = 3.3%. The diversity indexes (Shannon and Simpson) are shown in Table 1.
Table 1.
Distribution of the phylogenetic groups and diversity indexes among E. coli strains isolated from different domestic animals.
| Host | Phylogenetic group | Diversity indexes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B1 | B2 | C | D | E | F | Total | Shannon | Simpson | |
| Poultry | 64 | 51 | 3 | 10 | 6 | 6 | 9 | 149 | 0.6230 | 0.6878 |
| Cattle | 8 | 127 | 1 | 4 | 0 | 28 | 3 | 171 | 0.3689 | 0.4185 |
| Water buffalo | 2 | 45 | 5 | 4 | 5 | 9 | 1 | 71 | 0.5416 | 0.5681 |
| Total | 74 | 223 | 9 | 18 | 11 | 43 | 13 | 391 | — | — |
A chi-squared test checked the association between the host and phylogenetic group. Identification of phylogroups A (P < 0.001) and F (P = 0.018) was associated with E. coli strains isolated from poultry, while phylogroups B1 (P < 0.001) and E (P = 0.002) were associated with E. coli strains isolated from cattle. Phylogroups B2 (P = 0.003) and D (P = 0.017) were associated with E. coli strains isolated from water buffalo.
The CA was performed using the host and the phylogenetic group distribution. The bidimensional representation of phylogroup distribution in each of the three hosts is shown in Figure 1. The bidimensional representation explains 100% of the total variation with 85.20% explained by the 1st dimension and 14.80% by the 2nd dimension.
Figure 1.
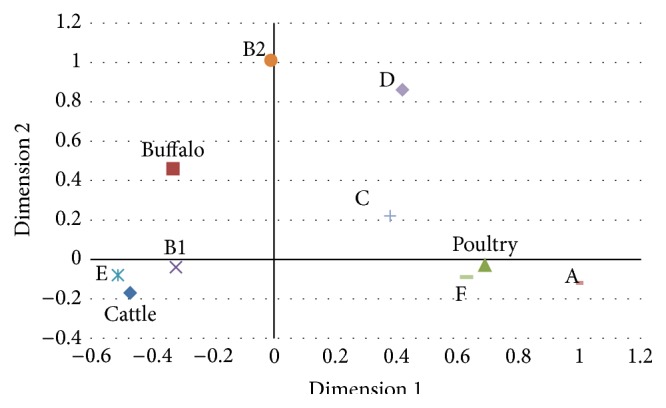
CA for the categories analyzed. Host and phylogroup that are similar fall close. This two-dimensional representation explains 100% of the total variation, with 85.20% explained by the 1st dimension and 14.80% by the 2nd dimension.
4. Discussion
This report determined the occurrence of phylogroup of Escherichia coli and demonstrated that some phylogroups are associated with the host analyzed. Phylogenetic studies help understand E. coli and its hosts and disease [6]. Moreover, food producing animals, such as cattle, water buffalo, and poultry, are an important source of EHEC in the food chain [9]. In Southern Brazil, beef and dairy cattle and water buffalos are important economically [10]. In addition, Brazil is the third largest producer of chicken meat and the largest exporter of this product [11].
The diversity indexes (Shannon and Simpson index) show that there is greater diversity in E. coli strains isolated from poultry than water buffalo and cattle (Table 1). The Shannon and Simpson index obtained for poultry were similar to those estimated by Carlos et al. [12], while cattle indices were lower than those obtained by Carlos et al. [12]. Cattle and water buffalo differ from poultry and share some characteristics such as diet and gut morphology; this may account for the differences in the diversity indexes.
Correspondence analysis examines the relationship between categorical nominal data using a contingency table of the categorical variables and transforms nonmetric data to metric data, allowing the mapping visualization, indicating that the higher the association is, the closer together the variables are in the maps [13]. Even though CA can be used to evaluate complex associations among variables, it is sometimes not sufficient to completely evaluate the associations of variables and it is suggested to use another simple (unconditional) analysis (e.g., chi-square) or multivariate analysis (e.g., logistic regression) in conjunction to complement the analytical procedures [14]. This is why we used both chi-square and CA to evaluate the relationship of E. coli phylogenetic group and the host from which the strains were isolated.
Our results indicate that B1 is the main phylogroup of E. coli isolated from domestic animals followed by phylogroup A. The results of the chi-square test and the CA agreed and showed that phylogroups B1 and E are associated with E. coli strains isolated from cattle and phylogroups A and F with poultry. E. coli strains from water buffalo were associated with phylogroups B2 and D in the chi-square but the CA showed no clear association, since these variables were not so close in the graph. Although CA did not indicate a strong association of E. coli strains of phylogroups B2 and D isolated from water buffalos, these two phylogroups were relatively closer to water buffalo than poultry and cattle and together with the chi-square results indicate that there is a tendency to detect E. coli strains of phylogroups B2 and D isolated from water buffalos.
The Extraintestinal pathogenic E. coli (ExPEC) strains are clustered mostly in groups B2 and D showing a link between phylogeny and virulence [15]. Our findings indicate that E. coli strains isolated from water buffalo calves may carry pathogenic characteristics of extraintestinal pathogenic E. coli. Studies have shown that phylogroup A is the most common phylogroup of strains obtained from ominivorous mammals and phylogroup B1 is prevalent in those isolated from herbivorous mammals [12, 16]. According to Gordon and Cowling [17], host habitat, diet, gut morphology, and body mass influence the distribution of the E. coli groups among the mammalian host. In the domestic animals analyzed here, diet and gut morphology seem to have influenced the distribution of the phylogroups. The CA can be used for molecular epidemiology studies to determine the phylogroup distribution among different hosts.
Molecular protocols such as sequencing the 16SrRNA gene and MLST are uneconomical for most screening purposes. Besides, MLST does not provide information concerning the phylogenetic group, and the phylogroup assignment depends on the “Clermont method” [5]. In our study, most phylogroups were detected in all three hosts studied; however, the chi-square test and the CA model indicate some host specificity. The PCR-based method to identify the seven phylogenetic groups was recently developed [4]; its use is rare in the literature. Our results provide knowledge of the phylogenetic composition of E. coli from domestic animals.
Acknowledgments
This work was supported by the CNPq (448357/2014-3) and Fapemig. Andrey Pereira Lage and Marcos Bryan Heinemann are indebted to CNPq for the fellowships. Fernanda Morcatti Coura thanks CNPq and Capes (13827/13-8) for her fellowship.
Conflict of Interests
The authors declare that they have no conflict of interests.
References
- 1.Mainil J. Escherichia coli virulence factors. Veterinary Immunology and Immunopathology. 2013;152(1-2):2–12. doi: 10.1016/j.vetimm.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 2.Nakazato G., de Campos T. A., Stehling E. G., Brocchi M., da Silveira W. D. Virulence factors of avian pathogenic Escherichia coli (APEC) Pesquisa Veterinaria Brasileira. 2009;29(7):479–486. doi: 10.1590/s0100-736x2009000700001. [DOI] [Google Scholar]
- 3.Bagheri M., Ghanbarpour R., Alizade H. Shiga toxin and beta-lactamases genes in Escherichia coli phylotypes isolated from carcasses of broiler chickens slaughtered in Iran. International Journal of Food Microbiology. 2014;177:16–20. doi: 10.1016/j.ijfoodmicro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Clermont O., Christenson J. K., Denamur E., Gordon D. M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental Microbiology Reports. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 5.Clermont O., Gordon D., Denamur E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology. 2015;161:980–988. doi: 10.1099/mic.0.000063. [DOI] [PubMed] [Google Scholar]
- 6.Tenaillon O., Skurnik D., Picard B., Denamur E. The population genetics of commensal Escherichia coli . Nature Reviews Microbiology. 2010;8(3):207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 7.Quinn P. J., Carter M. E., Markey B., Carter G. R. Enterobacteriaceae. In: Quinn P. J., Carter M. E., Markey B., Carter G. R., editors. Clinical Veterinary Microbiology. London, UK: Wolfe; 1994. [Google Scholar]
- 8.Pielou E. C. Ecological Diversity. New York, NY, USA: Wiley; 1975. [Google Scholar]
- 9.Martin A., Beutin L. Characteristics of Shiga toxin-producing Escherichia coli from meat and milk products of different origins and association with food producing animals as main contamination sources. International Journal of Food Microbiology. 2011;146(1):99–104. doi: 10.1016/j.ijfoodmicro.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira M. G., Brito J. R. F., Gomes T. A. T., et al. Diversity of virulence profiles of Shiga toxin-producing Escherichia coli serotypes in food-producing animals in Brazil. International Journal of Food Microbiology. 2008;127(1-2):139–146. doi: 10.1016/j.ijfoodmicro.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Alves J., Marques V. V., Pereira L. F. P., Hirooka E. Y., De Oliveira T. C. R. M. Multiplex pcr for the detection of Campylobacter spp. and Salmonella spp. in chicken meat. Journal of Food Safety. 2012;32(3):345–350. doi: 10.1111/j.1745-4565.2012.00386.x. [DOI] [Google Scholar]
- 12.Carlos C., Pires M. M., Stoppe N. C., et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiology. 2010;10, article 161 doi: 10.1186/1471-2180-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hair J. F., Black W. C., Babin B. J., Anderson R. E., editors. Multivariate Data Analysis. Upper Saddle River, NJ, USA: Pearson Prentice Hall; 2010. [Google Scholar]
- 14.Dohoo I. R., Ducrot C., Fourichon C., Donald A., Hurnik D. An overview of techniques for dealing with large numbers of independent variables in epidemiologic studies. Preventive Veterinary Medicine. 1997;29(3):221–239. doi: 10.1016/s0167-5877(96)01074-4. [DOI] [PubMed] [Google Scholar]
- 15.Escobar-Páramo P., Clermont O., Blanc-Potard A.-B., Bui H., Le Bouguénec C., Denamur E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli . Molecular Biology and Evolution. 2004;21(6):1085–1094. doi: 10.1093/molbev/msh118. [DOI] [PubMed] [Google Scholar]
- 16.Baldy-Chudzik K., Mackiewicz P., Stosik M. Phylogenetic background, virulence gene profiles, and genomic diversity in commensal Escherichia coli isolated from ten mammal species living in one zoo. Veterinary Microbiology. 2008;131(1-2):173–184. doi: 10.1016/j.vetmic.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Gordon D. M., Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149(12):3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]


