Abstract
Tibetan high-altitude adaptation (HAA) has been studied extensively, and many candidate genes have been reported. Subsequent efforts targeting HAA functional variants, however, have not been that successful (e.g., no functional variant has been suggested for the top candidate HAA gene, EPAS1). With WinXPCNVer, a method developed in this study, we detected in microarray data a Tibetan-enriched deletion (TED) carried by 90% of Tibetans; 50% were homozygous for the deletion, whereas only 3% carried the TED and 0% carried the homozygous deletion in 2,792 worldwide samples (p < 10−15). We employed long PCR and Sanger sequencing technologies to determine the exact copy number and breakpoints of the TED in 70 additional Tibetan and 182 diverse samples. The TED had identical boundaries (chr2: 46,694,276–46,697,683; hg19) and was 80 kb downstream of EPAS1. Notably, the TED was in strong linkage disequilibrium (LD; r2 = 0.8) with EPAS1 variants associated with reduced blood concentrations of hemoglobin. It was also in complete LD with the 5-SNP motif, which was suspected to be introgressed from Denisovans, but the deletion itself was absent from the Denisovan sequence. Correspondingly, we detected that footprints of positive selection for the TED occurred 12,803 (95% confidence interval = 12,075–14,725) years ago. We further whole-genome deep sequenced (>60×) seven Tibetans and verified the TED but failed to identify any other copy-number variations with comparable patterns, giving this TED top priority for further study. We speculate that the specific patterns of the TED resulted from its own functionality in HAA of Tibetans or LD with a functional variant of EPAS1.
Introduction
Tibetan highlanders have settled for more than 10,000 years in the world’s highest plateau, which has an average elevation of over 4,500 m, where the oxygen pressure is much lower (∼60%) than at sea level.1 The genetic adaptation to hypoxic environments contributes to their long-term inhabitation on the plateau. Facilitated by recent advances in genomic technologies and based on genome-wide SNP data, many studies have been conducted to search for candidate loci associated with high-altitude adaptation (HAA) in Tibetans.2–7 Among many reported HAA candidates, two hypoxia pathway genes (EPAS1 [MIM: 603349] and EGLN1 [MIM: 606425]) are the top two genes identified by most of the previous studies as having the most extreme signature of positive selection in Tibetans.
A major undertaking of the subsequent studies was to determine the functional genetic variants of the HAA candidate genes identified from previous genome-wide scans. One successful example is a high-frequency EGLN1 missense mutation that was identified by recent studies to contribute functionally to the Tibetan high-altitude phenotype.8,9 However, most efforts to study other genes with a similar purpose have not been successful, although several sequencing studies have been attempted. For instance, a previous sequencing study failed to identify any sequence variants in the exons, exon-intron boundaries, or promoter region of PPARA (MIM: 170998).10 The sequencing efforts on EPAS1 failed to identify any promising variants that might explain altered activity responsible for HAA in Tibetans.9,11
The patterns observed in many HAA genes, especially the signatures revealed by most studies on EPAS1, could not be explained by a random process. On the other hand, according to previous sequencing studies, functional variants in coding region do not exist. We thus suspect that other types of genetic variation, such as copy-number variation (CNV), probably play important roles, directly or indirectly, given that CNVs could alter gene expression.12,13 Taking advantage of the available genome-wide data of more than 120 Tibetan samples and a method developed in this study, we re-analyzed microarray image data with fluorescence-intensity information of more than one million probes and determined a Tibetan-enriched deletion (TED)—i.e., a 3.4-kb deletion 80 kb downstream of EPAS1 showed very striking differentiation between Tibetans and general worldwide populations. This deletion itself was observed by some previous studies but at a low frequency.14,15 We further validated this TED by using long PCR and Sanger sequencing in 70 additional Tibetan samples and more than 100 other diverse population samples, and all the deletion carriers had identical breakpoints (chr2: 46,694,276–46,697,683; UCSC Genome Browser hg19). Our analysis showed that the TED was in strong linkage disequilibrium (LD) with the EPAS1 coding region (r2 > 0.6) and with SNPs (r2 ≥ 0.8) that were reported previously to be associated with hemoglobin concentrations. Furthermore, our whole-genome deep sequencing (>60×) of seven Tibetan samples ranked this TED as the top HAA candidate and suggested it as a priority for further functional studies.
Material and Methods
Populations and Samples
We collected genome-wide microarray data of Tibetan samples from three published studies,4,6,7 and we refer to them here as TIB1 (GEO: GSE21661), TIB2, and TIB3 (GEO: GSE30481) (Table 1). All these samples were assayed with Affymetrix Genome-Wide Human SNP Array 6.0, which contains more than 1.8 million probes in total. Samples from the HapMap Project were also included in the analysis and consisted of the following populations: ASW (African ancestry in southwest USA), CEU (Utah residents with northern and western European ancestry from the CEPH collection), CHB (Han Chinese in Beijing, China), CHD (Chinese in Metropolitan Denver, CO), GIH (Gujarati Indians in Houston, TX), JPT (Japanese in Tokyo, Japan), LWK (Luhya in Webuye, Kenya), MXL (Mexican ancestry in Los Angeles, CA), MKK (Maasai in Kinyawa, Kenya), TSI (Toscani in Italia), and YRI (Yoruba in Ibadan, Nigeria). We also collected microarray data from other East and Southeast Asian populations. These data were also assayed with Affymetrix Genome-Wide Human SNP Array 6.0, and the samples included (1) 100 Korean (KOR) individuals from this study; (2) 18 Malay, 17 Senoi, and 12 Negritos from a previous study;16 and (3) 80 Han Chinese, 8 Yao, 6 Zhuang, 9 Dong, and 8 Li from a previous study.17
Table 1.
A Summary of the Tibetan Samples and Dataset
| Dataset | Sourcea(Location) | Sample Size | QC+ Samplesb | Methodc | Reference |
|---|---|---|---|---|---|
| TIB1 | Qinghai | 31 | 27 | microarray | Simonson et al.4 |
| TIB2 | Tibet, Qinghai, and Yunnan | 50 | 44 | microarray | Peng et al.6 |
| TIB3 | Tibet | 51 | 46 | microarray | Xu et al.7 |
| TIB4 | Tibet | 70 | 70 | LP and SS | this study |
| TIB-seq | Tibet | 7 | 7 | NGS | this study |
Abbreviations are as follows: LP, long PCR; NGS, next-generation sequencing; SS, Sanger sequencing.
Sample location: TIB1, Madou County; TIB2, randomly selected from Tibet, Qinghai, and Yunnan; TIB3 and TIB4, Lhasa, Nyingchi, Qamdo, Shannan, and Shigatse; TIB-seq, a subset from TIB4.
Those samples that passed quality control in this study.
Experimental methods for detecting or validating the CNV.
To verify the boundary and frequency of the deletion, we collected peripheral-blood samples of another set of 70 Tibetans (TIB4: 17 Lhasa [∼3,650 m], 5 Nyingchi [∼3,000 m], 19 Qamdo [∼3,240 m], 15 Shannan [∼3,700 m], and 14 Shigatse [∼3,837 m]; Table 1) from Tibet and collected blood samples of 182 non-Tibetans (50 Han Chinese, 50 Kazakhs, 50 Uyghur, 11 Hui, 8 Mongolian, 7 Khalkhas, 2 Ozbek, 2 Tatar, 1 Tujia, and 1 Xibe) from the surrounding regions of Tibet as the reference panel. Each individual was the offspring of a non-consanguineous marriage of members of the same nationality within three generations. Informed consent was acquired from the participants. All procedures were in accordance with the ethical standards of the Responsible Committee on Human Experimentation (approved by the Biomedical Research Ethics Committee of Shanghai Institutes for Biological Sciences) and the Helsinki Declaration of 1975 (revised in 2000).
CNV Detection from Microarray Data
We applied Birdsuite v.1.5.518 to detect CNVs in all Tibetan samples, as well as in the HapMap and other microarray samples. The software implemented two different methods to call CNVs from the microarray intensity data: (1) a clustering-based algorithm to determine genotype predefined loci (Canary) and (2) a hidden Markov model (HMM)-based algorithm to detect novel CNVs (Birdseye). We performed quality control at two levels: (1) samples with a CNV amount more than 5 SDs from the average were excluded, and (2) CNV calls (birdseye_canary_calls file) with a confidence score less than 5 were excluded. Then, we used the filtered results to generate a Tibetan CNV map. Because Birdsuite only provides coordinates in hg18, we used UCSC liftover to convert the coordinates into hg19. In this study, all the genomic coordinates were based on hg19, and the detected CNVs have been deposited in the Database of Genomic Structural Variation (dbVar: nstd111).
An Algorithm for Searching for Population-Differential CNVs in Microarray Intensity Data
To search for CNV regions (CNVRs) that are highly differentiated between Tibetans and Han Chinese, we developed the algorithm WinXPCNVer, which was particularly designed to search for CNVs that are highly differentiated between populations. The script of this software is available online. The basic idea of the algorithm is that the raw microarray intensity data will show detectable differences if a CNVR shows high differentiation between two populations. We calculated the VST19 for each probe set on the basis of the normalized intensity data (locus_summary file). Because the raw intensity of a single probe could be noisy, we set a non-overlapping sliding window with length L and calculated the statistic VST-w; that is, the average VST for the top n probes in each window (w). Because the Tibetan samples were from three different studies, we calculated the VST for each probe between each Tibetan group and CHB and subtracted the pairwise VST among Tibetans as follows:
We performed the analysis in different combinations of L and n: (1) L = 1 kb and n = 3; (2) L = 3 kb and n = 3; (3) L = 5 kb and n = 3; and (4) L = 5 kb and n = 5. We removed any window with probe number less than n. For each combination, we ranked all windows by VST-w and also manually checked the information in the top 20 windows. Further, we searched whether any genes with a functional annotation were located within 100 kb of the flanking regions.
Determination of Individual TED Genotype in Microarray Intensity Data
We determined individual TED genotypes in microarray intensity data by using the K-means method with further manual checks. We employed four probes (CN_839592, CN_839595, SNP_A-1898130, and SNP_A-4199859) with the highest VST in that window to assign the genotype (Figure S1). For Tibetan samples, the K-means method was good enough to distinguish different clusters; however, for the HapMap Asian samples (CHB, CHD, and JPT), the K-means method did not perform so well. Therefore, we manually checked their genotypes and referred to the Database of Genomic Variants (DGV),20 given that many HapMap samples have been well characterized by previous studies via different platforms.
A large number of HapMap samples were also sequenced by next-generation sequencing (NGS) and were included in the 1000 Genomes Project. We further referred to the deletion genotype and frequencies in the project report15 (DGV: estd199), which included 1,092 worldwide samples from 61 ASW, 85 CEU, 97 CHB, 100 CHS (Southern Han Chinese), 60 CLM (Colombians from Medellin, Colombia), 93 FIN (Finnish in Finland), 89 GBR (British in England and Scotland), 14 IBS (Iberian population in Spain), 89 JPT, 97 LWK, 66 MXL, 55 PUR (Puerto Ricans from Puerto Rico), 98 TSI, and 88 YRI.
Long-PCR Validation of the Deletion
We performed long PCR to detect and validate the zero-copy, one-copy, and two-copy samples in each population. Given that the previous studies14,15 reported the breakpoints of the same deletion in the 1000 Genomes Project samples in base-pair resolution, we amplified the region chr2: 46,693,938–46,697,928. The primers were designed with Primer3. A 20-μl mixture was prepared for each reaction with 1 μl template DNA. Amplification conditions consisted of an initial denaturation step at 94°C for 10 min, followed by 35 cycles of 94°C for 20 s, 68°C for 5 min, and 72°C for 2 min. The long-PCR products were observed by 1% agarose gel electrophoresis. The product size could be distinguished according to the number of copies in each sample: 583-bp products represented zero-copy and one-copy samples, and 3,991-bp products represented two-copy samples.
Determination of TED Breakpoints by Sanger Sequencing
We used Standard Sanger sequencing approaches to determine deletion regions in zero-copy and one-copy samples. PCR was performed with HotStarTaq DNA Polymerase (QIAGEN). A 20-μl mixture was prepared for each reaction and included 1 U HotStarTaq DNA Polymerase and 1 μl template DNA. 1 U SAP and 6 U Exo I were added into 8 μl PCR product for purification. The mixture was incubated at 37°C for 60 min, followed by incubation at 70°C for 10 min. Then, the purified PCR product was sequenced with the Big-Dye Terminator Cycle Sequencing Kit and an ABI 3130XL Genetic Analyzer (Applied Biosystems). With the information of breakpoints and the flanking sequences, we determined the mutation mechanism of the TED according to the pipeline from a previous study.21
Population Genetic Analysis
Geographic distribution of TED frequencies in Asia and worldwide were plotted onto a contour map with Surfer 10.0 (Golden Software), and the Kriging method was used for data interpolation. The p value for the frequency difference between Tibetans and worldwide populations was calculated with Fisher’s exact test, which was treated as a 2 × 2 table (TIB versus non-TIB and deletion-carrier versus non-deletion-carrier). FST was calculated as a reference22 in this study. We selected SNPs with FST larger than 0.5 between Tibetans and Han Chinese to infer the haplotype of deletion. The phase inference was performed by software PHASE v.2.1.23 The haplotypes inferred by PHASE analysis and with a frequency larger than 0.01 were used for building a haplotype network with Network 4.6.1.3.24 The haplotypes of chromosome 2 in Tibetans and CHB were inferred with software BEAGLE.25 When calculating LD between the TED and its flanking SNPs, we removed the SNPs with a minor allele frequency less than 0.2. Analysis of extended haplotype homozygosity (EHH)26 was performed with R package rehh.27 Selection age of the TED was estimated on the basis of the EHH results according to previous studies,6,28,29 which assumed a star genealogy of the haplotypes and that recombination happened independently in each genealogy. We assumed 25 years per generation. Under a soft-sweep model, we estimated the selection intensity according to a previous study.8 We estimated the confidence intervals (CIs) of selection intensity and the age of the TED by bootstrapping over haplotypes.
Whole-Genome Sequencing Analysis
Whole-genome deep sequencing (>60×) of seven Tibetan individuals (TIB-seq; Table 1) from TIB4 was performed in Wuxi AppTec in Shanghai with an Illumina HiSeq X according to Illumina-provided protocols. Whole-genome sequences (150-bp paired-end reads) were aligned to the human reference sequence (hg19) with bwa0.7.10-r78930 from the BWA-MEM algorithm. The aligned reads were sorted with SAMtools31 and then processed as suggested by the Genome Analysis Toolkit32,33 best practices. We performed mark duplicates, indel realignment, and base recalibration for the sorted BAM files to get the well-curated BAM files. The Korean whole-genome sequencing data were obtained and downloaded from the Korean Personal Genome Project (KPGP; see Web Resources). We used the same mapping procedure to align Korean sequences to hg19 and generated the BAM files.
To detect CNVs from Tibetan whole-genome sequence data, we used two algorithms, CNVnator34 and readDepth.35 The bin size was set to 100 bp for both algorithms. For the same individual, a segment was called as a CNV only if this segment had 50% overlap of length and the same variation type from both algorithms. The overlapped CNVs were merged into CNVRs. We re-genotyped these CNVRs in Tibetan and Korean samples with CNVnator and compared their frequency difference.
We used multiple sequentially Markovian coalescent (MSMC)36 to infer the change in effective population size from multiple genome sequences. To reduce the computational burden, we only used markers on chromosomes 1–10 of five Tibetan individuals. We set the autosomal mutation rate at 1.25 × 10−8 per base per generation and 25 years per generation.
Results
CNV Profiles in the Tibetan Populations
We first collected genome-wide microarray data of Tibetan samples from three studies,4,6,7 which we hereafter refer to as TIB1, TIB2, and TIB3 (Table 1). All these samples were assayed with an identical genotyping platform, Affymetrix Human Genome-wide SNP 6.0, which includes more than 946,000 probes designed for CNV detection. Birdsuite18 was employed for SNP and CNV calling from these data sets. Principal-component analysis based on the genome-wide SNPs showed that TIB2 and TIB3 samples clustered together, whereas TIB1 was separated (Figure 1A), which is consistent with the geographical location of those populations (Table 1). For the purpose of comparison, HapMap37 population samples with available Affymetrix SNP 6.0 raw intensity data were also included in our analysis. After quality control on both sample and CNV levels (see Material and Methods), a total of 15,516 CNV events were detected from 117 Tibetan samples. Whereas individuals from TIB3 had a smaller number of CNVs than did the other two Tibetan data sets on average, all three Tibetan groups carried fewer CNVs than did CHB (p < 10−5; Table S1). The median size of the CNV events was similar in both Tibetans and Han Chinese (8.5 and 67 kb for deletions and duplications, respectively; Figure S2). Furthermore, we merged the overlapping CNVs into CNVRs and estimated the allele frequency for each CNVR. To search for the CNVR with a significant allele-frequency difference between Tibetans and other populations, we calculated the pairwise FST between Tibetans and the HapMap populations. At this stage of the analysis, however, we failed to find any CNVs that were significantly different in frequency between Tibetans and the other populations, such as Han Chinese (see Discussion).
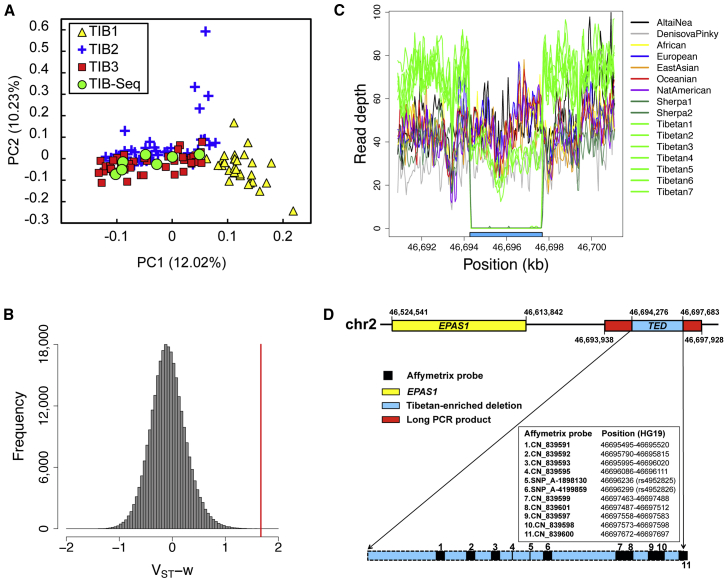
Figure 1.
TED Downstream of EPAS1
(A) Population structure of Tibetan samples from different sources (Qinghai: TIB1; Tibet: TIB2, TIB3 and TIB-seq). The principal-component analysis (PCA) plot was generated by 99,768 genome-wide random SNPs. Each dot represents one Tibetan individual. The x and y axes represent the first and second principal components (PCs), respectively, which explain 12.02% and 10.23% of the total variance, respectively.
(B) Genome-wide distribution of VST-w, calculated as the mean VST of the top three probes in each 3-kb sliding window. The red vertical line represents the TED downstream of EPAS1.
(C) Read depth (RD) of seven Tibetan, two Sherpa, one Neandertal, one Denisovan, and five modern human individuals. The deletion region is highlighted in the blue bar at the bottom. Samples with a homozygous or heterozygous deletion showed 0% or 50% of the normal (flanking) RD, respectively. Four Tibetan and two Sherpa individuals carried a homozygous deletion, and the other three Tibetan individuals carried a heterozygous deletion. No deletions were found in other individuals.
(D) Diagram of the locations of microarray probes, long-PCR primers, and EPAS1.
Searching for Population-Differential CNVs in Microarray Intensity Data
Despite the fact that the above routine analysis did not reveal any CNVs that were significantly different between populations, the raw intensity data we obtained from all the samples provided an opportunity to further deeply investigate the CNV architectures in both Tibetan and non-Tibetan samples. We suspected that some interesting signals showing a significant difference between the two populations might have been missed by CNV-calling algorithms developed for general purposes. Therefore, we developed a CNV-searching algorithm particularly for a two-population comparison. The basic idea was that the microarray raw intensity data would show detectable differences if there was a CNVR that was substantially differentiated between the two populations being compared. Because the raw intensity of a single probe could be noisy, we used a window-based measurement (i.e., VST-w) to decrease the effect of random noise while increasing the difference reflecting the true differentiation of the variants between two populations (see Material and Methods). We named our algorithm WinXPCNVer, a window-based cross-population differential-CNV detector. Our results demonstrated that this searching algorithm is much more powerful than the routine approach at identifying CNVs that differ between populations, especially at highly differentiated regions, where the routine calling algorithm fails to genotype CNVs correctly.
Using the HapMap Han Chinese (CHB) samples as a reference population, we calculated the VST-w under different conditions. We checked the top 20 windows with the largest VST-w values and searched for genes near each window. Surprisingly, we found that one signal (ranking at 7, 10, 13, and 17 at four conditions with different number of probes and window sizes; Figure 1B; see Material and Methods) fell in a CNVR located downstream of a previously identified hypoxia-inducing gene (EPAS1). However, we failed to find any other high-VST-w windows that were located in CNVRs or contained any genes in the 100-kb flanking regions.
Manual Check of Intensity Data and Experimental Validation of the CNVR
To confirm the signal identified by the above analysis, we first manually checked the raw intensity data. The target window contained three probes (CN_839592, CN_839595, and SNP_A-1898130) with a VST larger than 1.50. We plotted the intensity of the above three probes with the fourth-highest VST probe (SNP_A-4199859) in Tibetans and HapMap populations. Unlike other populations, which showed a typical biallelic SNP-clustering pattern (although a few samples were in a deletion state; Figures S1A–S1C), most Tibetan samples showed a typical deletion-like pattern (and only very few were in a normal two-copy state; Figure S1D). Because Birdsuite failed to call most of the deletions correctly, we re-genotyped this region in silico by using K-means clustering and manually corrected the copy number for each individual (Material and Methods).
Moreover, we whole-genome deeply sequenced (>60×) seven additional Tibetan individuals (TIB-seq) and found that reads were fully absent in four individuals and half absent in the other three individuals (Figure 1C), indicating that the four Tibetan individuals had a homozygous deletion and the other three had a heterozygous deletion.
This deletion has been included in DGV (e.g., DGV: dgv625n67,38 nsv44175739) but was only reported in non-Tibetan populations without precise breakpoints. In addition, no information of its frequency in Tibetans was available. Therefore, we conducted long PCR at the EPAS1 downstream region encompassing the deletion region by referring to DGV: esv266048015 (chr2: 46,694,273–46,697,681, hg19) and performed Sanger sequencing in an additional set of 70 Tibetan (TIB4) and 182 non-Tibetan samples from different Chinese ethnic groups (see Material and Methods and Figure 1D) to verify this deletion region. The results showed that the majority of the Tibetan samples carried the deletion but that most of the non-Tibetan samples did not (deletion frequency was 62/70 in TIB and 7/182 in non-TIB; Figure S3). The precise breakpoints of the deletion (chr2: 46,694,276–46,697,683) were determined by Sanger sequencing (Figure S4). Interestingly, the breakpoints were identical in all the deletion carriers. Therefore, we validated this deletion in both Tibetan and non-Tibetan samples and determined the boundaries of this deletion, hereafter referred to as the TED.
Frequency Distribution of the TED in Worldwide Populations
Tibetans carry a high frequency of the TED (88.6% in TIB4 and 94% in combined TIB1, TIB2, and TIB3 samples; Figure 2A). Among these deletion carriers, more than half have homozygous deletions. In contrast, in worldwide populations, these percentages are <10% for deletion carriers and 0% for homozygous deletion carriers (p < 10−15). In addition, no deletion was observed in either African or European populations (Figure S5; Table 2).
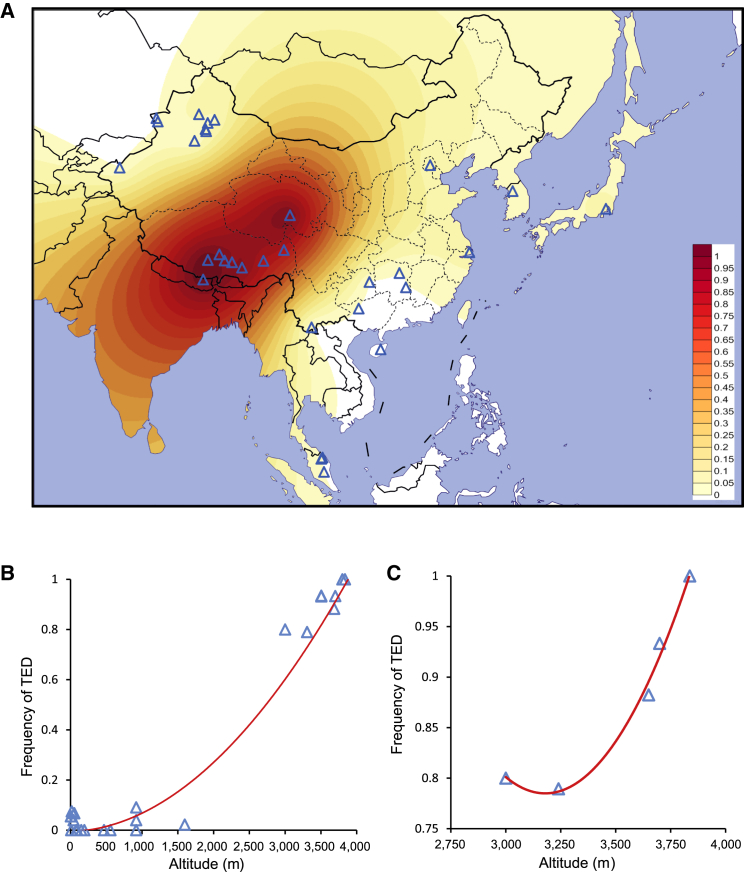
Figure 2.
Distribution of the TED Frequency among Populations and Its Correlation with Altitude
(A) Distribution of deletion frequency in Asian populations. Colors from yellow to red indicate the frequency from low to high, respectively. Each blue triangle represents a sampled population.
(B) Deletion frequency correlated (R2 = 0.958) with altitude in Asian populations (population information is listed in Table 2).
(C) Deletion frequency correlated (R2 = 0.989) with altitude in five Tibetan sub-groups (Lhasa, Nyingchi, Qamdo, Shannan, and Shigatse).
Table 2.
Frequency of the TED in Worldwide Populations
| Populationa | Sample Size | Zero-Copy Samples (Count) | One-Copy Samples (Count) | Two-Copy Samples (Count) | Methodb | Source |
|---|---|---|---|---|---|---|
| ASW | 87 | 0% (0) | 0% (0) | 100% (87) | microarray | HapMap |
| CEU | 177 | 0% (0) | 0% (0) | 100% (177) | microarray | HapMap |
| CHB | 89 | 0% (0) | 6.7% (6) | 93.3% (83) | microarray | HapMap |
| CHD | 90 | 0% (0) | 2.2% (2) | 97.8% (88) | microarray | HapMap |
| GIH | 90 | 0% (0) | 0% (0) | 100% (90) | microarray | HapMap |
| JPT | 91 | 0% (0) | 5.5% (5) | 94.5% (86) | microarray | HapMap |
| LWK | 90 | 0% (0) | 0% (0) | 100% (90) | microarray | HapMap |
| MXL | 84 | 0% (0) | 0% (0) | 100% (84) | microarray | HapMap |
| MKK | 179 | 0% (0) | 0% (0) | 100% (179) | microarray | HapMap |
| TSI | 90 | 0% (0) | 0% (0) | 100% (90) | microarray | HapMap |
| YRI | 180 | 0% (0) | 0% (0) | 100% (180) | microarray | HapMap |
| KOR | 100 | 0% (0) | 4.0% (4) | 96.0% (96) | microarray | this study |
| Malay | 18 | 0% (0) | 0% (0) | 100% (18) | microarray | Mokhtar et al.16 |
| Senoi | 17 | 0% (0) | 0% (0) | 100% (17) | microarray | Mokhtar et al.16 |
| Negrito | 12 | 0% (0) | 0% (0) | 100% (12) | microarray | Mokhtar et al.16 |
| Han Chinese | 80 | 0% (0) | 7.5% (6) | 92.5% (74) | microarray | Lou et al.17 |
| Yao | 8 | 0% (0) | 0% (0) | 100% (8) | microarray | Lou et al.17 |
| Zhuang | 6 | 0% (0) | 0% (0) | 100% (6) | microarray | Lou et al.17 |
| Dong | 9 | 0% (0) | 0% (0) | 100% (9) | microarray | Lou et al.17 |
| Li | 8 | 0% (0) | 0% (0) | 100% (8) | microarray | Lou et al.17 |
| TIB1 | 27 | 55.6% (15) | 40.7% (11) | 3.7% (1) | microarray | Simonson et al.4 |
| TIB2 | 44 | 43.2% (19) | 50.0% (22) | 6.8% (3) | microarray | Peng et al.6 |
| TIB3 | 46 | 54.3% (25) | 39.1% (18) | 6.5% (3) | microarray | Xu et al.7 |
| Han Chinese | 50 | 0% (0) | 4.0% (2) | 96.0% (48) | LP and SS | this study |
| Kazakh | 50 | 0% (0) | 4.0% (2) | 96.0% (48) | LP and SS | this study |
| Uyghur | 50 | 0% (0) | 4.0% (2) | 96.0% (48) | LP and SS | this study |
| Hui | 11 | 0% (0) | 9.1% (1) | 90.9% (10) | LP and SS | this study |
| Khirghiz | 7 | 0% (0) | 0% (0) | 100% (7) | LP and SS | this study |
| Mongolian | 8 | 0% (0) | 0% (0) | 100% (8) | LP and SS | this study |
| Ozbek | 2 | 0% (0) | 0% (0) | 100% (2) | LP and SS | this study |
| Tatar | 2 | 0% (0) | 0% (0) | 100% (2) | LP and SS | this study |
| Tujia | 1 | 0% (0) | 0% (0) | 100% (1) | LP and SS | this study |
| Xibe | 1 | 0% (0) | 0% (0) | 100% (1) | LP and SS | this study |
| TIB4 | 70 | 61.4% (43) | 27.1% (19) | 11.4% (8) | LP and SS | this study |
| ASW | 61 | 0% | 0% | 100% (61) | NGS | Abecasis et al.15 |
| CEU | 85 | 0% | 0% | 100% (85) | NGS | Abecasis et al.15 |
| CHB | 97 | 0% | 4.1% (4) | 95.9% (93) | NGS | Abecasis et al.15 |
| CHS | 100 | 0% | 1.0% (1) | 99.0% (99) | NGS | Abecasis et al.15 |
| CLM | 60 | 0% | 0% | 100% (60) | NGS | Abecasis et al.15 |
| FIN | 93 | 0% | 0% | 100% (93) | NGS | Abecasis et al.15 |
| GBR | 89 | 0% | 0% | 100% (89) | NGS | Abecasis et al.15 |
| IBS | 14 | 0% | 0% | 100% (14) | NGS | Abecasis et al.15 |
| JPT | 89 | 0% | 7.9% (7) | 92.1% (82) | NGS | Abecasis et al.15 |
| LWK | 97 | 0% | 0% | 100% (97) | NGS | Abecasis et al.15 |
| MXL | 66 | 0% | 0% | 100% (66) | NGS | Abecasis et al.15 |
| PUR | 55 | 0% | 0% | 100% (55) | NGS | Abecasis et al.15 |
| TSI | 98 | 0% | 0% | 100% (98) | NGS | Abecasis et al.15 |
| YRI | 88 | 0% | 0% | 100% (88) | NGS | Abecasis et al.15 |
| Dinka | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Mbuti | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| French | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Papuan | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Sardinian | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Karitians | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| San | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Mandenka | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Yoruba | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Dai | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Han Chinese | 1 | 0% (0) | 0% (0) | 100% (1) | NGS | Meyer et al.40 |
| Sherpa | 2 | 100% (2) | 0% (0) | 0% (0) | NGS | Jeong et al.41 |
| TIB-Seq | 7 | 57.1% (4) | 42.9% (3) | 0% (0) | NGS | this study |
Abbreviations are as follows: LP, long PCR; NGS, next-generation sequencing; SS, Sanger sequencing.
The full names of the abbreviated populations are listed in the Material and Methods.
Experimental methods for detecting or validating the CNV.
We further checked the status of this TED region in whole-genome sequence data of worldwide population samples. We did not observe the deletion in 11 individuals from seven diverse worldwide groups (Africans, Europeans, East Asians, Oceanians, Native Americans, Altai Neandertals, and Denisovans) that have been deeply sequenced40,42 (Figure 1C). However, two Sherpa who, like Tibetan people,41 lived in a high-altitude (>3,000 m) region carried the same homozygous breakpoint deletion that we validated in Tibetans.
We compared the frequency of the TED between Tibetans and Han Chinese (FST = 0.64) and confirmed that it was the most differential locus in the region encompassing EPAS1 (Figure S6). We inferred the haplotype of the TED with highly differentiated SNPs (FST > 0.5). Interestingly, one dominant haplotype occupied 88% of all the haplotypes with the deletion in Tibetans, and the deletion haplotype in the two Sherpa individuals was identical to the dominant one in the Tibetans (Figure S7). In addition, we investigated the relationship between TED frequency and the altitude of locations where Tibetan individuals live. Interestingly, we observed a strong correlation between the deletion frequency and the altitude in Asian populations (R2 = 0.958) and in five Tibetan sub-groups (TIB4, R2 = 0.989) (Figures 2B and 2C).
To search for other Tibetan-enriched CNVs whose pattern might be similar to that of this TED, we analyzed the seven deeply sequenced Tibetan samples by comparing them with high-coverage (∼30×) Korean samples from the KPGP (see Material and Methods). However, we failed to identify a second CNV showing a comparable pattern from the available data.
LD between the TED and Its Flanking SNPs Associated with HAA
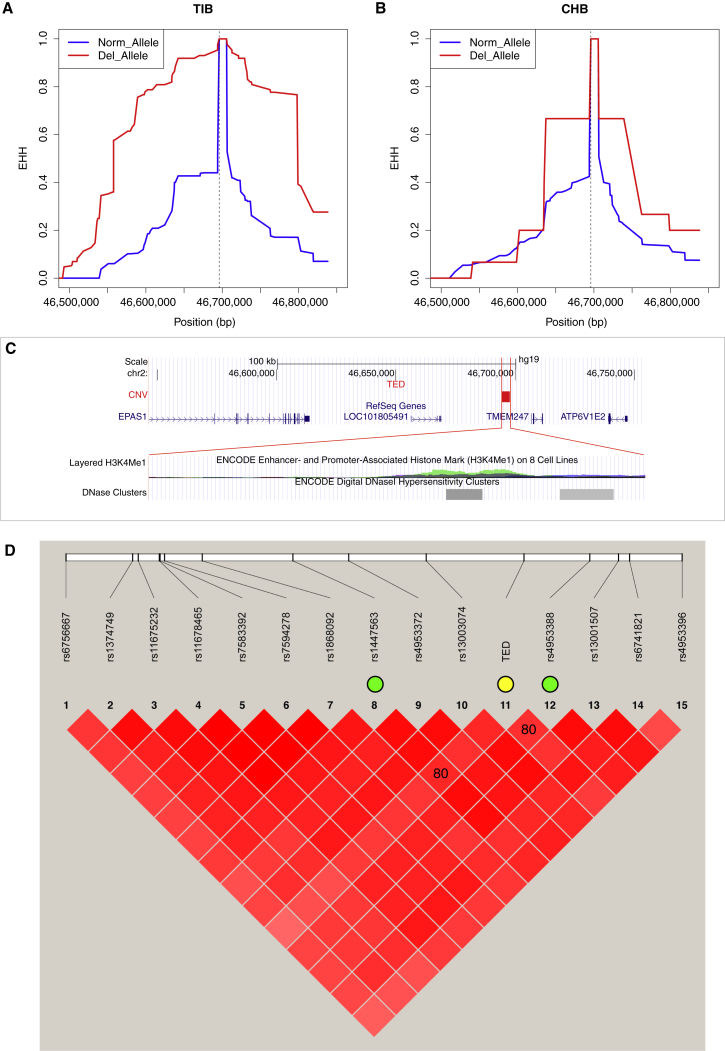
We examined the LD between the TED and its flanking SNPs. Upstream of the TED, LD of the deletion allele and its flanking region in Tibetans (r2 > 0.5) extended over nearly 100 kb and overlapped ten exons of EPAS1. The highest LD (r2 ≥ 0.8) was observed in the SNPs (rs13003074, rs4953388, rs1447563, and rs6741821) of the EPAS1 downstream region, in which previous studies have reported the highest FST between Tibetans and Han Chinese.2,7 In contrast to Tibetans, Han Chinese showed much shorter LD, which decayed substantially at both 5′ and 3′ regions (Figure S8). We further analyzed EHH, and the results indicated that EHH was longer in the deletion allele in Tibetans than in the normal allele (Figure 3A); in contrast, both the deletion allele and the normal allele in Han Chinese only showed limited EHH (Figure 3B), which was consistent with the LD pattern. This strong EHH signal suggests that this region could be a target of natural selection. Furthermore, we estimated the selection intensity and the age of the selected deletion allele (see Material and Methods). The age of selection on the TED was estimated to be 12,803 (95% CI = 12,075–14,725) years. Under a model assuming selection on a standing variant and using the deletion allele frequency in CHB (0.034) as an approximated variant frequency in ancestral Tibetans before selection, we estimated the selection intensity as 0.0084 (95% CI = 0.0073–0.0089).
Figure 3.
EHH and Functional Annotation of the TED
(A and B) EHH plot of the deletion in (A) TIB and (B) CHB. The dashed line indicates the deletion position; the blue and red curves represent the normal allele and the deletion allele, respectively.
(C) Functional annotation generated by the UCSC Genome Browser. Blue bars represent RefSeq genes, and the red bar represents the deletion. A regulation signal (H3K4Me1) from the regulation database ENCODE overlapped the deletion (bottom panel). Colors in the regulation track represent different cell lines: Gm12878 (red), H1 ES (yellow), HMEC (green), HSMM (aqua), HUVEC (blue), K562 (cyan), NHEK (purple), and NHLF (pink). The gray tracks are the digital DNaseI hypersensitivity clusters from ENCODE.
(D) The strong LD (r2 = 0.80) between the TED and two identified SNPs associated with hemoglobin concentrations in Tibetans2. The color in the red squares represents the strength of the LD (i.e., the darker, the stronger). The TED is highlighted with a yellow circle, and the other 14 markers are the SNPs with the highest FST (>0.5) between TIB and CHB in this study. The SNPs in the green circle are the ones found to be associated with hemoglobin concentrations in a previous study.2 The relative positions of these two SNPs encompassing the deletion suggest an association between the TED and hemoglobin concentrations in the Tibetan population.
Functional Annotation of the TED
By searching in the regulation database ENCODE, we found an enhancer- and promoter-associated histone mark (H3K4me1) and two digital DNaseI hypersensitivity clusters overlapping the TED. The H3K4me1 histone mark is the mono-methylation of lysine 4 of the H3 histone protein, and it is associated with enhancers and with DNA regions downstream of transcription start sites. The regulation signals were found in human mammary epithelial cells (HMECs), human epidermal keratinocyte (NHEK) cells, and K562 cells, and they were most pronounced in HMECs (Figure 3C). Consistently, the histone mark region is associated with the DNaseI hypersensitivity clusters, which have been regarded as indicators of regulatory elements.43 These signals suggest a regulatory role for the TED in nearby genes, especially in EPAS1, LOC101805491, TMEM247, and ATP6V1E2. However, except for those of EPAS1 and ATP6V1E2, the functions of the other genes (LOC101805491 and TMEM247) are unknown. We also used weight-matrix-based software Match44 to predict the transcription factor binding site in the sequence of the deletion region. Detailed information is listed in Table S2.
At the phenotypic level, one previous study identified that eight highly differentiated SNPs near EPAS1 were in strong LD with the SNPs associated with hemoglobin concentration in Tibetans.2 Interestingly, among the highly differentiated SNPs in that study, the two top SNPs (rs1447563 and rs4953388, which were located to the left and right of the TED, respectively) were in strong LD with the TED (r2 = 0.8) (Figure 3D), suggesting an association between the TED and hemoglobin concentrations.
Discussion
In this study, we conducted genome-wide studies to search for signals of HAA in Tibetans on the basis of raw genome-wide microarray data and whole-genome deep sequencing data. With a much larger sample size, we were able to replicate most of the HAA signals on the basis of SNP data reported by previous studies. Notably, a genome-wide search of CNV data allowed us to identify a TED causing more than 90% of Tibetan individuals to lose at least one copy (i.e., a heterozygous deletion) and 50% to lose both copies (i.e., a homozygous deletion). This 3.4-kb TED was prevalent in Tibetans and Sherpa but had a low frequency or was absent in other Asian (<10%) and worldwide populations (0%; p < 10−15).
The deletion itself was previously reported in some non-Tibetan populations. For example, several studies, including the HapMap and 1000 Genomes Projects, have already reported this deletion,14,38,39,45 and they were mainly derived from the Han Chinese (CHB) and the Japanese (JPT) samples. Only one study reported the deletion in Tibetans, but it did not provide frequency information.3 Moreover, the deletion has not been experimentally validated by any previous studies. Whereas two CEU individuals (NA12146 and NA10847, from a trio) with one copy were reported in some studies,39,46 we considered them as copy normal because neither the 1000 Genomes Project15 detected this deletion nor could any deletion pattern from the intensity plot of the microarray of the two individuals be observed. We also found that the genotyping of this deletion by Birdsuite was not reliable in Tibetans. Most of the genotypes in Tibetans were called “missing data,” which was probably due to the parameters that did not fit the Tibetan samples in the Canary package, and the HMM-based package Birdseye was not sensitive enough to detect the probe-intensity changes in such a small segment. This was also the reason why it was missed in many previous studies, including one of our own.7 The method developed in this study (WinXPCNVer) allows searching for differential CNVs between populations on the basis of raw microarray intensity data. It contributed to the successful identification of this particular TED. However, HAA in Tibetans could be associated with more structural variants, especially those smaller than 1 kb, which are difficult for algorithms to detect or are not covered by microarray probes.
A recent sequencing study of EPAS1 found that Tibetans have a unique 5-SNP motif that differs from that of other worldwide populations; this motif was suspected and reported to be introgressed by Denisovans.11 We also observed this 5-SNP motif in our data and found it in complete LD (r2 = 1) with the TED in seven whole-genome-sequenced Tibetan samples. However, we did not observe the deletion in the Denisovan sequence (Figure 1C), which indicates that the LD between the TED and the 5-SNP motif was established in modern humans or in Tibetans after the genetic introgression, if it did occur.
The mechanism of the TED was characterized as a non-homologous event that had limited homology14 and was non-recurrent,47 and we also confirmed this with our data (Figure S4). Consistently, the deletion was observed with the same breakpoints in all the deletion carriers of Tibetan and non-Tibetan Chinese samples. Considering the fact that the deletion is overrepresented in Tibetans and Sherpa and is present at a very low frequency exclusively in a heterozygous state in other East Asian populations, the deletion is likely to have occurred before the separation of Tibetans and other East Asian populations. Its high frequency in Tibetans and Sherpa is probably due to the hitchhiking effect as a result of strong LD with EPAS1 under natural selection or a consequence of being directly selected because of its own functional role in the HAA of Tibetans. Under the latter scenario, the estimated selection age of the TED allele was less than the age of the previously reported EPAS1 selected allele.6 This is because the deletion’s EHH was longer than that of the previous SNP (with the largest FST between Tibetans and CHB) used as a surrogate of the selected mutant. Moreover, we used MSMC36 to infer the effective population size (Figure S9). Interestingly, on the basis of the curve of the effective population size inferred from sequencing data, the estimated age coincided with the peak of Tibetan expansion at 13,000 years ago (Figure S9).
EPAS1, the top HAA signal identified by almost all of the previous studies in Tibetans, encodes the transcription factor involved in the induction of genes when oxygen levels fall. The LD between the TED and EPAS1 reached r2 > 0.6 at the 3′ gene region. If the function of the TED is involved in HAA, it is reasonable to speculate that EPAS1 could be a target gene regulated by the TED either directly or indirectly. Many studies have demonstrated not only that a coding-region CNV could affect gene expression but also that a non-coding-region CNV could be functional. For example, a disease-associated duplication was reported to affect the function of PMP22 (MIM: 601097)48 even though it is located in the regulatory region about 34 kb away from the coding region of PMP22. Similarly, a deletion was reported to be more than 1 Mb away from SOX9 (MIM: 608160).49 In addition, it is unexpected that a 3.4-kb deletion such as the TED would exist in the coding region of EPAS1 given the important function of the gene, and a knockout of EPAS1 in mice would result in pancytopenia.50 Therefore, we believe that the function of EPAS1, as the gene in the strongest LD with this TED, could be substantially affected. Nevertheless, this does not exclude the possibility that the TED could influence other flanking genes, given that other than EPAS1, three more RefSeq genes are located in the 100-kb flanking region of the TED (Figure 3C; Table S3). The closest genes are TMEM247 (transmembrane protein 247) and LOC101805491 (an RNA gene), which are downstream and upstream of the TED, respectively, and whose LD is r2 = 0.8. However, the functions of these two genes are largely unknown. The third gene is ATP6V1E2 (ATPase, H+ transporting, lysosomal 31 kDa, V1 subunit E2), which is downstream of the TED and has moderate LD (0.39 < r2 < 0.68). This gene is related to H+ATPase activity, but it is not clear whether the gene is involved in any HAA-related pathway. Additionally, it is also possible that the TED could affect other distant genes (e.g., via trans-regulation).
In summary, although the function of the TED has not yet been fully characterized, many lines of evidence support that the TED is a promising candidate that might have played a critical role in HAA of Tibetans. Accordingly, here we propose two hypotheses that are both supported by our current data but need further experimental investigation. Hypothesis 1 is that the TED itself functionally and directly contributes to the HAA of the Tibetan people. A good amount of evidence supports this: (1) the TED is ranked highly across the whole genome and has a frequency that is extremely differentiated between the Tibetans and all the other lowland populations; (2) the TED’s frequency is strongly correlated with altitude; (3) it has available functional annotation, including that it is close to EPAS1 and overlaps the H3K4me1 histone mark and two DNaseI signals; and (4) it shows an apparent signature of natural selection. Hypothesis 2 suggests that the TED is a simple tag of a functional variant and that the outstanding patterns we observed simply resulted from strong LD between the TED and the cryptic functional SNPs outside or inside EPAS1. Some evidence also supports this: (1) the TED showed extended homozygosity and strong LD with the region overlapping EPAS1, and (2) the TED was in strong LD with EPAS1 SNPs associated with a reduced blood concentration of hemoglobin, as reported in previous study.2
To test the two hypotheses, we suggest that human cell lines with the TED and without the TED should be cultured under hypoxia conditions and that the direction and magnitude of the expression changes of the two genes upstream and downstream of the TED (EPAS1 and TMEM247, respectively) should then be measured. The role of the TED in regulating EPAS1 expression can be largely confirmed if significant changes are observed in EPAS1 expression. Indirect evidence from previous studies has indicated that the effect of the TED is most likely to downregulate EPAS1 expression. For instance, the TED is in strong LD with the allele of the SNP rs13006131, which was reported to be associated with reduced hemoglobin concentration.2 Furthermore, the H3K4me1 histone mark within the TED is an enhancer, and EPAS1 expression was observed to be lower in Tibetans than in Han Chinese.51 Taken together, if the TED is a causal variant (hypothesis 1), the deletion would cause the loss of the enhancer and decrease EPAS1 expression, eventually reducing the hemoglobin concentration. For further distinguishing the two hypotheses (i.e., disassociating the TED from other variants inside or outside EPAS1), many more cell lines with different combinations (haplotypes) of the TED and the other variants in LD with the TED should be examined and compared for gene-expression changes, although a considerable amount of labor is expected to be required. In either case, we believe that the TED we identified in this study is worthy of further functional investigation. Such efforts would open a window into understanding the functional role of EPAS1 and provide a significant increase in knowledge about the molecular basis of HAA in Tibetans.
Acknowledgments
We thank three anonymous reviewers for their helpful comments on the manuscript. We thank Dr. Yundi Chen and his colleagues from Wuxi AppTec for their technical assistances during whole-genome sequencing. These studies were supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS; XDB13040100), by National Natural Science Foundation of China grants (91331204, 31171218, 31260263, and 31260252), and by the Science and Technology Commission of Shanghai Municipality (14YF1406800). S.X. is a Max-Planck Independent Research Group Leader and a member of the CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges the support of the National Program for Top-Notch Young Innovative Talents of the “Wanren Jihua” Project.
Published: June 11, 2015
Footnotes
Supplemental Data include nine figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.05.005.
Accession Numbers
The Database of Genomic Structural Variation (dbVar) accession number for the CNVs reported in this paper is dbVar: nstd111.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org
Database of Genomic Structural Variation (dbVar), http://www.ncbi.nlm.nih.gov/dbvar/
Database of Genomic Variants, http://dgv.tcag.ca/dgv/app/home
ENCODE, http://genome.ucsc.edu/ENCODE/
Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/
HapMap data, http://hapmap.ncbi.nlm.nih.gov/
HGDP-CEPH Genome Diversity panel database, http://www.cephb.fr/hgdp/index.php
HGDP SNP data, http://www.hagsc.org/hgdp/files.html
Korean Personal Genome Project (KPGP), http://opengenome.net/index.php/Main_Page
OMIM, http://www.omim.org/
Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
UCSC Genome Browser, http://genome.ucsc.edu/
WinXPCNVer, http://www.picb.ac.cn/PGG/resource.php
Supplemental Data
References
- 1.Beall C.M. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. USA. 2007;104(1):8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigham A., Bauchet M., Pinto D., Mao X., Akey J.M., Mei R., Scherer S.W., Julian C.G., Wilson M.J., López Herráez D. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 5.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng Y., Yang Z., Zhang H., Cui C., Qi X., Luo X., Tao X., Wu T., Ouzhuluobu, Basang Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 7.Xu S., Li S., Yang Y., Tan J., Lou H., Jin W., Yang L., Pan X., Wang J., Shen Y. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- 8.Xiang K., Ouzhuluobu, Peng Y., Yang Z., Zhang X., Cui C., Zhang H., Li M., Zhang Y., Bianba Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol. Biol. Evol. 2013;30:1889–1898. doi: 10.1093/molbev/mst090. [DOI] [PubMed] [Google Scholar]
- 9.Lorenzo F.R., Huff C., Myllymäki M., Olenchock B., Swierczek S., Tashi T., Gordeuk V., Wuren T., Ri-Li G., McClain D.A. A genetic mechanism for Tibetan high-altitude adaptation. Nat. Genet. 2014;46:951–956. doi: 10.1038/ng.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pineda Torra I., Jamshidi Y., Flavell D.M., Fruchart J.C., Staels B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol. Endocrinol. 2002;16:1013–1028. doi: 10.1210/mend.16.5.0833. [DOI] [PubMed] [Google Scholar]
- 11.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weischenfeldt J., Symmons O., Spitz F., Korbel J.O. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 13.Schlattl A., Anders S., Waszak S.M., Huber W., Korbel J.O. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. 2011;21:2004–2013. doi: 10.1101/gr.122614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mokhtar S.S., Marshall C.R., Phipps M.E., Thiruvahindrapuram B., Lionel A.C., Scherer S.W., Peng H.B. Novel population specific autosomal copy number variation and its functional analysis amongst Negritos from Peninsular Malaysia. PLoS ONE. 2014;9:e100371. doi: 10.1371/journal.pone.0100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou H., Li S., Yang Y., Kang L., Zhang X., Jin W., Wu B., Jin L., Xu S. A map of copy number variations in Chinese populations. PLoS ONE. 2011;6:e27341. doi: 10.1371/journal.pone.0027341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korn J.M., Kuruvilla F.G., McCarroll S.A., Wysoker A., Nemesh J., Cawley S., Hubbell E., Veitch J., Collins P.J., Darvishi K. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam H.Y., Mu X.J., Stütz A.M., Tanzer A., Cayting P.D., Snyder M., Kim P.M., Korbel J.O., Gerstein M.B. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat. Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weir B.S., Hill W.G. Estimating F-statistics. Annu. Rev. Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 23.Stephens M., Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandelt H.J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 25.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 27.Gautier M., Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 28.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tishkoff S.A., Reed F.A., Ranciaro A., Voight B.F., Babbitt C.C., Silverman J.S., Powell K., Mortensen H.M., Hirbo J.B., Osman M. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;11 doi: 10.1002/0471250953.bi1110s43. 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller C.A., Hampton O., Coarfa C., Milosavljevic A. ReadDepth: a parallel R package for detecting copy number alterations from short sequencing reads. PLoS ONE. 2011;6:e16327. doi: 10.1371/journal.pone.0016327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiffels S., Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 38.Park H., Kim J.I., Ju Y.S., Gokcumen O., Mills R.E., Kim S., Lee S., Suh D., Hong D., Kang H.P. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat. Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I., Maller J.B., Kirby A. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 40.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong C., Alkorta-Aranburu G., Basnyat B., Neupane M., Witonsky D.B., Pritchard J.K., Beall C.M., Di Rienzo A. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat. Commun. 2014;5:3281. doi: 10.1038/ncomms4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kel A.E., Gössling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P., Wellcome Trust Case Control Consortium Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Seeman P., Liu P., Weterman M.A., Gonzaga-Jauregui C., Towne C.F., Batish S.D., De Vriendt E., De Jonghe P., Rautenstrauss B. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am. J. Hum. Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon C.T., Tan T.Y., Benko S., Fitzpatrick D., Lyonnet S., Farlie P.G. Long-range regulation at the SOX9 locus in development and disease. J. Med. Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- 50.Scortegagna M., Morris M.A., Oktay Y., Bennett M., Garcia J.A. The HIF family member EPAS1/HIF-2alpha is required for normal hematopoiesis in mice. Blood. 2003;102:1634–1640. doi: 10.1182/blood-2003-02-0448. [DOI] [PubMed] [Google Scholar]
- 51.Petousi N., Croft Q.P., Cavalleri G.L., Cheng H.Y., Formenti F., Ishida K., Lunn D., McCormack M., Shianna K.V., Talbot N.P. Tibetans living at sea level have a hyporesponsive hypoxia-inducible factor system and blunted physiological responses to hypoxia. J. Appl. Physiol. 2014;116:893–904. doi: 10.1152/japplphysiol.00535.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.