Abstract
Orange carotenoid protein (OCP) is the photoactive protein that is responsible for high light tolerance in cyanobacteria. We studied the kinetics of the OCP photocycle by monitoring changes in its absorption spectrum, intrinsic fluorescence, and fluorescence of the Nile red dye bound to OCP. It was demonstrated that all of these three methods provide the same kinetic parameters of the photocycle, namely, the kinetics of OCP relaxation in darkness was biexponential with a ratio of two components equal to 2:1 independently of temperature. Whereas the changes of the absorption spectrum of OCP characterize the geometry and environment of its chromophore, the intrinsic fluorescence of OCP reveals changes in its tertiary structure, and the fluorescence properties of Nile red indicate the exposure of hydrophobic surface areas of OCP to the solvent following the photocycle. The results of molecular-dynamics studies indicated the presence of two metastable conformations of 3′-hydroxyechinenone, which is consistent with characteristic changes in the Raman spectra. We conclude that rotation of the β-ionylidene ring in the C-terminal domain of OCP could be one of the first conformational rearrangements that occur during photoactivation. The obtained results suggest that the photoactivated form of OCP represents a molten globule-like state that is characterized by increased mobility of tertiary structure elements and solvent accessibility.
Introduction
Carotenoids belong to a special class of organic compounds that are responsible for the physiological functions of many living systems. An important role of carotenoids is to protect the photosynthetic apparatus from reactive oxygen species (1,2). The xanthophyll cycle was found to play a key role in energy dissipation within the chlorophyll-containing antennae of algae and higher plants by nonphotochemical quenching (NPQ) (3–5). Cyanobacteria perform light harvesting by means of supramolecular complexes, called phycobilisomes, that consist of several types of phycobiliproteins (6–9). The utilization of water-soluble pigment-protein complexes (e.g., phycocyanin and allophycocyanin) for the construction of antennae allows cyanobacteria to effectively absorb light in the spectral region, where absorption of chlorophyll molecules is insignificant. However, this requires additional mechanisms that protect the cyanobacteria from excess excitation energy in antennae complexes. One mechanism introduces strong nonphotochemical quenching of the phycobilisome fluorescence (10–13). A specific 35 kDa water-soluble, carotenoid-binding protein, orange carotenoid protein (OCP), plays an essential role in this process (14–17). Numerous experiments performed both in vivo and in vitro have shown that OCP is a photoactive protein that is capable of reversibly changing its spectral and structural properties initiated by photon absorption (18–23). OCP acts as a light-intensity sensor in initiating the protective photocycle. Upon absorption of a blue-green light quantum OCP switches to the active form, which interacts with the phycobilisome and effectively quenches its fluorescence, thus reducing the amount of excitation energy that is transferred to the chlorophylls of the reaction centers. In addition, OCP has been found to be an effective quencher of singlet oxygen (24).
Results from x-ray crystallography have provided valuable information about the structure of OCP in its stable (noninteracting with phycobilisomes) form, which is referred to as the orange (OCPO) form (19,25–27). It was shown that OCP consists of two structural domains (the N- and C-terminal domains, hereafter referred to as the N- and C-domains), between which the 3′-hydroxyechinenone (hECN) carotenoid chromophore is bound (28–30). This chromophore is responsible for the protein’s absorption in the visible spectral region (10), which exhibits a red shift upon photon absorption and consequent OCP photoswitching (31). The photoactivated form of OCP is referred to as the red (OCPR) form. It was proposed that the photoactivity of OCP is due to specific carotenoid-protein interactions (32). Site-directed mutagenesis experiments have shown the essential role of tyrosine (Tyr) and tryptophan (Trp) residues in the photoactivity of OCP, as well as of a salt bridge between arginine and glutamic acid that stabilizes the interaction between the N- and C-domains (28,33). The facts that a 16 kDa red carotenoid protein (RCP) can be obtained from OCPO by removal of the C-domain, and that the resultant RCP is spectrally and functionally similar to OCPR, including its ability to effectively quench the fluorescence of phycobilisomes, raised the idea that OCP exhibits structural and functional modularity (29,34).
Interestingly, the structure of OCP was found to be similar to other photoactive proteins with a multistep reversible photocycle (21), such as photoactive yellow protein (PYP) (35) and light, oxygen, and voltage (LOV)-sensitive domain proteins. For instance, the mixed α-helical/β-sheet C-domain resembles the fold of the nuclear transport factor 2 (NTF2) and blue light using FAD (BLUF) domain-containing photosensors (32). However, the photocycle of OCP is significantly less studied compared with that of other photoactive proteins (36–43). This is especially notable regarding the conformational dynamics of OCP that follows its photoactivation, and the structural properties of OCPR.
Ultrafast spectroscopy techniques have enabled investigators to study the dynamics of the photoactivation and relaxation processes in OCP and to explore the pathways of energy dissipation in its orange and red forms (23,34,44–46). Excitation of OCP leads to the population of new carotenoid excited states. One of these states, which is populated shortly after excitation, is characterized by a pronounced charge transfer character with a 0.6 ps lifetime (23). It was assumed that π-stacking between aromatic amino acids in OCP and the rings of the hECN carotenoid act as a zipper that is opened upon photon absorption. Several works also indicated a release of the hECN carotenoid, i.e., a weakening of its interaction with the protein matrix after photoactivation of OCP, suggesting strong conformational rearrangements of the protein’s globule (29,47). However, no changes in the protein secondary structure of OCP were observed upon its photoactivation (29). Using a Hofmeister series of ions, King et al. (48) obtained OCP’s red form in vitro without constant illumination. This allowed them to measure the circular dichroism (CD) spectrum of the red form, which was characterized by significantly lower CD activity than the orange form, suggesting a lowering of asymmetry upon activation and weakening of protein-chromophore interactions.
In this work, we studied the kinetics of the photocycle of OCP in vitro by probing both the parameters of its hECN chromophore and the protein matrix. We performed the latter by using fluorescence spectroscopy, with the excitation of Trp residues in OCP as well as the hydrophobic probe Nile red (NR), bound to OCP, serving as a fluorescence reporter. The obtained results led us to conclude that the active signaling state of OCP (OCPR) is a molten globule-like structure that is characterized by high flexibility due to the lack of tertiary structure. We also performed resonance Raman spectroscopy experiments complemented by quantum-chemistry/molecular-dynamics (QC/MD) calculations to evaluate changes in the hECN chromophore geometry of the first conformational rearrangement of OCP upon its photoactivation.
Materials and Methods
Materials
Cells of the cyanobacterium Arthrospira maxima were grown in Zarrouk’s medium (49) at 30°C under luminescent lamps with continuous white-light illumination at 100 μmol photons m−2s−1. The cells with expressed OCP were collected by centrifugation for 15 min at 7000 g and then washed once and resuspended in 30 mL of 40 mM Tris-HCl buffer, pH 8.8 (buffer A). The suspension obtained was sonicated twice in an Ultrasonics Processor VCX130 (Sonics & Materials) at 40% intensity and centrifuged for 20 min at 12,000 g and 4°C to remove cell debris and other insoluble material. The supernatant was subjected to ammonium sulfate fractionation. The green protein pellet at 0–20% saturation of ammonium sulfate and membrane fragments were removed by 1 h of ultracentrifugation at 105,000 g. Ammonium sulfate was added to the blue supernatant for up to 40% of saturation and the dark-blue pellet was collected by centrifugation for 30 min at 12,000 g and 4°C. The supernatant was further supplied with ammonium sulfate for up to 60% of saturation, and after 30 min on ice was centrifuged to obtain a blue iridescent pellet and a rose-tinted supernatant. The blue pellet was dissolved in 15 mL of 1.2 M ammonium sulfate solution in buffer A. The resultant solution was clarified by 30 min centrifugation at 12,000 g and 4°C, and then subjected to hydrophobic interaction chromatography on a 5 mL HiTrap Phenyl-Sepharose (GE Healthcare) column using 1.2–0 M ammonium sulfate gradient on buffer A (10 column volumes) developed at a 2 mL/min flow rate at ambient temperature. Intense orange fractions, free from cytochrome C absorbance (maximum at ∼416 nm) and eluted before the blue peak, were collected and then dialyzed overnight against 2 L of buffer A, followed by dialysis against 1 L of 40 mM potassium phosphate buffer, pH 7.3, to obtain the final OCP solution, which exhibited the characteristic dark-adapted absorbance spectrum (Fig. 1 A).
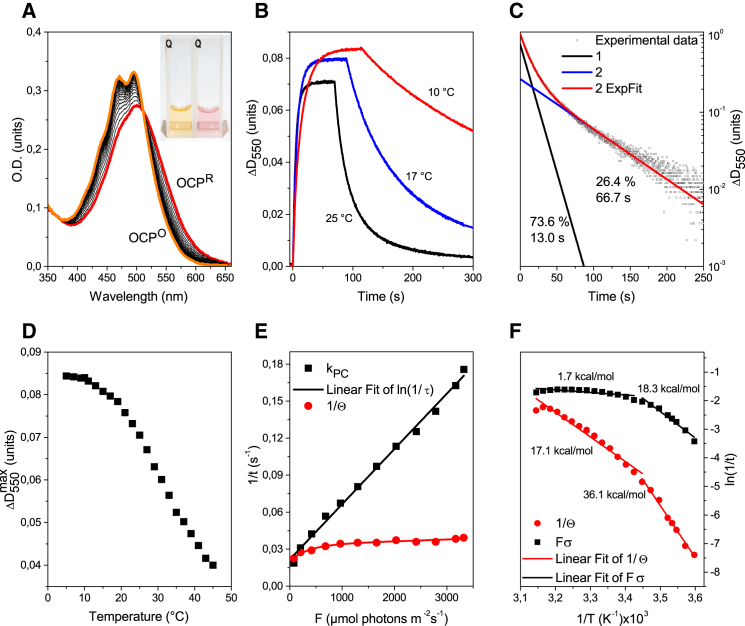
Figure 1.
(A) Visible absorption spectra of A. maxima OCPO, OCPR, and a set of intermediate states measured in a 1-cm path length cuvette at 20°C during OCPR → OCPO relaxation. Photoconversion was induced by illumination with 3000 μmol photons m−2s−1 of blue-green light. The inset shows a change in the sample’s color due to photoconversion. (B) Typical OCPO → OCPR → OCPO conversion curves measured as changes in optical density at 550 nm at 10°C, 17°C, and 25°C. (C) Approximation of OCPR → OCPO conversion measured at 28°C with a biexponential function, . (D) Temperature dependence of the photoconversion amplitude, measured as the maximum absorbance changes at 550 nm. (E) Dependency of OCPO → OCPR (kPC) and OCPR → OCPO rates on photon flux density . (F) Arrhenius plots for the calculated rate of OCPO → OCPR activation under 3000 μmol photons m−2s−1 of blue-green light (squares) and the subsequent relaxation rate in darkness (circles). Low- and high-temperature regions were approximated by linear functions to estimate the activation energies. For further explanation, see text below and Supporting Material. To see this figure in color, go online.
NR and α-lactalbumin (LALBA) from bovine milk were purchased from Sigma-Aldrich. Bovine serum albumin (BSA) was obtained from Thermo Scientific. All solutions for OCP purification were prepared using Milli-Q quality water (Millipore) and were filtrated before use.
Conditions of OCP illumination used to monitor photoconversion
In all experiments, a set of 2 W light-emitting diodes (LEDs) with maximum emission at 460 nm, delivering up to 10,000 μmol photons m−2s−1, was used for blue-green illumination of the samples (i.e., for OCPO → OCPR photoconversion). The OCPR → OCPO conversion occurs in darkness and consequently can be observed after the end of the illumination of the sample with blue-green light. Five independent preparations of the OCP sample were carried out. All experiments were performed in vitro in aqueous solutions of isolated OCP. Each experiment was repeated at least five times.
Absorption measurements
Absorption spectra were recorded using a Perkin Elmer Lambda-25 spectrophotometer. The kinetics of OCP photoconversion was measured as the change of optical density at 550 nm with 0.1 s time resolution, and the precision of the optical density measurement was 5 × 10−4. A band-pass filter FB450-40 (Thorlabs) was installed in the spectrophotometer to eliminate a possible influence of the blue-green illumination on the values of optical density measured at 550 nm.
Fluorescence measurements
Steady-state fluorescence measurements were performed using a FluoroMax-4 spectrofluorometer (Horiba Jobin Yvon). Samples were diluted to an optical density of ∼0.1 units at 280 nm to avoid inner filter effects and reabsorption. The Trp fluorescence of OCP was measured upon 295 nm excitation, and emission spectra were recorded in the 305–500 nm wavelength region. We also measured the kinetics of Trp fluorescence after the photocycle of OCP (OCPO → OCPR and OCPR → OCPO conversion). The kinetics was measured at the fixed wavelength (360 nm) upon 295 nm excitation with 0.1 s time resolution, and the widths of the excitation and emission slits were set to 1 and 15 nm, respectively.
The fluorescence emission of the hydrophobic probe NR was measured in OCP solution during the OCPO → OCPR and OCPR → OCPO conversion. The excitation wavelength was set to 560 nm and the emission was recorded in the 590–730 nm region. The fluorescence kinetics of NR after the OCP conversion was measured at a fixed wavelength (640 nm) upon 560 nm excitation with a 0.1 s time resolution, and the widths of the excitation and emission slits were set to 1 and 15 nm, respectively.
Fluorescence decay kinetics with a picosecond time resolution were collected with the use of a time- and wavelength-correlated single-photon counting setup (Becker & Hickl, Germany). Excitation was performed with 1) a 280 nm laser diode EPLED-280 (Edinburgh Instruments, Scotland) with a 700 ps pulse full width at half-maximum (FWHM), and 2) a 405 nm laser diode (InTop, Russia) delivering 13 pJ, 26 ps (FWHM) pulses, driven at a repetition rate up to 50 MHz. Excitation 1 was used for Trp fluorescence measurements, and excitation 2 was used for experiments with NR. Fluorescence decay curves were approximated by a sum of exponential decay functions with the SPCImage (Becker and Hickl, Germany) software package. To compare different decay curves, we calculated the average decay time according to the following expression:
where and are the lifetime and amplitude (normalized to unity: ) of the i-th fluorescence decay component, respectively. To obtain the time-integrated fluorescence spectra, the number of photons in each spectral channel was summed up.
Raman spectroscopy measurements
Raman spectra were obtained under continuous excitation at 532 nm. The laser beam was focused on a 0.1 mm glass capillary connected to a peristaltic pump (Heidolph, Germany). Raman scattered light was collected and subsequently imaged using a microscope-based system (NTMDT, Ntegra Spectra, Russia). OCPO Raman spectra were collected under rapid-flow conditions (2 mL/min) to minimize the effect of OCPO → OCPR photoconversion in the registered volume of the sample. The laser beam (532 nm, 5 mW) was focused in a 100 μm spot. Turning off the peristaltic pump led to an immediate conversion of the illuminated sample into the red form. Similar spectra were obtained under flow conditions with additional illumination of the sample with 460 nm blue LED (10,000 μmol photons m−2s−1) before Raman probing was conducted.
QC and MD modeling
We calculated the equilibrium conformation of an NR molecule using QC. For QC calculations, we used a FireFly software package (50) based on the source code of GAMESS US (51). We used the B3LYP5 functional for DFT calculations and the 6-311+G∗ basis set. We searched for the geometrical attachment site of NR to OCP using the Hex Protein Docking online service with a standard set of parameters (52). In this procedure, OCP and NR were considered as rigid bodies interacting through electrostatic and dispersion forces. The highest-scored OCP-NR complex structure represented one of the most likely arrangements for this protein-dye interaction. The initial coordinates of OCP were obtained from the Protein Data Bank (PDB ID: 1M98 for OCP from Arthrospira maxima (26)). In the set of the 50 highest-scored models, there were only a few in which the NR molecule was not in the central cavity of OCP.
MD simulations were performed using the GROMACS software package (53) and the OPLS-AA (54) force field with an integration step of 1 fs and no constraints of hydrogen atoms. The initial coordinates were taken from the PDB (ID: 3MG1 (19)), and electrostatic interactions were determined using the PME method with Fourier spacing of 0.12 nm. The cutoff for electrostatic and dispersion interactions was 1.25 nm for water molecules, and a TIP4P model was exploited. A model of the cofactor molecule (hECN) was assembled from the atoms of standard for OPLS-AA types, and no partial charges corrections were made. The model of OCP was dissolved in 27,639 water molecules in a 9 × 9 × 11 nm rectangular box, and a 100 ns MD simulation was performed in periodic boundary conditions at 300 K and 1 bar on the Lomonosov supercomputer at the Research Computing Center of Moscow State University.
Results and Discussion
Photocycle of OCP as revealed by absorption measurements
We studied the OCPO → OCPR → OCPO photoconversion using absorption spectroscopy according to well-established procedures (18). The absorption of OCP in the visible region is due to the presence of an hECN carotenoid chromophore, namely, the S0→S2 transition. In the stable OCPO form, the absorption band of hECN exhibits a vibronic structure, whereas upon photoactivation it becomes red shifted by ∼10 nm and loses vibronic structure (Fig. 1 A). This was interpreted as a consequence of the carotenoid-protein interaction, which is weakened in OCPR compared with OCPO (31). The kinetics of OCP photoactivation and relaxation is usually studied by measuring changes in optical density at ∼550 nm, where the absorption is mainly due to OCPR. Typical OCPO → OCPR → OCPO conversion curves measured at 10°C, 17°C, and 25°C are presented in Fig. 1 B. In this figure, the gradual increase of optical density represents the accumulation of OCPR caused by blue-green light illumination, and the decrease of optical density after switching off the illumination represents the spontaneous relaxation of OCPR to OCPO in darkness. The latter process is considered to be light independent (18). It should be noted that the absorption spectrum of OCPR is also characterized by a slightly increased optical density in the 300–320 nm region (Supporting Material), which may correspond to an all-trans to 15-cis isomerization of the carotenoid according to Saito et al. (55).
Essentially, the OCPR → OCPO relaxation in darkness was biexponential in all of the experiments, which is clearly seen on a semilogarithmic scale (Fig. 1 C). The characteristic relaxation times were 13 and 66 s with an amplitude ratio of 2:1 at room temperature. Interestingly, all works to date that have dealt with the measurement of OCP conversion using absorption spectroscopy reported a monoexponential character of the OCPR → OCPO relaxation kinetics, although they did not provide the results of the approximation. Biexponential relaxation may be specific to A. maxima OCP and should be tested for OCP from Synechocystis, since despite the great similarity of these species, they differ in certain properties (56). Another intriguing fact is that the ratio of the amplitudes of these two components in the OCP relaxation kinetics in darkness was not dependent on temperature (Supporting Material). It was recently shown that hECN binds to only 81% of A. maxima OCP, and ECN binds to the remaining 19% (56). Therefore, the presence of two different OCP fractions can probably explain the biexponential shape of the OCPR → OCPO conversion.
We also measured the concentration dependency of the kinetics of the optical density changes that occurred during the OCP conversion cycle. Neither the rates of OCPR relaxation nor the amplitudes of the components showed a dependence on the total concentration of OCP in solution in the 0.05–1 μM range. This indicates that bimolecular interactions have no influence on the kinetics of OCP relaxation as revealed by the changes in hECN absorption.
Gwizdala et al. (14) and Wilson et al. (18) reported several interesting features of the OCP conversion as revealed by changes in ΔD at 550 nm: 1) a dramatic dependence of the OCPR → OCPO relaxation rate on temperature (3000 s)−1 at 5°C versus (20 s)−1 at 30°C, which gives an activation energy for the relaxation process of ∼40 kcal/mol); 2) an absence of the temperature dependence of the OCPO → OCPR photoactivation rate (at constant blue-green light intensity); and 3) a significant dependence of the OCPO → OCPR photoactivation rate on the ionic strength, whereas no dependence on ionic strength was observed for the OCPR → OCPO relaxation rate. These facts clearly demonstrate the difference between the processes involved in the OCPO → OCPR and OCPR → OCPO transitions.
Wilson et al. (18) deduced the absence of a temperature dependence of the OCPO → OCPR photoconversion rate from the similar slopes of during the initial stage of photoconversion at high temperatures; however, according to a recent work (56), at low temperatures those rates will change. We applied a different procedure to estimate the rates of the OCPO → OCPR and OCPR → OCPO transitions, as described below.
The rate of photoconversion of OCP upon light absorption is equal to the product of light intensity (in cm−2s−1) and the photoconversion cross-section σ, which in turn is the product of the absorption cross-section σexc (in cm2) and photoconversion quantum yield β. Assuming = 3000 μmol photons m−2s−1, and taking the hECN absorption cross-section σexc = 4.6 × 10−16 cm2 (at 470 nm) (11), the photoconversion rate of ∼(10 s)−1 (the value obtained in our experiments at room temperature; see Fig. 1 E) results in β ≈ 10−3, which is in good agreement with the value published in Gorbunov et al. (11). Then, in darkness, OCPR spontaneously converts to OCPO with the characteristic rate .
The kinetics of OCPR accumulation (and consequently the changes in optical density at 550 nm) can be described by the following equation (see Supporting Material for details):
| (1) |
Hence, the rate of absorption changes that occur during the OCPO → OCPR conversion can be described by a specific rate , which depends on both and . It can be seen that Eq. 2 predicts the dependence of the maximum photoconversion level ΔDmax on the rate of OCPR relaxation , which exhibits a strong temperature dependence (Fig. 1 B):
| (2) |
This fact is illustrated in Fig. 1 D, where the dependence of the maximum optical density at 550 nm is shown as a function of temperature. Fig. 1 D clearly shows that the amplitude of the absorbance changes at low temperatures is significantly higher than that at high temperatures. This is in good agreement with experimental data obtained for OCP from Synechocystis (18).
We followed a standard procedure to determine the activation energy for the OCPO → OCPR photoconversion (at a constant photon flux density of 3000 μmol photons m−2s−1) and the subsequent OCPR → OCPO relaxation in darkness by measuring the changes in OCP absorbance at 550 nm in the temperature range of 5–45°C (Fig. 1 F). The time courses were approximated by a biexponential function, , to determine the average time constants for photoactivation and relaxation , respectively. Then, to obtain the value, we subtracted from the rate according to Eq. 2.
The temperature dependencies of and are presented in the form of Arrhenius plots in Fig. 1 F. It is known that a single rate-limited, thermally activated process results in a straight line within an Arrhenius plot, from which both the activation energy and the preexponential factor can be determined. However, our results clearly demonstrate that both the OCPO → OCPR and OCPR → OCPO transition rates show a positive convexity of their Arrhenius plots. Such a phenomenon can be explained by a decrease of activation energy at high temperatures (57). From the data obtained, we determined that the OCPO → OCPR photoactivation rate has a low activation energy of 1.7 kcal/mol at high temperatures and ∼18.3 kcal/mol at low temperatures, whereas the OCPR → OCPO relaxation rate has a strong temperature dependency, resulting in a 36.1 kcal/mol energy barrier at low temperatures (Fig. 1 F). Nonlinear Arrhenius plots of rate constants are typical for protein-folding processes in solution, where the heat capacity increment correlates with the hydrophobic surface exposure during the conformational transition (58). It should be noted that such nonlinear behavior was observed for NPQ and consequent fluorescence recovery in vivo (59).
Although the proposed model allows us to describe many experimental facts, some phenomena are outside of its framework; however, they should be mentioned. At a constant temperature (25°C), the rate of the OCPO → OCPR photoconversion depends linearly on the photon flux density of actinic illumination (Fig. 1 E), which is in agreement with Eq. 2. From Eq. 2, it is apparent that the rate of the OCPO→OCPR conversion is kpc = Fσ + 1/, and kpc should be equal to the rate of OCPR → OCPO relaxation 1/ at low photon flux density . A linear fit of the data (see Fig. 1 E) indicates that the kpc0 value is equal to ∼(50 s)−1, which is almost twofold faster than the rate of the OCPR → OCPO relaxation measured experimentally at this temperature at high F values. This discrepancy is due to the fact that the rate of OCPR → OCPO relaxation 1/ also depends on (Fig. 1 E). This fact cannot be described by the model presented in the Supporting Material, which suggests that the OCP relaxation in darkness is light independent (i.e., it does not depend on the initial conditions).
The intrinsic fluorescence of OCP exhibits the photocycle
Upon excitation at 295 nm of dark-adapted OCP, we observed a fluorescence emission with a maximum at 343 nm (Fig. 2 A) corresponding to the Trp fluorescence of OCP, which has five Trp residues. The position of the maximum of Trp fluorescence is strongly influenced by the polarity of its microenvironment (60–62): the more polar the environment, the more red shifted is the fluorescence emission. The maximum position at 343 nm observed for OCP (Fig. 2 A) is characteristic for Trp contacting with the structured water molecules (63). This is in good agreement with the results of x-ray crystallography (19) indicating that several water molecules (including structurally conserved) are in the immediate proximity of four Trp residues in the N-domain.
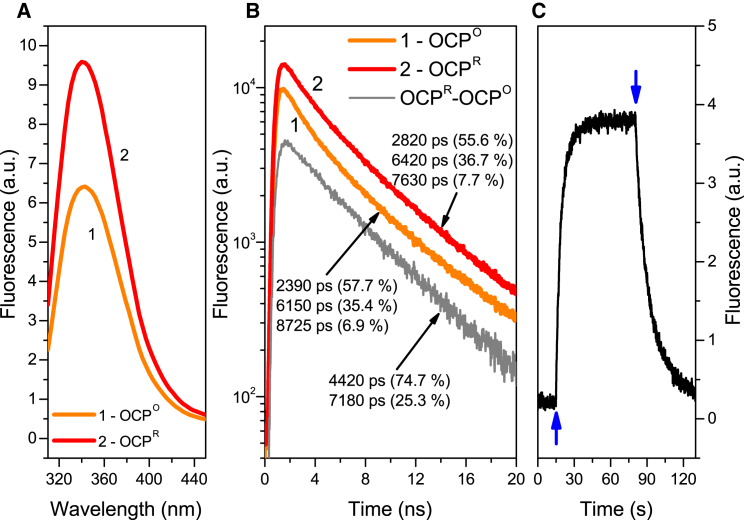
Figure 2.
(A) Trp fluorescence spectra of A. maxima OCPO (1) and OCPR (2), each measured in a quartz cuvette with a 1 cm path length. (B) Fluorescence decay kinetics of OCPO and OCPR registered at 340 nm, obtained under 280 nm excitation by a 700 ps FWHM laser diode at 2°C; the gray line represents the difference between OCPR and OCPO fluorescence kinetics. (C) OCPO → OCPR → OCPO photoconversion measured as Trp fluorescence changes at 340 nm using 3000 μmol photons m−2s−1 blue-green light intensity at 25°C. The arrows indicate the beginning and end of blue-green illumination. To see this figure in color, go online.
Exposure of the OCP aqueous solution to 3000 μmol photons m−2s−1 of the actinic 460 nm light caused a 30% increase of Trp fluorescence intensity, and the spectral band shape became blue shifted by ∼3 nm (Fig. 2 A; Supporting Material). The blue shift may be indicative of changes in the local Trp environment, as negatively charged residues near the pyrrole end or positive charges near the benzene end of the Trp ring system will shift the fluorescence maximum to shorter wavelengths (i.e., produce a blue shift), with the opposite configuration producing a red shift (60). The observed rise of the Trp fluorescence intensity could be described by a monoexponential function, with a characteristic time constant of (6 s)−1, which is in good agreement with the results obtained from absorption spectroscopy with the same actinic light intensity (Fig. 2 C). The relaxation kinetics of the Trp fluorescence changes (Fig. 2 C) after switching off the actinic light exhibited a biexponential character consistent with the results obtained from the absorption measurements (Fig. 1 C). In particular, the kinetics of OCP relaxation at room temperature could be fitted by a sum of two exponential decays with time constants = 12 s and = 60 s, which are in agreement with the OCPR → OCPO relaxation rates observed in absorption measurements (Fig. 1).
Trp fluorescence is characteristic of changes in a protein’s tertiary structure. Hence, the observed cyclic behavior of the Trp fluorescence intensity, which follows the photoactivation and relaxation of OCP, clearly indicates the conformational rearrangements of OCP. Interestingly, the kinetics of conformational changes monitored via Trp fluorescence changes (Fig. 2 C) was similar to the kinetics of OCP’s hECN spectral changes (Fig. 1 B). This fact demonstrates the principal role of carotenoid-protein interactions in the formation of OCP photocycle.
The increase of Trp fluorescence after the photoactivation of OCP indicates that the reduction of the quenching efficiency of the Trp excited state is likely to be caused by changes in the protein conformation. A possible explanation for this is that a reduction in the overall efficiency of excitation energy transfer (EET) from Trp residues to the hECN chromophore occurs during photoactivation. For instance, the Trp-290 and Trp-110 residues, which are required for OCP’s photoactivity, are in close proximity to the hECN carotenoid (33). One can assume that due to the OCPO → OCPR transition, the distance (or orientation) between hECN and Trp-290/Trp-110 changes significantly, resulting in a decrease of EET efficiency.
Previously, an EET from Trp to the chromophore was shown for another photoactive protein, photoactive yellow protein (PYP) (64). In that work, a gradual increase in the fluorescence lifetime of the single Trp residue in PYP upon photobleaching of its cofactor (apo-PYP) was demonstrated. It was found that although the fluorescence lifetime of Trp in the native (stable) form of PYP was 0.18 ns, it reached 4.8 ns upon bleaching of the chromophore. Upon photoactivation by 470 nm illumination, PYP was converted to its intermediate (signaling) state, for which the fluorescence lifetime of Trp was 0.82 ns. Notably, Gensch et al. (65) studied the photocycle in PYP using Trp fluorescence. We also observed the energy transfer from Trp residues to the chromophore of the red fluorescent protein mRFP1 using time-resolved and nonlinear fluorimetry (66). The EET rate in this case was k = 3.7 × 109 s−1. The energy transfer between Trp and chromophores with lower-lying energy levels is a typical situation in protein photophysics, and this effect could be expected in the case of OCP, where the Trp and hECN chromophore interaction, as well as its relative positions, changes significantly upon photoactivation.
To investigate EET, we performed time-resolved measurements of Trp fluorescence of OCP in the picosecond time domain (Fig. 2 B). The obtained results showed characteristic variations in the intrinsic fluorescence lifetime of OCP upon photoactivation with 460 nm actinic light. In both cases we observed fluorescence decay kinetics characterized by three components: 2390 ps (57.7%), 6150 ps (35.4%), and 8725 (6.9%)) in OCPO, and 2820 ps (55.6%), 6420 ps (36.7%), and 7630 (7.7%) in OCPR. The corresponding average fluorescence lifetimes are 4.15 and 4.51 ns for OCPO and OCPR, respectively, so the overall changes are on the order of 10%. However, the amplitudes of the fluorescence decay curves exhibited the same 30% increase as the intensity of the Trp emission in steady-state experiments (Fig. 2 A). A possible reason for this is the fast rate of EET, which might exceed the time resolution of our setup (∼100 ps). As the distances between certain Trp residues and hECN in OCP are significantly lower than typical values of Foerster radii for macromolecules (∼2 nm), we believe this explanation could be reasonable. On the other hand, the Trp-290 residue, which forms a hydrogen bond with the β-ring of hECN in the C-domain of OCPO, can be expected to be statically quenched due to ground-state interactions of Trp and hECN, whereas in OCPR, this residue may exhibit fluorescence. The fact that the difference between the fluorescence decay kinetics of the red and orange forms (Fig. 2 B) can be sufficiently described by a monoexponential function with a 5120 ps lifetime suggests that only one type of new fluorescing centers appears during photoconversion.
Transient changes in the hydrophobic properties of OCP as probed by NR fluorescence
In addition to the intrinsic Trp fluorescence, extrinsic fluorescence labels are extensively used to monitor conformational changes in proteins. One such fluorescent label is NR dye, which is a hydrophobic probe. This fluorescent label is bound into the hydrophobic cavities of a protein, and its photophysical properties (both absorption and fluorescence) are affected by the parameters of its microenvironment (67). In DMSO, NR exhibits a high fluorescence quantum yield and its fluorescence decays monoexponentially with a lifetime τ = 4.2 ns (Fig. 3 B). An almost identical fluorescence decay kinetics was observed upon addition of NR to an aqueous solution of the 66.5 kDa protein BSA, which is characterized by the presence of several hydrophobic sites that enable binding of the dye (data not shown). In contrast, NR fluorescence is significantly quenched in PBS buffer. As a result, the fluorescence lifetime of NR in this medium is almost 10-fold shorter than that in BSA or DMSO. Almost the same low fluorescence quantum yield of NR was detected in a solution of the 14.1 kDa protein LALBA (Supporting Material), which possesses a low number of hydrophobic regions. Hence, NR could serve as a good tool for monitoring conformational changes associated with alterations of the protein’s hydrophobicity (67,68). This approach was previously applied to investigate the photocycle in PYP (35).
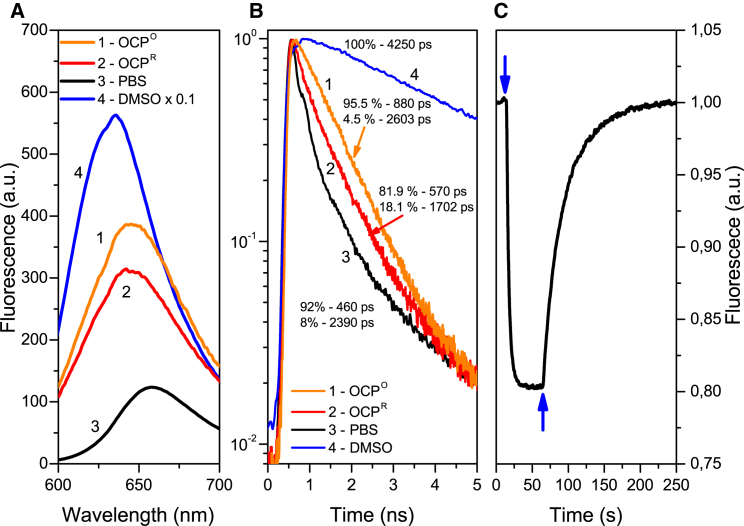
Figure 3.
(A and B) NR fluorescence spectra (A) and fluorescence decay kinetics (B) in A. maxima OCPO (1) and OCPR (2). The molar ratio of NR to OCP was equal to one. Corresponding curves for NR fluorescence in DMSO (3) and PBS buffer (4) are presented in blue and black, respectively. Note that the fluorescence spectrum of NR in DMSO was normalized (×0.1) to fit in the graph. (C) OCPO → OCPR → OCPO photoconversion measured as NR fluorescence changes at 640 nm using 3000 μmol photons m−2s−1 blue-green light intensity at 25°C. The arrows indicate the beginning and end of blue-green illumination. To see this figure in color, go online.
Here, we sought to determine whether NR can bind to OCP, and whether its fluorescence is sensitive to photoconversion of OCP. After adding OCPO to an NR solution in PBS buffer, we observed an increase of NR fluorescence intensity, indicating that NR was interacting with OCPO, and presumably binding to the hydrophobic site(s) of OCP. We found that the OCPO-NR complexes exhibit a biexponential fluorescence decay, with decay time constants = 880 ps (95.5%) and = 2600 ps (4.5%) (Fig. 3 B, 1). The presence of two components in the fluorescence decay kinetics may indicate the existence of multiple NR binding sites in OCP or partial binding of NR to OCP.
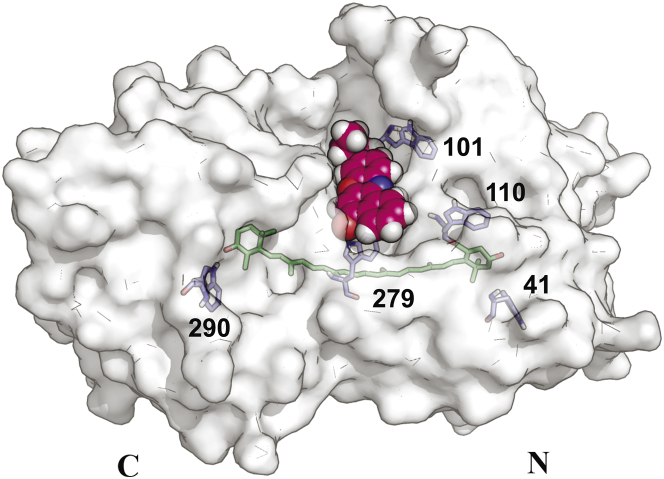
To find the most probable structure of the OCPO-NR complex, we used molecular docking methods. The docking results obtained from these calculations are presented in Fig. 4. It can be seen from this figure that NR most likely binds inside the cavity between the C- and N-domains of OCP, close to several hydrophobic amino acid residues (Trp-101, -110, and -279) and Arg-155-Glu-246, which form a salt bridge between the N- and C-domains of OCP and are simultaneously accessible for bulk water. Upon binding of NR, OCP remained photoactive, as confirmed by absorption spectroscopy and Trp fluorescence measurements.
Figure 4.
Probable position of NR dye inside the central cavity of OCPO from A. maxima OCP (PDB ID: 1M98) according to the docking procedure (see Materials and Methods). The protein is shown in surface representation, the NR molecule is shown as spheres, and Trp residues are labeled corresponding to A. maxima amino acid numbering. To see this figure in color, go online.
As in the case of intrinsic Trp fluorescence, reversible changes of NR fluorescence occurred upon blue-green illumination and subsequent relaxation in darkness. During OCP photoactivation, we observed a decrease of NR fluorescence accompanied by a significant decrease of the two aforementioned fluorescence lifetimes to = 570 ps (81.9%) and = 1700 ps (18.1%) (Fig. 3 B, 2). After switching off the actinic blue-green light, we observed a slow increase of NR fluorescence intensity, which reached the initial level measured before the blue-green illumination (Fig. 3 C). It should be noted that no photo-induced changes were detected in NR solutions in DMSO, or in mixtures of NR-BSA or NR-LALBA. Moreover, in the OCP solution, the kinetics of the NR fluorescence recovery in darkness was biexponential (with the same time constants) as in the case of the OCPR-OCPO conversion measured using absorption spectroscopy and Trp fluorescence. We can conclude that the observed correlation of NR fluorescence with the kinetic parameters of the photocycle of OCP indicates changes in the hydrophobicity of the NR environment. These changes observed upon photoactivation most likely suggest disassembly of the tentative NR-accepting cavity (Fig. 4) and concomitant exposure of multiple OCP hydrophobic areas that do not bind NR.
Hendriks et al. (35) observed similar effects for another photoactive protein, PYP, and concluded that the signaling state of PYP is molten-globule like. The term “molten globule” refers to protein folding intermediates, which have a secondary structure similar to that observed in the native state, whereas the tertiary structure is not established (69). The molten-globule state is usually characterized by an increase of the protein’s effective volume and an exposure of hydrophobic regions to solution due to a folding disorder (58).
Using gel filtration, Leverenz et al. (29) recently showed that although OCPR has a larger volume than OCPO, they have almost identical CD spectra in the far-UV, which is indicative of an unaltered secondary structure. Moreover, it was shown that in the visible region, the CD spectra of the orange and red forms are different, indicating a less ordered structure of the chromophore in the red form (48). Considering these facts together with the hydrophobicity changes reported by NR, one can hypothesize that the red form of OCP itself represents a molten-globule structure. This is in line with the fact that the signaling state of PYP is also considered to be a molten globule (35).
The destruction of the Arg-155-Glu-246 salt bridge and other photo-induced changes can lead to specific movements of the N- and C-domains of OCP and to an increase in OCP’s flexibility. The cyclic changes in the hydrophobicity and overall geometry of OCP seem to be crucial for providing a microenvironment around the hECN cofactor, which is required for implementing NPQ of phycobilisome fluorescence.
Resonance Raman spectra and MD of OCP
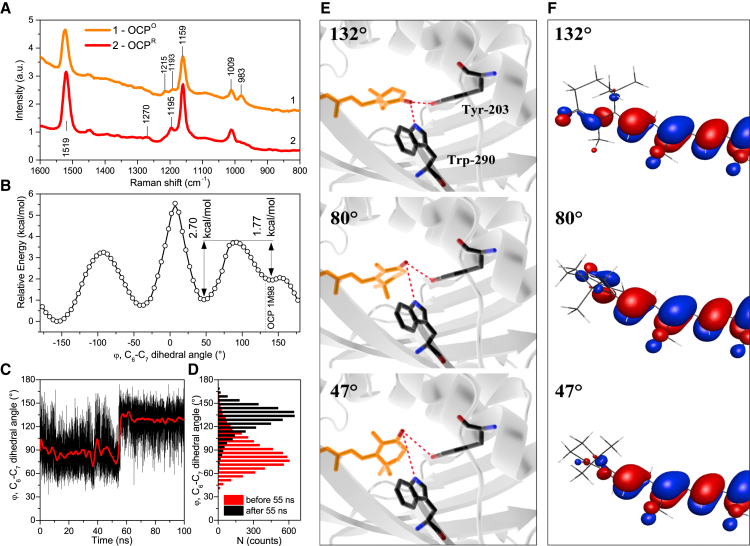
Resonance Raman spectroscopy provides important information about the structure and conformation of the OCP chromophore; therefore, we used this method to study photoconversion. Resonance Raman spectra of OCPO and OCPR are presented in Fig. 5 A. The most striking feature that distinguishes the Raman spectra of OCPO and OCPR (Fig. 5) is the large difference in the intensity of the 983 cm−1 band, which is related to a hydrogen-out-of-plane (HOOP) wagging mode, as shown previously (29). Application of the density functional theory (DFT) demonstrated that the intensity of the ν4 vibration (983 cm−1) is associated with distortion of the molecular conformation from a planar configuration. Moreover, it was previously shown that ν4 is sensitive to configuration changes induced by a β-ring rotation around the C6-C7 bond (70–72). For β-carotene, it was shown that an increase of the ν4 band intensity indicates that the conjugation between the β-ionylidene rings and the polyene chain is moderately strong (71). It should be noted that the hydroxyl substitution has almost no effect on the rotational energy profile of the β-ionylidene ring (70); therefore, these kinds of ν4 band intensity changes are probably not specific to β-carotene and could also apply to hECN. According to Schlucker et al. (72), β-carotene is characterized by the maximum intensity of the ν4 band at C6-C7 dihedral angle (φ) values equal to 40° and 135°, and the minimum intensity of the ν4 oscillations corresponds to 90° and 180° of β-ionylidene ring rotation. From the OCP crystal structure (PDB ID: 1M98), one can determine that the dihedral angle (φ) between the polyene chain and the β-ring within the C-domain of OCP is equal to 132°. According to theoretical calculations (71), rotation from 132° to 80–90° can result in an approximately fivefold decrease of the 983 cm−1 band intensity for β-carotene. Such changes in the 983 cm−1 band intensity were observed in our experiments (Fig. 5 A) and in a previous study (29).
Figure 5.
(A) Resonance Raman spectra of OCPO (1) and OCPR (2) recorded upon excitation by a 532 nm laser. Spectra are shifted along the y axis for better comparison. (B) Relation between the C6-C7 dihedral angle (φ) and the potential energy of hECN. (C) Results of the OCP MD simulations are presented as changes in the dihedral angle (φ) between the polyene chain and 4-keto-β-ionylidene ring of the hECN molecule in the C-domain of OCP in time. (D) Distributions of C6-C7 dihedral-angle (φ) values during the first 55 ns and after 45 ns of MD simulation. (E) Different hECN conformations corresponding to dihedral angles (φ) equal to 132°, 80°, and 47° between the polyene chain and β-ring of hECN in the C-domain of OCPO (based on data from PDB ID: 1M98). Note the difference between the distances of hECN’s carbonyl oxygen, Trp-290, and Tyr-203. (F) Schematic representation of the highest occupied molecular orbital corresponding to C6-C7 dihedral angles (φ) of 132°, 80°, and 47°. To see this figure in color, go online.
According to our calculations (Fig. 5 B), the hECN molecule is characterized by a local minimum of the potential energy for rotation around C6-C7 at 130°. A low (1.77 kcal/mol) energy barrier exists for a β-ring rotation from 130° to 50° (Fig. 5 B), which is in good agreement with the value of activation energy (1.7 kcal/mol at high temperatures) estimated from the absorption changes during the OCPO → OCPR photoconversion (Fig. 1 F). This value is lower than the energy of a single hydrogen bond (3–5 kcal/mol) (73). In fact, hydrogen bonds between Trp-290/Tyr-203 and the carbonyl oxygen of the hECN molecule exist (25,26,33), which explains the absence of a free β-ring rotation in OCPO and the pronounced vibronic structure of its absorption spectrum. However, the energy of 532 nm quanta is equal to 53.5 kcal/mol, which is sufficient to rotate the β-ring and consequently break the hydrogen bonds. Such a rotation may instantly increase the distance between hECN’s carbonyl oxygen and Trp-290 and Tyr-203 (Fig. 5) from approximately 2.9 to 6.6 Å, and from 2.6 to 4.6 Å, respectively. This can lead to significant changes in the efficiency of EET from Trp to hECN, resulting in an increase in Trp fluorescence (see Fig. 2) and a blue shift of Trp fluorescence (Supporting Material) due to changes in the local charge distribution of Trp-290 (Fig. 5 E). In addition, such a rotation changes the conjugation of the π-orbitals in the ionylidene ring and the π-orbitals in the polyene chain. The observed decrease in the conjugation length (π-electron system; Fig. 5 F) could explain the changes in the 983 cm−1 band intensity, in good agreement with the data presented in Liu et al. (71). However, such a decrease in the conjugation length cannot explain the red shift of OCPR hECN S0→S2 absorption, as it occurs in other carotenoid-binding proteins (74,75). Changes in S0→S2 absorption probably are induced by other factors, such as carotenoid environment and bending of the polyene chain. Thus, it seems probable that the rotation of the β-ring in the C-domain is the first conformational step in the reaction sequence that leads toward the red form of OCP.
Another distinction between the Raman spectra of OCPO and OCPR was found in the 1100–1300 cm−1 spectral range, the so-called fingerprint region of the carotenoid, where weak lines that are sensitive to both the nature of the end groups and the chain conformation are concentrated. Upon photoactivation of OCP, we observed an increase in the ratio between the bands at 1195 cm−1 and 1159 cm−1, a parameter that is characteristic for an s-trans to 7-cis isomerization, i.e., a rotation of the β-ring (76).
However, Raman spectroscopy only allows one to detect changes in certain steady-state configurations of hECN; it does not allow one to determine the C6-C7 dihedral angle (φ) between the polyene chain and the β-ring, because several equivalent configurations can result in a low intensity of the 983 cm−1 band. To solve this problem and find these angles, we used MD simulations and studied the behavior of the end groups of the hECN molecule in OCP. hECN in the N-domain of OCP exhibited one metastable configuration (at least on a 100 ns timescale) with a dihedral angle between the polyene chain and β-ring equal to −50°, which is close to its conformation in the crystal (Supporting Material). Simultaneously, in the C-domain we observed two metastable positions of the 4-keto-β-ionylidene ring corresponding to φ values equal to 80° and 130° (Fig. 5 D). Surprisingly, a 130° dihedral angle was obtained after a 60 ns period and subsequently remained constant (Fig. 5 C). This conformation probably corresponds to OCPO. Another conformation (φ = 80°) was obtained almost immediately after the onset of the calculations and remained stable for 60 ns. This indicates that in the beginning of the in silico experiment, the structure of OCP was somehow destabilized, probably due to hydration, which caused a rotation of the 4-keto-β-ionylidene ring in the C-domain of OCP. Definitely, one cannot claim that the configuration of OCP characterized by the 80° dihedral angle between the polyene chain and the 4-keto-β-ionylidene ring directly corresponds to the OCPR. It is obvious that the red form should be characterized by a wide range of structural changes not only in hECN but also in the OCP apoprotein. However, such small rearrangements of hECN (acting as the zipper) upon photon absorption can induce the appearance of new degrees of freedom, which are crucial for formation of the red signaling state.
Conclusions
OCP is known as a photoactive protein whose active form is capable of binding specifically to phycobiliproteins present in the light-harvesting antennae of cyanobacteria (i.e., phycobilisomes) (19,21,33,56,59,77). This suggests that the photoactivation process must be accompanied by significant structural rearrangements of the OCP structure. To date, however, most studies have focused on light-induced conformational changes of OCP in vitro and changes in the properties of the hECN chromophore (i.e., its electronic structure as revealed by ultrafast (23,31,34,44–46) and absorption spectroscopy (18), and its geometry as revealed by Raman (18,29) and CD (48) spectroscopy), and only a few experiments have dealt with the conformational changes of the apoprotein part of OCP (47). It has been frequently argued that one of the structural determinants underlying the photoactivity of OCP is the interaction between the hECN chromophore and aromatic amino acid residues in OCP. Hence, the parameters of OCP’s structural changes could provide important insights into the formation of its photocycle.
The kinetics of the photocycle of OCP is usually studied by examining the changes in its absorption in the visible area of the spectrum, which characterize changes in the electronic structure, environment, and geometry of the hECN chromophore (Fig. 1). Here, we obtained the kinetic characteristics of OCP’s photocycle using two other methods, which revealed the changes in the protein’s structure. It was shown that both the intrinsic (Trp) fluorescence of OCP (Fig. 2) and the fluorescence of NR (Figs. 3 and 4), incorporated into OCP’s structure, exhibit cyclic behavior similar to that observed in OCP’s (i.e., hECN’s) absorption spectrum. The changes in Trp fluorescence that occur after photoconversion and relaxation of OCP are indicative of changes in the protein’s tertiary structure, and the changes in NR fluorescence reveal the transient exposure of hydrophobic regions of OCP to solvent. Together with the facts that 1) the far-UV CD and Raman spectra of OCP indicate an unchanged secondary structure of the protein in both stable and active forms, and 2) the gel-filtration experiments demonstrate an enhanced volume of OCP in its active form (29), our results suggest that the active OCPR form assumes a molten-globule-like state. The molten-globule state is the intermediate of protein folding, which has a secondary structure similar to the native form, whereas the tertiary structure in this state is not established. A molten globule is typically characterized by an enhanced volume of the protein and an increased exposure of hydrophobic regions to solvent (58). A molten globule-like structure was previously suggested for another photoactive protein, PYP, in a study that used NR as a fluorescent probe (35). Hence, we demonstrated that the photocycle of OCP has some features in common with that of PYP. Moreover, a recent work (52) showed that the addition of chaotropic ions to OCP solution can lead to its photoactivation due to conformational changes and increased entropy, which is in fair agreement with the molten globule-like character of OCPR. Notably, a similar effect was also observed for PYP, in that the addition of chaotropes promoted the photocycle (78). In addition, it was shown that photo-induced changes disrupt the interactions of an α-helix from the N-domain with a β-sheet from the C-domain, leading to destabilization of the central interface of the latter (47). We assume that the results of our experiments with NR are associated with such rearrangements of the cavity between the N- and C-domains, and provide direct evidence of disorganization of the tertiary structure.
The measurements of the changes in the absorption spectrum of OCP, as well as its intrinsic fluorescence and NR fluorescence, all of which follow its photocycle in time, clearly demonstrate that the relaxation (OCPR → OCPO conversion) process in darkness exhibits two temporal components with a 2:1 amplitude ratio (Fig. 1 C) independently of temperature. These two components in the relaxation kinetics of OCP could be attributed to either 1) the presence of an intermediate state in the OCPR → OCPO conversion, or 2) the existence of two subpopulations of OCP forms in the solution. The absence of a temperature dependence of the amplitude ratio makes assumption 1 improbable, because temperature changes should result in changes of the populations of OCPR and the intermediate state, leading to an alteration of amplitude ratio. According to mass-spectroscopy data (77), the stable OCPO form appears as a homodimer, whereas upon photoactivation, a dimer-monomer transition takes place. Hence, the two subpopulations in assumption 2 could hypothetically be the monomeric and dimeric forms of OCP. However, the absence of a dependence of the OCPR-OCPO conversion rates on the OCP concentration in solution, as shown by absorption spectroscopy data, suggests that the dimerization process is not manifested in the changes of OCP’s absorption spectrum. Therefore, the formation of OCPO dimers should occur on timescales exceeding those of spectral changes due to the OCPR-OCPO transition. Another possible explanation may reflect multiple subpopulations of OCP with different chromophores, which are due to the ability of apoprotein to bind both hECN and ECN (56).
The Arrhenius plot of the rates of two components in the OCPR-OCPO relaxation kinetics demonstrated a nonlinear character with positive convexity. Such behavior is typical for protein folding rates and could be explained by the change of the rate-limiting step at high temperatures, which could be due to the increased role of an entropy factor. We also measured the temperature dependence of the OCP photoconversion quantum yield, which provided an ∼1.7 kcal/mol activation barrier at high temperatures, consistent with the barrier of the β-ring rotation in hECN obtained from DFT calculations. The results of the MD simulations indicate the presence of two metastable conformations of hECN, which is consistent with the characteristic changes of the Raman spectra (Fig. 5). Rotation of the 4-keto-β-ionylidene ring may cause a disruption of a hydrogen bond between the oxygen of hECN and nitrogen in the pyrrole ring of Trp-290, which may explain the observed increase in Trp fluorescence (Fig. 2). Those observations suggest that the first step in OCP conversion is the rotation of the β-ionylidene ring in the hECN chromophore in the C-domain after photon absorption, which serves to initiate subsequent larger protein-assisted dynamics due to continuous pigment-protein interactions.
Author Contributions
E.G.M., E.A.S., and N.N.S. performed research and wrote the manuscript. D.V.Z., E.Y.P., G.V.T., K.E.K., and G.S.B. performed research. F.-J.S. and T.F. analyzed data. V.V.F. and A.B.R. designed research. V.Z.P. designed research, analyzed data, and wrote the manuscript.
Acknowledgments
The authors thank Professor N.B. Gusev for help with OCP purification and valuable advice.
This work was supported by the Russian Foundation for Basic Research (14-04-01536A, 15-04-01930A, and 14-05-91177-Gfen-A to V.Z.P., E.G.M., and VV.F.), the Russian Scientific Foundation (14-17-00451 to V.V.F., E.A.S., G.S.B., and E.G.M.), and the Russian Ministry of Education and Science (MK-5949.2015.4 to E.G.M., G.V.T., and K.E.K.). E.M. received a fellowship from the Dynasty Foundation. This work was also supported by the German Federal Ministry of Education and Research within the framework of RUS 10/026 and 11/014, and COST within the framework of COST action MP1205.
Editor: Elizabeth Rhoades.
Footnotes
Supporting Materials and Methods, seven figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00662-1.
Supporting Material
References
- 1.Frank H.A., Cogdell R.J. Carotenoids in photosynthesis. Photochem. Photobiol. 1996;63:257–264. doi: 10.1111/j.1751-1097.1996.tb03022.x. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki S., Mizuguchi K., Shimizu H. A novel astaxanthin-binding photooxidative stress-inducible aqueous carotenoprotein from a eukaryotic microalga isolated from asphalt in midsummer. Plant Cell Physiol. 2013;54:1027–1040. doi: 10.1093/pcp/pct080. [DOI] [PubMed] [Google Scholar]
- 3.Härtel H., Lokstein H., Rank B. Kinetic studies on the xanthophyll cycle in barley leaves (influence of antenna size and relations to nonphotochemical chlorophyll fluorescence quenching) Plant Physiol. 1996;110:471–482. doi: 10.1104/pp.110.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horton P., Ruban A.V., Walters R.G. Regulation of light harvesting in green plants (indication by nonphotochemical quenching of chlorophyll fluorescence) Plant Physiol. 1994;106:415–420. doi: 10.1104/pp.106.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbonera D., Gerotto C., Morosinotto T. NPQ activation reduces chlorophyll triplet state formation in the moss Physcomitrella patens. Biochim. Biophys. Acta. 2012;1817:1608–1615. doi: 10.1016/j.bbabio.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Liu H., Zhang H., Blankenship R.E. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science. 2013;342:1104–1107. doi: 10.1126/science.1242321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian L., Gwizdala M., van Amerongen H. Picosecond kinetics of light harvesting and photoprotective quenching in wild-type and mutant phycobilisomes isolated from the cyanobacterium Synechocystis PCC 6803. Biophys. J. 2012;102:1692–1700. doi: 10.1016/j.bpj.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maksimov E.G., Schmitt F.J., Rubin A.B. Anomalous temperature dependence of the fluorescence lifetime of phycobiliproteins. Laser Phys. Lett. 2013;10:055602. [Google Scholar]
- 9.Maksimov E.G., Kuzminov F.I., Paschenko V.Z. Photosystem 2 effective fluorescence cross-section of cyanobacterium Synechocystis sp. PCC6803 and its mutants. J. Photochem. Photobiol. B. 2011;104:285–291. doi: 10.1016/j.jphotobiol.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Rakhimberdieva M.G., Stadnichuk I.N., Karapetyan N.V. Carotenoid-induced quenching of the phycobilisome fluorescence in photosystem II-deficient mutant of Synechocystis sp. FEBS Lett. 2004;574:85–88. doi: 10.1016/j.febslet.2004.07.087. [DOI] [PubMed] [Google Scholar]
- 11.Gorbunov M.Y., Kuzminov F.I., Falkowski P.G. A kinetic model of non-photochemical quenching in cyanobacteria. Biochim. Biophys. Acta. 2011;1807:1591–1599. doi: 10.1016/j.bbabio.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Stadnichuk I.N., Yanyushin M.F., Pashchenko V.Z. Quenching of phycobilisome fluorescence by orange carotenoid protein. Dokl. Biochem. Biophys. 2011;439:167–170. doi: 10.1134/S1607672911040053. [DOI] [PubMed] [Google Scholar]
- 13.Tian L., van Stokkum I.H., van Amerongen H. Site, rate, and mechanism of photoprotective quenching in cyanobacteria. J. Am. Chem. Soc. 2011;133:18304–18311. doi: 10.1021/ja206414m. [DOI] [PubMed] [Google Scholar]
- 14.Gwizdala M., Wilson A., Kirilovsky D. In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the orange carotenoid protein in synechocystis PCC 6803. Plant Cell. 2011;23:2631–2643. doi: 10.1105/tpc.111.086884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirilovsky D., Kerfeld C.A. The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta. 2012;1817:158–166. doi: 10.1016/j.bbabio.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Boulay C., Abasova L., Kirilovsky D. Occurrence and function of the orange carotenoid protein in photoprotective mechanisms in various cyanobacteria. Biochim. Biophys. Acta. 2008;1777:1344–1354. doi: 10.1016/j.bbabio.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Boulay C., Wilson A., Kirilovsky D. Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2010;107:11620–11625. doi: 10.1073/pnas.1002912107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson A., Punginelli C., Kirilovsky D. A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl. Acad. Sci. USA. 2008;105:12075–12080. doi: 10.1073/pnas.0804636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson A., Kinney J.N., Kerfeld C.A. Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria. J. Biol. Chem. 2010;285:18364–18375. doi: 10.1074/jbc.M110.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadnichuk I.N., Yanyushin M.F., Paschenko V.Z. Fluorescence quenching of the phycobilisome terminal emitter LCM from the cyanobacterium Synechocystis sp. PCC 6803 detected in vivo and in vitro. J. Photochem. Photobiol. B. 2013;125:137–145. doi: 10.1016/j.jphotobiol.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Kirilovsky D., Kerfeld C.A. The orange carotenoid protein: a blue-green light photoactive protein. Photochem. Photobiol. Sci. 2013;12:1135–1143. doi: 10.1039/c3pp25406b. [DOI] [PubMed] [Google Scholar]
- 22.Stadnichuk I.N., Yanyushin M.F., Paschenko V.Z. Site of non-photochemical quenching of the phycobilisome by orange carotenoid protein in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta. 2012;1817:1436–1445. doi: 10.1016/j.bbabio.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 23.Berera R., van Stokkum I.H., van Grondelle R. The photophysics of the orange carotenoid protein, a light-powered molecular switch. J. Phys. Chem. B. 2012;116:2568–2574. doi: 10.1021/jp2108329. [DOI] [PubMed] [Google Scholar]
- 24.Sedoud A., López-Igual R., Kirilovsky D. The cyanobacterial photoactive orange carotenoid protein is an excellent singlet oxygen quencher. Plant Cell. 2014;26:1781–1791. doi: 10.1105/tpc.114.123802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerfeld C.A., Wu Y.P., Yeates T.O. Crystals of the carotenoid protein from Arthrospira maxima containing uniformly oriented pigment molecules. Acta Crystallogr. D Biol. Crystallogr. 1997;53:720–723. doi: 10.1107/S0907444997006999. [DOI] [PubMed] [Google Scholar]
- 26.Kerfeld C.A., Sawaya M.R., Yeates T.O. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure. 2003;11:55–65. doi: 10.1016/s0969-2126(02)00936-x. [DOI] [PubMed] [Google Scholar]
- 27.Sutter M., Wilson A., Kerfeld C.A. Crystal structure of the FRP and identification of the active site for modulation of OCP-mediated photoprotection in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2013;110:10022–10027. doi: 10.1073/pnas.1303673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson A., Gwizdala M., Kirilovsky D. The essential role of the N-terminal domain of the orange carotenoid protein in cyanobacterial photoprotection: importance of a positive charge for phycobilisome binding. Plant Cell. 2012;24:1972–1983. doi: 10.1105/tpc.112.096909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leverenz R.L., Jallet D., Kerfeld C.A. Structural and functional modularity of the orange carotenoid protein: distinct roles for the N- and C-terminal domains in cyanobacterial photoprotection. Plant Cell. 2014;26:426–437. doi: 10.1105/tpc.113.118588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punginelli C., Wilson A., Routaboul J.M., Kirilovsky D. Influence of zeaxanthin and echinenone binding on the activity of the orange carotenoid protein. Biochim. Biophys. Acta. 2009;1787::280–288. doi: 10.1016/j.bbabio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Polívka T., Kerfeld C.A., Sundström V. Spectroscopic properties of the carotenoid 3′-hydroxyechinenone in the orange carotenoid protein from the cyanobacterium Arthrospira maxima. Biochemistry. 2005;44:3994–4003. doi: 10.1021/bi047473t. [DOI] [PubMed] [Google Scholar]
- 32.Kerfeld C.A. Structure and function of the water-soluble carotenoid-binding proteins of cyanobacteria. Photosynth. Res. 2004;81:215–225. doi: 10.1023/B:PRES.0000036886.60187.c8. [DOI] [PubMed] [Google Scholar]
- 33.Wilson A., Punginelli C., Kirilovsky D. Essential role of two tyrosines and two tryptophans on the photoprotection activity of the orange carotenoid protein. Biochim. Biophys. Acta. 2011;1807:293–301. doi: 10.1016/j.bbabio.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Chábera P., Durchan M., Polívka T. Excited-state properties of the 16kDa red carotenoid protein from Arthrospira maxima. Biochim. Biophys. Acta. 2011;1807:30–35. doi: 10.1016/j.bbabio.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Hendriks J., Gensch T., van Thor J.J. Transient exposure of hydrophobic surface in the photoactive yellow protein monitored with Nile red. Biophys. J. 2002;82:1632–1643. doi: 10.1016/S0006-3495(02)75514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer A.E., Qin Y., McCombs J.E. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29:144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scanziani M., Häusser M. Electrophysiology in the age of light. Nature. 2009;461:930–939. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- 38.Okumoto S. Imaging approach for monitoring cellular metabolites and ions using genetically encoded biosensors. Curr. Opin. Biotechnol. 2010;21:45–54. doi: 10.1016/j.copbio.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorostiza P., Isacoff E.Y. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lórenz-Fonfría V.A., Resler T., Heberle J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA. 2013;110:E1273–E1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F., Vierock J., Deisseroth K. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miesenböck G. The optogenetic catechism. Science. 2009;326:395–399. doi: 10.1126/science.1174520. [DOI] [PubMed] [Google Scholar]
- 43.Jung A., Reinstein J., Schlichting I. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J. Mol. Biol. 2006;362:717–732. doi: 10.1016/j.jmb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.De Re, E., G. S. Schlau-Cohen, …, G. R. Fleming. 2013. Comparing the photophysics of the two forms of the orange carotenoid protein using 2D electronic spectroscopy. XVIIIth Int. Conf. Ultrafast Phenomena, Lausanne, Switzerland. EPJ Web of Conferences, Vol. 41, id.08008.
- 45.De Re E., Schlau-Cohen G.S., Fleming G.R. Insights into the structural changes occurring upon photoconversion in the orange carotenoid protein from broadband two-dimensional electronic spectroscopy. J. Phys. Chem. B. 2014;118:5382–5389. doi: 10.1021/jp502120h. [DOI] [PubMed] [Google Scholar]
- 46.Niedzwiedzki D.M., Liu H., Blankenship R.E. Excited state properties of 3′-hydroxyechinenone in solvents and in the orange carotenoid protein from Synechocystis sp. PCC 6803. J. Phys. Chem. B. 2014;118:6141–6149. doi: 10.1021/jp5041794. [DOI] [PubMed] [Google Scholar]
- 47.Liu H., Zhang H., Blankenship R.E. Mass spectrometry footprinting reveals the structural rearrangements of cyanobacterial orange carotenoid protein upon light activation. Biochim. Biophys. Acta. 2014;1837:1955–1963. doi: 10.1016/j.bbabio.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 48.King J.D., Liu H., Blankenship R.E. Chemical activation of the cyanobacterial orange carotenoid protein. FEBS Lett. 2014;588:4561–4565. doi: 10.1016/j.febslet.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 49.Zarrouk, C. 1966. Contribution à l’étude d’une cyanophycée: influence de divers facteurs physiques et chimiques sur la croissance et la photosynthèse de Spirulina maxima (Setch et Gardner) Geitler. PhD thesis, Université de Paris, Paris, France.
- 50.Granovsky, A. A. Firefly version 8.0.0. http://classic.chem.msu.su/gran/firefly/index.html. Accessed December 2014.
- 51.Schmidt M.W., Baldridge K.K., Montgomery J.A. General atomic and molecular electronic structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- 52.Ritchie D.W., Kozakov D., Vajda S. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics. 2008;24:1865–1873. doi: 10.1093/bioinformatics/btn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jorgensen W.L., Maxwell D.C., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 55.Saito S., Tasumi M., Eugster C.H. Resonance Raman spectra (5800–40 cm−1) of all-trans and 15-cis isomers of β-carotene in the solid state and in solution. Measurements with various laser lines from ultraviolet to red. J. Raman Spectrosc. 1983;14:299–309. [Google Scholar]
- 56.Jallet D., Thurotte A., Kirilovsky D. Specificity of the cyanobacterial orange carotenoid protein: influences of orange carotenoid protein and phycobilisome structures. Plant Physiol. 2014;164:790–804. doi: 10.1104/pp.113.229997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Truhlar D., Kohen A. Convex Arrhenius plots and their interpretation. Proc. Natl. Acad. Sci. USA. 2001;98:848–851. doi: 10.1073/pnas.98.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Privalov P.L. Stability of proteins: small globular proteins. Adv. Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 59.Rakhimberdieva M.G., Bolychevtseva Y.V., Karapetyan N.V. Protein-protein interactions in carotenoid triggered quenching of phycobilisome fluorescence in Synechocystis sp. PCC 6803. FEBS Lett. 2007;581:2429–2433. doi: 10.1016/j.febslet.2007.04.056. [DOI] [PubMed] [Google Scholar]
- 60.Vivian J.T., Callis P.R. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lakowicz J.R. 2nd ed. Kluwer Academic/Plenum Publishers; New York: 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 62.Permyakov E.A., Burstein E.A. Some aspects of studies of thermal transitions in proteins by means of their intrinsic fluorescence. Biophys. Chem. 1984;19:265–271. doi: 10.1016/0301-4622(84)87009-x. [DOI] [PubMed] [Google Scholar]
- 63.Hixon J., Reshetnyak Y.K. Algorithm for the analysis of tryptophan fluorescence spectra and their correlation with protein structural parameters. Algorithms. 2009;2:1155–1176. [Google Scholar]
- 64.Otto H., Hoersch D., Heyn M.P. Time-resolved single tryptophan fluorescence in photoactive yellow protein monitors changes in the chromophore structure during the photocycle via energy transfer. Biochemistry. 2005;44:16804–16816. doi: 10.1021/bi051448l. [DOI] [PubMed] [Google Scholar]
- 65.Gensch T., Hendriks J., Hellingwerf K.J. Tryptophan fluorescence monitors structural changes accompanying signalling state formation in the photocycle of photoactive yellow protein. Photochem. Photobiol. Sci. 2004;3:531–536. doi: 10.1039/b401600a. [DOI] [PubMed] [Google Scholar]
- 66.Banishev A.A., Shirshin E.A., Fadeev V.V. Determination of photophysical parameters of red fluorescent protein mRFP1 under ultraviolet excitation by methods of laser fluorimetry. Appl. Opt. 2010;49:6637–6644. doi: 10.1364/AO.49.006637. [DOI] [PubMed] [Google Scholar]
- 67.Sackett D.L., Wolff J. Nile red as a polarity-sensitive fluorescent probe of hydrophobic protein surfaces. Anal. Biochem. 1987;167:228–234. doi: 10.1016/0003-2697(87)90157-6. [DOI] [PubMed] [Google Scholar]
- 68.Polverini E., Cugini G., Gensch T. Molten globule formation in apomyoglobin monitored by the fluorescent probe Nile Red. Biochemistry. 2006;45:5111–5121. doi: 10.1021/bi051905y. [DOI] [PubMed] [Google Scholar]
- 69.Ptitsyn O.B. Molten globule and protein folding. Adv. Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 70.Landrum J.T., Chatfield D.C., Fernandez M.V. The conformation of end-groups is one determinant of carotenoid topology suitable for high fidelity molecular recognition: a study of beta- and epsilon-end-groups. Arch. Biochem. Biophys. 2010;493:169–174. doi: 10.1016/j.abb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 71.Liu W.-L., Wang Z.-G., Su W.-H. Effect of β-ring rotation on the structures and vibrational spectra of β-carotene: density functional theory analysis. J. Phys. Chem. A. 2008;112:10580–10585. doi: 10.1021/jp802024v. [DOI] [PubMed] [Google Scholar]
- 72.Schlucker S., Szeghalmi A., Kiefer W. Density functional and vibrational spectroscopic analysis of OI-carotene. J. Raman Spectrosc. 2003;34:413–419. [Google Scholar]
- 73.Ben-Tal N., Sitkoff D., Honig B. Free energy of amide hydrogen bond formation in vacuum, in water, and in liquid alkane solution. J. Phys. Chem. B. 1997;101:450–457. [Google Scholar]
- 74.Maruta S., Kosumi D., Hashimoto H. The dependence of excitation energy transfer pathways on conjugation length of carotenoids in purple bacterial photosynthetic antennae. Physica Status Solidi (b) 2011;248:403–407. [Google Scholar]
- 75.Cianci M., Rizkallah P.J., Helliwell J.R. The molecular basis of the coloration mechanism in lobster shell: β-crustacyanin at 3.2-A resolution. Proc. Natl. Acad. Sci. USA. 2002;99:9795–9800. doi: 10.1073/pnas.152088999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Merlin J.C. Resonance Raman spectroscopy of carotenoids and carotenoid-containing systems. Pure Appl. Chem. 1985;57:785–792. [Google Scholar]
- 77.Zhang H., Liu H., Blankenship R.E. Molecular mechanism of photoactivation and structural location of the cyanobacterial orange carotenoid protein. Biochemistry. 2014;53:13–19. doi: 10.1021/bi401539w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khoroshyy P., Dér A., Zimányi L. Effect of Hofmeister cosolutes on the photocycle of photoactive yellow protein at moderately alkaline pH. J. Photochem. Photobiol. B. 2013;120:111–119. doi: 10.1016/j.jphotobiol.2012.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.