Abstract
Ablepharon macrostomia syndrome (AMS) and Barber-Say syndrome (BSS) are rare congenital ectodermal dysplasias characterized by similar clinical features. To establish the genetic basis of AMS and BSS, we performed extensive clinical phenotyping, whole exome and candidate gene sequencing, and functional validations. We identified a recurrent de novo mutation in TWIST2 in seven independent AMS-affected families, as well as another recurrent de novo mutation affecting the same amino acid in ten independent BSS-affected families. Moreover, a genotype-phenotype correlation was observed, because the two syndromes differed based solely upon the nature of the substituting amino acid: a lysine at TWIST2 residue 75 resulted in AMS, whereas a glutamine or alanine yielded BSS. TWIST2 encodes a basic helix-loop-helix transcription factor that regulates the development of mesenchymal tissues. All identified mutations fell in the basic domain of TWIST2 and altered the DNA-binding pattern of Flag-TWIST2 in HeLa cells. Comparison of wild-type and mutant TWIST2 expressed in zebrafish identified abnormal developmental phenotypes and widespread transcriptome changes. Our results suggest that autosomal-dominant TWIST2 mutations cause AMS or BSS by inducing protean effects on the transcription factor’s DNA binding.
Introduction
Ablepharon macrostomia syndrome (AMS [MIM: 200110]) and Barber Say syndrome (BSS [MIM: 209885]) are congenital ectodermal dysplasias.1–26 AMS is a disorder defined by absent eyelids, macrostomia, microtia, redundant skin, sparse hair, dysmorphic nose and ears, variable abnormalities of the nipples, genitalia, fingers, and hands, largely normal intellectual and motor development, and poor growth (Table 1).5,6,11,19,20,24 BSS is characterized by ectropion, macrostomia, ear abnormalities, bulbous nose with hypoplastic alae nasi, redundant skin, hypertrichosis, and variable other features.14,16,21,23,25,26 Several instances of parent-to-child transmission suggest that both AMS and BSS are inherited in an autosomal-dominant fashion,2,8,9,21,22 but no specific gene defect has been associated with these disorders. The substantial phenotypic overlap between AMS and BSS, as well as a shared mode of inheritance, supports the hypothesis that the two disorders are caused by dominant mutations in the same gene.10,15,16
Table 1.
Clinical Features of Individuals Harboring TWIST2 Mutations
| Subject | TWIST2 Alteration | Gender | Eyelids | Mouth | Nose | Ears | Skin | Hair | Nipples | Genitalia | Hands | Other | Development |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMS-1.120 | p.Glu75Lys | M | severe hypoplastic eyelids bilateral | macrostomia | depressed nasal bridge | microtia first degree, cryptotia, low-set | wrinkled | sparse | normal | ambiguous | normal | hypertelorism | normal |
| AMS-2.19 | p.Glu75Lys mosaic | M | right upper eyelid defect, absent eyelashes | normal | normal | microtia first degree, increased posterior angulation | normal | sparse | normal | normal | normal | – | normal |
| AMS-2.29 | p.Glu75Lys paternally inherited | F | ablepharon bilateral | macrostomia | depressed nasal bridge, under-developed ala ansi | microtia first degree, low-set | redundant | sparse, absent lanugo | hypoplastic | hypoplastic labia majora | cutaneous syndactyly | hypertelorism | – |
| AMS-2.39 | p.Glu75Lys paternally inherited | F | ablepharon bilateral | macrostomia | depressed nasal bridge, anteverted nostrils | microtia first degree | thin, redundant | sparse, absent lanugo | absent | hypoplastic labia majora | small nails | omphalocele, anteriorly placed anus | – |
| AMS-3.1 (case 2 in Stevens and Sargent24) | p.Glu75Lys de novo | M | ablepharon bilateral | macrostomia | depressed nasal bridge, cleft ala nasi | microtia first degree, low-set | thin, wrinkled | sparse, absent lanugo | hypoplastic | ambiguous genitalia, micropenis, cryptorchidsim | cutaneous syndactyly, camptodactyly | single umbilical artery, lipoma overlying metopic suture | mild gross and fine motor delay, articulation errors with speech |
| AMS-4.1 (case 3 in Stevens and Sargent24) | p.Glu75Lys de novo | F | ablepharon bilateral | macrostomia | depressed nasal bridge, cleft ala nasi | microtia first degree, low-set, unilateral hearing loss | thin, wrinkled | sparse, absent lanugo | normal | hypoplastic labia majora, urethral opening in vagina | camptodactyly | hemiparesis due to cerebral hemorrhage, anteriorly placed anus | mild delays, receives PT, OT, and speech therapy |
| AMS-5.1 (case 4 in Stevens and Sargent24) | p.Glu75Lys de novo | F | ablepharon bilateral | macrostomia | cleft ali nasi | microtia first degree, mild hearing loss | thin, wrinkled | sparse, absent lanugo | hypoplastic | normal | camptodactyly | zygomatic hypoplasia | normal |
| AMS-6.15 | p.Glu75Lys mosaic | F | severe hypoplastic eyelids bilateral, absent eyelashes | macrostomia | under-developed ala nasi | microtia first degree, high-frequency hearing loss | thin, wrinkled | sparse | hypoplastic nipples and breast | normal | cutaneous syndactyly, camptodactyly | aplastic zygomatic arches | normal |
| AMS-7.122 | p.Glu75Lys mosaic | M | severe hypoplastic eyelids bilateral | macrostomia? | normal | microtia first degree | redundant | variable follicle density on posterior scalp | normal | ambiguous genitalia | clinodactyly radial f5 bilateral | low anterior hairline, omphalocele | normal |
| AMS-7.222 | p.Glu75Lys paternally inherited | M | ablepharon bilateral | macrostomia | under-developed ala nasi | microtia first degree | thin, redundant | sparse, absent lanugo | normal | hypoplasia of labia majora | cutaneous syndactyly | omphalocele, anteriorly placed anus, absent zygomatic arches | mild gross motor delay, mild receptive language delay, significant early expressive language delay |
| BSS-1.110 | Gln77_Arg78dup de novo | F | severe hypoplastic eyelids bilateral, ectropion | macrostomia | bulbous nose | cup-shaped, hypoplastic external auditory canals, hearing loss | wrinkled, dry | marked hypertrichosis | inverted | “snout-shaped” labia majora | normal | velopharyngeal incompetence | delayed language development, dyslalia, dysgrammatism |
| BSS-2.114 | p.Glu75Gln de novo | F | ectropion | macrostomia | broad nasal width, bulbous nose, hypoplastic ala nasi | microtia first degree, hypoplastic external auditory canals | wrinkled, translucent | marked hypertrichosis | normal | ambiguous | normal | parental consanguinity | – |
| BSS-3.1 (this paper) | p.Glu75Gln de novo | F | ectropion | macrostomia | broad nasal width, bulbous and prominent nose | small, hypoplastic external auditory canals | wrinkled, visible veins over thorax | marked hypertrichosis | hypoplastic | normal | normal | low anterior hairline, sparse eyebrows, hypertelorism, hypoplastic maxilla, gum hypertrophy, widely spaced teeth | mild delay |
| BSS-4.121 | p.Glu75Gln mosaic | M | coarse eyebrows, telecanthus | macrostomia | bulbous nose, broad nasal width | normal | redundant, dry skin | marked hypertrichosis | inverted, hypoplastic | normal | normal | low anterior hair line | normal |
| BSS-4.221 | p.Glu75Gln paternally inherited | F | ectropion, telecanthus, epiblepharon | macrostomia, mild micrognathia | bulbous nose, broad nasal width | low-set ears, small external canal, concha, extra fold | redundant, dry skin, lipodystrophy | marked hypertrichosis | inverted, hypoplastic | normal | normal | low anterior hair line, thin vermillion of lips | normal |
| BSS-4.321 | p.Glu75Gln paternally inherited | F | ectropion, ocular telecanthus, epiblepharon | macrostomia, mild micrognathia | bulbous nose, broad nasal width | low-set ears, microtia first degree, concha extra fold | redundant, dry skin, lipodystrophy | marked hypertrichosis | inverted, hypoplastic | normal | normal | low anterior hair line, thin vermillion of lips | normal |
| BSS-5.125 | p.Glu75Gln | M | ectropion bilateral, sparse lashes | macrostomia | bulbous nose | microtia first degree | lax, redundant skin | marked hypertrichosis | hypoplastic | shawl scrotum | normal | clubfeet, reduced elastic fibers on skin biopsy | – |
| BSS-6.1 (this paper) | p.Glu75Ala de novo | F | hypoplasia, microblepharon, ectropion (bilateral) | macrostomia | broad nasal width, bulbous nose, hypoplastic ala nasi | microtia first degree | thin (general), redundant (trunk) | marked hypertrichosis (back and limbs) | hypoplastic | mild hypoplasia of labia majora | brachydacytly and clinodactyly f5 prominent digit pads | high palate, lumbar flat angioma | normal |
| BSS-7.115 | p.Glu75Gln de novo | F | microblepharon, ectropion | macrostomia | broad nasal width, bulbous nose | microtia first degree, low-set | thin, redundant | marked hypertrichosis, lanugo hair, sparse eyebrows | absent | normal | normal | parental consanguinity, telengectasias, hypdontia malocculusion | normal |
| BSS-8.17 | p.Glu75Gln | M | ectropion bilateral | macrostomia | bulbous nose, anteverted nares | microtia first degree, hypoplasia external auditory canals | redundant | marked hypertrichosis | hypoplastic | bilateral cryptorchidism | normal | hypertelorism | normal |
| BSS-9.1 (this paper) | p.Glu75Gln de novo | F | ectropion/bilateral “lagophthalmos” as described by ophthalmologist | macrostomia | bulbous nose | microtia first degree | thin, wrinkled | marked hypertrichosis over the back | hypoplastic | normal | normal | hypertelorism | normal |
| BSS-10.116 | p.Glu75Ala | F | ectropion bilateral, sparse lashes | macrostomia | bulbous nose, broad nasal width | microtia first degree, narrow auditory canal | lax, redundant skin | marked hypertrichosis | hypoplastic | hypoplastic labia minora | normal | hypertelorism, delayed eruption of teeth | mild language delay |
Shown are the clinical features of individuals with ablepharon macrostomia syndrome (AMS) and Barber Say syndrome (BSS) and mutation included in this study. Description of signs and symptoms adhere to the published guidelines according to the Elements for Morphology recommendations.27–34 Mutations in TWIST2 (GenBank: NM_057179.2) account for all cases in the study.
We employed extensive clinical phenotyping, exome sequencing, and expression studies to determine the genetic basis for AMS and BSS. We show that both AMS and BSS are due to dominant mutations in TWIST2 (MIM: 607556), affecting a highly conserved residue. TWIST2 (also called Dermo-1), which binds to E-box DNA motifs (5′-CANNTG-3′) as a heterodimer with other bHLH proteins such as the ubiquitously expressed protein E12, is thought to act as a negative regulator of transcription.35–40 TWIST2 expression is temporally restricted and tissue specific. During embryonic development, TWIST2 is highly expressed in the craniofacial mesenchyme and in chondrogenic precursors. Previous studies suggest that TWIST2 regulates mesenchymal stem cell differentiation and directs the development of dermal and chondrogenic tissues.35,38,40,41 Disturbance of these processes due to dominant mutations in TWIST2 could, therefore, cause the distinctive clinical features and facial patterning defects observed in AMS and BSS. Molecular analyses suggest that these mutations alter the DNA-binding activity of TWIST2, leading to both dominant-negative and gain-of-function effects.
Methods
Individuals Included in the Study
The original six family members of a pedigree including AMS-7.1 and AMS-7.2 were enrolled in the NIH Undiagnosed Diseases Program and admitted to the National Institutes of Health Clinical Center (NIH-CC). That family, together with AMS-6.1, were enrolled in protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders,” approved by the National Human Genome Research Institute (NHGRI) Institutional Review Board (IRB). Targeted genetic testing, karyotype analysis, and chromosomal microarray analysis revealed no significant findings. SNP array analysis showed no anomalous regions of homozygosity or significant copy-number variants. Studies of 7 additional AMS-affected individuals from 5 families and 12 BBS-affected individuals from 10 families were approved by the institutional review boards of the Policlinico Tor Vergata University Hospital, the University of Magdeburg, University Medical Center Utrecht, Ghent University Hospital, and Radboud University Medical Center Nijmegen. Written informed consent was obtained from all affected individuals or parents.
Electron Microscopy
Skin biopsies were fixed for 48 hr at 4°C in 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) and washed with cacodylate buffer three times. The tissues were fixed with 2% OsO4 for 2 hr, washed again with 0.1 M cacodylate buffer three times, washed with water, and placed in 1% uranyl acetate for 1 hr. The tissues were subsequently serially dehydrated in ethanol and embedded in Spurr’s (Electron Microscopy Sciences). Semi-thick (∼1,000 nm) and thin (∼80 nm) sections were obtained by utilizing the Leica ultracut-UCT ultramicrotome (Leica) and placed either on glass slides for toluidine blue staining or onto 300 mesh copper grids and stained with saturated uranyl acetate in 50% methanol and then with lead citrate. The grids were viewed in the JEM-1200EXII electron microscope (JEOL Ltd) at 80 kV and images were recorded on the XR611M, mid mounted, 10.5 Mpixel, CCD camera (Advanced Microscopy Techniques).
Fibroblast Culture
Primary dermal fibroblasts were cultured from forearm skin-punch biopsies as described.42 Genomic DNA was extracted from AMS-3.1, -6.1, -7.1 (hyperpigmented, affected), and -7.2 and from BSS-4.2 and -6.1 dermal fibroblasts as well as ATCC adult control, AMS-7.1 (hypopigmented, unaffected), and the mother of AMS-7.2 dermal fibroblasts as unaffected control.
Genetic Analysis
Genomic DNA was extracted from whole blood of AMS-7.1 and AMS-7.2 and their unaffected family members via the Gentra Puregene Blood Kit (QIAGEN). SNP analyses were performed with an Illumina Omni Express 12 (hg18) SNP array and the Genome Studio software program. Whole-exome sequencing was performed with the Illumina HiSeq2000 platform and the TrueSeq capture kit (Illumina) by the NIH Intramural Sequencing Center (NISC). Sequence data were aligned to the human reference genome (hg19) via Novoalign (Novocraft Technologies). Variants were filtered based on allele frequencies in the NIH Undiagnosed Diseases Program43–45 cohort (<0.06) and were confirmed by Sanger sequencing.
Protein Modeling
A computerized model of TWIST2 bound to DNA was generated by YASARa and WHAT IF Twinset via standard parameters with PDB: 1NKP (Myc-Max Recognizing DNA) as template. A homodimeric model was produced by superimposing two TWIST2 models on the original PDB: 1NKP file.
ChIP-Seq
Stably transfected T-REx-HeLa cells, treated for 24 hr with 1 μg/ml tetracycline to induce recombinant WT, p.Glu75Lys, p.Glu75Gln, p.Glu75Ala, and p.Gln77_Arg78dup TWIST2 overexpression, were fixed, pelleted, and frozen according to a cell fixation protocol provided by Active Motif. Sheared chromatin from T-REx HeLa cells without recombinant TWIST2 served as a negative control and was similarly fixed, pelleted, and frozen as a negative control. Chromatin shearing, ChIP, and DNA sequencing were performed by Active Motif. ChIP was performed with a monoclonal anti-FLAG M2 antibody (Sigma Aldrich). ChIPed DNA was sequenced on the Illumina NextSeq 500 platform. Short reads were aligned to human reference genome (hg19) with BWA.46 Binding peaks were identified with MACS using standard parameters.47 Peaks shared between different samples as well as the Jaccard coefficients representing the correlations between samples were calculated with BedTools.48 Peaks were annotated and summarized with CEAS.49 The consensus-binding motif for the WT TWIST2 sample peaks was determined with GEM2.5 with a minimum k-mer length of 6 and a maximum k-mer length of 20.50
Plasmids and Transfection
pCMV6 plasmid containing human TWIST2 cDNA was purchased from Origene Technologies. TWIST2 cDNA was amplified by PCR with Platinum Taq DNA Polymerase High Fidelity (Life Technologies) using pCMV6/TWIST2 plasmid as template. PCR amplification was performed with a forward primer containing FLAG-HA tags. The PCR product was then cloned into the Gateway entry vector pENTR/D-TOPO (Life Technologies) according to the manufacturer’s protocol. Mutations associated with AMS and BSS (c.223G>A [p.Glu75Lys], c.223G>C [p.Glu75Gln], c.224A>C [p.Glu75Ala], and c.229_234dupCAGCGC [p.Gln77_Arg78dup]) were introduced into the TWIST2 containing pENTR/D-TOPO plasmid with the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent) according to manufacturer’s protocol. All primer sequences are listed in Table S1. TWIST2 inserts were then recombined into the Gateway mammalian expression plasmid pT-REx-DEST30 (Life Technologies) according to the manufacturer’s protocol.
T-REx-HeLa cells (Life Technologies) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% (v/v) heat-inactivated fetal bovine serum, 1% (v/v) penicillin-streptomycin, and 5 μg/ml blasticidin. T-REx-HeLa cells were transfected with 2.5 μg pT-REx-DEST30 vector with Lipofectamine 2000 reagent (Life Technologies) and selected with 400 μg/ml Geneticin. Individual clones were screened for TWIST2 expression before and after 24 hr treatment with 1 μg/ml tetracycline. Clones with the highest expression after 24 hr and with minimal leaky expression were chosen for use in further assays.
Zebrafish Experiments
Zebrafish (Danio rerio) were maintained under an approved animal study protocol, in accordance with the Zebrafish Book.51 The human wild-type and mutant TWIST2 (p.Glu75Lys and p.Glu75Gln) were cloned into pCS2GW by Gateway (Life Technologies). Subcloned cDNAs were linearized and used for in vitro synthesis of capped mRNA via mMESSAGE mMACHINE SP6 Ultra Kit (Life Technologies). Wild-type embryos of Tupfel long fin (TL) strain embryos were injected at the one-cell stage with 10 pg (phenotypical analysis) or 2 pg (RNA sequencing) mRNA. Embryos were raised at 28°C in Embryo Medium (E3).
For RNA sequencing, total RNA was extracted from approximately 70 non-injected, wild-type hTWIST2 mRNA-injected, p.Glu75Lys mRNA-injected, and p.Glu75Gln mRNA-injected zebrafish embryos at shield stage in triplicate. Each embryo was microinjected with approximately 10 pg mRNA. RNA extraction was carried out with TRizol (Life Technologies BV) according to the manufacturer’s recommendations. For all samples, total RNA was re-suspended in MQ water prior to sequencing. RNA-seq libraries were prepared according the TruSeq Stranded Total RNA Sample Preparation, Low Sample (LS) protocol. The 12 RNA-seq samples were run on a single HiSeq2500 flow cell. Quality control in FastQ files was performed with FastQC, followed by alignment to the zebrafish genome via STAR.52 A second-quality control step was performed on the generated BAM files via Picards collectRNAMetrics. The read count per gene was determined by HTSeq-count and normalized with DESeq.53 Differential expression analysis was done on the normalized read count tables. GO term analysis was carried out with Gorilla in fast mode54,55 on the expression profiles of the top 10,000 protein-coding genes ranked on adjusted p value from zebrafish embryos overexpressing hTWIST2 variants. The analysis was carried out on datasets of 26,457 protein-coding genes for p.Glu75Lys and p.Glu75Gln after comparison with wild-type hTWIST2-overexpressing samples.
Results
Clinical Characteristics
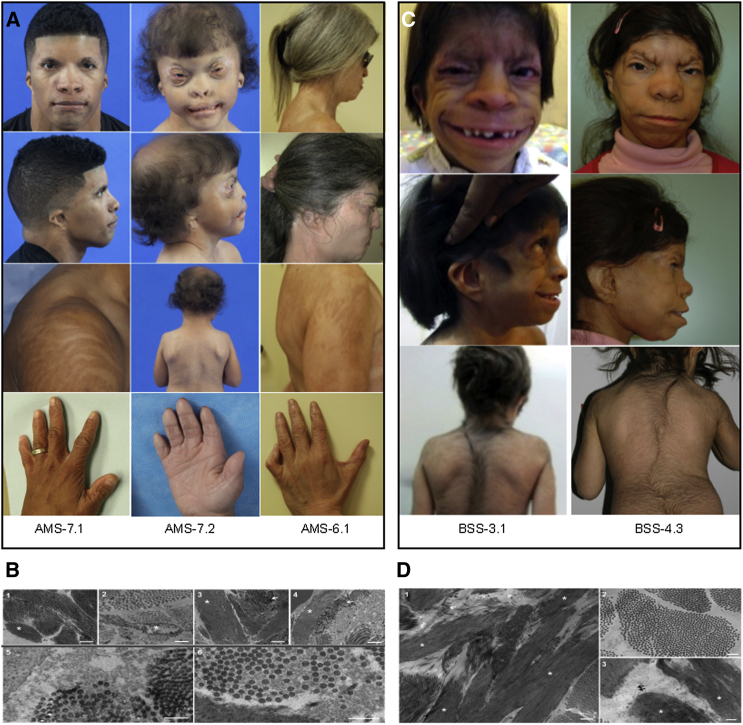
The clinical features of the AMS-affected cohort were previously described in the literature, and their findings and the original citations are listed in Table 1. Overall, the persons with AMS exhibited ablepharon or microblepharon, macrostomia, varying anomalies of the nose and ears, lax redundant skin, sparse hair, variable abnormalities of the nipples, genitalia, fingers, and hands, and largely normal intellectual and motor development. AMS-6.1, AMS-7.1, and AMS-7.2 have typical facial, extremity, and adipose features typical of AMS (Figure 1A). Extensive craniofacial phenotyping of AMS-7.2 also identified undescribed features of harlequin-shaped eyebrows with absence of the distal third, a hypoplastic nasal dorsum with no projection of the nasal tip, absent columella, hypoplastic ala nasi, macrostomia, CL II malocclusion with 50% overbite, a Brody bite, conical shaped teeth, and a long uvula. The macrostomia was characterized by deficient lateral development of the vermillion border and an inability to raise the oral commissures upon smiling, suggesting a discontinuity of the orbicularis oris muscle. Electron microscopy of skin biopsies of AMS-7.1 and AMS-7.2 showed thin, disrupted elastic fibers with areas of amorphous deposits along abnormally oriented collagen fibers and adjacent areas of microfibrillar proliferation (Figure 1B). Masson-Trichrome staining showed abnormal reticulodermal collagen patterns in AMS-7.1 and AMS-7.2 (Figure S1A), whereas elastic fiber (Elastic Van Geison) staining appeared within normal limits (Figure S1B).
Figure 1.
Clinical, Histological, and Molecular Defects in Ablepharon-Macrostomia Syndrome and Barber-Say Syndrome
(A) Face, shoulders, and hands of AMS-6.1, AMS-7.1, and AMS-7.2, demonstrating dysmorphic features detailed in Table 1. Shoulder photographs of AMS-6.1 and AMS-7.1 highlight Blaschko-like hyperpigmented banding indicative of mosaicism. Hands show mild cutaneous syndactyly and clinodactyly.
(B) Electron microscopy of skin of unaffected control (1), AMS-6.1 (2), AMS-7.1 (3), and AMS-7.2. In (1), the elastin (asterisk) is ovoid in shape, whereas in (2), (3), and (4), the elastin appears elongated and, in some areas, fractured. Collagen fibers in (1) appear organized and are oriented in bundles. In (3) and (4), some collagen fibers appear in disarray (arrows) and show curved edges. In addition, some collagen fibers show variable diameters (6). Surrounding the elastin in (5) (AMS-7.1) and (6) (AMS-7.2) are flocculent and amorphous deposits that disrupt organization of collagen bundles. Scale bars represent 1,000 nm (1–4) and 500 nm (5 and 6).
(C) Face, back, and hands of BSS-3.1 and BSS-4.2 showing dysmorphic features detailed in Table 1. Note hypertrichosis.
(D) Electron microscopy of skin of BSS-3.1. Note the long and thin elastin fibers (asterisk) and collagen fibers in disarray (arrow) in (1). Similar to AMS, some collagen fibers show variable diameters (2). Surrounding the elastin in (3) are flocculent and amorphous deposits that disrupt organization of elastin and collagen bundles. Scale bars represent 1,000 nm (1) and 500 nm (2 and 3).
The BSS-affected individuals exhibited ectropion, macrostomia, bulbous noses, malformed ears in the spectrum of microtia first degree, thin, redundant skin, hypertrichosis, hypoplastic nipples, and normal hands and development, together with other variable features (Table 1; Figure 1C). Electron microscopy of the skin biopsy of BSS-3.1 showed findings similar to those of AMS, i.e., thin and long elastic fibers, abnormally oriented collagen fibers, and areas of microfibrillar proliferation and amorphous deposits (Figure 1D). BSS-3.1 and BSS-9.1 have not been previously reported; their clinical features are detailed in Table 1.
DNA Studies
Initial studies were performed on two individuals with AMS (7.1 and 7.2), members of a three-generation pedigree. Targeted genetic testing, karyotype analysis, and chromosomal microarray analysis revealed no significant findings. SNP array analysis showed no anomalous regions of homozygosity or significant copy-number variants. Exome sequencing on all family members revealed a single nonsynonymous heterozygous mutation in TWIST2, c.223G>A (GenBank: NM_057179.2), encoding the predicted deleterious protein alteration p.Glu75Lys. Targeted sequencing revealed the same TWIST2 mutation in ten AMS-affected individuals from seven independent families (Table 1). Targeted sequencing of 11 BSS-affected individuals (Figure 1B) identified heterozygous missense mutations; nine had a c.223G>C (p.Glu75Gln) mutation (GenBank: NM_057179.2) and two had c.224A>C (p.Glu75Ala) mutations (GenBank: NM_057179.2). A 12th individual with BSS, BSS-1.1, carried a heterozygous c.229_234dupCAGCGC (p.Gln77_Arg78dup) mutation (GenBank: NM_057179.2) in TWIST2 (Table 1).
In all instances in which DNA was available from both unaffected parents, the TWIST2 mutation occurred de novo in the first generation of individuals affected with AMS or BSS and was heritable in the third generation. Three disease-transmitting fathers with mild AMS or BSS and variable skin pigmentation were mosaic for a TWIST2 mutation, based upon next-generation sequencing of peripheral blood DNA (AMS-6.1) and Sanger sequencing of DNA from affected and unaffected skin (AMS-7.1) (Figure S2). None of the disease-causing TWIST2 mutations were present in the NIH Undiagnosed Diseases Program’s exome cohort, and none were reported in public variant databases such as NHGRI CLINSEQ,36 dbSNP 142,35 1000 Genomes Project Database, NHLBI Exome Sequencing Project EVS v.0.0.30, or the ExAC database (see Web Resources).
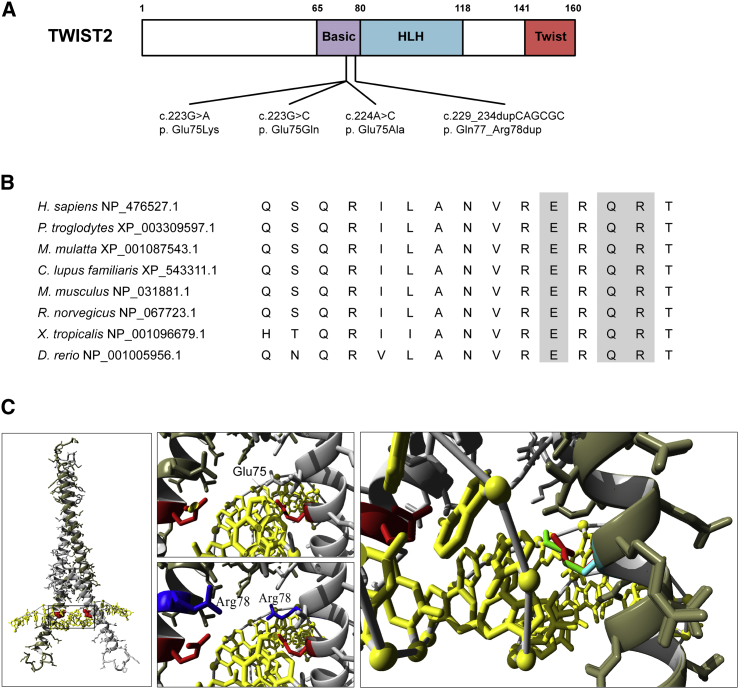
Protein Modeling
TWIST2 contains three functional domains: basic, helix-loop-helix, and twist box (Figure 2A). The TWIST2 alterations associated with AMS and BSS (p.Glu75Lys, p.Glu75Gln, p.Glu75Ala, and p.Gln77_Arg78dup) all fall within the basic domain of the protein, which mediates DNA binding. The residues affected in AMS and BSS are conserved from zebrafish to human (Figure 2B). In silico TWIST2 modeling suggests that mutations affecting the p.Glu75 residue do not alter global protein structure but could alter DNA binding (Figure 2C). The p.Glu75 residue is putatively oriented toward the major groove of bound DNA and in close proximity to the first two nucleotides of the E-box motif. Therefore, the mutations associated with AMS and BSS (as well as p.Gln77_Arg78dup) could alter the DNA-binding activity of TWIST2.
Figure 2.
Mutations in the Basic Domain of TWIST2 Associated with AMS and BSS
(A) Schematic of TWIST2 (GenBank: NP_476527.1) with locations of de novo missense variants identified in individuals with AMS and BSS.
(B) Protein sequence alignment of vertebrate TWIST2 homologs. Residues in the basic domain affected by de novo variants are shaded gray.
(C) Dimeric TWIST2 bHLH protein (gray) with bound DNA (yellow) model with inset. The p.Glu75 residue (red) is oriented toward the DNA major grove; this residue could be involved in hydrogen bonding with the first two nucleotides of the consensus E-box motif or positioning residue p.Arg78.
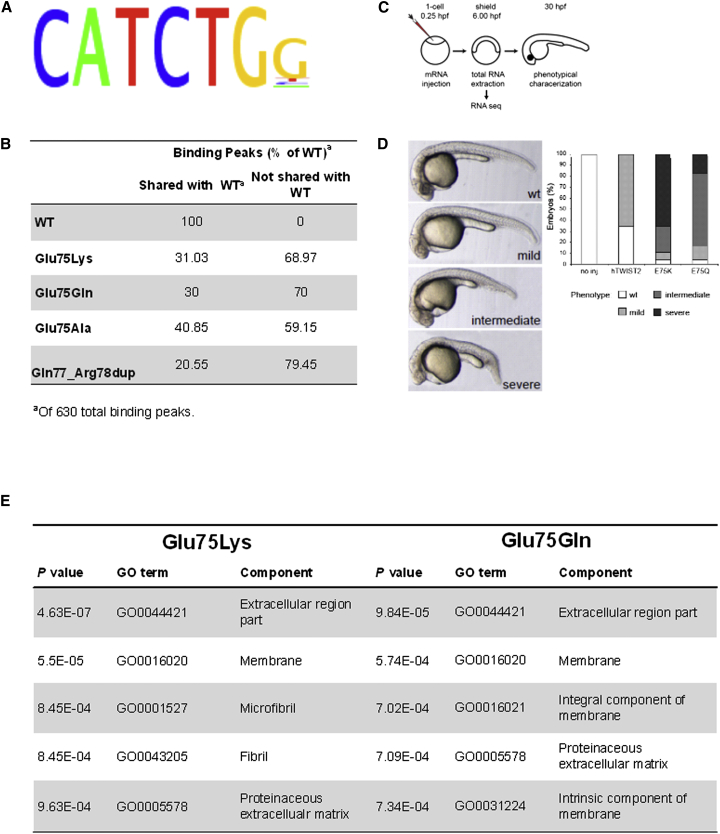
Wild-Type and Mutant TWIST2 Binding Sites
We characterized the binding pattern of both wild-type and mutant TWIST2 in an agnostic and genome-wide fashion. Specifically, we performed ChIP-seq on sheared cross-linked chromatin from T-REx HeLa cells overexpressing recombinant FLAG-HA-tagged TWIST2 proteins; sheared chromatin from T-REx HeLa cells without recombinant TWIST2 served as a negative control. We identified 630 binding peaks associated with wild-type TWIST2. Events associated with wild-type TWIST2 binding were significantly enriched (p < 0.05) near promoters (<1,000 bp, <2,000 bp, and <3,000 bp) as well as in 5′ UTRs (data not shown). The consensus binding motif determined for wild-type TWIST2 was 5′-CATCTGG-3′ (Figure 3A), which represents a canonical E-box. The TWIST2 alterations (p.Glu75Lys, p.Glu75Gln, p.Glu75Ala, and p.Gln77_Arg78dup) shared only a fraction of their binding peaks with WT TWIST2 (Figure 3B); p.Glu75Ala and p.Gln77_Arg78dup TWIST2, both associated with BSS, shared only 25 binding peaks in common with the wild-type. A significant number of binding peaks detected for the mutant TWIST2 proteins were not detected for the wild-type protein (Figure 3B).
Figure 3.
Effect of TWIST2 Mutations on HeLa Cell DNA Binding Sites and on Zebrafish Development and Gene Expression
(A) Chromatin from HeLa cells overexpressing wild-type TWIST2 was subjected to ChIP-seq, identifying 630 DNA binding sites with a consensus sequence typical of an E-box motif.
(B) ChIP-seq showed that the numbers of binding sites for p.Glu75Lys, p.Glu75Gln, p.Glu75Ala, and p.Gln77_Arg78dup TWIST2 were reduced compared to WT TWIST2 and that the mutants bound to many sites not shared with WT TWIST2.
(C) Schematic of the zebrafish studies, involving embryos microinjected with mRNA at the 1-cell stage and either used at shield stage (6 hpf) for RNA-seq or left to grow until approximately 30 hpf for phenotypic characterization.
(D) Appearance of mild, moderate, and severely affected embryos, and quantification of the phenotypes induced at 30 hpf by overexpression of WT, p.Glu75Lys, and p.Glu75Gln hTWIST2. Embryos were injected at the 1-cell stage with 10 pg mRNA. hTWIST2 variants induced defects in head structures and failure of the posterior end of the embryo to extend properly. The p.Glu75Lys and p.Glu75Gln mutants induced stronger developmental defects than wild-type hTWIST2 mRNA.
(E) GO term analysis on ranked gene lists from RNA-seq for p.Glu75Lys and p.Glu75Gln mRNA. Extracellular matrix, membrane, and cytoskeleton proteins are downregulated.
TWIST2 and Zebrafish Development
To elucidate the effect of TWIST2 mutations in vivo, we assessed the functional consequences of injecting wild-type and mutant (p.Glu75Lys and p.Glu75Gln) human TWIST2 (hTWIST2) mRNA into zebrafish at the 1-cell stage (Figure 3C). The introduction of wild-type hTWIST2 led to mild developmental defects (mainly mild brain hypoplasia) in approximately 65% of injected zebrafish (Figure 3D). The injection of p.Glu75Lys and p.Glu75Gln hTWIST2 RNA, however, led to predominantly intermediate and severe developmental defects, including severe head hypoplasia, unclear midbrain-hindbrain boundary, dysmorphic body trunk, and pericardial edema. These results were confirmed in stable transgenic zebrafish lines that express human wild-type and mutant TWIST2 (p.Glu75Lys and p.Glu75Gln) under the control of a Cre-loxP inducible system (Figure S3A).
To gain insight into the genetic processes underlying the developmental phenotypes, we performed RNA sequencing on injected embryos at shield stage. Compared to injection of wild-type hTWIST2, injection of p.Glu75Lys or p.Glu75Gln hTWIST2 caused differential expression of 162 genes from a total of 26,457 datasets (adjusted p value < 0.05); of these, 28 were common to both p.Glu75Lys and p.Glu75Gln hTWIST2 injection (Figure S3B) and all but one were up- or downregulated in the same fashion in both mutants (Figure S3C). The majority of expression changes of these target genes induced by overexpression of p.Glu75Lys and p.Glu75Gln were confirmed by analyzing the stable transgenic zebrafish embryos (Figure S3D). Gene ontology (GO) analyses revealed the greatest reduction in the expression of genes related to extracellular matrix (ECM), membrane components, and cytoskeleton (fibrils) (Figure 3E).
Discussion
We have shown that recurrent dominant mutations in the DNA binding domain of TWIST2 are responsible for two ectodermal dysplasias associated with congenital malformations and dysmorphic facial features, i.e., AMS and BSS. All 10 AMS-affected and 11 of 12 BSS-affected individuals carried a mutation in the highly conserved p.Glu75 amino acid (either p.Glu75Lys, p.Glu75Gln, or p.Glu75Ala). Mutations in the basic domain of TWIST2 drastically altered the spectrum of DNA binding, reducing normal binding and increasing binding to off-target sites. The dominant nature of the mutations, then, could be explained by the abnormal 50% of TWIST2 homodimers and bHLH heterodimers that either reduced binding to the normal contingent of DNA binding sites or conferred a neomorphic function by binding to other sites. Based upon the multitude of DNA binding sites affected by AMS and BSS mutations, the phenotypic manifestations of both AMS and BSS probably result from transcriptional effects on more than a single gene. Our findings support the importance of the DNA binding domain of bHLH transcription factors: autosomal-dominant mutations in the DNA binding domain of TWIST1 (GenBank: NM_000474.3; MIM: 601622), another member of the bHLH transcriptional regulators, have been associated with Saethre-Chotzen syndrome (MIM: 101400), which is characterized with craniosynostosis and limb abnormalities.56,57
Development appears to be exquisitely sensitive to the influence of TWIST2. Simple overexpression of the wild-type protein in zebrafish caused a mild developmental phenotype. In addition, although AMS and BSS result from missense mutations of the same amino acid, p.Glu75, the phenotypes depend on the substituting amino acid. A lysine at TWIST2 residue 75 results in AMS, whereas a glutamine or alanine yields BSS. This suggests that a single amino acid alteration, yielding two phenotypically distinct disorders, dictates specific groups of targeted developmental genes, some shared and some not shared between AMS and BSS (Figure S4B). Those genes might well encode proteins of the extracellular matrix, whose expression in zebrafish was downregulated by the mutations that cause AMS and BSS (Figure S3). In fact, the phenotypes of those zebrafish are reminiscent of sly and bal mutants,58 which are loss-of-function alleles of gamma-1- and alpha-1-laminins, key components of the extracellular matrix. TWIST2 is recognized as a key regulator of mesenchymal cell fate during embryonic development and of epithelial-mesenchymal transition in human cancers;59–63 therefore, TWIST2 mutations might alter the ECM by causing aberrant gene expression.
We conclude that recurrent dominant mutations in the DNA binding domain of TWIST2 are responsible for AMS and BSS, ectodermal dysplasias with congenital malformations and dysmorphic facial features. All 10 AMS-affected and 11 of 12 BSS-affected individuals had an alteration in p.Glu75 of TWIST2. The zebrafish developmental anomalies arising from injection of mutant TWIST2 RNA and ChIP studies suggest two possible mechanisms: a dominant-negative effect due to loss of binding to the normal contingent of TWIST2 DNA binding sites or a neomorphic mechanism due to binding of the mutant TWIST2 to extraneous promoter sites. Supporting a contribution by the first mechanism is the phenotypic overlap with Setleis syndrome (MIM: 227260), a less severe ectodermal dysplasia characterized by bitemporal lesions as well as eyelash and eyebrow defects,64–68 that has been associated with homozygous loss-of-function mutations in TWIST2.
Acknowledgments
We thank the patients, their families, and the treating physicians for their cooperation, encouragement, and interest. This work was supported by the Intramural Research Program of the NHGRI, Netherlands Organization for Health Research and Development grant 319 912-12-109 (B.B.A.d.V.), Wilhelmina Children’s Hospital fund (G.v.H.), and Netherlands Organization for Health Research and Development veni grant 916-12-095 (A.H.). One AMS fibroblast cell line was from NICHD Brain and Tissue Bank for Developmental Disorders (N01-HD-4-3368/N01-HD-4-3383). We are grateful to the following: Dr. Michael Wright (Northern Genetics Service, Newcastle upon Tyne Hospitals, UK), Dr. Salmo Raskin (Laboratorio Genetika, Alameda Augusto Stellfeld, Curitiba Parana, Brazil), Dr. Donna M. McDonald-McGinn (Clinical Genetics Center, Children’s Hospital of Philadelphia), Dr. Edward Cowan (NCI/NIH, Bethesda, MD), Dr. Maria Luisa Martínez-Frías (University Complutense and CIAC/ISCIII, Madrid, Spain), Dr. Elena Campione (Dermatology Unit of Tor Vergata University, Rome, Italy), Dr. Frederic Pérez-Álvarez (Hospital Universitari de Girona Dr. Josep Trueta, Spain), Dra. Beatriz López-García (Hospital Quirón-La Floresta, Zaragoza, Spain), Dr. Anthony Liu and Dr. K.Y. Wong (QMH, Hong Kong), Dr. T.Y. Tan (Royal Children’s Hospital, Melbourne, Australia), Prof. Marina Gallottini (Department of Stomatology, Dental School of USP, Brazil), Prof. Raoul C.M. Hennekam (Academic Medical Center, University of Amsterdam, the Netherlands), Prof. Wilson Araujo Silva, Jr. (Department of Genetics and Center of Genomic Medicine, Riberao Preto University/Clinical Hospital, Brazil), and Dr. Luis Rohena (San Antonio Military Medical Center, TX). The views expressed in this article are those of the authors and do not reflect the official policy of the Department of Army/Navy/Air Force, Department of Defense, or U.S. Government.
Published: June 25, 2015
Footnotes
Supplemental Data include three figures, one table, and Supplemental Methods and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.05.017.
Contributor Information
May Christine V. Malicdan, Email: malicdanm@mail.nih.gov.
William A. Gahl, Email: gahlw@helix.nih.gov.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project (11_2010 data release), ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20100804/
CLINSEQ, http://www.genome.gov/20519355
ExAC Browser (accessed 11, 2015), http://exac.broadinstitute.org/
FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Gene Ontology Consortium, http://geneontology.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
Supplemental Data
References
- 1.Cruz A.A.V., Guimarães F.C., Obeid H.N., Ferraz V.E.F., Noce T.R., Martinez F.E. Congenital shortening of the anterior lamella of all eyelids: the so-called ablepharon macrostomia syndrome. Ophthal. Plast. Reconstr. Surg. 1995;11:284–287. doi: 10.1097/00002341-199512000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Cruz A.A., Souza C.A., Ferraz V.E., Monteiro C.A., Martins F.A. Familial occurrence of ablepharon macrostomia syndrome: eyelid structure and surgical considerations. Arch. Ophthalmol. 2000;118:428–430. [PubMed] [Google Scholar]
- 3.Amor D.J., Savarirayan R. Intermediate form of ablepharon-macrostomia syndrome with CNS abnormalities. Am. J. Med. Genet. 2001;103:252–254. doi: 10.1002/ajmg.1540. [DOI] [PubMed] [Google Scholar]
- 4.Barber N., Say B., Bell R.F., Merveille O.C. Macrostomia, ectropion, atrophic skin, hypertrichosis and growth retardation. Syndr. Ident. 1982;8:6–9. [Google Scholar]
- 5.Brancati F., Mingarelli R., Sarkozy A., Dallapiccola B. Ablepharon-macrostomia syndrome in a 46-year-old woman. Am. J. Med. Genet. A. 2004;127A:96–98. doi: 10.1002/ajmg.a.20658. [DOI] [PubMed] [Google Scholar]
- 6.Cavalcanti D.P., Matejas V., Luquetti D., Mello M.F., Zenker M. Fraser and Ablepharon macrostomia phenotypes: concurrence in one family and association with mutated FRAS1. Am. J. Med. Genet. A. 2007;143A:241–247. doi: 10.1002/ajmg.a.31426. [DOI] [PubMed] [Google Scholar]
- 7.David A., Gordeeff A., Badoual J., Delaire J. Macrostomia, ectropion, atrophic skin, hypertrichosis: another observation. Am. J. Med. Genet. 1991;39:112–115. doi: 10.1002/ajmg.1320390124. [DOI] [PubMed] [Google Scholar]
- 8.Dinulos M.B., Pagon R.A. Autosomal dominant inheritance of Barber-Say syndrome. Am. J. Med. Genet. 1999;86:54–56. doi: 10.1002/(sici)1096-8628(19990903)86:1<54::aid-ajmg10>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Ferraz V.E., Melo D.G., Hansing S.E., Cruz A.A., Pina-Neto J.M. Ablepharon-macrostomia syndrome: first report of familial occurrence. Am. J. Med. Genet. 2000;94:281–283. doi: 10.1002/1096-8628(20001002)94:4<281::aid-ajmg3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 10.Haensel J., Kohlschmidt N., Pitz S., Keilmann A., Zenker M., Ullmann R., Haaf T., Bartsch O. Case report supporting that the Barber-Say and ablepharon macrostomia syndromes could represent one disorder. Am. J. Med. Genet. A. 2009;149A:2236–2240. doi: 10.1002/ajmg.a.32993. [DOI] [PubMed] [Google Scholar]
- 11.Hornblass A., Reifler D.M. Ablepharon macrostomia syndrome. Am. J. Ophthalmol. 1985;99:552–556. doi: 10.1016/s0002-9394(14)77956-5. [DOI] [PubMed] [Google Scholar]
- 12.Jackson I.T., Shaw K.E., del Pinal Matorras F. A new feature of the ablepharon macrostomia syndrome: zygomatic arch absence. Br. J. Plast. Surg. 1988;41:410–416. doi: 10.1016/0007-1226(88)90084-7. [DOI] [PubMed] [Google Scholar]
- 13.Kallish S., McDonald-McGinn D.M., van Haelst M.M., Bartlett S.P., Katowitz J.A., Zackai E.H. Ablepharon-Macrostomia syndrome--extension of the phenotype. Am. J. Med. Genet. A. 2011;155A:3060–3062. doi: 10.1002/ajmg.a.34287. [DOI] [PubMed] [Google Scholar]
- 14.Martínez Santana S., Pérez Alvarez F., Frías J.L., Martínez-Frías M.L. Hypertrichosis, atrophic skin, ectropion, and macrostomia (Barber-Say syndrome): report of a new case. Am. J. Med. Genet. 1993;47:20–23. doi: 10.1002/ajmg.1320470105. [DOI] [PubMed] [Google Scholar]
- 15.Martins F., Ortega K.L., Hiraoka C., Ricardo P., Magalhães M. Oral and dental abnormalities in Barber-Say syndrome. Am. J. Med. Genet. A. 2010;152A:2569–2573. doi: 10.1002/ajmg.a.32898. [DOI] [PubMed] [Google Scholar]
- 16.Mazzanti L., Bergamaschi R., Neri I., Perri A., Patrizi A., Cacciari E., Forabosco A. Barber-Say Syndrome: report of a new case. Am. J. Med. Genet. 1998;78:188–191. [PubMed] [Google Scholar]
- 17.McCarthy G.T., West C.M. Ablepheron macrostomia syndrome. Dev. Med. Child Neurol. 1977;19:659–663. doi: 10.1111/j.1469-8749.1977.tb07999.x. [DOI] [PubMed] [Google Scholar]
- 18.Ng J.D., Rajguru D.S. Oculoplastic approach to treating Barber-Say syndrome. Ophthal. Plast. Reconstr. Surg. 2006;22:232–234. doi: 10.1097/01.iop.0000217575.01061.00. [DOI] [PubMed] [Google Scholar]
- 19.Pellegrino J.E., Schnur R.E., Boghosian-Sell L., Strathdee G., Overhauser J., Spinner N.B., Stump T., Grace K., Zackai E.H. Ablepharon macrostomia syndrome with associated cutis laxa: possible localization to 18q. Hum. Genet. 1996;97:532–536. doi: 10.1007/BF02267081. [DOI] [PubMed] [Google Scholar]
- 20.Price N.J., Pugh R.E., Farndon P.A., Willshaw H.E. Ablepharon macrostomia syndrome. Br. J. Ophthalmol. 1991;75:317–319. doi: 10.1136/bjo.75.5.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roche N., Houtmeyers P., Janssens S., Blondeel P. Barber-Say syndrome in a father and daughter. Am. J. Med. Genet. A. 2010;152A:2563–2568. doi: 10.1002/ajmg.a.33622. [DOI] [PubMed] [Google Scholar]
- 22.Rohena L., Kuehn D., Marchegiani S., Higginson J.D. Evidence for autosomal dominant inheritance of ablepharon-macrostomia syndrome. Am. J. Med. Genet. A. 2011;155A:850–854. doi: 10.1002/ajmg.a.33900. [DOI] [PubMed] [Google Scholar]
- 23.Sod R., Izbizky G., Cohen-Salama M. Macrostomia, hypertelorism, atrophic skin, severe hypertrichosis without ectropion: milder form of Barber-Say Syndrome. Am. J. Med. Genet. 1997;73:366–367. [PubMed] [Google Scholar]
- 24.Stevens C.A., Sargent L.A. Ablepharon-macrostomia syndrome. Am. J. Med. Genet. 2002;107:30–37. doi: 10.1002/ajmg.10123. [DOI] [PubMed] [Google Scholar]
- 25.Tenea D., Jacyk W.K. What syndrome is this? Barber-Say syndrome. Pediatr. Dermatol. 2006;23:183–184. doi: 10.1111/j.1525-1470.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 26.Cortes F.M., Troncoso L.A., Alliende A.R., Curotto B.L. Barber-Say syndrome: further delineation of the clinical spectrum. Genet. Mol. Biol. 2000;23:265–267. [Google Scholar]
- 27.Allanson J.E., Cunniff C., Hoyme H.E., McGaughran J., Muenke M., Neri G. Elements of morphology: standard terminology for the head and face. Am. J. Med. Genet. A. 2009;149A:6–28. doi: 10.1002/ajmg.a.32612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biesecker L.G., Aase J.M., Clericuzio C., Gurrieri F., Temple I.K., Toriello H. Elements of morphology: standard terminology for the hands and feet. Am. J. Med. Genet. A. 2009;149A:93–127. doi: 10.1002/ajmg.a.32596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carey J.C., Cohen M.M., Jr., Curry C.J., Devriendt K., Holmes L.B., Verloes A. Elements of morphology: standard terminology for the lips, mouth, and oral region. Am. J. Med. Genet. A. 2009;149A:77–92. doi: 10.1002/ajmg.a.32602. [DOI] [PubMed] [Google Scholar]
- 30.Hall B.D., Graham J.M., Jr., Cassidy S.B., Opitz J.M. Elements of morphology: standard terminology for the periorbital region. Am. J. Med. Genet. A. 2009;149A:29–39. doi: 10.1002/ajmg.a.32597. [DOI] [PubMed] [Google Scholar]
- 31.Hennekam R.C., Allanson J.E., Biesecker L.G., Carey J.C., Opitz J.M., Vilain E. Elements of morphology: standard terminology for the external genitalia. Am. J. Med. Genet. A. 2013;161A:1238–1263. doi: 10.1002/ajmg.a.35934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennekam R.C., Cormier-Daire V., Hall J.G., Méhes K., Patton M., Stevenson R.E. Elements of morphology: standard terminology for the nose and philtrum. Am. J. Med. Genet. A. 2009;149A:61–76. doi: 10.1002/ajmg.a.32600. [DOI] [PubMed] [Google Scholar]
- 33.Hunter A., Frias J.L., Gillessen-Kaesbach G., Hughes H., Jones K.L., Wilson L. Elements of morphology: standard terminology for the ear. Am. J. Med. Genet. A. 2009;149A:40–60. doi: 10.1002/ajmg.a.32599. [DOI] [PubMed] [Google Scholar]
- 34.Klinger G., Merlob P. Elements of morphology: standard terminology for the ear--additional features. Am. J. Med. Genet. A. 2009;149A:1606. doi: 10.1002/ajmg.a.32893. author reply 1607. [DOI] [PubMed] [Google Scholar]
- 35.Li L., Cserjesi P., Olson E.N. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev. Biol. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- 36.Barnes R.M., Firulli A.B. A twist of insight - the role of Twist-family bHLH factors in development. Int. J. Dev. Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong X.Q., Li L. Dermo-1, a multifunctional basic helix-loop-helix protein, represses MyoD transactivation via the HLH domain, MEF2 interaction, and chromatin deacetylation. J. Biol. Chem. 2002;277:12310–12317. doi: 10.1074/jbc.M110228200. [DOI] [PubMed] [Google Scholar]
- 38.Sosic D., Olson E.N. A new twist on twist--modulation of the NF-kappa B pathway. Cell Cycle. 2003;2:76–78. [PubMed] [Google Scholar]
- 39.Šošić D., Richardson J.A., Yu K., Ornitz D.M., Olson E.N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 40.Isenmann S., Arthur A., Zannettino A.C., Turner J.L., Shi S., Glackin C.A., Gronthos S. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 41.Lee M.S., Lowe G., Flanagan S., Kuchler K., Glackin C.A. Human Dermo-1 has attributes similar to twist in early bone development. Bone. 2000;27:591–602. doi: 10.1016/s8756-3282(00)00380-x. [DOI] [PubMed] [Google Scholar]
- 42.Cullinane A.R., Vilboux T., O’Brien K., Curry J.A., Maynard D.M., Carlson-Donohoe H., Ciccone C., Markello T.C., Gunay-Aygun M., Huizing M., Gahl W.A., NISC Comparative Sequencing Program Homozygosity mapping and whole-exome sequencing to detect SLC45A2 and G6PC3 mutations in a single patient with oculocutaneous albinism and neutropenia. J. Invest. Dermatol. 2011;131:2017–2025. doi: 10.1038/jid.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markello T.C., Han T., Carlson-Donohoe H., Ahaghotu C., Harper U., Jones M., Chandrasekharappa S., Anikster Y., Adams D.R., Gahl W.A., Boerkoel C.F., NISC Comparative Sequencing Program Recombination mapping using Boolean logic and high-density SNP genotyping for exome sequence filtering. Mol. Genet. Metab. 2012;105:382–389. doi: 10.1016/j.ymgme.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gahl W.A., Markello T.C., Toro C., Fajardo K.F., Sincan M., Gill F., Carlson-Donohoe H., Gropman A., Pierson T.M., Golas G. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet. Med. 2012;14:51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gahl W.A., Tifft C.J. The NIH Undiagnosed Diseases Program: lessons learned. JAMA. 2011;305:1904–1905. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- 46.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 50.Machanick P., Bailey T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westerfield M. Eugene: University of Oregon Press; 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 52.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eden E., Lipson D., Yogev S., Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.el Ghouzzi V., Le Merrer M., Perrin-Schmitt F., Lajeunie E., Benit P., Renier D., Bourgeois P., Bolcato-Bellemin A.L., Munnich A., Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat. Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 57.Howard T.D., Paznekas W.A., Green E.D., Chiang L.C., Ma N., Ortiz de Luna R.I., Garcia Delgado C., Gonzalez-Ramos M., Kline A.D., Jabs E.W. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat. Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 58.Stemple D.L., Solnica-Krezel L., Zwartkruis F., Neuhauss S.C., Schier A.F., Malicki J., Stainier D.Y., Abdelilah S., Rangini Z., Mountcastle-Shah E., Driever W. Mutations affecting development of the notochord in zebrafish. Development. 1996;123:117–128. doi: 10.1242/dev.123.1.117. [DOI] [PubMed] [Google Scholar]
- 59.Floc’h N., Kolodziejski J., Akkari L., Simonin Y., Ansieau S., Puisieux A., Hibner U., Lassus P. Modulation of oxidative stress by twist oncoproteins. PLoS ONE. 2013;8:e72490. doi: 10.1371/journal.pone.0072490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu J., Qin L., He T., Qin J., Hong J., Wong J., Liao L., Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasparotto D., Polesel J., Marzotto A., Colladel R., Piccinin S., Modena P., Grizzo A., Sulfaro S., Serraino D., Barzan L. Overexpression of TWIST2 correlates with poor prognosis in head and neck squamous cell carcinomas. Oncotarget. 2011;2:1165–1175. doi: 10.18632/oncotarget.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang T., Li Y., Wang W., Tuerhanjiang A., Wu Z., Yang R., Yuan M., Ma D., Wang W., Wang S. Twist2, the key Twist isoform related to prognosis, promotes invasion of cervical cancer by inducing epithelial-mesenchymal transition and blocking senescence. Hum. Pathol. 2014;45:1839–1846. doi: 10.1016/j.humpath.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Teng Y., Li X. The roles of HLH transcription factors in epithelial mesenchymal transition and multiple molecular mechanisms. Clin. Exp. Metastasis. 2014;31:367–377. doi: 10.1007/s10585-013-9621-6. [DOI] [PubMed] [Google Scholar]
- 64.Cervantes-Barragán D.E., Villarroel C.E., Medrano-Hernández A., Durán-McKinster C., Bosch-Canto V., Del-Castillo V., Nazarenko I., Yang A., Desnick R.J. Setleis syndrome in Mexican-Nahua sibs due to a homozygous TWIST2 frameshift mutation and partial expression in heterozygotes: review of the focal facial dermal dysplasias and subtype reclassification. J. Med. Genet. 2011;48:716–720. doi: 10.1136/jmedgenet-2011-100251. [DOI] [PubMed] [Google Scholar]
- 65.Franco H.L., Casasnovas J.J., Leon R.G., Friesel R., Ge Y., Desnick R.J., Cadilla C.L. Nonsense mutations of the bHLH transcription factor TWIST2 found in Setleis Syndrome patients cause dysregulation of periostin. Int. J. Biochem. Cell Biol. 2011;43:1523–1531. doi: 10.1016/j.biocel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosti R.O., Uyguner Z.O., Nazarenko I., Bekerecioglu M., Cadilla C.L., Ozgur H., Lee B.H., Aggarwal A.K., Pehlivan S., Desnick R.J. Setleis syndrome: clinical, molecular and structural studies of the first TWIST2 missense mutation. Clin. Genet. 2014 doi: 10.1111/cge.12539. Published online December 11, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tukel T., Šošić D., Al-Gazali L.I., Erazo M., Casasnovas J., Franco H.L., Richardson J.A., Olson E.N., Cadilla C.L., Desnick R.J. Homozygous nonsense mutations in TWIST2 cause Setleis syndrome. Am. J. Hum. Genet. 2010;87:289–296. doi: 10.1016/j.ajhg.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Girisha K.M., Bidchol A.M., Sarpangala M.K., Satyamoorthy K. A novel frameshift mutation in TWIST2 gene causing Setleis syndrome. Indian J. Pediatr. 2014;81:302–304. doi: 10.1007/s12098-013-1253-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.