Abstract
In spite of the success of genome-wide association studies (GWASs), only a small proportion of heritability for each complex trait has been explained by identified genetic variants, mainly SNPs. Likely reasons include genetic heterogeneity (i.e., multiple causal genetic variants) and small effect sizes of causal variants, for which pathway analysis has been proposed as a promising alternative to the standard single-SNP-based analysis. A pathway contains a set of functionally related genes, each of which includes multiple SNPs. Here we propose a pathway-based test that is adaptive at both the gene and SNP levels, thus maintaining high power across a wide range of situations with varying numbers of the genes and SNPs associated with a trait. The proposed method is applicable to both common variants and rare variants and can incorporate biological knowledge on SNPs and genes to boost statistical power. We use extensively simulated data and a WTCCC GWAS dataset to compare our proposal with several existing pathway-based and SNP-set-based tests, demonstrating its promising performance and its potential use in practice.
Keywords: aSPU test, genome-wide association studies (GWASs), GRASS, PLINK, SNP, SPU, SSU tests
Introduction
Genome-wide association studies (GWASs) have been successful in identifying many genetic variants, mainly SNPs, associated with complex and common disease (see, for example, the online Catalog of Published Genome-Wide Association Studies). However, only a small proportion of the estimated heritability for most human complex traits can be explained by the identified genetic variants. One possible reason is that, due to small effect sizes and genetic heterogeneity (i.e., multiple causal variants), the standard single-SNP-based analysis might not have enough power to identify many causal variants. Although many human genetic diseases are caused by variants in multiple genes, it has been increasingly recognized that, because genomic variants of these genes lead to the same or similar phenotypes, these genes are likely to be functionally related, and such functional relatedness can be exploited to identify novel genes containing variants related to disease. One way to organize functionally related genes is through biological pathways, such as annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.1 Association analysis of multiple genes with related functions is here generically called pathway analysis (or gene set analysis), which might improve power over testing on single SNPs or single genes one by one. One convincing source of evidence is from tumor sequencing studies, e.g., The Cancer Genome Atlas (TCGA).2 Although a few genes (e.g., TP53 [MIM: 191170]) harbor many mutations related to cancer, most harbor few mutations in a tumor-dependent way. For example, a tumor might contain mutations in PTEN (MIM: 601728), not in NF1 (MIM: 613113), whereas another tumor contains mutations in NF1, not in PTEN. Individually, each of the genes in a related pathway has only a low mutation frequency, but collectively, they have a much higher mutation frequency. Hence, for a disease (e.g., cancer) involving a few pathways, a pathway analysis by aggregating information across multiple genes in a relevant pathway will boost statistical power, and thus is preferred. For example, among the 316 ovarian cancer (MIM: 167000) tumors studied by TCGA, 45% of them had genomic alterations (somatic mutations and DNA copy-number changes) in the PI3K/RAS signaling pathway. This pathway contains seven genes—PTEN, PIK3CA (MIM: 171834), AKT1 (MIM: 164730), AKT2 (MIM: 164731), NF1, KRAS (MIM: 190070), and BRAF (MIM: 164757)—each with only low to moderate genomic alterations in 7%, 18%, 3%, 6%, 12%, 11%, and 0.5% of the tumors, respectively; hence, it should be more powerful to detect genomic alterations at the pathway level than at the individual gene level.
The importance of pathway analysis and many existing approaches have been reviewed by several authors.3–5 Many pathway-based analysis methods for GWAS data are evolved from those for gene expression data;6,7 however, higher-dimensional data are involved in the former with up to hundreds to thousands of SNPs, compared to only tens to hundreds of genes in the latter. On the other hand, because it is known that not all the SNPs in any gene or any pathway are related to a disease, statistically it is most important and challenging to adaptively aggregate information over multiple unknown causal SNPs while minimizing the effects of non-causal SNPs. Existing approaches have some limitations. For example, a popular approach8 used the minimum p value of the multiple SNPs in a gene to summarize association information for the gene, which is not efficient if there are multiple weakly associated SNPs inside the gene. Two other methods, GATES-Simes9 and HYST,10 combine gene-level p values based on GATES,11 a gene-based test using an extended Simes procedure to correct multiple testing while calculating the p value quickly and possibly based on SNP summary statistics (instead of individual-level SNP and phenotype data); GATES-Simes uses an extended Simes procedure to extract the most significant gene-level p value for a pathway, whereas HYST uses Fisher’s method to combine multiple genes’ p values. Hence, as to be confirmed later, GATES-Simes behaves like the minimum p value method, losing power if there are multiple SNPs and/or multiple genes with only weak association strengths; in contrast, HYST, as Fisher’s method, is expected to be low powered if an increasing number of the genes in a pathway are not associated with the trait. A very recent approach12 uses a variance-component test to aggregate information across multiple SNPs non-adaptively, which will lose power in the presence of many non-associated genes. The fundamental problem is the non-adaptive nature of these methods at both the SNP and gene levels. Our proposal is based on a highly adaptive test called adaptive sum of powered score (aSPU) test originally proposed for analysis of rare variants (RVs).13 The main idea of the aSPU test is that, because we do not know which and how many SNPs in the given set are associated with a trait, we first construct a class of tests over-weighting a sequence of increasingly smaller sets of the top-ranked (i.e., most statistically significant) SNPs, then select the test with the most significant result (with a proper adjustment for multiple testing). For relatively small sets of RVs, the aSPU test often outperforms other tests.13 Here we extend the aSPU test to pathway analysis of either common variants (CVs) or RVs. One change we made is that, because the analysis unit of a pathway analysis is a gene but genes can contain quite different numbers of SNPs, we need to modify the aSPU test to treat each gene equally a priori. More importantly, the proposed test is adaptive with respect to both genes and SNPs, which is critical because we do not know a priori how many genes in a pathway are associated and how many SNPs in an associated gene are associated with the given trait. We will compare our proposal with two aforementioned pathway-analysis methods, GATES-Simes and HYST, and two other popular ones, one based on penalized regression (called GRASS) and the other as a representative two-step approach based on SNP screening then combining as implemented in the software PLINK (called simply Plink in the sequel),14,15 largely because the latter two methods have been widely applied to GWASs in practice.16–18
Material and Methods
Data and Notation
We consider the most popular case-control study design as adopted in GWASs, though the methods can be extended to other study designs, e.g., with a quantitative or survival trait. Suppose that for subject i = 1,…,n, Yi = 0 or 1 is a binary trait, e.g., an indicator of disease, and Xi = (Xi1,…, Xik)′ is the vector of the genotype scores for k SNPs, possibly drawn from multiple genes in a pathway. We use additive coding for each SNP; that is, Xij is the number of the copies of an allele at SNP j for subject i. It is possible to include other covariates, but for simplicity we ignore them. We consider a logistic regression model:
| (Equation 1) |
We’d like to test the null hypothesis H0 : β = (β1,…, βk)′ = 0; that is, there is no association between any SNPs and the trait under H0. The score vector U = (U1,…, Uk)′ for β and its covariance matrix are
where and are the sample means of Yis and Xis, respectively. The classic score test statistic is TScore = U′V−1U, which, however, in the current context with a large k, relative to the sample size n, might be low powered, as its asymptotically equivalent Wald test and likelihood ratio test. As shown theoretically,19 as the dimension k increases, the power of the score test might diminish, tending to the type I error rate α. The most popular univariate single SNP-based test, call UminP here, is with Vjj = Var(Uj), which might also be low powered if we have many small . Two alternatives, called the Sum and SSU tests, are
The Sum test is powerful when all or most with the same sign, but not otherwise. As shown by Pan,20 the SSU test can be regarded as a variance-component test21,22 and is closely related to an empirical Bayes test for high-dimensional data23 and a nonparametric MANOVA test.24 In particular, variance-component tests, including kernel machine regression (KMR), have been advocated for SNP set analysis and empirically shown to be powerful in many cases.21,22,25 Nevertheless, as shown in Pan et al.,13,26 because a variance-component test is not adaptive, in the presence of many non-associated SNPs as anticipated in the current context of pathway analysis, it might lose power. Accordingly, a more powerful and adaptive test was proposed as reviewed next.
Review: The Data-Adaptive aSPU Test
Pan et al.13 proposed a class of sum of powered score (SPU) tests in a different context for analysis of RVs:
| (Equation 2) |
The SPU tests cover the Sum and SSU tests as two special cases with a corresponding γ = 1 and γ = 2, respectively. Importantly, as γ → ∞ (and as an even integer), then the SPU test would approach the UminP test if the variances of the score components are a constant (or if their varying variances are ignored, which might be advantageous in certain cases); the reason is simple:
Without covariates, we propose using permutations to obtain p values. More generally, to adjust for covariates, the parametric bootstrap (or, alternatively, permuting residuals) can be used for inference. Specifically, we will first fit a null model under H0, then simulate a new set of traits Y(b)s from the fitted null model for b = 1,…,B; we calculate the test statistic based on each set of simulated Y(b); finally, we calculate the p value as . We used B = 500 in our simulations for a nominal significance level at 5%.
There is no uniformly most powerful test in multilocus association testing; on the other hand, it has been found empirically that the Sum, SSU, and UminP tests performed well under different situations. For a given dataset, to adaptively choose the value of γ for the SPU tests, Pan et al.13 propose an adaptive SPU (aSPU) test that simply combines the results of multiple SPU tests: suppose that we have some candidate values of γ in Γ, e.g., Γ = {1, 2, 3, …, 8} as used in our later experiments, and suppose that the p value of the SPU(γ) test is pγ, then the aSPU test simply takes the minimum p value:
Of course, TaSPU is no longer a genuine p value; we recourse to the parametric bootstrap to estimate its p value. As before, first, we simulate B independent copies Y(b) from the null distribution of Y and obtain the null score vectors U(b) for b = 1,2,…,B. We then calculate the corresponding SPU test statistics and their p values . Thus, we have , and the final p value of the aSPU test .
A Data-Adaptive Pathway-Based Test: aSPUpath
Given a pathway S with |S| genes, we partition the score vector according to the genes as with the score subvector for gene g (with kg SNPs) as based on the logistic regression model (or other generalized linear models or proportional hazards model). The gene-specific SPU statistic and the pathway-based SPU statistic are, respectively,
| (Equation 3) |
| (Equation 4) |
where two scalars γ > 0 and γG > 0, gene-specific weights for SNPs and , and gene-specific weights for genes are pre-specified. wg is used to incorporate prior information on SNPs, e.g., to up-weight SNPs associated with gene expression, whereas wG can be based on gene functional annotations or gene expression data to represent prior likelihoods of their being functional (and associated with the trait); without prior knowledge or data, or for simplicity, we can simply use wg = 1 and wG = 1, which are to be used by default unless specified otherwise in this paper. Note that SPU (γ, wg; g) is standardized by the gene-specific number of SNPs, kg, so that large genes will not dominate a pathway analysis (since the genes in a pathway are the analysis units and are thus treated equally a priori if no weighting is desired). The intuition behind using γG is like that for γ: in general, a larger γG (or γ) is more effective if there are fewer associated genes (or SNPs) with larger effects in a pathway (or in a gene), but not otherwise. Two extreme examples are the following: (1) γG = 1 (or γ = 1), treating all genes (or SNPs) equally, which is most powerful if all the genes (or SNPs) are associated with the trait with similar effect sizes and in the same direction (i.e., all positive or all negative); (2) γG = ∞ (or γ = ∞), using only the most significant gene (or SNP) as the evidence against the null hypothesis, which is most powerful if there are only one or few genes (or SNPs) associated with the trait with a large effect size. Between the two extremes, other values of γG (or γ) might be more powerful. For example, if only a subset of the genes (or SNPs) are associated with different effect sizes and different directions, using γG = 2 (or γ = 2) might be more powerful, as variance-component tests (e.g., KMR); on the other hand, if the proportion of the associated genes (or SNPs) decreases, a larger value, say γG = 4 (or γ = 4), might lead to a more powerful test; often γG = 8 or 16 (or γ = 8 or 16) gives the results similar to using γG = ∞ (or γ = ∞). We also note that, if the association directions of (most) associated genes (or SNPs) are in the same direction, using an odd integer of γG (or γ) might be more powerful; otherwise, using an even integer is more promising. These points have been empirically verified for RV analysis13 and polygenic testing in GWASs.26 In practice, because an optimal value of γG (or γ) is unknown, depending on the unknown genetic association patterns, one has to conduct a grid search over a wide range of possible values for γG (or γ), but searching over too many will introduce extra variability and thus lead to power loss. Based on our experience coupled with the goal of a pathway-based analysis, to take advantage of possibly multiple associated genes (and SNPs), we suggest trying (and ) as shown in the results below, though this needs to be further studied.
For any given (γ,γG), as for SPU(γ), we recourse to resampling to calculate its p value . Its power depends on the choice of (γ,γG). A pathway-based aSPU test is defined as
| (Equation 5) |
aiming to select from multiple PathSPU tests the most powerful one. Similar to that for the aSPU test, we propose using a single layer of the permutation or parametric bootstrap to calculate the p values.
For the possible situation where multiple genes in a pathway might contain quite different proportions of causal SNPs, we might use a more general pathway-based test with a gene-specific γg for each gene g. Denote , we can modify the tests as
| (Equation 6) |
| (Equation 7) |
The corresponding aSPUpath2 test is computationally more demanding in searching for suitable values of more parameters in γ and γG, which will also introduce more variability to the results and thus might lead to loss of power. This needs to be studied further.
Other Modifications
We also considered single-gene-based approaches and those based on dimension reduction. Because they did not outperform the proposed aSPUpath, we will present just a summary that might be interesting.
As a representative of single gene-based approaches, we considered applying SPU and aSPU tests to each gene in a pathway, then using the minimum gene-level p value as a final test statistic for the pathway. It is easy to see that the pathway-based SPU(∞) (after ignoring the inverse weighting by the number of SNPs and the possible use of weights) and single gene-based SPU(∞) are almost the same; hence, our proposed aSPUpath test is more adaptive and thus expected to be more flexible and powerful.
For dimension reduction, as in GRASS, for each gene we replaced its individual SNP genotype scores by their top few principal components (PCs) that accounted for at least 95% of total variation, and then we applied the pathway-based aSPU test to these PCs. Perhaps due to the adaptivity of the original aSPUpath test and possible loss of information by PCs, we did not find improvement by the use of PCs in our simulations. However, given that PC-based tests27,28 are viable competitors to variance-component tests as discussed in Schaid et al.,12 we had an interesting, perhaps surprising, observation: applying the SPU(2) (i.e., SSU) test (that is equivalent to a variance-component test) to the original genotypes or the PCs gave almost the same result; an explanation is offered below.
Suppose that X is the n × k matrix of the original genotype scores. We apply a singular value decomposition: , where we assume that the eigen values have been put in descending order as the diagonal elements of . The first L PCs are the columns of , where VL is an n × L matrix containing the first L columns of V and is an L × L diagonal matrix containing the first L eigen values. Now we can compare the two SSU statistics when applied to X and PL, respectively:
But for other γ ≠ 2, we would expect that, in general, SPU(γ) would give different results when applied to the original genotype scores X and its top PCs PL, respectively.
Simulation Set-ups
We conducted extensive simulation studies to evaluate and compare the performance of the aSPUpath test with several alternative methods. Our general set-ups were similar to those (set-ups A–D) in Chen et al.14 except that we simulated SNPs, not PCs (called eigenSNPs therein) of SNPs, to mimic real data. Specifically, set-up A was the null case with no causal SNP, while the other three set-ups contained causal SNPs in 1, 5, and 10 genes, respectively. We considered one pathway containing 20 genes, each of which might contain 1–20 SNPs, or 3–100 SNPs; there was at most one causal SNP inside each gene. To cover possible situations with more than one causal SNP inside a gene, we added set-ups B′–D′, in which we randomly selected 1–3 causal SNPs in a gene. Furthermore, to mimic real pathways as in KEGG, we also considered cases E and F with 40 and 80 genes, respectively, in a pathway while all other aspects were similar to set-up D′. The SNPs inside each gene might or might not be correlated whereas the SNPs from different genes were always independent, and the causal SNPs might or might not be included in the data.
The simulated genotypes were generated as in Wang and Elston.29 First, we generated a latent vector Z = (Z1,…,Zk)′ from a multivariate Normal distribution with a first-order auto-regressive (AR1) covariance structure: between any latent components i and j; ρ = 0 and ρ > 0 randomly chosen from a uniform distribution U(0, 0.8) were used to generate (neighboring) SNPs in linkage equilibrium and in linkage disequilibrium (LD), respectively. The number of SNPs inside each gene, kg, was randomly chosen between 1 and 20, or between 3 and 100. Second, the latent vector was dichotomized to yield a haplotype with MAFs each randomly selected uniformly between 0.05 and 0.4 for CVs or between 0.001 and 0.01 for RVs. Third, we combined two independent haplotypes and obtained genotype data: Xi = (Xi1,…,Xik)′ for subject i. Fourth, for a non null case, the first SNP inside the first k1 = 1 or 5 or 10 genes, corresponding to set-ups B–D, was chosen to be causal with βj = logOR ≠ 0, and all other βj = 0; we also tried set-ups B′–D′, E, and F with 1–3 randomly chosen causal SNPs. For the null case, all βj = 0. Fifth, the disease status Yi of subject i was generated from the logistic regression model (Equation 1). We used β0 = −log(0.05 / 0.95) for a 5% background disease probability; that is, . Sixth, as in a case-control study, we sampled n/2 = 500 cases and n/2 = 500 controls in each dataset.
Throughout the simulations, we fixed the test significance level at α = 0.05. We used the R package SNPath implementing GRASS and Plink;14 we implemented other methods in R package aSPU. Because the program for Plink was quite slow, we ran only 100 independent replicates for Plink, but 1,000 replicates for others in each set-up.
Results
Simulation Results for CVs
For comparison, we included the SSU (i.e., SPU(2)) and UminP tests; the former is equivalent to a global pathway-based test of Goeman et al.6 as shown in Pan,30 and the latter is the most popular single SNP-based test in GWASs. The UminP test often performed similarly to SPU(∞) (data not shown).
Type I Error
As shown in Table 1, it appears that each test could control its type I error rate satisfactorily around or within 0.05.
Table 1.
Empirical Type I Error Rates of the Tests for CVs
| Set-up | aSPUpath | GRASS | Plink | aSPU | SSU | UminP | GATES-Simes | HYST |
|---|---|---|---|---|---|---|---|---|
| 200 indep SNPs | .055 | .057 | .02 | .053 | .046 | .057 | .047 | .022 |
| 1,000 indep SNPs | .048 | .067 | .03 | .050 | .052 | .040 | .040 | .028 |
| 200 corr SNPs | .054 | .064 | .05 | .048 | .040 | .062 | .050 | .042 |
Comparison of the aSPUpath Test with Other Tests
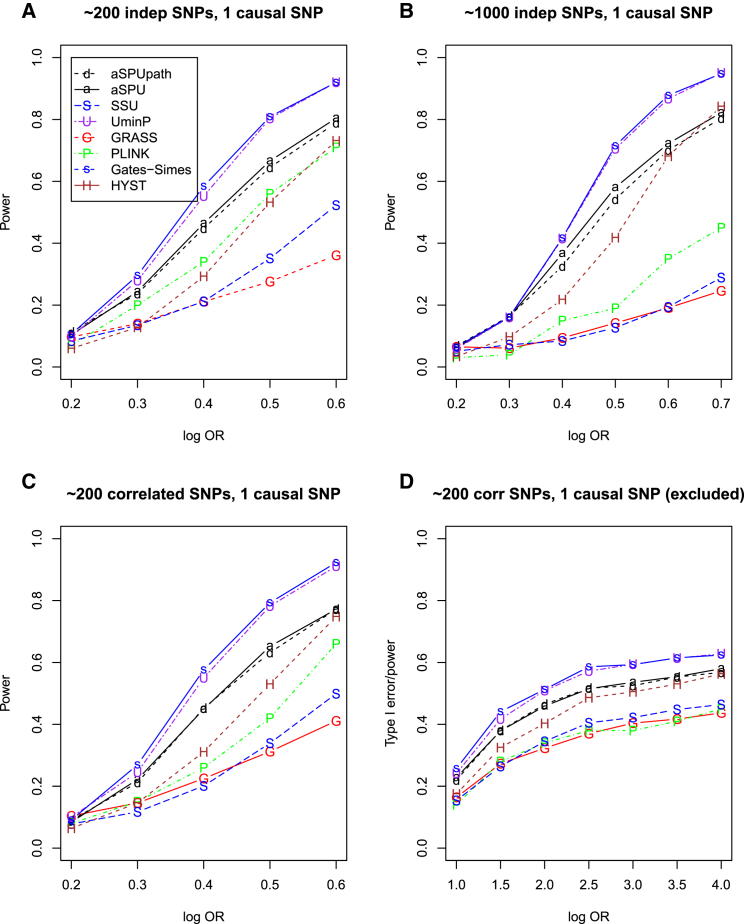
We first consider set-up B, an extreme scenario that is least favorable to pathway or SNP set analysis: because there was only one causal SNP, single SNP-based analysis as implemented in the UminP test was expected to be most powerful, which was confirmed as shown in Figure 1; GATES-Simes also achieved the highest power as UminP. Nevertheless, the aSPU and aSPUpath tests performed similarly and were the next most powerful. As shown in Figure 1A with about 200 independent SNPs, besides UminP/GATES-Simes and aSPU/aSPUpath, Plink was most powerful, closely followed by HYST, then by SSU, and finally GRASS. Figure 1B shows that, with about 1,000 independent SNPs, the aSPU and aSPUpath tests showed even a more striking advantage over the other pathway- or SNP set-based tests except GATES-Simes, suggesting the former two’s (and the latter four’s) robustness (and lack of robustness) to an increasing number of SNPs. In particular, the performance of SSU deteriorated with its power close to that of GRASS. Figure 1C shows that, with about 200 correlated SNPs (with the causal SNP included), the power trend was similar to that with 200 independent SNPs, though GRASS performed better than Plink and SSU with smaller ORs. As shown in Figure 1D, with about 200 correlated SNPs with the causal SNP excluded, again we found GATES-Simes and UminP, closely followed by the aSPU and aSPUpath tests, then by HYST, to be the top performers, while the other three tests were similarly low powered.
Figure 1.
Empirical Power for Simulation Set-up B with a Pathway of 20 Genes
One gene included one causal SNP.
(A and C) Each gene contained 1–20 independent (A) or correlated (C) SNPs.
(B) Each gene contained 3–100 independent SNPs.
(D) Each gene contained 1–20 correlated SNPs and the causal SNP was excluded in analysis.
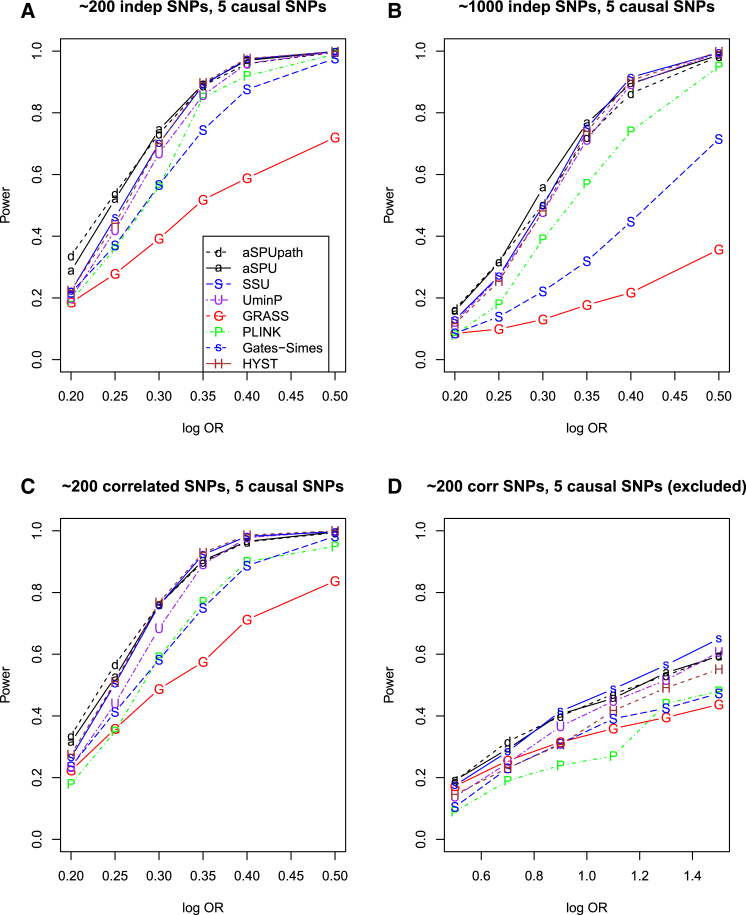
In set-up C with five causal SNPs (Figure 2), again the aSPU and aSPUpath tests performed similarly and now they had an edge over the UminP test, especially for smaller ORs, since the latter uses only the single SNP with the strongest signal while ignoring the signals from other four causal SNPs. HYST also performed well, especially for large ORs, and the power of GATES-Simes was close to or slightly higher than that of the UminP test. However, differing from set-up B, we notice that the SSU test and Plink performed similarly, shown in Figures 2A and 2C, and one was more powerful than the other in Figures 2B and 2D, respectively. Figure 2D showed that, with the five causal SNPs excluded, GRASS could perform well when the causal effect size was small (and the power was low).
Figure 2.
Empirical Power for Simulation Set-up C with a Pathway of 20 Genes
Five genes each included one causal SNP.
(A and C) Each gene contained 1–20 independent (A) or correlated (C) SNPs.
(B) Each gene contained 3–100 independent SNPs.
(D) Each gene contained 1–20 correlated SNPs and the causal SNP was excluded in analysis.
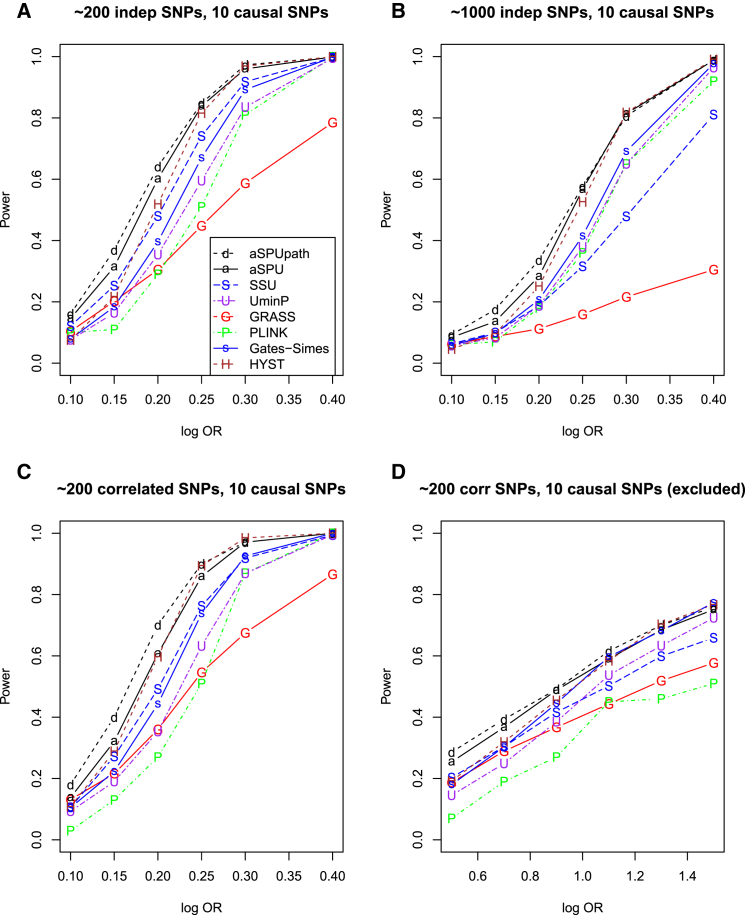
Now consider a case favoring pathway or SNP set analysis in set-up D with ten causal SNPs (Figure 3). The aSPUpath test was the sole winner, having an edge over the aSPU test; in particular, the two tests could be much more powerful than the UminP test and GATES-Simes, although HYST performed well for large effect sizes. As shown in Figures 3A and 3C, even the SSU test was much more powerful than the UminP test, confirming the advantage of combining information across multiple causal SNPs. On the other hand, in Figure 3B with about 1,000 SNPs, GATES-Simes, UminP, and Plink were tied (after aSPUpath, aSPU, and HYST) as the next tier of the most powerful, followed by SSU, then by GRASS; the low power of SSU test was due to its non-robustness to a large number of non-associated SNPs: it did not down-weight enough the larger number of non-associated SNPs; in contrast, the two adaptive tests, aSPU and aSPUpath, did not suffer much from the presence of a large number of non-associated SNPs. GRASS could beat Plink when the causal effect size was small with (Figures 3A and 3C) or without (Figure 3D) the presence of the causal SNPs.
Figure 3.
Empirical Power for Simulation Set-up D with a Pathway of 20 Genes
Ten genes each included one to three causal SNPs.
(A and C) Each gene contained 1–20 independent (A) or correlated (C) SNPs.
(B) Each gene contained 3–100 independent SNPs.
(D) Each gene contained 1–20 correlated SNPs and the causal SNP was excluded in analysis.
In all the above three situations, each gene contained either no or only one causal SNP, which might be too restrictive. To cover possible situations with more than one causal SNP inside a gene, we considered set-ups B′–D′, in which we randomly selected 1–3 causal SNPs in 1, 5, and 10 genes, respectively (while other genes contained no causal SNPs). The main results remained the same as before except the following as shown in Figure 4 for set-up D′. First, there was a larger power advantage of the aSPUpath over the aSPU test for a larger number of SNPs (Figure 4B). Second, there was improved performance of GRASS: for example, for small effect sizes, GRASS was consistently more powerful than Plink, though it was still outperformed by aSPUpath. It is clear that GATES-Simes behaved like, albeit a little more powerful than, the UminP test, and HYST was more powerful than the other two but less powerful than aSPUpath.
Figure 4.
Empirical Power for Simulation Set-up D′ with a Pathway Containing 20 Genes
Ten genes each included one to three causal SNPs.
(A and C) Each gene contained 1–20 independent (A) or correlated (C) SNPs.
(B) Each gene contained 3–100 independent SNPs.
(D) Each gene contained 1–20 correlated SNPs and the causal SNP was excluded in analysis.
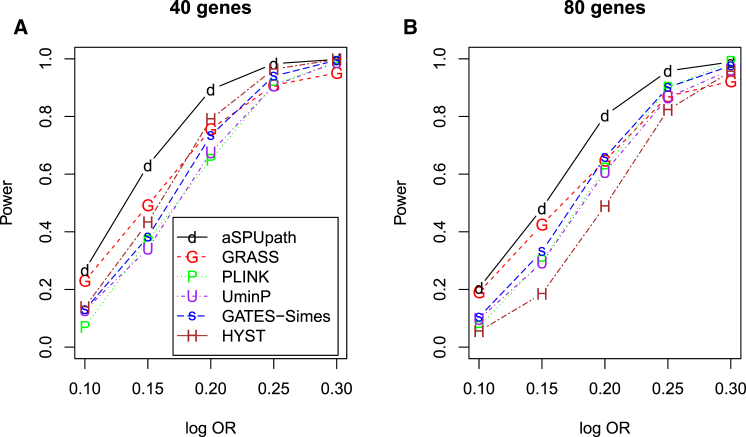
To mimic KEGG pathways, most of which contain more than 20 genes (e.g., Table 4), we considered two set-ups similar to set-up D′ but with 40 or 80 genes in each pathway and each gene with 1–20 correlated SNPs. As shown in Figure 5, aSPUpath remained the most powerful in most situations, especially with relatively small ORs as in typical GWASs for complex traits, under which GRASS performed second best. Again, GATES-Simes performed similarly as the UminP test, and HYST lost power as the number of the non-associated genes in the pathway increased.
Table 4.
Results of the WTCCC CD GWAS Data Application: KEGG Pathways with p Values < 0.00001 by Any of aSPUpath, GRASS, GATES-Simes, and HYST
| KEGG ID | Pathway Names | No. of Genes | No. of SNPs |
p Values |
|||
|---|---|---|---|---|---|---|---|
| aSPUpath | GRASS | GATES-Simes | HYST | ||||
| hsa04630 | Jak-STAT signaling pathway∗ | 145 | 1,410 | <0.00001 | <0.00001 | <0.00001 | <0.00001 |
| hsa04060 | cytokine-cytokine receptor interaction∗ | 247 | 2,506 | <0.00001 | <0.00001 | <0.00001 | .00001 |
| hsa04660 | T cell receptor signaling pathway∗ | 105 | 1,373 | <0.00001 | <0.00001 | .00081 | .00021 |
| hsa04310 | Wnt signaling pathway | 143 | 2,087 | <0.00001 | <0.00001 | .00089 | .00238 |
| hsa05310 | asthma | 27 | 271 | <0.00001 | <0.00001 | .00071 | .00002 |
| hsa05330 | allograft rejection | 34 | 466 | <0.00001 | <0.00001 | .00089 | <0.00001 |
| hsa05414 | dilated cardiomyopathy (DCM) | 89 | 2,605 | <0.00001 | <0.00001 | .00382 | .02188 |
| hsa05416 | viral myocarditis | 67 | 1,263 | <0.00001 | <0.00001 | .00148 | <0.00001 |
| hsa04972 | pancreatic secretion | 93 | 2,187 | <0.00001 | .00003 | .00072 | .00211 |
| hsa04621 | NOD-like receptor signaling pathway∗ | 57 | 502 | <0.00001 | .00542 | <0.00001 | .01012 |
| hsa04062 | chemokine signaling pathway∗ | 174 | 2,714 | <0.00001 | .00061 | .00131 | .00119 |
| hsa04810 | regulation of actin cytoskeleton | 201 | 3,347 | <0.00001 | .00108 | .00156 | .00962 |
| hsa05131 | shigellosis | 60 | 784 | <0.00001 | .00434 | <0.00001 | .00159 |
| hsa00230 | purine metabolism | 154 | 2,810 | .00759 | <0.00001 | .05376 | .02156 |
| hsa04144 | endocytosis | 180 | 2,575 | .00190 | <0.00001 | .00139 | .01397 |
| hsa04145 | phagosome | 136 | 1,469 | .00101 | <0.00001 | .00314 | .00272 |
| hsa04270 | vascular smooth muscle contraction | 113 | 2,887 | .00025 | <0.00001 | .00086 | .00566 |
| hsa04350 | TGF-beta signaling pathway | 82 | 831 | .00080 | <0.00001 | .00060 | .01381 |
| hsa04514 | cell adhesion molecules (CAMs) | 122 | 3,312 | .00120 | <0.00001 | .00311 | .00043 |
| hsa04612 | antigen processing and presentation | 63 | 543 | .00129 | <0.00001 | .00146 | .00016 |
| hsa04650 | natural killer cell mediated cytotoxicity | 124 | 1,464 | .00199 | <0.00001 | .02586 | .00336 |
| hsa04672 | intestinal immune network for IgA production | 45 | 393 | .00073 | <0.00001 | .00105 | .00009 |
| hsa04940 | type I diabetes mellitus | 39 | 714 | .00031 | <0.00001 | .00102 | <0.00001 |
| hsa05332 | graft-versus-host disease | 33 | 440 | .00036 | <0.00001 | .00086 | .00001 |
| hsa04622 | RIG-I-like receptor signaling pathway | 65 | 474 | .00004 | .0318 | <0.00001 | .30502 |
Asterisks (∗) indicate positive control pathways.
Figure 5.
Empirical Power for Simulation Set-ups E and F with a Pathway Containing 40 and 80 Genes, Respectively
Ten genes each included 1–3 causal SNPs, and each gene contained 1–20 correlated SNPs. Set-up E with a pathway of 40 genes (A) and set-up F with a pathway of 80 genes (B) are shown.
In summary, we found that the aSPUpath and aSPU tests were much more powerful than pathway-based GRASS, HYST, and Plink, and the SSU test for SNP set analysis, across all the simulation set-ups considered. In the presence of multiple causal SNPs or of multiple genes containing causal SNPs, as anticipated for pathway analysis, they also outperformed the single SNP-based UminP test, which often operated like GATES-Simes; between the two adaptive tests, the aSPUpath test had an edge over the aSPU test in some situations, especially for a larger number of non-associated SNPs and for casual SNPs with small effect sizes.
Comparison of the aSPUpath Test with Its Other Variants
For set-up B with only one causal SNP, the single-gene-based aSPU and pathway-based aSPU tests had almost identical power and were much more powerful than the PC-based aSPU test. The reason was the following. First, because there was only one single causal SNP, a single-gene-based approach would not lose power as compared to a pathway-based approach aiming to combine information across multiple genes; at the same time, a pathway-based approach in general would not gain either under this situation. Second, note that the aSPU test could realize effective SNP selection by adaptively choosing the tuning parameter γ to down-weight non-associated SNPs; however, each PC is a linear combination of all the SNPs, a mixture of both associated and non-associated SNPs, hindering the ability of the PC-based aSPU test to select SNPs effectively.
For set-up C with five causal SNPs, the pathway-based aSPU test was more powerful than the gene-based aSPU test, and the PC-based aSPU test was still the least powerful.
For set-up D with ten causal SNPs, the pathway-based aSPU test was by far the most powerful. For 200 SNPs, the PC-based aSPU test was more powerful than the single-gene-based aSPU; however, with about 1,000 SNPs, the single-gene-based aSPU was more powerful than the PC-based aSPU, presumably due to the fact that each PC contained too many non-associated SNPs, diluting the association effects.
As in GRASS, we also tried to first construct gene-specific SPU test statistics before combining them across a pathway but did not find it working better than the simple aSPUpath test discussed here.
In summary, we found that overall our proposed aSPUpath test performed better than the single-gene-based aSPU and PC-based aSPU tests.
Incorporating Prior Knowledge by Weighting
As discussed earlier, our proposed method can incorporate biological knowledge or prior data on the likelihood of SNPs and genes being functional through weighting them differentially. We did a preliminary study to explore the use of informative weighting in set-up D with ten genes, each containing one causal SNP and with a total of about 200 correlated SNPs. We applied our proposed test with wg = 1, but with wG = 1 or wG ≠ 1 to assess the effects of some correctly specified and some mis-specified gene weights (while the effects of SNP weighting could be explored similarly). We generated wG,g ∼ U (0.2, 0.6), a uniform distribution between 0.2 and 0.6, for genes containing no causal SNPs, but for other genes (containing causal SNPs) wG,g ∼ U (0.2 + δ, 0.6 + δ) for several values of δ ≥ 0. Increasing values of δ reflected increasing informativeness of the weights, while δ = 0 represented completely random and non-informative weighting. Note that, with the overlapping weights for the genes containing causal SNPs and for those without any causal SNP, although the weights might be informative, strictly speaking they were mis-specified. As shown in Table 2, it is clear that our proposed aSPUpath test was most powerful; its weighted version was robust to mis-specified and completely random weights (with δ = 0) with only small power loss, while gaining higher power with more informative weights.
Table 2.
Empirical Type I Error logOR = 0 and Power at logOR ≠ 0 of Various Tests for about 200 Correlated SNPs in a 20-Gene Pathway for Set-up D
| logOR |
aSPUpath |
GRASS | Plink | aSPU | SSU | UminP | GATES-Simes | HYST | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wG= 1 | δ = 0 | δ = .1 | δ = .2 | δ = .3 | δ = .4 | ||||||||
| 0 | .054 | .052 | .051 | .050 | .048 | .044 | .064 | .05 | .048 | .040 | .062 | .050 | .042 |
| 0.15 | .400 | .397 | .430 | .468 | .489 | .517 | .216 | .13 | .321 | .272 | .190 | .223 | .289 |
| 0.2 | .701 | .656 | .713 | .747 | .769 | .791 | .360 | .27 | .607 | .492 | .353 | .443 | .597 |
| 0.25 | .900 | .873 | .907 | .926 | .931 | .936 | .546 | .51 | .859 | .763 | .632 | .738 | .894 |
Simulation Results for RVs
With the increasing availability of sequencing data, it has become more important and urgent to develop and apply pathway-based analysis of RVs; there have been few such studies. For this purpose, we did a simulation study to assess the performance and show the potential of our proposed test for pathway analysis of RVs. To save space, we present results only for a simulation set-up similar to set-up D: a pathway contained 20 genes, 0 or 10 of which each contained one causal RV among 1–20 RVs for the null or non null cases, respectively. The MAFs for the RVs were randomly drawn between 0.1% and 1% for the control samples. We considered both independent and correlated RVs within each gene.
For comparison, we also included several existing popular or competitive tests. In particular, we included the Sum (i.e., SPU(1)) as a representative burden test, the SSU (i.e., SPU(2)) test that was shown by Basu and Pan31 to be competitive and closely related to several other association tests such as C-alpha test32 and kernel machine regression or SKAT,33 and three adaptive tests that appeared recently, a kernel-based adaptive clustering (KBAC) test,34 a p value weighted sum test (PWSU),35 and an estimated regression coefficient (EREC) test.36
As shown in Table 3, all the methods seem to have type I error rates around the nominal level of 0.05.
Table 3.
Empirical Type I Error Rates of the Tests for RVs
| Set-up | aSPUpath | GRASS | aSPU | Sum | SSU | UminP | SKAT | KBAC | PWST | EREC |
|---|---|---|---|---|---|---|---|---|---|---|
| 200 indep SNPs | .059 | .058 | .060 | .048 | .051 | .068 | .050 | .054 | .053 | .048 |
| 200 corr SNPs | .058 | .065 | .047 | .051 | .060 | .045 | .058 | .048 | .054 | .052 |
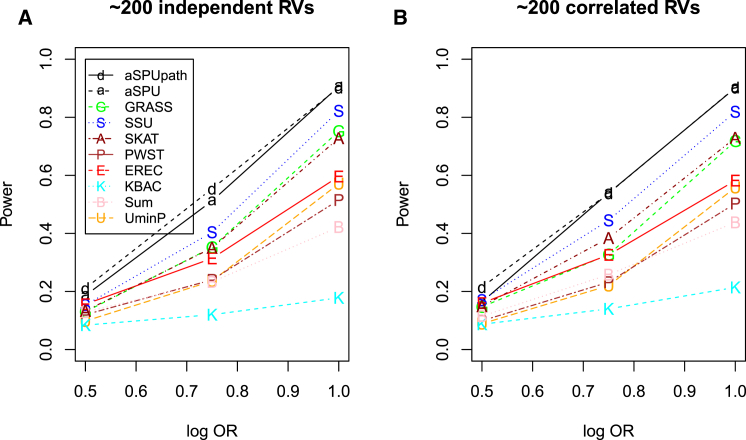
As shown in Figure 6, the relative performance of the various tests did not strongly depend on whether there were within-gene correlations among the RVs. Clearly, the aSPUpath test was the most powerful, closely followed by the usual aSPU test, then followed by the SSU test, then GRASS, SKAT, and EREC tests. Although the SSU and SKAT are closely related, because SKAT over-weights rare variants with smaller MAFs, which was not a correct assumption in our simulations, here the SSU test was more powerful than SKAT. It is worth noting that here GRASS was much more powerful than Plink, perhaps due to the latter’s ineffective screening on each individual RV, which contained only a quite limited association information content with a low MAF.
Figure 6.
Empirical Power for RVs in Simulation Set-up D2 with a Pathway Containing 20 Genes
Ten genes each included one causal RV. Each gene contained 1–20 independent (A) or correlated (B) RVs.
The PWST and the single RV-based UminP test performed similarly. The KBAC had lowest power. Note that here all the causal RVs had an equal association strength (and direction), which was supposed to be ideal for the Sum test (or other burden tests); however, due to the presence of many non-associated RVs, the Sum test and several other adaptive tests did not perform well due to their non- or not-so-good selection or down-weighting of the many non-associated RVs, as discussed in Pan et al.13
Example
We applied the proposed aSPUpath test, as well as the GRASS test, to the Wellcome Trust Case Control Consortium (WTCCC) GWAS data for Crohn disease (CD [MIM: 266600]).37 CD, a type of inflammatory bowel disease, is also considered an autoimmune disease with a strong genetic component.3 The WTCCC GWAS dataset contains 2,000 CD-affected case subjects and 3,000 control subjects with a total of 500,568 SNPs. Following the WTCCC’s quality control (QC) recommendations, we removed subjects and SNPs that did not pass the QC criteria, resulting in 469,612 SNPs in 1,748 case subjects and 2,938 control subjects. We further restricted the pathway analysis to SNPs with MAF of at least 1%. We retrieved a total of 214 human biological pathways from the KEGG database.1 Because a too-small pathway can give results not too different from a gene-based analysis, whereas the annotated function of a large pathway is likely to be non-specific, many authors restricted their analyses to pathways of certain sizes. For example, Chen et al.14 and Wang et al.3 considered pathways with at least 10 genes, whereas Gui et al.9 included only pathways containing between 10 and 300 genes. Following the previous authors, to facilitate interpretation of the results, we excluded too-small (<10 genes) and too-big (>500 genes) pathways, which resulted in 197 pathways. We obtained the genomic coordinates of SNPs and genes according to human reference genome hg19 and assigned a SNP to a gene if it is located within 20,000 base pairs (20 kb) upstream or downstream of the gene to include SNPs in regulatory regions. A total of 64,557 SNPs were mapped to the 197 pathways including 4,572 unique genes. The median number of genes in a pathway was 47 with the first and third quartiles being 27 and 76, and the median number of SNPs in a gene was 8 with the first and third quartiles being 4 and 17, respectively. We employed a stage-wise permutation strategy for both aSPUpath and GRASS tests: we first performed 5,000 permutations for all pathways and then increased to 100,000 permutations for those pathways with p values < 0.01 in the first stage. We set the significance threshold at 0.00025 to control the family-wise error rate (FWER) at 0.05 based on the Bonferroni correction for 197 pathways.
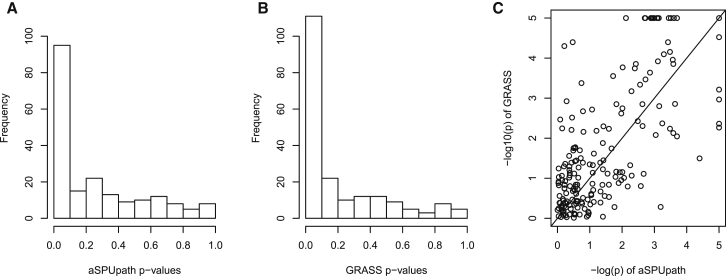
Figure 7 shows the histograms of the p values across the 197 KEGG pathways by the new method and GRASS; their distributions were similar, though GRASS gave a larger number of more significant p values. Overall, the two methods gave similar and complementary results: although many common pathways were identified to be significant by both methods, each also detected some unique pathways. For example, at the significance threshold of 0.00025, aSPUpath and GRASS identified 18 and 35 significant pathways, respectively, among which 11 were common. The Spearman’s rank correlation coefficient between the p values of the two methods was 0.65.
Figure 7.
Distributions of the p Values from aSPUpath and GRASS and Their Comparison in the Log10 Scale for the WTCCC CD Data
Shown are aSPUpath (A) and GRASS (B) and comparison (C).
Table 4 shows 24 KEGG pathways with p values less than 0.00001 by either method, i.e., none of the permuted test statistics exceeded the observed one based on 100,000 permutations. Interestingly, five pathways that have been confirmed to be associated with susceptibility to CD by meta-analysis and replication studies3,38,39 are all among the 24 pathways. Three of them had p values less than 0.00001 by both methods. Of note, the JAK-STAT signaling pathway (hsa04630) has been identified in quite a few previous pathway analyses.9,10,40 This pathway has 145 genes, including IL23R (MIM: 607562) with a cluster of genome-wide significant SNPs in the WTCCC GWAS of CD, and nine additional genes, for example, JAK2 (MIM: 147796) and STAT3 (MIM: 102582), which were found to be associated with CD in a large-scale meta-analysis.38 Therefore, it is relatively easy to be identified by several pathway analysis methods.3,9 On the other hand, two positive control pathways, namely, NOD-like receptor signaling pathway (hsa04621) and Chemokine signaling pathway (hsa04062), had p values < 0.00001 only by aSPUpath, but were not significant by GRASS (p values > 0.00025). It is noteworthy that SNPs in NOD2 (MIM: 605956) in the NOD-like receptor signaling pathway were the first to be identified to be associated with CD and confer the highest risk for CD development among all CD-susceptibility SNPs discovered thus far.39,41 The NOD-like receptor signaling pathway includes not only NOD2, but also several other CD-associated genes, including TNF (MIM: 191160), CCL2 (MIM: 158105), and CCL7 (MIM: 158106), making it one of the most-well-understood pathways underlying CD susceptibility.42 The data application here demonstrates that our proposed aSPUpath test is a competitive and complementary approach to the GRASS test.
For comparison, we also ran GATES-Simes and HYST, yielding 5 and 4 pathways with p values < 0.00001, respectively, all but one of which had a p value < 0.00001 by either aSPUpath or GRASS. The only exception was pathway hsa04622 “RIG-I-like receptor signaling;” the four methods, aSPUpath, GRASS, GATES-Simes, and HYST, gave p values of 0.00004, 0.0318, <0.00001, and 0.3050, respectively.
Discussion
We have proposed a powerful adaptive test for pathway analysis of genetic SNP data as arising in GWASs.3,4,43 Because any pathway analysis involves multiple genes, each containing multiple SNPs, it is desirable to apply a test that can maintain high power with a large number of non-associated SNPs (or genes) and multiple only weakly associated SNPs (or genes), an ideal case for our proposed test. On the other hand, because the genes in a pathway can contain different numbers of SNPs, to avoid undue influence from a large (or small) gene, we modify the tests to take account of varying gene lengths. Our proposed test introduces two parameters (γ and γG) to achieve the objective. For example, if there are only few genes, each containing many associated SNPs (e.g., due to LD), a large value of γ and a small value of γG would yield a more powerful test; because the truth is unknown, we use data to adaptively estimate their optimal values. The adaptivity of the proposed test at the gene level and/or at the SNP level is missing from many existing tests for pathway or SNP set analysis, such as the SSU and SKAT tests. As supported by our numerical examples, the proposed test can gain power in many situations and serve as a tool complementary to existing methods like GRASS.
Our proposed test is general and applicable to CVs or RVs. It can be modified, e.g., via suitable weighting on SNPs, for analysis of both CVs and RVs, as shown for the SSU test in Basu and Pan.31 In addition, we can also introduce some weights at the gene and SNP levels to incorporate biological knowledge on which genes or SNPs are more likely to be causal. We have focused on testing on a single pathway; an alternative is to take account of possible overlapping or hierarchical structures of some pathways as discussed in Schaid et al.12 These topics warrant future investigation.
Finally, we note that our proposed approach is in the category of “self-contained tests,” not “competitive tests,” because the null hypothesis to be tested here fits the former better than the latter: we are interested in detecting any pathways with any SNPs associated with a trait, not in detecting ones that are over-enriched with associated SNPs. Furthermore, as argued by Goeman and Buhlmann,44 the same test on the former is necessarily more powerful than on the latter. Following Zhou et al.,45 we can extend our aSPUpath to competitive testing. Our goal also differs from that of Newton et al.,46 which goes beyond only identifying significant pathways, but also aims to uncover the common theme shared among the identified significant pathways.
Acknowledgments
The authors are grateful to the reviewers for helpful and constructive comments. This research was supported by NIH grant R01HL116720. W.P. was also supported by NIH grants R01GM113250, R01HL105397, and R01GM081535, and P.W. by R01CA169122 and R21HL126032. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the WTCCC data is available from http://www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under award 076113.
Published: June 25, 2015
Contributor Information
Wei Pan, Email: weip@biostat.umn.edu.
Peng Wei, Email: peng.wei@uth.tmc.edu.
Web Resources
The URLs for data presented herein are as follows:
References
- 1.Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K., Li M., Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 4.Fridley B.L., Biernacka J.M. Gene set analysis of SNP data: benefits, challenges, and future directions. Eur. J. Hum. Genet. 2011;19:837–843. doi: 10.1038/ejhg.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L., Jia P., Wolfinger R.D., Chen X., Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goeman J.J., van de Geer S.A., de Kort F., van Houwelingen H.C. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 7.Newton M.A., Quintana F.A., den Boon J.A., Sengupta S., Ahlquist P. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann. Appl. Stat. 2007;1:85–106. [Google Scholar]
- 8.Wang K., Li M., Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gui H., Li M., Sham P.C., Cherny S.S. Comparisons of seven algorithms for pathway analysis using the WTCCC Crohn’s Disease dataset. BMC Res. Notes. 2011;4:386. doi: 10.1186/1756-0500-4-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M.-X., Kwan J.S.H., Sham P.C. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am. J. Hum. Genet. 2012;91:478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M.-X., Gui H.-S., Kwan J.S.H., Sham P.C. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaid D.J., Sinnwell J.P., Jenkins G.D., McDonnell S.K., Ingle J.N., Kubo M., Goss P.E., Costantino J.P., Wickerham D.L., Weinshilboum R.M. Using the gene ontology to scan multilevel gene sets for associations in genome wide association studies. Genet. Epidemiol. 2012;36:3–16. doi: 10.1002/gepi.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan W., Kim J., Zhang Y., Shen X., Wei P. A powerful and adaptive association test for rare variants. Genetics. 2014;197:1081–1095. doi: 10.1534/genetics.114.165035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L.S., Hutter C.M., Potter J.D., Liu Y., Prentice R.L., Peters U., Hsu L. Insights into colon cancer etiology via a regularized approach to gene set analysis of GWAS data. Am. J. Hum. Genet. 2010;86:860–871. doi: 10.1016/j.ajhg.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deelen J., Uh H.-W., Monajemi R., van Heemst D., Thijssen P.E., Böhringer S., van den Akker E.B., de Craen A.J.M., Rivadeneira F., Uitterlinden A.G. Gene set analysis of GWAS data for human longevity highlights the relevance of the insulin/IGF-1 signaling and telomere maintenance pathways. Age (Dordr.) 2013;35:235–249. doi: 10.1007/s11357-011-9340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verschuren J.J.W., Trompet S., Sampietro M.L., Heijmans B.T., Koch W., Kastrati A., Houwing-Duistermaat J.J., Slagboom P.E., Quax P.H.A., Jukema J.W. Pathway analysis using genome-wide association study data for coronary restenosis--a potential role for the PARVB gene. PLoS ONE. 2013;8:e70676. doi: 10.1371/journal.pone.0070676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei P., Tang H., Li D. Insights into pancreatic cancer etiology from pathway analysis of genome-wide association study data. PLoS ONE. 2012;7:e46887. doi: 10.1371/journal.pone.0046887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J. Test of significance based on wavelet thresholding and Neyman’s truncation. J. Am. Stat. Assoc. 1996;91:674–688. [Google Scholar]
- 20.Pan W. Relationship between genomic distance-based regression and kernel machine regression for multi-marker association testing. Genet. Epidemiol. 2011;35:211–216. doi: 10.1002/gepi.20567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzeng J.Y., Zhang D., Pongpanich M., Smith C., McCarthy M.I., Sale M.M., Worrall B.B., Hsu F.C., Thomas D.C., Sullivan P.F. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. Am. J. Hum. Genet. 2011;89:277–288. doi: 10.1016/j.ajhg.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu M.C., Kraft P., Epstein M.P., Taylor D.M., Chanock S.J., Hunter D.J., Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goeman J.J., van de Geer S., van Houwelingen H.C. Testing against a high dimensional alternative. J. R. Stat. Soc., B. 2006;68:477–493. [Google Scholar]
- 24.Wessel J., Schork N.J. Generalized genomic distance-based regression methodology for multilocus association analysis. Am. J. Hum. Genet. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwee L.C., Liu D., Lin X., Ghosh D., Epstein M.P. A powerful and flexible multilocus association test for quantitative traits. Am. J. Hum. Genet. 2008;82:386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan W., Chen Y.M., Wei P. Testing for polygenic effects in genome-wide association studies. Genet. Epidemiol. 2015;39:306–316. doi: 10.1002/gepi.21899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Abbott D. A principal components regression approach to multilocus genetic association studies. Genet. Epidemiol. 2008;32:108–118. doi: 10.1002/gepi.20266. [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Wang L., Hu B., Guo M., Barnard J., Zhu X. Pathway-based analysis for genome-wide association studies using supervised principal components. Genet. Epidemiol. 2010;34:716–724. doi: 10.1002/gepi.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang T., Elston R.C. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am. J. Hum. Genet. 2007;80:353–360. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan W. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genet. Epidemiol. 2009;33:497–507. doi: 10.1002/gepi.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu S., Pan W. Comparison of statistical tests for disease association with rare variants. Genet. Epidemiol. 2011;35:606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D.J., Leal S.M. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q., Irvin M.R., Arnett D.K., Province M.A., Borecki I. A data-driven method for identifying rare variants with heterogeneous trait effects. Genet. Epidemiol. 2011;35:679–685. doi: 10.1002/gepi.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin D.Y., Tang Z.Z. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng G., Luo L., Siu H., Zhu Y., Hu P., Hong S., Zhao J., Zhou X., Reveille J.D., Jin L. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet. 2010;18:111–117. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strober W., Asano N., Fuss I., Kitani A., Watanabe T. Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn’s disease. Immunol. Rev. 2014;260:249–260. doi: 10.1111/imr.12193. [DOI] [PubMed] [Google Scholar]
- 42.Billmann-Born S., Lipinski S., Böck J., Till A., Rosenstiel P., Schreiber S. The complex interplay of NOD-like receptors and the autophagy machinery in the pathophysiology of Crohn disease. Eur. J. Cell Biol. 2011;90:593–602. doi: 10.1016/j.ejcb.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 43.Torkamani A., Topol E.J., Schork N.J. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goeman J.J., Bühlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y.H., Barry W.T., Wright F.A. Empirical pathway analysis, without permutation. Biostatistics. 2013;14:573–585. doi: 10.1093/biostatistics/kxt004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newton M.A., He Q., Kendziorski C. A model-based analysis to infer the functional content of a gene list. Stat. Appl. Genet. Mol. Biol. 2012;11:9. doi: 10.2202/1544-6115.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]