Abstract
Three multiplex PCR assays were developed to identify the 11 most common Listeria monocytogenes clones in clinical and food samples; 270 (95.7%) of 282 strains of serogroups IVb, IIb, IIa, and IIc were identified accurately. This novel tool is a rapid and efficient alternative to multilocus sequence typing for identification of L. monocytogenes clones.
TEXT
Human and food isolates of the food-borne pathogen Listeria monocytogenes belong predominantly to two phylogenetic lineages (1–3). Lineage I isolates belong mostly to serotypes 4b and 1/2b, whereas lineage II isolates mostly belong to serotypes 1/2a and 1/2c. A multiplex PCR assay (4) allows identification of PCR serogroups IVb (serotypes 4b and rare variants 4d and 4e), IIa (serotypes 1/2a and 3a), IIb (serotypes 1/2b, 3b, and 7), IIc (serotypes 1/2c and 3c), and L (serotypes 4a, 4ab, and 4c). Unfortunately, serogrouping has limited discriminatory power, thus providing limited resolution for epidemiological typing. Multilocus sequence typing (MLST) can discriminate isolates within the same serogroup (5). The large-scale application of MLST led to the identification of numerically predominant and internationally widespread clonal complexes (CCs), which are considered clones, i.e., sets of isolates descending from a single common ancestor (5–9). Most major clones have been involved in outbreaks of listeriosis (7, 10–12). Rapid identification of MLST clones is important for epidemiological surveillance and outbreak investigation. However, MLST is time-consuming and is too expensive for routine use in most laboratories involved in L. monocytogenes typing. The objective of this work was to enable rapid affordable identification of major clones of L. monocytogenes by multiplex PCR.
Clone-specific target genes for PCR were identified based on comparative analyses of 104 genomes that were representative of the diversity of clones of L. monocytogenes (our unpublished results). We determined the pan-genome, i.e., the full complement of protein-coding gene families of the species, as described previously (13). Gene families specific for major clones, lineages, or serogroups were identified and represented putative target genes for PCR identification. Among these, target genes that were of sufficient size and were not present on mobile genetic elements were selected. Primers were then carefully designed (see Table S1 in the supplemental material), using Primer3 (14), to produce amplification products with different sizes and to amplify DNA at a unique annealing temperature. Target genes were grouped into three “clonogrouping” multiplex PCR assays, which were intended to be used downstream of the widely used PCR serogrouping assay. First, the IVb clonogrouping PCR assay was designed to identify serogroup IVb clones CC1, CC2, CC4, and CC6 or any other serogroup IVb isolate. Second, the IIb PCR assay was designed to identify serogroup IIb clones CC3 and CC5 or any other serogroup IIb isolate. Third, the IIaIIc PCR assay was designed to identify either serogroup IIa clones CC7, CC8, CC121, and CC155 or clone CC9, most isolates of which are in serogroup IIc. Expected amplification results are given in Table S2 in the supplemental material. PCR mixture details are given in Table S3 in the supplemental material. The three multiplex PCRs were performed with an initial denaturation step of 94°C for 30 s, 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 75 s, and a final elongation step of 72°C for 10 min. Eight microliters of the PCR products was mixed with 3 μl of loading buffer, and the products were separated on a 1.5% agarose gel, in 1× Tris-borate-EDTA (TBE) buffer, at 120 V for 90 min.
A total of 282 L. monocytogenes strains were used to validate the method (see Table S4 in the supplemental material). Among these, 185 were analyzed previously by MLST (6, 7, 15), whereas the MLST genotypes of the 97 remaining strains were determined in the present study. The latter strains were selected from the Collection of Listeria of the Institut Pasteur (CLIP) maintained at the French National Reference Centre and the WHO Collaborating Centre for Listeria. The strains were isolated in 33 countries, with 57% of the strains from France. There were 202 strains (72%) from human infections and 42 (15%) from food, and the remaining strains were from animals and the environment. DNA extractions were performed using the Promega Wizard genomic DNA purification kit. PCR serogrouping and MLST were performed as described previously (4, 5). Novel alleles and sequence types (STs) were submitted to the international MLST database (http://www.pasteur.fr/mlst). Clonal complexes were defined as groups of allelic profiles sharing at least 6 of 7 alleles with at least one other member of the group (5). Allelic profiles sharing less than 6 alleles with all other allelic profiles were designated singletons; these groups were defined based on MLST data from this study and previous studies (5–8, 15). The discrimination index was calculated using an online tool (http://darwin.phyloviz.net/ComparingPartitions).
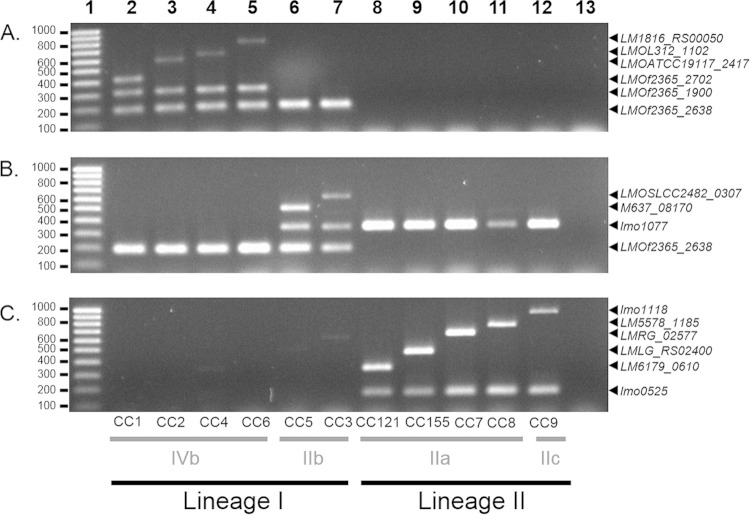
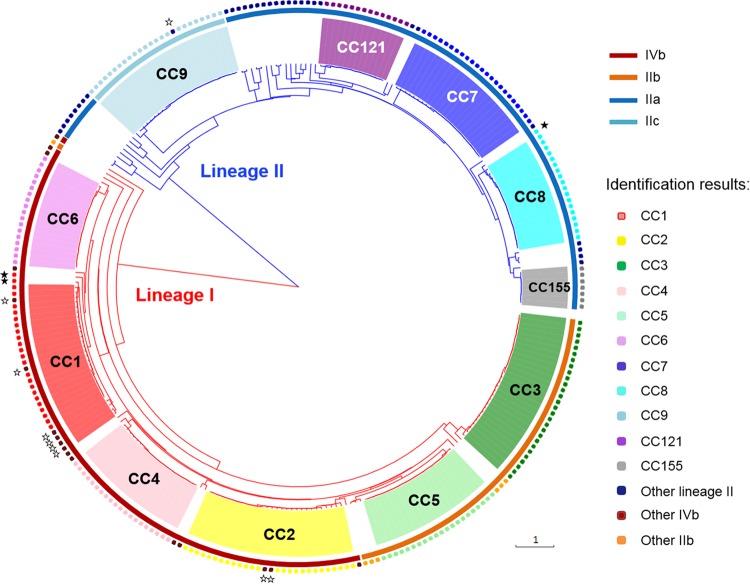
The 282 strains were analyzed using the three multiplex PCR assays (see Table S4 in the supplemental material). Figure 1 shows PCR amplification patterns obtained for representatives of the 11 target clones. Amplification results were highly consistent with the expected profiles of the presence or absence of clone-specific genes and lineage- or serogroup-specific control genes (see Table S2 in the supplemental material). Overall, 223 (96.1%) of 232 strains that belonged to target clones were correctly identified (Table 1 and Fig. 2); 100% of strains belonging to CC3, CC4, CC5, CC6, CC7, CC8, CC121, and CC155 were successfully identified, and correct identification rates were 78.6% (22 of 28 strains) for CC1, 93.1% (27 of 29 strains) for CC2, and 96% (24 of 25 strains) for CC9. No isolate of a target clone was misidentified as belonging to another clone. For the nine strains that were not successfully identified, the identification was correct at the level of lineage and PCR serogroup but was incomplete because the expected clone-specific PCR product was not amplified (Fig. 2; see also Table S4 in the supplemental material). To understand these discordances, genome sequencing was performed, confirming the absence of the gene for five strains, i.e., LM235, LM68, LM71, LM12-01748, and SLCC63 (data not shown). For the remaining strains (LM12980, LM86256, LM18354, and LM19700), the gene was present without priming site mismatches, which was confirmed by simplex PCR.
FIG 1.
Agarose gel electrophoresis of PCR products generated by IVb clonogrouping multiplex PCR (A), IIb clonogrouping multiplex PCR (B), and IIaIIc clonogrouping multiplex PCR (C). Lanes 2 to 12, L. monocytogenes strains of CC1 (CLIP2013/00068), CC2 (CLIP2012/00799), CC4 (CLIP2011/00181), CC6 (CLIP2010/01520), CC5 (CLIP2011/00140), CC3 (CLIP2011/00787), CC121 (CLIP2012/00189), CC155 (2010/01410), CC7 (CLIP2012/01275), CC8 (CLIP2012/01049), and CC9 (CLIP2010/01314), respectively; lane 13, negative control; lane 1, Smart Ladder small fragment (Eurogentec). Genes corresponding to the amplified fragments are indicated on the right. Molecular sizes (in base pairs) are given on the left.
TABLE 1.
Identification results
| CC | No. of isolates | No. (%) of correct identifications | No. (%) of incomplete identifications | No. (%) of incorrect identifications |
|---|---|---|---|---|
| CC1 | 28 | 22 (78.6) | 6 (21.4) identified as other serogroup IVb | |
| CC2 | 29 | 27 (93.1) | 2 (6.9) identified as other serogroup IVb | |
| CC3 | 28 | 28 (100) | ||
| CC4 | 20 | 20 (100) | ||
| CC5 | 21 | 21 (100) | ||
| CC6 | 19 | 19 (100) | ||
| CC7 | 23 | 23 (100) | ||
| CC8 | 18 | 18 (100) | ||
| CC9 | 25 | 24 (96) | 1 (4.0) identified as other lineage II | |
| CC121 | 14 | 14 (100) | ||
| CC155 | 7 | 7 (100) | ||
| Other serogroup IVb | 11 | 9 (81.8) | 2 (18.2) identified as CC1 | |
| Other serogroup IIb | 7 | 7 (86) | ||
| Other lineage II | 32 | 31 (96.9) | 1 (3.1) identified as CC8 | |
| Total | 282 | 270 (95.7) | 9 (3.2) | 3 (1.1) |
FIG 2.
Phylogenetic tree based on concatenation of the seven MLST gene sequences of the 282 strains that were tested using the three multiplex PCRs. Branches of lineage I are in red and branches of lineage II are in blue. Clones defined based on MLST data are indicated with colored areas at the leaves. Colored areas around the tree indicate the serogroups defined with the PCR serogrouping method. Identification results based on the clonogrouping multiplex PCR method are indicated by rounded squares on the outer circle (see key). White stars, incomplete identifications; black stars, incorrect identifications.
Of the 50 strains that did not belong to target clones, 47 (94%) yielded the expected lineage- and serogroup-specific PCR fragments, and three were misidentified as either CC1 (LM84790 and LM10-01572) or CC8 (LM10-01432), from which they differed by only two or three MLST alleles. Genomic sequencing of the three strains confirmed the presence of the unexpected genes (data not shown). Given the close phylogenetic relatedness of these strains to the target clones, these results suggest that the clone-specific gene was acquired ancestrally, rather than horizontally. Strains that were identified by the multiplex PCR only at the lineage or serogroup level could be characterized more precisely using classic MLST (5).
In conclusion, we have developed a set of three multiplex PCRs enabling clonogrouping, i.e., determining whether a strain belongs to one of the major clonal groups of L. monocytogenes. Overall, 270 (95.7%) of 282 strains were identified at the clone level, whereas nine (3.2%) were identified correctly above the clone level and only three (1.1%) were misidentified. This novel tool is intended to be used in a two-step strategy, i.e., first, use of the PCR serogrouping method (4) and, second, depending on the PCR serogroup determined, use of the appropriate clonogrouping PCR assay (IVb, IIb, or IIaIIc) for identification at the MLST clone level. Unlike MLST, this strategy can provide clone identification in less than 1 day starting from a bacterial culture. Based on the data set of this study, the discriminatory power (Simpson's index) of clonogrouping was 92.3% (95% confidence interval [CI], 91.8 to 92.8%), whereas PCR serogrouping had a discrimination level of only 77.5% (95% CI, 76.3 to 78.7%). Therefore, this widely applicable new tool will significantly refine, beyond PCR serogrouping, the genotypic characterization of L. monocytogenes strains. It will thus provide improved resolution for molecular surveillance while rapidly providing information to risk managers during investigations of outbreaks. In investigations of putative epidemiologically linked cases, it may rapidly provide information that strains that belong to the same PCR serogroup actually belong to different CCs, in a more-timely manner than time-consuming but more discriminatory methods such as pulsed-field gel electrophoresis (PFGE) and genome-based typing. Finally, the wide use of this novel method should contribute to better defining the distribution of major clonal groups in clinical, food, and environmental sources.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported financially by the Institut Pasteur, INSERM, French Government Investissement d'Avenir grant ANR-10-LABX-62-IBEID, and Institut de Veille Sanitaire.
A patent application describing the clonogrouping method was submitted (V. Chenal-Francisque, M. M. Maury, M. Lavina, M. Touchon, A. Leclercq, M. Lecuit, and S. Brisse, patent application EP 15 306 154.4, 10 July 2015, European Patent Office).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00738-15.
REFERENCES
- 1.Piffaretti JC, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser JM, Selander RK, Rocourt J. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A 86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun 65:2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, Lecuit M, Brisse S. 2011. Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis 17:1110–1112. doi: 10.3201/eid/1706.101778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantinelli T, Chenal-Francisque V, Diancourt L, Frezal L, Leclercq A, Wirth T, Lecuit M, Brisse S. 2013. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J Clin Microbiol 51:3770–3779. doi: 10.1128/JCM.01874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haase JK, Didelot X, Lecuit M, Korkeala H, L. monocytogenes MLST Study Group, Achtman M. 2014. The ubiquitous nature of Listeria monocytogenes clones: a large-scale multilocus sequence typing study. Environ Microbiol 16:405–416. doi: 10.1111/1462-2920.12342. [DOI] [PubMed] [Google Scholar]
- 9.Adgamov R, Zaytseva E, Thiberge JM, Brisse S, Ermolaeva S. 2012. Genetically related Listeria monocytogenes strains isolated from lethal human cases and wild animals, p 235–250. In Caliskan M. (ed), Genetic diversity in microorganisms. InTech, Rijeka, Croatia. [Google Scholar]
- 10.Kathariou S. 2003. Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes, p 243–256. In Torrence ME, Isaacson RE (ed), Microbial food safety in animal agriculture. Iowa State University Press, Ames, IA. [Google Scholar]
- 11.Knabel SJ, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon CA, Gilmour MW. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J Clin Microbiol 50:1748–1751. doi: 10.1128/JCM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EP. 2014. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol 6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3: new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chenal-Francisque V, Diancourt L, Cantinelli T, Passet V, Tran-Hykes C, Bracq-Dieye H, Leclercq A, Pourcel C, Lecuit M, Brisse S. 2013. Optimized multilocus variable-number tandem-repeat analysis assay and its complementarity with pulsed-field gel electrophoresis and multilocus sequence typing for Listeria monocytogenes clone identification and surveillance. J Clin Microbiol 51:1868–1880. doi: 10.1128/JCM.00606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.