Abstract
BK virus (BKV) infection causing end-organ disease remains a formidable challenge to the hematopoietic cell transplant (HCT) and kidney transplant fields. As BKV-specific treatments are limited, immunologic-based therapies may be a promising and novel therapeutic option for transplant recipients with persistent BKV infection. Here, we describe a whole-genome, deep-sequencing methodology and bioinformatics pipeline that identify BKV variants across the genome and at BKV-specific HLA-A2-, HLA-B0702-, and HLA-B08-restricted CD8 T-cell epitopes. BKV whole genomes were amplified using long-range PCR with four inverse primer sets, and fragmentation libraries were sequenced on the Ion Torrent Personal Genome Machine (PGM). An error model and variant-calling algorithm were developed to accurately identify rare variants. A total of 65 samples from 18 pediatric HCT and kidney recipients with quantifiable BKV DNAemia underwent whole-genome sequencing. Limited genetic variation was observed. The median number of amino acid variants identified per sample was 8 (range, 2 to 37; interquartile range, 10), with the majority of variants (77%) detected at a frequency of <5%. When normalized for length, there was no statistical difference in the median number of variants across all genes. Similarly, the predominant virus population within samples harbored T-cell epitopes similar to the reference BKV strain that was matched for the BKV genotype. Despite the conservation of epitopes, low-level variants in T-cell epitopes were detected in 77.7% (14/18) of patients. Understanding epitope variation across the whole genome provides insight into the virus-immune interface and may help guide the development of protocols for novel immunologic-based therapies.
INTRODUCTION
BK virus (BKV) is the leading viral cause of premature kidney transplant failure in kidney transplant recipients and hemorrhagic cystitis in hematopoietic cell transplant (HCT) recipients. In kidney transplantation, BKV-associated nephropathy affects 1 to 10% of recipients with allograft failure as high as 80% (1). Similarly, BKV-associated hemorrhagic cystitis occurs in 5 to 40% of HCT recipients and results in the need for blood transfusions and prolonged hospitalization (2). Despite the serious posttransplantation morbidity associated with persistent BKV replication, treatment options remain limited, and BKV remains a significant challenge to the transplant field.
BKV is a polyomavirus with a 5.1-kbp closed circular DNA genome. The genome consists of a noncoding control region (NCCR) that contains the origin of replication and bidirectional promoters, the early genes encoding small tumor (ST) and large tumor (LT) antigens, and the late genes encoding viral capsid proteins 1 to 3 (VP1 to VP3) and agnoprotein (Fig. 1) (3). BKV infection is ubiquitous with a peak age of infection of 2 to 5 years and an 80% seroprevalence by adulthood (4). After primary infection by respiratory or oral routes, BKV establishes a persistent infection in renal tubular epithelial cells and uroepithelium, but can replicate to high levels and cause significant pathology in immunocompromised patients.
FIG 1.

Organization of the BKV genome. Four long-range, inverse primer pairs (black arrowheads) were used to generate overlapping PCR amplicons for whole-genome sequencing. agno, agnoprotein; ori, origin.
BKV-specific T cells have an important role in controlling BKV replication. As no effective antivirals currently exist, treatment is based on restoring the underlying cellular immune defect by reducing immunosuppression when high levels of BKV in plasma or urine are detected (5, 6). Several studies have confirmed that a robust cellular immune response directed at BKV peptides is correlated with resolution of viremia (7–9). However, a reduction in immunosuppressive therapy is not always possible and must be carefully balanced with maintaining adequate immunosuppression to prevent allograft rejection. A promising treatment option that infuses virus-specific T cells to boost the host immune response to combat persistent viral infections has shown encouraging results in transplant recipients with Epstein-Barr virus and cytomegalovirus infections (10–12). Given the limited treatment options against BKV, adoptive T-cell transfer is an attractive and novel therapeutic option for HCT and kidney transplant recipients with persistent BKV infection.
Effective adoptive T-cell therapy requires a firm understanding of the virus-immune interface. In particular, infused T cells must recognize BKV peptides displayed by host cells. Currently, genetic variation in regions of the BKV genome that encode T-cell epitopes is not known. Notably, T-cell epitopes have been identified in nearly all BKV proteins, including VP1 to VP3, LT, and ST (7, 13). We therefore developed a whole-genome, deep-sequencing methodology and informatics pipeline to detect amino acid variants in previously described T-cell epitopes in pediatric HCT and kidney transplant recipients with active BKV infection. Such knowledge may provide insight into optimization of cell-mediated immunotherapy.
(Part of this research was presented at IDWeek 2014, Philadelphia, PA, 8 to 12 October 2014.)
MATERIALS AND METHODS
Ethics.
This study was approved by the Stanford University Institutional Review Board.
Study samples.
Pediatric HCT and kidney transplant recipients with active BK infection were identified during routine, preemptive monitoring for BK disease with weekly quantitative PCR testing. BKV whole-genome sequencing was performed on 65 samples collected during 2011-2013 from 18 patients local to our institution with quantifiable BKV DNAemia in plasma or urine. Demographic and clinical characteristics, including age, sex, HLA-match status, and diagnosis of BK nephropathy by histopathologic criteria or BK hemorrhagic cystitis by clinical criteria, were obtained through chart review.
Nucleic acid extraction, BKV quantitation, and standard VP1 sequencing.
Viral nucleic acids were extracted from frozen plasma or urine samples as previously described (14). BKV DNA was quantitated by real-time PCR (14). The VP1 region of one specimen from each patient was amplified using forward primer 5′-TGGACTTAAGAAATCAAC-AAAGTGTACATTCAGGA-3′ and reverse primer 5′-CTGCTGAAGATTCCCAAAGGTCAGAC-3′. Each reaction included 1× PCR master mix (2×; Thermo Fisher Scientific Inc., Waltham, MA), 500 nM each primer, and 5 μl nucleic acid. The reactions were run in a DNA Engine PTC-200 (Bio-Rad, Hercules, CA), with the following cycling conditions: 95°C for 2 min; 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min 40 s; and final extension for 10 min at 72°C. The PCR products were sequenced by bidirectional dideoxynucleotide termination sequencing using the same forward and reverse primers (Elim Biopharmaceuticals, Inc., Hayward, CA).
Long-range PCR.
BKV genomes were amplified by inverse long-range PCR with four primer sets (Fig. 1 and Table 1) using the SequalPrep Long PCR kit (Invitrogen, Grand Island, NY). The 20-μl reactions were composed of 2 μl 10× reaction buffer, 0.4 μl DMSO, 1 μl 10× enhancer A, 1.8 U enzyme, 250 nM each primer, and 10 μl nucleic acid extract. The reactions were run in a DNA engine PTC-200 (Bio-Rad, Hercules, CA), using the following cycling parameters: 94°C for 2 min; 5 cycles of 94°C for 10 s, 60°C for 30 s, and 68°C for 5 min; 35 cycles of 94°C for 10 s, 60°C for 30 s, and 68°C for 5 min +20 s/cycle; followed by a final extension at 68°C for 10 min. For each sample, all four PCR products were pooled, the column was purified using the GeneJET PCR purification kit (Thermo Fisher Scientific Inc., Waltham, MA), and the concentration of DNA was determined using the Quant-iT PicoGreen dsDNA assay kit (Life Technologies, Carlsbad, CA), according to the manufacturer's recommendations. Purified and quantitated PCR products were stored at −20°C until library preparation.
TABLE 1.
Primers used for BKV amplification and sequencing
| Primer name | Sequence (5′ to 3′)a | Positionb | Gene |
|---|---|---|---|
| Long-range primer | |||
| LR 1-F | AATTTAAATGAGGACCTAACCTGTGGAAATCT | 1855–1886 | VP1 |
| LR 1-R | TCTGGGTTTAGGAAGCATTCTACCTCT | 1700–1727 | VP1 |
| LR 2-F | CTGCTTTTGTATAAGCCACTTTTAAGCTTGTGT | 2690–2722 | Large T antigen |
| LR 2-R | AGGCCTCTCCACTGTTGTGTTCCAG | 2387–2411 | VP1 |
| LR 3-F | ATTAGCAGTAGCAACAAGGTCATTCCACTTTGTA | 3065–3098 | Large T antigen |
| LR 3-R | TGAATGGAAGGAAAGGCTGGATTCTGA | 2987–3013 | Large T antigen |
| LR 4-F | GCTTTAGATCTCTGAAGGGAGTTTCTCCAATT | 4640–4671 | Small T antigen |
| LR 4-R | CTTGCCTGCTTTGCTGTGTATAC | 4316–4338 | Large T antigen |
| VP1 primer | |||
| VP1-F | TGGACTTAAGAAATCAACAAAGTGTACATTCAGGA | 1408–1440 | VP2/VP3 |
| VP1-R | CTGCTGAAGATTCCCAAAGGTCAGAC | 2797–2822 | Large T antigen |
Each forward (F) primer is preceded by 5′-TACGGTAGCAGAGACTTGGTCT-3′ and each reverse (R) primer is preceded by 5′-GCGGCCGCTAATACGACTCACTATAGG-3′.
Relative to the Dunlop reference sequence (GenBank accession no. V01108).
Plasmid controls.
The long-range PCR product of primer set 1 (LR 1-F and LR 1-R), using BKV control DNA extracted from a NATtrol BK Virus Linearity Panel (ZeptoMetrix Corporation, Buffalo, NY) as the template, was cloned into the pJET1.2 cloning vector using the CloneJET PCR cloning kit (Thermo Fisher Scientific Inc., Waltham, MA), according to the manufacturer's instructions. Plasmids were propagated in Escherichia coli NEB10 beta cells and isolated using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific Inc.). Plasmid sequences were confirmed by bidirectional dideoxynucleotide termination sequencing (Elim Biopharmaceuticals, Inc., Hayward, CA) and used for estimation of error in the library generation and sequencing steps.
Library preparation.
Sequencing libraries were prepared from approximately 500 ng of purified long-range PCR products using the Ion Xpress Plus fragment library kit (Life Technologies, Carlsbad, CA). DNA was fragmented using Ion Shear Plus reaction buffer and 10 μl enzyme mix in a 50-μl reaction volume followed by incubation at 37°C for 2 min. Fragmentation reactions were stopped by addition of 5 μl Ion Shear stop buffer, and fragments were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA). Purified fragments were nick repaired, adaptor ligated, and barcoded in a 100-μl reaction mixture containing 1× ligase buffer, 3 μl Ion P1 adaptor, 3 μl Ion Xpress barcode, 2 μl dNTP mix, 2 μl DNA ligase, and 8 μl nick repair polymerase. The reaction mixtures were incubated at 25°C for 15 min and then at 72°C for 5 min. The nick-repaired and adaptor-ligated fragments were immediately purified using Agencourt AMPure XP magnetic beads and size selected in E-Gel SizeSelect 2% agarose gels (Life Technologies, Carlsbad, CA), targeting fragments of ∼350 bp. The size-selected library was amplified in a PCR containing 100 μl Platinum PCR SuperMix High Fidelity (Invitrogen, Grand Island, NY), 5 μl Library Amplification Primer Mix, and 25 μl unamplified library. The following cycling conditions were used: 95°C for 5 min and 8 cycles of 95°C for 15 s, 58°C for 15 s, and 70°C for 1 min. The resulting PCR products were purified using AMPure XP magnetic beads and checked for quality on the Agilent 2100 expert bioanalyzer using a high-sensitivity DNA kit (Agilent Technologies, Santa Clara, CA). The bioanalyzer concentration estimate was used for normalization of each sample library in the final pool of up to 16 samples for a single sequencing run. The pooled library concentration was estimated using the Quant-iT PicoGreen dsDNA assay kit (Life Technologies, Carlsbad, CA), and the concentration was calculated using the following equation: library concentration (nanomolar) = [(sample DNA concentration in nanograms per microliter) × 106/[656.6 × (amplicon length in base pairs)]. The pooled library was adjusted to a 450 pM concentration before sequencing.
Ion Torrent sequencing.
An emulsion PCR was performed on 45 pM library fragments mixed with capture beads on the Ion OneTouch system supplemented with the Ion Torrent Personal Genome Machine (PGM) template OT2 200 kit. The template libraries were sequenced on the Ion Torrent PGM using the Ion 314 chip kit v2 and the Ion Torrent PGM Sequencing 200 kit v2, according to the manufacturer's instructions (all from Life Technologies, Carlsbad, CA).
Alignment of sequencing reads.
The full-length BKV subtype Ib1 sequence was used as the reference (isolate WW; GenBank accession no. AB211371.1). The WW sequence contains an intact, nonrearranged NCCR. The WW sequence was renumbered to match the numbering of BKV Dunlop (V01108/NC_001538.1). The reads in FASTQ files from the Ion Torrent suite were quality filtered by trimming out bases from the 3′ end until the moving average quality score of a 30-bp window was >15. The sequence reads were then aligned to the reference using Burrows-Wheeler Aligner Smith-Waterman Alignment (BWA-SW) with default parameters (15). The BAM alignment files were checked visually using Tablet for coverage and read counts (16). Consensus sequences were generated using the pileup function in SAMTools (17).
Phylogenetic analysis, evaluation of recombination.
Consensus sequences obtained via Ion Torrent and VP1 sequences obtained via Sanger sequencing, as well as reference BKV sequences representing each of the known BKV subtypes (Table 2), were analyzed by multiple-sequence alignment using Clustal X 2.0.12 (18). Phylogenetic distances were calculated by the Neighbor joining (NJ) method and exported in the PHYLIP format to generate a phylogenetic tree using MEGA 5.2 (19).
TABLE 2.
BKV whole-genome reference sequences
Error modeling and variant calling.
In order to identify all variants before any noise filter was applied, a custom python script using pysam was used for variant calling. A pileup table containing nucleotide base counts, as well as deletion and insertion events at each position along the reference was generated from the BAM file after poor quality bases as represented by N in Tablet were filtered out. For each position, any nonreference base with a count above zero was called and listed in the raw variants file.
All variants found in the plasmid clones were considered noise, except where the plasmid differed from the reference sequences based on confirmatory Sanger sequencing. At nucleotide positions where no variant was detected, the noise value was considered to be zero (20). Error rates for the plasmid clones were calculated as the mean percent noise reported for all sequenced positions (21). The noise data were analyzed in R using packages vcd, ggplot2, and MASS. A generalized linear (Poisson) model was used to estimate the dependence of the number of noise reads on the covariates (20, 22). The homopolymer covariate was defined as a repeat of 3 or more of the same nucleotide along with nonidentical flanking nucleotides (23). This definition generated 2,045 homopolymer and 3,096 nonhomopolymer positions on the reference sequence.
Evaluation of T-cell epitopes.
The consensus whole-genome sequences were evaluated for the presence of variants in 27 BKV-specific HLA-A2-restricted peptide sequences and 1 HLA-B0702- and HLA-B08-restricted peptide sequence (24, 25) with NCBI bl2seq using TBLASTN 2.2.30+ (26). Genotype-specific amino acid changes were excluded from the epitope variation analysis. Minor variants in these peptide sequences were also quantified for each sample.
RESULTS
BKV detection and subtyping.
A total of 65 samples from 18 pediatric patients (10 HCT and 8 kidney transplant recipients), including 30 urine and 35 plasma samples, were sequenced. Patient demographics are shown in Table 3. A median of 45,466 reads per sample aligned to the BKV subtype Ib1 reference sequence (range, 5,469 to 457,910; interquartile range [IQR], 46,084). In 92% (60/65) of these samples at least 100× coverage was obtained over 95% of the complete BKV genome. In the remaining 8% (5/65), long-range PCR amplification occurred with only one of the four inverse primer pairs, leaving a short sequence gap that lacked coverage. However, in these samples, 100× coverage was achieved over 95% of the target amplicon, and the sequences were analyzed downstream (see Fig. S1 in the supplemental material).
TABLE 3.
Characteristics of pediatric kidney and hematopoietic cell transplant recipients with BK viremia or viruria
| Characteristic | No. of patients |
|---|---|
| Kidney transplant (n = 8) | |
| Age (mean [range]) (yr) | 14 (3–19) |
| Sex (no.) | |
| Male | 5 |
| Female | 3 |
| HLA matches per patient (no. [range]) | 2.25 (0–3) |
| Donor (no.) | |
| Living | 3 |
| Cadaveric | 5 |
| Hematopoietic cell transplant (n = 10) | |
| Age (mean [range]) (yr) | 12 (9–21) |
| Sex (no.) | |
| Male | 5 |
| Female | 5 |
| Conditioning regimen (no.) | |
| Myeloablative | 5 |
| Reduced intensity | 5 |
| HLA match (no.) | |
| Matched related | 3 |
| Matched unrelated | 7 |
| Stem cell source (no.) | |
| Bone marrow | 5 |
| Umbilical | 5 |
The median plasma BK viral load was 107,000 copies/ml (range, <1,000 to 6.24 × 106; IQR, 268,945) and 22/35 had >10,000 copies/ml plasma. The median plasma viral load was higher in HCT recipients than in kidney transplant recipients (112,000 versus 43,250 copies/ml). All urine samples had a viral load of >50,000/ml except one at 9,950/ml, and 25/30 had viral loads of >6 × 107 copies/ml. The median urine BK viral load was >6 × 107 copies/ml in both HCT and kidney transplant recipients.
Phylogenetic analysis of whole-genome sequences from each patient showed 22% (4/18) subtype Ia, 50% (9/18) subtype Ib1, 22% (4/18) subtype Ib2, and 6% (1/18) subtype III (see Fig. S2 in the supplemental material). Genotypes were similarly represented between HCT and kidney transplant recipients. Subtypes Ic, II, and IV were not identified. Phylogenetic analysis of VP1 sequences obtained via Sanger sequencing confirmed the subtype identified by whole-genome sequencing (data not shown). Neither mixed subtype infections nor intersubtype recombinants were identified (data not shown). Furthermore, no rearrangements were observed in the NCCR (27).
Determining the error threshold.
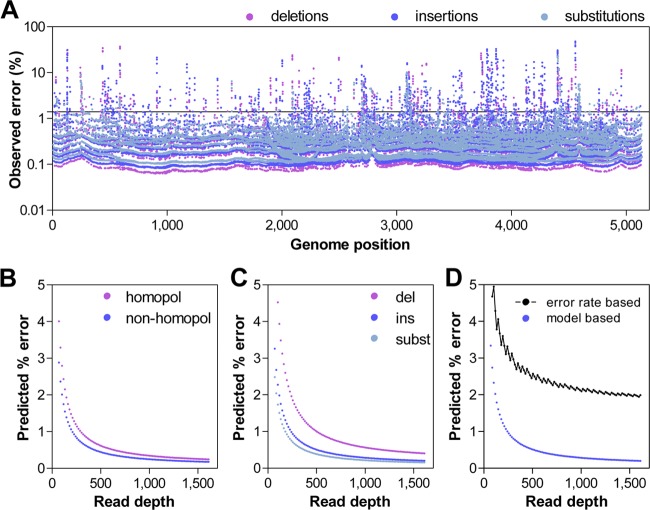
To determine the error in our sequencing approach, we subjected cloned BKV subtype III to the same sequencing protocol utilized for the patient plasma and urine samples. The variants detected from the plasmid sequencing data were considered noise and used to set an error threshold that limits variant false discovery. Based on these experiments, the overall observed per base error rate was determined to be 0.014, and the error was distributed across the genome (Fig. 2A).
FIG 2.
Determination of the error threshold. (A) Error events observed across the genome in a plasmid clone. Insertions, deletions, and substitutions are color coded. The solid black line represents the overall per base error rate of 0.014. (B) Prediction of error rates in homopolymer and nonhomopolymer regions. (C) Prediction of insertion (ins), deletion (del), and substitution (subst) error rates based on the sequence depth using a general linear model (Poisson) with observed error data. (D) Overall error using a general linear model. The black curve represents the error threshold utilized in this study. The threshold was calculated from the overall per base error rate using the Poisson distribution at P < 0.05.
To further characterize the error in our sequencing protocol, we utilized the observed error to model the central tendencies of the method noise using a Poisson model. This analysis demonstrated that the error rate was higher at lower read depths, and the errors were more likely to be deletions and insertions, rather than substitutions (Fig. 2C). The error rate was also higher in homopolymer sequences (Fig. 2B).
BKV variant analysis.
Based on these error data, we took a conservative approach for rare variant calling. Variants from the raw variant call files were evaluated via the following series of sequential steps. First, variants with fewer than 20 total reads or fewer than 3 nonreference reads were discarded. Next, variants showing bias of >0.85 for the percent variant detected in the forward and reverse reads were discarded [bias = 1 − (minimum/maximum)]. Thresholds were then dynamically set based on the read depth according to the Poisson function using the observed plasmid error rate as the probability of error at a P value of <0.05 (23). Variants present at levels below this threshold were discarded (Fig. 2D).
For further refinement, genomic positions defining subtype-specific variants were ignored. The remaining variants were translated to predict an amino acid change in their respective gene context. Variants resulting in synonymous and nonsense changes, as well as variants detected in the BKV plasmid control, were also discarded.
The median number of amino acid variants identified per sample was 8 (range, 2 to 37; IQR, 10). The distribution of variants by gene is shown in Fig. 3A. When normalized for length, there was no statistical difference in the median number of variants across all genes. Similarly, the median number of variants in plasma was 12 (range, 2 to 37; IQR, 13) and not statistically different from the median number of variants found in urine (6; range, 2 to 27; IQR, 6).
FIG 3.
BKV variant analysis. (A) Distribution of amino acid variants in the BKV protein coding genes. (B) Distribution of amino acid variant abundance across the BKV genome. Gray bars, medians.
Of all variants, 77% (566/735) were detected at a frequency of <5%. The median number of variants identified at <5% was 7 (range, 0 to 34; IQR, 9), which was statistically different from the median number of higher frequency variants: 5 to 20% (1; range, 0 to 6; IQR, 2), 20 to 50% (0; range, 0 to 3; IQR, 0), 50 to 90% (0; range, 0 to 2; IQR, 0), >90% (1; range, 0 to 4; IQR, 2) (Fig. 3B).
Conservation of T-cell epitopes.
A total of 27 BKV HLA-A2-restricted peptide sequences and 1 HLA-B0702- and HLA-B08-restricted peptide sequence, as previously described, were evaluated for nonsynonymous mutations in patient samples (24, 25). The consensus BKV sequence among specimens revealed limited variability in these T-cell epitopes. After the exclusion of genotype-specific amino acid changes, only one kidney transplant recipient was found to have a nonsynonymous mutation (S405C in BKV peptide T p398 CLLPKMDSV) in all six plasma specimens obtained weekly over a 1.5-month period of BK viremia. Despite limited variability in the consensus BKV sequence, low-level variants in T-cell epitopes accounting for <10% of the viral population were detected in 77.7% (14/18) of patients (Table 4). The frequencies of low-level variants in patient samples ranged from 1.4 to 9.3%. Select changes, particularly in the LT gene, occurred in multiple patients (K180N, 5 patients; M178I, 3 patients; D409E, 3 patients).
TABLE 4.
Low-level variants of BKV HLA-A2-restricted peptide sequencesa detected in clinical samples of pediatric kidney and hematopoietic cell transplant recipients
| Antigen position | Peptide sequence | Mutation | No. of patients | Variant frequency (%)b |
|---|---|---|---|---|
| VP1 antigen | ||||
| BKV VP1 p62 | NLRGFSLKL | F66L | 1 | 1.87 |
| BKV VP1 p84 | KMLPCYSTA | L86P | 1 | 1.87 |
| BKV VP1 p108 | LLMWEAVTV | E112G | 1 | 2.38 |
| BKV VP1 p128 | NLHAGSQKV | L129F | 1 | 1.72 |
| Q134P | 1 | 4.49 | ||
| K135Q | 1 | 5.34 | ||
| BKV VP1 p245 | LLDEQGVGPL | L254F | 1 | 1.67 |
| BKV VP1 p259 | SLYVSAADI | L260P | 1 | 3.62 |
| Large T antigen | ||||
| BKV T p157 | TLACFAVYT | V163A | 1 | 3.03 |
| BKV T p176 | KLMEKYSVT | M178I | 3 | 2.26; 2.30; 3.53 |
| E179G | 2 | 2.25; 2.27 | ||
| K180N | 5 | 1.92; 2.34; 2.34; 2.49; 3.27 | ||
| BKV T p199 | FLTPHRHRV | H203R | 1 | 1.96 |
| BKV T p216 | KLCTFSFLI | K216R | 1 | 1.88 |
| C218R | 1 | 1.79 | ||
| T219A | 1 | 1.93 | ||
| T219N | 1 | 2.39 | ||
| F220L | 1 | 4.03 | ||
| S221R | 1 | 3.31 | ||
| S221C | 1 | 3.3 | ||
| S221N | 1 | 3.18 | ||
| I224F | 2 | 2.00; 2.37 | ||
| BKV T p313 | PYHFKYHEKHFANAT | K321E | 1 | 2.04 |
| T327N | 1 | 1.97 | ||
| BKV T p362 | MLTERFNHIL | T364A | 1 | 1.94 |
| F367S | 2 | 1.98; 3.22 | ||
| BKV T p395 | WLHCLLPKM | H397R | 1 | 2.07 |
| BKV T p398 | CLLPKMDSV | S405F | 1 | 4.02 |
| BKV T p406 | VIFDFLHCI | D409E | 3 | 1.44; 1.65; 2.21 |
| D409G | 2 | 1.56; 2.61 | ||
| D409Y | 2 | 1.55; 2.74 | ||
| BKV T p410 | FLHCIVFNV | L411S | 1 | 4.03 |
| BKV T p558 | SLQNSEFLL | N561T | 1 | 2.1 |
| S562P | 1 | 3.36 | ||
| BKV T p570 | ILQSGMTLL | I570F | 2 | 2.05; 2.19 |
| Small T antigen | ||||
| BKV T p4 | VLNREESME | R7G | 1 | 1.8 |
| BKV T p10 | SMELMDLLGL | E12V | 1 | 9.28 |
| BKV T p125 | FLRKEPLVWI | R127G | 2 | 1.51; 1.86 |
| K128R | 1 | 1.85 |
Low-level variants of BKV HLA-B0702- and HLA-B08-restricted peptide sequences were not identified.
Variants that appeared in multiple specimens for a patient were averaged. Frequencies for each patient are listed.
DISCUSSION
We present a whole-genome, deep-sequencing methodology and informatics pipeline that provides comprehensive BKV sequence information to detect genetic variations in plasma and urine samples. This method offers excellent coverage and the ability to detect low-level variants within a virus population. The informatics pipeline and error model can accurately distinguish variants from noise, resulting in a sensitive, quantitative approach for detection of nucleotide and amino acid variants across the BKV genome.
Genetic variations of BKV have been evaluated in several studies, although most have characterized variations in specific BKV genes, particularly VP1 and the NCCR. In these studies, variants were described, but an association with end-organ disease was not always found. In one study, single base-pair mutations were detected more frequently in samples from patients with BK nephropathy, but amino acid changes were randomly distributed in BK nephropathy patients and controls (28). Other studies have identified amino acid changes in the external loops of VP1, which mediate BKV attachment to target cells but did not find an association with viral load or incidence of BK nephropathy (29, 30). Similarly, amplifications, deletions, and rearrangements in the NCCR have not been shown to influence the development of hemorrhagic cystitis or BK nephropathy in either HCT or kidney transplant recipients (27, 31). Several studies have defined genetic variation over the entire BKV genome via traditional Sanger sequencing, although these studies primarily assessed phylogenetic relationships between BKV strains for optimal classification schemas and characterization of circulating genotypes (32–34).
Using whole-genome, deep-sequencing we obtained complete BKV sequence information for pediatric HCT and kidney transplant recipients with a BKV infection. An analysis of the whole genome revealed no significant differences in amino acid variants across genes. The total number of variants detected was minimal, with most variants present at a frequency of <5%. Unlike prior studies, no NCCR rearrangements were noted in our samples. Given the limited frequency of BKV variants and the few patients in the study who developed hemorrhagic cystitis or BK nephropathy, no associations between variants and BK disease was made in our study.
We also evaluated for variants in CD8 T-cell epitopes and found limited variability in these peptide sequences. The predominant virus population in urine and plasma samples harbored T-cell epitopes identical to those of the reference BKV strain that was matched for the BKV genotype. Such limited variability may reflect the minimal selection pressure on BKV due to the lack of an effective antiviral immune response in an immunocompromised host. This is consistent with findings in studies evaluating intrahost diversity of other viruses which report increased viral diversity in response to immune pressure and decreased diversity with immunosuppression (35–37). However, despite the conservation of epitopes, we detected low levels of variants that resulted in amino acid changes in the majority of patient samples. Such BKV variants may encode peptides that escape immune recognition and subsequently emerge in the presence of selection pressure from immunotherapy.
The limited BKV epitope variability identified in this study supports the use of cytotoxic T cells that target consensus BKV peptides for immunotherapy. Consideration of the BKV genotype and infusion of T cells that recognize genotype-specific epitopes may optimize the effectiveness of T-cell therapy. Furthermore, an analysis of low-level BKV variants before and after T-cell therapy may provide insight on viral evolution and detection of escape variants, particularly if therapy fails to control persistent BKV infection.
Limitations exist in the whole-genome next-generation sequencing methodology used for this study. Higher error rates have been observed with the Ion Torrent PGM than with the Illumina MiSeq, highlighting the importance of the error model in distinguishing variants from error (38). Similar to other studies, we found that sequencing errors were more often insertions and deletions and primarily located in homopolymer regions (38, 39). While the use of long-range, inverse PCR specifically enriched samples for virus with intact genomes, fragmented genomes would not have amplified and, ultimately, were not evaluated. The relevance of such sequence data is unclear. Additionally, the read lengths and method of library preparation limits our ability to determine whether variants are present in the same virus. Thus, the frequencies of variants in a sample reflect the total viral population.
Although much work has been done to characterize BKV T-cell epitopes, knowledge of the full repertoire of epitopes remains incomplete and limited to certain HLA types. While we evaluated genetic variation in HLA-A2-, HLA-B0702-, and HLA-B08-restricted peptide sequences as reported in previous studies (24, 25), this is likely to represent a minority of existing T-cell epitopes, and further characterization of epitopes is needed. Moreover, genetic variation in epitopes may not necessarily result in peptides that escape cytotoxic T-cell recognition. Understanding the relationship between epitope variants and T cells that may have broad peptide recognition requires further study.
Here, we used next-generation sequencing to elucidate variants over the entire BKV genome and at CD8 T-cell epitopes in pediatric HCT and kidney transplant recipients with BKV infection. We describe a whole-genome, deep-sequencing methodology and informatics pipeline that can yield unique information on BKV population dynamics. A detailed understanding of the virus and its relationship with the host immune response promises to provide insights into novel therapeutic approaches that have the potential to more effectively control BKV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staffs of the Stanford Clinical Virology Laboratory, the Stanford Blood Center Histocompatibility, Immunogenetics and Disease Profiling Laboratory, and the Pediatric Kidney Transplant Clinic at Lucile Packard Children's Hospital for their continued diligent work and dedication to patient care. We also thank summer student Kory Wilkinson for his contributions to this project.
This research was supported by NIH training grant 5T32AI007502-19 (S.K.T.) and a generous gift from Beta Sigma Phi (S.K.T.), an international women's service organization. The Tissue Engineering Center of Excellence Program Endowment Fund, Lucile Packard Children's Hospital, also provided support for this work.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01385-15.
REFERENCES
- 1.Balba GP, Javaid B, Timpone JG Jr. 2013. BK polyomavirus infection in the renal transplant recipient. Infect Dis Clin North Am 27:271–283. doi: 10.1016/j.idc.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Funk GA, Gosert R, Comoli P, Ginevri F, Hirsch HH. 2008. Polyomavirus BK replication dynamics in vivo and in silico to predict cytopathology and viral clearance in kidney transplants. Am J Transplant 8:2368–2377. doi: 10.1111/j.1600-6143.2008.02402.x. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch HH. 2005. BK virus: opportunity makes a pathogen. Clin Infect Dis 41:354–360. doi: 10.1086/431488. [DOI] [PubMed] [Google Scholar]
- 4.Knowles WA, Pipkin P, Andrews N, Vyse A, Minor P, Brown DW, Miller E. 2003. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J Med Virol 71:115–123. doi: 10.1002/jmv.10450. [DOI] [PubMed] [Google Scholar]
- 5.Ginevri F, Azzi A, Hirsch HH, Basso S, Fontana I, Cioni M, Bodaghi S, Salotti V, Rinieri A, Botti G, Perfumo F, Locatelli F, Comoli P. 2007. Prospective monitoring of polyomavirus BK replication and impact of pre-emptive intervention in pediatric kidney recipients. Am J Transplant 7:2727–2735. doi: 10.1111/j.1600-6143.2007.01984.x. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch HH, Randhawa P. 2013. BK polyomavirus in solid organ transplantation. Am J Transplant 13(Suppl 4):179–188. doi: 10.1111/ajt.12110. [DOI] [PubMed] [Google Scholar]
- 7.Binggeli S, Egli A, Schaub S, Binet I, Mayr M, Steiger J, Hirsch HH. 2007. Polyomavirus BK-specific cellular immune response to VP1 and large T-antigen in kidney transplant recipients. Am J Transplant 7:1131–1139. doi: 10.1111/j.1600-6143.2007.01754.x. [DOI] [PubMed] [Google Scholar]
- 8.Schachtner T, Muller K, Stein M, Diezemann C, Sefrin A, Babel N, Reinke P. 2011. BK virus-specific immunity kinetics: a predictor of recovery from polyomavirus BK-associated nephropathy. Am J Transplant 11:2443–2452. doi: 10.1111/j.1600-6143.2011.03693.x. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt T, Adam C, Hirsch HH, Janssen MW, Wolf M, Dirks J, Kardas P, Ahlenstiel-Grunow T, Pape L, Rohrer T, Fliser D, Sester M, Sester U. 2014. BK polyomavirus-specific cellular immune responses are age-dependent and strongly correlate with phases of virus replication. Am J Transplant 14:1334–1345. doi: 10.1111/ajt.12689. [DOI] [PubMed] [Google Scholar]
- 10.Heslop HE, Slobod KS, Pule MA, Hale GA, Rousseau A, Smith CA, Bollard CM, Liu H, Wu MF, Rochester RJ, Amrolia PJ, Hurwitz JL, Brenner MK, Rooney CM. 2010. Long-term outcome of EBV-specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood 115:925–935. doi: 10.1182/blood-2009-08-239186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Micklethwaite KP, Clancy L, Sandher U, Hansen AM, Blyth E, Antonenas V, Sartor MM, Bradstock KF, Gottlieb DJ. 2008. Prophylactic infusion of cytomegalovirus-specific cytotoxic T lymphocytes stimulated with Ad5f35pp65 gene-modified dendritic cells after allogeneic hemopoietic stem cell transplantation. Blood 112:3974–3981. doi: 10.1182/blood-2008-06-161695. [DOI] [PubMed] [Google Scholar]
- 12.Gerdemann U, Katari UL, Papadopoulou A, Keirnan JM, Craddock JA, Liu H, Martinez CA, Kennedy-Nasser A, Leung KS, Gottschalk SM, Krance RA, Brenner MK, Rooney CM, Heslop HE, Leen AM. 2013. Safety and clinical efficacy of rapidly-generated trivirus-directed T cells as treatment for adenovirus, EBV, and CMV infections after allogeneic hematopoietic stem cell transplant. Mol Ther 21:2113–2121. doi: 10.1038/mt.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller K, Schachtner T, Sattler A, Meier S, Friedrich P, Trydzenskaya H, Hinrichs C, Trappe R, Thiel A, Reinke P, Babel N. 2011. BK-VP3 as a new target of cellular immunity in BK virus infection. Transplantation 91:100–107. doi: 10.1097/TP.0b013e3181fe1335. [DOI] [PubMed] [Google Scholar]
- 14.Kapusinszky B, Chen SF, Sahoo MK, Lefterova MI, Kjelson L, Grimm PC, Kambham N, Concepcion W, Pinsky BA. 2013. BK polyomavirus subtype III in a pediatric renal transplant patient with nephropathy. J Clin Microbiol 51:4255–4258. doi: 10.1128/JCM.01801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahoo MK, Lefterova MI, Yamamoto F, Waggoner JJ, Chou S, Holmes SP, Anderson MW, Pinsky BA. 2013. Detection of cytomegalovirus drug resistance mutations by next-generation sequencing. J Clin Microbiol 51:3700–3710. doi: 10.1128/JCM.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flaherty P, Natsoulis G, Muralidharan O, Winters M, Buenrostro J, Bell J, Brown S, Holodniy M, Zhang N, Ji HP. 2012. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res 40:e2. doi: 10.1093/nar/gkr861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullagh P, Nelder JA. 1989. Generalized linear models, 2nd ed Chapman and Hall, London, United Kingdom. [Google Scholar]
- 23.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res 17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mani J, Jin N, Schmitt M. 2014. Cellular immunotherapy for patients with reactivation of JC and BK polyomaviruses after transplantation. Cytotherapy, in press. doi: 10.1016/j.jcyt.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Melenhorst J, Hensel N, Rezvani K, Sconocchia G, Kilical Y, Hou J, Curfman B, Major E, Barrett AJ. 2006. T-cell responses to peptide fragments of the BK virus T antigen: implications for cross-reactivity of immune response to JC virus. J Gen Virol 87:2951–2960. doi: 10.1099/vir.0.82094-0. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr MJ, McCormack GP, Mutton KJ, Crowley B. 2006. Unique BK virus non-coding control region (NCCR) variants in hematopoietic stem cell transplant recipients with and without hemorrhagic cystitis. J Med Virol 78:485–493. doi: 10.1002/jmv.20566. [DOI] [PubMed] [Google Scholar]
- 28.Boldorini R, Allegrini S, Miglio U, Paganotti A, Veggiani C, Mischitelli M, Monga G, Pietropaolo V. 2009. Genomic mutations of viral protein 1 and BK virus nephropathy in kidney transplant recipients. J Med Virol 81:1385–1393. doi: 10.1002/jmv.21520. [DOI] [PubMed] [Google Scholar]
- 29.Tremolada S, Delbue S, Castagnoli L, Allegrini S, Miglio U, Boldorini R, Elia F, Gordon J, Ferrante P. 2010. Mutations in the external loops of BK virus VP1 and urine viral load in renal transplant recipients. J Cell Physiol 222:195–199. doi: 10.1002/jcp.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krautkrämer E, Klein TM, Sommerer C, Schnitzler P, Zeier M. 2009. Mutations in the BC-loop of the BKV VP1 region do not influence viral load in renal transplant patients. J Med Virol 81:75–81. doi: 10.1002/jmv.21359. [DOI] [PubMed] [Google Scholar]
- 31.Olsen GH, Hirsch HH, Rinaldo CH. 2009. Functional analysis of polyomavirus BK non-coding control region quasispecies from kidney transplant recipients. J Med Virol 81:1959–1967. doi: 10.1002/jmv.21605. [DOI] [PubMed] [Google Scholar]
- 32.Luo C, Bueno M, Kant J, Martinson J, Randhawa P. 2009. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol 83:2285–2297. doi: 10.1128/JVI.02180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chehadeh W, Nampoory MR. 2013. Genotypic diversity of polyomaviruses circulating among kidney transplant recipients in Kuwait. J Med Virol 85:1624–1631. doi: 10.1002/jmv.23639. [DOI] [PubMed] [Google Scholar]
- 34.Drew RJ, Walsh A, Laoi BN, Crowley B. 2012. Phylogenetic analysis of the complete genome of 11 BKV isolates obtained from allogenic stem cell transplant recipients in Ireland. J Med Virol 84:1037–1048. doi: 10.1002/jmv.23240. [DOI] [PubMed] [Google Scholar]
- 35.Grad YH, Newman R, Zody M, Yang X, Murphy R, Qu J, Malboeuf CM, Levin JZ, Lipsitch M, DeVincenzo J. 2014. Within-host whole-genome deep sequencing and diversity analysis of human respiratory syncytial virus infection reveals dynamics of genomic diversity in the absence and presence of immune pressure. J Virol 88:7286–7293. doi: 10.1128/JVI.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni YH, Chang MH, Chen PJ, Hsu HY, Lu TW, Lin KH, Lin DT. 1999. Decreased diversity of hepatitis C virus quasispecies during bone marrow transplantation. J Med Virol 58:132–138. doi:. [DOI] [PubMed] [Google Scholar]
- 37.Renzette N, Gibson L, Jensen JD, Kowalik TF. 2014. Human cytomegalovirus intrahost evolution-a new avenue for understanding and controlling herpesvirus infections. Curr Opin Virol 8:109–115. doi: 10.1016/j.coviro.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Hoecke S, Verhelst J, Vuylsteke M, Saelens X. 2015. Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing. BMC Genomics 16:79. doi: 10.1186/s12864-015-1284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.