Abstract
The currently available diagnostics for Clostridium difficile infection (CDI) have major limitations. Despite mounting evidence that toxin detection is paramount for diagnosis, conventional toxin immunoassays are insufficiently sensitive and cytotoxicity assays too complex; assays that detect toxigenic organisms (toxigenic culture [TC] and nucleic acid amplification testing [NAAT]) are confounded by asymptomatic colonization by toxigenic C. difficile. We developed ultrasensitive digital enzyme-linked immunosorbent assays (ELISAs) for toxins A and B using single-molecule array technology and validated the assays using (i) culture filtrates from a panel of clinical C. difficile isolates and (ii) 149 adult stool specimens already tested routinely by NAAT. The digital ELISAs detected toxins A and B in stool with limits of detection of 0.45 and 1.5 pg/ml, respectively, quantified toxins across a 4-log range, and detected toxins from all clinical strains studied. Using specimens that were negative by cytotoxicity assay/TC/NAAT, clinical cutoffs were set at 29.4 pg/ml (toxin A) and 23.3 pg/ml (toxin B); the resulting clinical specificities were 96% and 98%, respectively. The toxin B digital ELISA was 100% sensitive versus cytotoxicity assay. Twenty-five percent and 22% of the samples positive by NAAT and TC, respectively, were negative by the toxin B digital ELISA, consistent with the presence of organism but minimal or no toxin. The mean toxin levels by digital ELISA were 1.5- to 1.7-fold higher in five patients with CDI-attributable severe outcomes, versus 68 patients without, but this difference was not statistically significant. Ultrasensitive digital ELISAs for the detection and quantification of toxins A and B in stool can provide a rapid and simple tool for the diagnosis of CDI with both high analytical sensitivity and high clinical specificity.
INTRODUCTION
The recent increases in the global incidence and severity of Clostridium difficile infection (CDI) (1–3) are of major concern, and CDI now consumes substantial resources for diagnosis, treatment, and infection control (2, 4). A recent U.S. prevalence survey of health care-associated infections (HAI) (5) found that C. difficile was the most commonly reported pathogen, causing 12.1% of HAI. Despite available therapies, treatment failure and recurrence are common (1, 2, 6). The emergence of epidemic strains capable of toxin hyperproduction and increased disease severity/mortality (2, 7, 8) has further increased the urgency to improve methods for diagnosis and treatment. Accurate diagnosis remains the cornerstone of effective management.
Disease caused by C. difficile infection is due to the effects of two large protein exotoxins, toxins A and B (9, 10). While the presence of toxin is necessary to cause disease, the optimal method for diagnosing CDI remains controversial. Appropriate patient selection can improve diagnostic accuracy for all assays (2, 11). The classic gold standard stool assay, toxigenic culture (TC), is complex, lengthy, and unstandardized; furthermore, it is increasingly recognized that detection of a toxigenic organism, rather than the toxin itself, is suboptimal. Another classical method, the cell culture cytotoxicity assay, has also been used as a gold standard in comparisons of diagnostic assays (12). The cytotoxicity assay primarily detects toxin B, which is a far more potent cytotoxin than toxin A (13). Despite arguments that the detection of toxin in stool has highest clinical specificity and predictive value (2, 11, 12, 14), nucleic acid amplification testing (NAAT) for ultrasensitive detection of toxigenic organisms is increasingly employed for diagnosis. However, studies have repeatedly reported on NAAT-positive individuals who clinically would not be diagnosed with CDI (12, 15–18). Thus, by detecting C. difficile organisms, rather than toxins A and B, NAAT may lack specificity for clinical disease (2).
Despite the importance of toxin detection for CDI diagnosis, the current methods for toxin detection remain inadequate. While a cell cytotoxicity assay can have good analytical sensitivity (limit of detection [LOD], 1 to 10 pg/ml for toxin B spiked into buffer [19]), this assay (like TC) is laborious, slow, and unstandardized. Conventional qualitative toxin immunoassays are widely used (2) but have high LODs (e.g., ∼0.8 to 2.5 ng/ml) (20, 21) and poor sensitivity (52 to 75% versus TC and 72 to 83% versus cytotoxicity assay [22, 23]). Given these limitations, the field is poised for the development of a simple toxin detection test that combines high analytical sensitivity with the clinical specificity of toxin detection. Disease severity has been correlated with fecal toxin levels in some preliminary studies (24, 25); hence, a sensitive and quantitative toxin assay may be ideal for both diagnosis and the identification of those with more severe disease who need aggressive therapy.
We describe here the development and validation of ultrasensitive and quantitative digital ELISAs for the detection of toxins A and B based on single-molecule array (Simoa) technology (26, 27). Simoa technology is based on the high-efficiency capture and labeling of single protein molecules on paramagnetic beads and their detection in arrays of femtoliter-sized wells (26, 27). The ability to isolate and detect single protein molecules leads to dramatic improvements in sensitivity such that concentrations of proteins in femtograms per milliliter can be detected, typically 1,000-fold more sensitive than conventional ELISA. Digital ELISAs have been shown to be highly robust and reproducible for the detection of low levels of proteins in highly complex matrices, including cell culture supernatant, serum, plasma, and cerebrospinal fluid (CSF) (e.g., 26–29). Our assays represent the first application of Simoa for the measurement of proteins in stool specimens. These assays may provide clinical sensitivity equal to that of NAAT and TC, with higher clinical specificity, and thus offer the potential for improved approaches to diagnosis and management.
MATERIALS AND METHODS
Experimental design.
The objectives of this research project were to develop digital ELISAs for C. difficile toxins A and B with an analytical LOD of ∼1 pg/ml in stool and to clinically validate those digital ELISAs using clinical specimens originally submitted for C. difficile testing by NAAT, comparing digital ELISA results to the results of NAAT, toxigenic culture, and cytotoxicity assay. A second exploratory objective was to evaluate correlations between stool toxin concentrations and disease severity and outcomes.
Toxigenic culture (TC) is the most widely recognized laboratory gold standard for the diagnosis of CDI (2, 4), and thus our sample size calculations were built around the TC results. Our goal was to estimate digital ELISA sensitivities and specificities with a margin of error (determined by 95% confidence interval) for each of <10%. The target sensitivity and specificity for each digital ELISA were determined to be ≥95% for both, matching the reported sensitivity (90 to 95%) and specificity (94 to 96%) of NAAT compared to toxigenic culture (30–32). Assuming sensitivity in the range of ≥95% for each optimized digital ELISA, we determined that a sample size of 70 positive samples would give us a margin of error of ±5%, and 50 samples would give a margin of error of ±6%. However, we recognized that it was probable that toxigenic culture would overestimate actual toxin production in the host, and thus that the actual sensitivity of digital ELISA versus toxigenic culture might be lower. If the sensitivity of digital ELISA versus toxigenic culture were instead 85%, 70 true-positive samples would still give us a margin of error of ±8%. The same calculations applied to the needed numbers of negative specimens. The number of samples required for testing was further adjusted based on the expected yield of toxigenic culture in NAAT-positive versus NAAT-negative specimens.
The handling of outliers and numbers of replicates performed are described below and in the Results. Operators performing digital ELISAs, cytotoxicity assays, and TC were blinded to all other assay results for the stool specimens; those reviewing clinical data were blinded to all assay results other than the clinical result (NAAT).
Conventional ELISA.
Conventional plate- and bead-based ELISAs were used to screen antibodies for assay development and to test culture filtrates (CF). We ultimately selected one pair of monoclonal antibodies against toxin A (Meridian Life Sciences, Memphis, TN) and another pair against toxin B (bioMérieux, Lyon, France). For the bead-based ELISA, paramagnetic beads (5 × 106/ml) coated in capture antibodies were incubated with buffer containing purified native toxin A or B (prepared from C. difficile strain VPI 10463 in C.P.K's laboratory using established methods [33]) or CF in the wells of a microtiter plate for 2 h at room temperature. Captured toxin proteins were labeled with a biotinylated detection antibody (0.1 μg/ml) and an enzyme conjugate (0.5 nM streptavidin-β-galactosidase). Following washes, enzyme substrate was added to the microtiter plate wells, and fluorescent signals were measured using a Tecan plate reader.
Simoa assays (digital ELISAs).
Frozen aliquots of stool specimens were completely thawed at room temperature and mixed thoroughly either by vortexing or using a wooden applicator stick. Stool samples were then diluted and filtered to remove particulates before testing by digital ELISA.
Details of the Simoa technology used to develop and perform digital ELISAs have been described previously (26, 27). Antibody-coated capture beads and biotinylated detection antibodies were prepared using standard methods (26, 27). Digital ELISAs were performed on the Simoa HD-1 analyzer (Quanterix Corporation) (34) by automating the following steps. Capture beads (2.5 × 106/ml for both toxin A and B assays) were incubated with diluted stool samples for 15 min at 23°C. The beads were washed three times with 5× phosphate-buffered saline (PBS) plus 0.1% Tween 20. Captured toxin proteins were labeled with a biotinylated detection antibody (0.4 μg/ml for the A assay and 0.2 μg/ml for the B assay) and an enzyme conjugate (250 pM streptavidin-β-galactosidase). Following the addition of enzyme substrate, beads were loaded into arrays of femtoliter-sized wells for the isolation and detection of bound molecules. The total assay time was 69 min.
Simoa signals were quantified as the average enzymes per bead (AEB) (26, 27). Signals from the digital ELISAs were calibrated by spiking a series of known concentrations of purified native toxin A or B (prepared as previously described [33]) into either buffer or NAAT-negative stool samples processed as described above. Calibration curves were used to calculate toxin concentrations (in picograms per milliliter) in stool specimens. For each digital ELISA, all calibrators and samples were assayed in triplicate. For the calibrators, the average AEB, standard deviation (SD), and percent coefficient of variation (%CV) were calculated. For samples, the average, SD, and %CV were calculated for both AEB and measured toxin concentrations.
To evaluate assay accuracy, NAAT-negative stool samples were spiked (after dilution) with or without a known concentration of purified native toxin A or B, processed as described above, and tested by digital ELISA. The percent recovery of spiked toxins was calculated as follows: (measured concentration in spiked stool − measured concentration in nonspiked stool)/expected concentration of spiked toxins × 100.
Interfering factor analysis.
A panel of potential interfering substances was evaluated to determine the effect of their presence in stool specimens on the detection of toxin A or B using digital ELISA. Briefly, each of the potential interfering substances was individually spiked into diluted stool samples at targeted concentrations. These stool samples were then processed, and the concentration of toxin A or B was measured and compared to that of controls without added interferent. The tested substances included (concentrations noted are maximum final concentrations tested) vancomycin (100 mg/ml), metronidazole (40 mg/ml), loperamide HCl, salicylate, bismuth subsalicylate, Imodium (tablets or liquid; McNeil-PPC), Pepto-Bismol (tablets or liquid; Proctor & Gamble), barium sulfate, and 50% (vol/vol) human whole blood.
Culture filtrates.
A panel of clinical strains previously typed by restriction endonuclease analysis (REA) (provided by D.N.G.) were each cultured in chopped meat broth for 24 h. CFs were prepared by centrifugation at 3,000 × g to obtain supernatant, followed by passage through a 0.2-μm syringe filter. CFs were tested by a conventional bead-based ELISA, as described above.
Clinical stool specimens.
Clinical stool specimens (each collected from the patient <72 h prior and stored at 4°C) that had been tested for toxigenic C. difficile by NAAT (illumigene; Meridian Bioscience, Inc.) were aliquoted and frozen at −80°C. This study was approved by the Beth Israel Deaconess Medical Center (BIDMC) institutional review board (IRB) with a waiver of informed consent. Samples were obtained from adult inpatients and outpatients of both genders and were collected based on specimen volume/age and NAAT result. Fifty-two samples (11 NAAT positive and 41 NAAT negative) were used for assay development, and 149 additional samples (65 NAAT positive and 84 NAAT negative) were collected for assay validation. Stool consistency was scored at the time of collection by study staff as solid (n = 22), semisolid (n = 35), or liquid (n = 92). Separate aliquots of each validation specimen were subsequently thawed and tested by digital ELISA, cytotoxicity assay, and toxigenic culture (TC). Relevant clinical and laboratory data corresponding to each sample were collected by chart review. The presence or absence of diarrhea as documented at the time of sample collection was determined using the following definition: three or more unformed bowel movements during any 24-h period during the 48 h before the time of stool collection, or diarrhea specifically mentioned in medical doctor/registered nurse (MD/RN) progress notes as being present in the 48 h before stool collection. CDI was in turn defined as diarrhea plus positive results from one or more of the assays evaluated in this study: NAAT, toxin B digital ELISA, cytotoxicity assay, or toxigenic culture. A severe outcome potentially attributable to CDI was defined as intensive care unit (ICU) admission, colectomy, or death within 40 days of stool testing.
Cytotoxicity assay.
Cytotoxicity assays were performed using standard methods (19) in C.P.K.'s laboratory using stool samples diluted 1:100 in PBS, which were added to NIH 3T3 fibroblast cell monolayers and incubated for 24 h. A positive result required a typical cell rounding effect in >50% of the cells that was inhibited using specific antiserum to toxins A and B.
Toxigenic culture.
Toxigenic culture was performed using standard well-established methods (35) in D.N.G.'s laboratory. Stool samples were directly inoculated onto selective taurocholate-cefoxitin-cycloserine-fructose-agar (TCCFA) plates (36). If initial TCCFA culture failed to yield colonies with characteristic C. difficile morphology, an alcohol shock protocol (37) was performed on the stool sample to enhance the recovery of C. difficile spores. All C. difficile isolates were evaluated for their ability to produce toxin in vitro by culturing in brain heart infusion (BHI) medium for 48 to 72 h and testing supernatants in the Bartels cell cytotoxicity assay (MarDx Diagnostics, Inc., Carlsbad, CA). Samples that yielded a C. difficile isolate that did not produce toxin were scored as TC negative. All C. difficile isolates recovered from C. difficile culture were REA typed in D.N.G.'s laboratory using established methods (38, 39).
Statistical analysis.
During sample analysis, measured toxin levels for stool specimens were extrapolated from the calibration curve based upon a 4-parameter logistic (4PL) nonlinear regression model (1/y2 weighted). The analytical LOD, an interpolated toxin concentration corresponding to the Simoa signal (AEB) for blank (buffer) plus 3 SD, was reported as the toxin level for samples that were not detectable by Simoa assays. For samples that gave saturated Simoa signals (out of the dynamic range of the assay), a toxin concentration of 12,000 pg/ml (postdilution correction) corresponding to the upper dynamic range of both assays was reported. Clinical sensitivity was determined by measuring the percentage of positives by a reference method (TC, cytotoxicity assay, or NAAT) that were identified as positive by Simoa assays, and clinical specificity was determined by measuring the percentage of negatives by a reference method (TC, cytotoxicity assay, or NAAT) that were identified as negative by Simoa assays. A 95% confidence interval (CI) was also calculated for each estimation using this equation: P ± 1.96 [P × (1 − P)/N]1/2, where P is sensitivity (or specificity) measured as a proportion, and N is the number of true-positive or -negative samples by a given reference method. Statistical analysis for clinical correlation was performed using Stata (version 12; StataCorp LP, TX, USA). Continuous variables were expressed as the mean ± standard deviation (SD). Mean toxin levels ([toxin A], [toxin B], or [toxin A + B]) in subjects with CDI-attributable severe outcomes were compared to mean toxin levels in subjects without CDI-attributable severe outcomes by Student's t test.
RESULTS
Digital ELISA development and limits of detection in buffer.
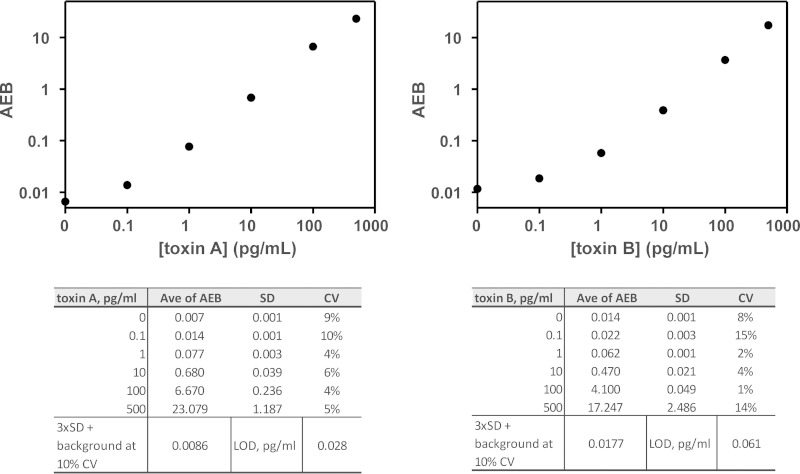
We first screened a panel of antibodies by ELISA to find pairs that detected purified native toxin A or B with highest analytical sensitivity and specificity (optimal signal/background). We then used culture filtrates (CF) prepared (see Materials and Methods) from a panel of 12 clinical C. difficile isolates representing the major strains in circulation (provided by D.N.G.) to confirm that our toxin A and B monoclonal antibody pairs were able to detect the toxins produced by all 12 isolates (Table 1). After optimization of the digital ELISAs, the typical limit of detection (LOD) for native toxin spiked into calibration buffer was 0.028 pg/ml for toxin A and 0.061 pg/ml for toxin B (Fig. 1).
TABLE 1.
Detection of C. difficile toxins A and B by conventional bead-based ELISA in culture filtrates prepared from a panel of C. difficile isolates
| Culture filtrate | REA type/strain informationa | Expected toxin production | Toxin A detection by bead-based ELISA | Toxin B detection by bead-based ELISA |
|---|---|---|---|---|
| 1 | B1 | A+/B+ | + | + |
| 2 | J9 | A+/B+ | + | + |
| 3 | K14 | A+/B+ | + | + |
| 4 | Y2 | A+/B+ | + | + |
| 5 | CF2 | A−/B+ | NDb | + |
| 6 | ATCC BAA-1801 | None | ND | ND |
| 7 | Medium | None | ND | ND |
| 8 | VPI10463 | A+/B+ | + | + |
| 9 | R23 | Sequenced A+/B+ strain | + | + |
| 10 | BI1 (historic NAP1) | A+/B+/binary+ | + | + |
| 11 | BI6 (epidemic NAP1) | A+/B+/binary+ | + | + |
| 12 | BI17 (epidemic NAP1) | A+/B+/binary+ | + | + |
| 13 | BK1 (ribotype 078) | A+/B+/binary+ | + | + |
REA, restriction endonuclease analysis.
ND, not detected.
FIG 1.
Representative calibration curves for toxin A and B assays. The LOD, defined as a concentration corresponding to the average enzymes per bead (AEB) from the zero calibrator plus 3 SD, was 0.028 pg/ml for toxin A (left) and 0.061 pg/ml for toxin B (right) from the presented curves, respectively, using a 10% CV for the zero calibrator.
Limits of detection in stool.
To measure the LOD in stool, we spiked native purified toxins A and B into clinical stool samples that had tested negative by NAAT (illumigene; Meridian Bioscience) during routine clinical testing. Stool samples were diluted prior to spiking and then processed as described in Materials and Methods. The LOD in stool was defined as an interpolated concentration corresponding to an average AEB from NAAT-negative stools plus 3 SD, corrected for the dilution factor. Our assays detected native toxins in stool with LODs of 0.45 pg/ml (toxin A) and 1.50 pg/ml (toxin B), respectively. The percent recovery ranged from 50 to 125% (mean, 84%). The mean %CV of Simoa signal (AEB) was 8%; this imprecision translated into a mean imprecision of 15% in measured toxin concentrations for all stool specimens analyzed during this study, including those close to the LOD that have intrinsically higher imprecision. Of note, in ∼6% of the samples, we observed aggregation of the paramagnetic beads (after incubation in the stool samples) that prevented measurement using Simoa. This limitation was overcome by using a modified dilution buffer for those samples that caused aggregation.
Potential interfering factors.
A panel of potential interfering substances (at concentrations at or above those tested for commercial enzyme immunoassays [EIAs] [see, e.g., references 40, 41]) was tested in our assay (see Materials and Methods). We saw no evidence of interference from excess levels of vancomycin, metronidazole, loperamide HCl, salicylate, bismuth subsalicylate, Imodium (tablets or liquid), Pepto-Bismol (tablets or liquid), barium sulfate, or 50% (vol/vol) whole blood.
Clinical validation.
One hundred forty-nine clinical specimens (<72 h old) that had tested either positive (n = 65) or negative (n = 84) by NAAT during routine clinical testing were tested with the two digital ELISAs (each run in triplicate and averaged), cytotoxicity assay, and TC (with TC followed by REA typing of all C. difficile isolates). Of the 149 patients providing these samples, 71 were male and 78 were female; age ranged from 19 to 97 years (mean, 58 years; median, 58 years), and 109 (73%) were inpatients.
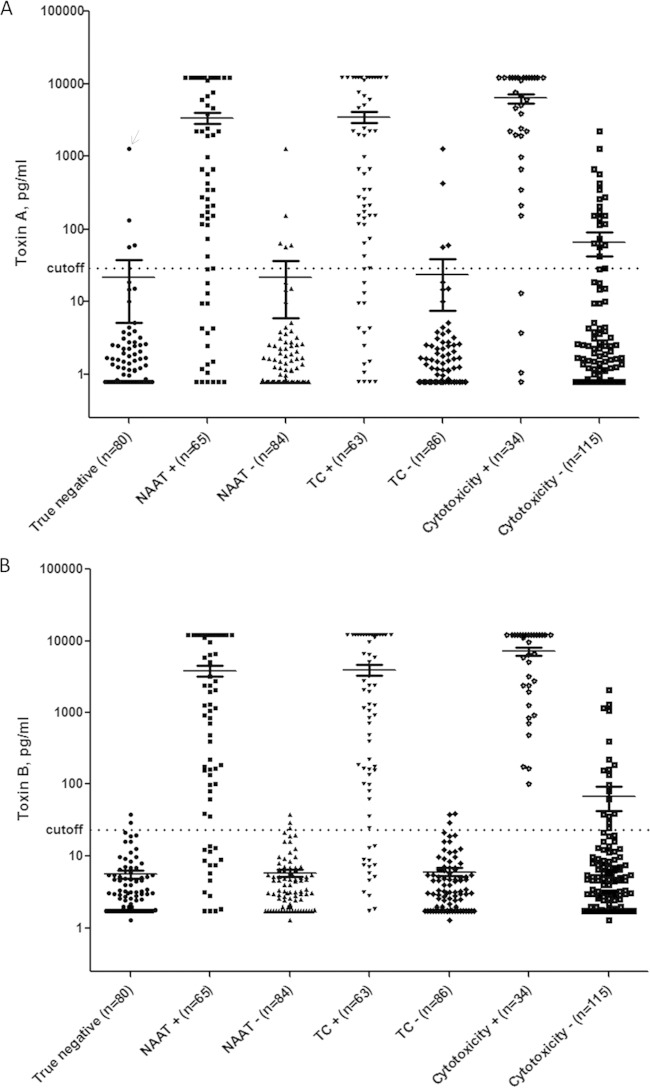
A clinical cutoff for a positive result for each digital ELISA was calculated by averaging the Simoa signal (AEB) for true-negative stool samples (n = 80; negative by NAAT, TC, and cytotoxicity assay; Fig. 2A and B) plus 3 standard deviations (SD) of that mean, and then converting AEB into picograms per milliliter using a comprehensive calibration curve. We excluded one of the 80 true-negative samples from our calculation of the cutoff because it was an extreme outlier on the toxin A assay (Fig. 2A, marked with arrow) and substantially distorted the mean for that assay. Using the remaining 79 true negatives, the calculated cutoff for a positive result was 29.4 pg/ml for the toxin A assay and 23.3 pg/ml for the toxin B assay; with these cutoffs, the toxin A and B digital ELISA specificities in the true-negative group were 96% and 98%, respectively.
FIG 2.
Toxin A and B digital ELISA results. Toxin A (A) and toxin B (B) digital ELISA results for groups of samples testing positive versus negative on other assays (i.e., NAAT positive versus NAAT negative, TC positive versus TC negative, and cytotoxicity positive versus cytotoxicity negative). The mean signals in each group are indicated by horizontal lines. The calculated clinical cutoffs for each digital ELISA (29.4 pg/ml for the toxin A assay and 23.3 pg/ml for the toxin B assay) are shown as dotted lines spanning each figure. (A) Arrow indicates a sample that was excluded from calculation of the cutoff because it was an extreme outlier and substantially distorted the mean for that assay. NAAT, nucleic acid amplification testing; TC, toxigenic culture.
Toxin concentrations (in picograms per milliliter) measured by digital ELISA spanned a >4-log dynamic range (Fig. 2A and B). Plots of the toxin A and B digital ELISA results for groups of samples testing positive versus negative on other assays (i.e., NAAT positive versus NAAT negative, TC positive versus TC negative, and cytotoxicity positive versus cytotoxicity negative) are shown in Fig. 2A and B. As expected, for the toxin B digital ELISA, 16/65 (25%) samples that were positive by NAAT and 14/63 (22%) samples that were positive by TC were negative by the toxin B digital ELISA, consistent with the presence of organism (but minimal or no toxin) (Fig. 2B). In contrast, 34/34 (100%) samples positive by the cytotoxicity assay were positive by the toxin B digital ELISA. As expected based on the LODs, we noted some overlap between the toxin B digital ELISA signals in cytotoxicity-positive versus cytotoxicity-negative samples in the range of ∼102 to 2,078 pg/ml (Fig. 2B). The results from the toxin A digital ELISA (Fig. 2A) revealed four samples for which the cytotoxicity assay and TC were positive but toxin A digital ELISA was negative. REA typing of isolates obtained from TC confirmed that all four isolates were REA type CF, a group of isolates that produce toxin B but not toxin A. TC and the cytotoxicity assay were used as alternative laboratory gold standards for calculations of digital ELISA sensitivity and specificity (Table 2); for comparison, we also calculated the sensitivity and specificity of each digital ELISA versus NAAT (Table 2).
TABLE 2.
Sensitivity and specificity of the toxin A and B digital ELISAs as compared to three alternative reference standardsa
| Digital ELISA for toxin: | Sensitivity vs reference standard shown |
Specificity vs reference standard shown |
||||
|---|---|---|---|---|---|---|
| Toxigenic culture | Cytotoxicity assay | NAAT | Toxigenic culture | Cytotoxicity assay | NAATb | |
| A | 75 ± 10.7 | 88 ± 10.9 | 71 ± 11.0 | 95 ± 4.6 | 84 ± 6.9 | 94 ± 4.2 |
| B | 78 ± 10.2 | 100 ± 0.0 | 75 ± 10.5 | 97 ± 3.6 | 87 ± 6.3 | 96 ± 4.2 |
Data are presented as % ± 95% CI.
NAAT, nucleic acid amplification testing.
Correlation of toxin detection with disease and disease severity.
Among the 73 patients whose stool tested positive by any of the assays used (toxin A and B digital ELISAs, NAAT, TC, or cytotoxicity assay), there were 8 subjects who had a severe outcome potentially attributable to CDI, as defined by ICU admission, colectomy, or death within 40 days of stool testing. A detailed chart review (by two clinicians blinded to stool assay results other than NAAT) indicated that 5/8 severe outcomes (3 ICU admissions, 1 colectomy, and 1 death) were likely attributable to CDI. Mean toxin levels ([toxin A], [toxin B], or [toxin A + B]) in the 5 subjects with CDI-attributable severe outcomes were higher (1.7-fold, 1.5-fold, and 1.6-fold, respectively) than the mean toxin levels in the 68 subjects without CDI-attributable severe outcomes, although these trends did not reach statistical significance (P = 0.10, 0.18, and 0.08, respectively). No significant correlations between toxin concentration and peak white blood cell (WBC) count (P = 0.965) or peak creatinine (Cr) level (P = 0.966) (each measured as peak in the window from 5 days before to 2 days after stool sample collection date) were observed.
DISCUSSION
Within the controversial, complex, and rapidly shifting C. difficile diagnostic landscape, one major theme is emerging: that detection of toxins, rather than of bacteria capable of producing those toxins, is central to the accurate diagnosis of CDI (2, 12, 14, 42). Qualitative enzyme immunoassays (EIAs) that detect C. difficile toxins in stool were, for many years, the mainstay of diagnosis, used by >90% of laboratories in the United States (2). Compared to TC, however, these assays show limited sensitivity (52 to 75% [22, 23]); consistent with this finding, the analytical LODs in stool for some of the highest-performing EIAs (23) are ∼1 ng/ml (0.8 to 2.5 ng/ml) (20, 21). Ryder et al. (25) described a cell-based assay (that has not progressed to clinical use) for the quantification of toxin in stool and calculated stool toxin concentrations as low as 30 pg/ml. Their data indicated that almost half of the toxin-positive specimens would not be detected by EIAs with LODs of ∼1 ng/ml. However, EIAs have been shown to have high specificity versus TC (96 to 98% [22, 23]) and high clinical specificity (42, 43).
The cell culture cytotoxicity assay for the direct detection of toxin in stool is reemerging as a reference assay that is favored over TC (12). Cytotoxicity assays have LODs below those of EIA (e.g., 1.5 pg/ml for toxin B in buffer [19]) but are complex and lengthy, making them unsuitable for routine diagnostic use. Moreover, in contrast to its low LOD in buffer, we observed that the cytotoxicity assay appeared to have varied sensitivity for the detection of toxin B in stool (Fig. 2B). It is possible that toxins in the samples, while detectable by digital ELISA, may not have been biologically active at the time of testing (further, we acknowledge that the clinical significance of toxin detection by the digital ELISA in cytotoxicity assay-negative samples is unknown). One large comparison study found the cytotoxicity assay to be approximately 86% sensitive compared to TC (23), and the question has been raised as to whether this difference is due to the lower sensitivity of the cytotoxicity assay or the detection, by TC, of C. difficile organisms in the absence of toxin production (14). Consistent with the second possibility, it was reported recently that C. difficile toxin production is regulated by quorum sensing, whereby toxin synthesis may be absent at low bacterial concentrations (44). Hence, highly sensitive methods to detect C. difficile spores and DNA may yield positive results in the absence of toxin production. A recent United Kingdom-based study (12) compared TC with cytotoxicity testing on >12,000 specimens and correlated the results with clinical data. While positive cytotoxicity assay results correlated with increased mortality, TC-positive/cytotoxicity assay-negative results did not, indicating that the presence of appreciable amounts of toxin (and not just the presence of toxigenic C. difficile) was of primary importance. The authors concluded that the “detection of toxin is an essential step in the diagnosis of C. difficile infection” (12).
Given the limitations of the existing toxin detection assays, there has been a shift toward diagnosis by NAAT for the tcdA and tcdB genes, with its potential for high sensitivity and rapid turnaround time (but with higher expense). Frustratingly, despite high sensitivity and specificity versus TC (90 to 95% and 94 to 96%, respectively [30–32]), NAAT is confounded by its inability to distinguish disease from colonization or even transient contamination with environmental spores (14). The positive predictive values for the commercial NAATs (even versus TC) are low (71 to 79% for prevalence ≤20% [32, 45]), in contrast to their high negative predictive values (98 to 99% in the same studies). The problem remains that, like TC, NAAT indicates the presence of C. difficile organisms capable of producing toxin but not the presence of actual fecal toxin. Importantly, a strain's capacity for toxin production in vitro may not reflect that strain's production of toxins in the highly variable in vivo environment. Akerlund et al. (24) demonstrated no correlation between fecal toxin levels and toxin yields in vitro for given isolates or between in vitro yields and disease severity. Moreover, the toxin genes are under complex regulatory control, and the expression of toxin proteins is impacted by numerous environmental factors, including temperature, carbon source/amino acid availability, and antibiotic concentration (46, 47). While the detection of toxigenic organisms may be appropriate for determining the need for infection control measures (45), it is not necessarily optimal for deciding whether or not toxigenic C. difficile is the cause of a patient's symptoms (16). Notably, multiple studies have reported NAAT-positive individuals who clinically did not have CDI (12, 15–18).
Until now, no highly sensitive assay existed that could rapidly detect and quantify both toxins A and B in stool samples at the time of diagnosis. Our assays have high analytical sensitivity and specificity and, given that we are detecting toxin directly, high clinical specificity—cumulative advantages that may provide higher diagnostic, and even prognostic, accuracy over that of existing assays. As expected, our analytical sensitivity (∼1 pg/ml, determined using NAAT-negative samples with uniformly low background) was lower than our final clinical cutoffs (∼20 pg/ml, determined using a large number of NAAT-negative samples with varied background, i.e., more representative of “real-life” sample quality). Using our clinical cutoff, our toxin B assay has 100% sensitivity versus cytotoxicity assay, 97% specificity versus TC, and 98% specificity in samples negative by NAAT, TC, and cytotoxicity assay. As expected, 22 to 25% of NAAT-positive and TC-positive samples were negative for toxin B by our ultrasensitive digital ELISA, suggesting that these samples contained toxigenic organisms but minimal or no toxin (see Table 2). These patients would potentially fall into the category of “C. difficile excretor” suggested by Planche et al. (12); however, our study design was such that we cannot prove that these patients (NAAT positive/TC positive and digital ELISA negative) did not have CDI. Future studies should focus on determining the clinical diagnostic and prognostic value of ultrasensitive detection and quantification of stool toxin (by digital ELISA) in symptomatic patients as well as the clinical significance of NAAT or TC positivity in the absence of detectable toxin.
A final advantage of these assays over existing methods is the ability to separately detect and quantify both toxins A and B. While each toxin has been shown to be independently capable of causing disease, their relative contributions remain unclear (e.g. 9, 10, 48, 49). Our results indicate a trend for patients with severe CDI to have higher stool toxin levels. However, our study was not designed or powered to draw firm conclusions on this issue; larger studies are needed to answer this important question. If toxin levels are shown to correlate with disease severity, response to treatment, and/or risk for recurrence, the quantification of toxin might provide critical information to guide management decisions.
In conclusion, we have successfully developed ultrasensitive and quantitative digital ELISAs for C. difficile toxins A and B. Our assays detect native toxins A and B in stool with analytical LODs that are ∼1,000-fold more sensitive than those of current EIAs, can quantify toxin across a 4-log range, and detect toxins from all major clinical isolates tested. This is the first application of Simoa technology to the measurement of proteins in stool specimens; our results indicate that this novel technology is likely to prove valuable for other applications involving the measurement of fecal proteins. Many complex and important questions remain regarding these toxins and the overall pathogenesis of CDI, and our assays provide a new tool with which to address these questions. Moreover, our assays offer the potential for a future paradigm shift in how CDI is diagnosed and managed.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (grant 5 R21AI103612-02, to N.R.P., C.P.K., and D.C.D.). Monoclonal antibodies to toxin B were provided by bioMérieux.
C.P.K. is a consultant of MedImmune, Stellar Biotechnologies, and Synthetic Biologics, a member of the advisory boards of Cubist/Merck, OpenBiome, Sanofi Pasteur, and Seres Health and holds a subcontract through Claremont BioSolutions for research funding from the National Institutes of Health. D.N.G holds patents for the prevention of CDI licensed to ViroPharma/Shire, is a consultant for ViroPharma/Shire, MedImmune, Sanofi Pasteur, and Pfizer, and is a member of the advisory boards of Rebiotix, Merck, Summit, and Actelion. L.S., M.Z., and D.C.D. are paid employees of Quanterix Corporation. D.C.D. owns stock in Quanterix Corporation. N.R.P., J.H., K.S., M.W., X.C., H.X., D.A.L., and S.P.S. declare no conflicts of interest.
REFERENCES
- 1.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH, Society for Healthcare Epidemiology of America, Infectious Diseases Society of America. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 3.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 4.Schutze GE, Willoughby RE, Committee on Infectious Diseases. 2013. Clostridium difficile infection in infants and children. Pediatrics 131:196–200. doi: 10.1542/peds.2012-2992. [DOI] [PubMed] [Google Scholar]
- 5.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamboj M, Khosa P, Kaltsas A, Babady NE, Son C, Sepkowitz KA. 2011. Relapse versus reinfection: surveillance of Clostridium difficile infection. Clin Infect Dis 53:1003–1006. doi: 10.1093/cid/cir643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald LC, Killgore GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 8.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 9.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 11.Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CA, Dunne WM Jr. 2011. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol 49:2887–2893. doi: 10.1128/JCM.00891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O'Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. 2013. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect Dis 13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X, Wang J, Steele J, Sun X, Nie W, Tzipori S, Feng H. 2009. An ultrasensitive rapid immunocytotoxicity assay for detecting Clostridium difficile toxins. J Microbiol Methods 78:97–100. doi: 10.1016/j.mimet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilcox MH, Planche T, Fang FC, Gilligan P. 2010. What is the current role of algorithmic approaches for diagnosis of Clostridium difficile infection? J Clin Microbiol 48:4347–4353. doi: 10.1128/JCM.02028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ota KV, McGowan KL. 2012. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile Nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol 50:1185–1188. doi: 10.1128/JCM.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koo HL, Van JN, Zhao M, Ye X, Revell PA, Jiang ZD, Grimes CZ, Koo DC, Lasco T, Kozinetz CA, Garey KW, DuPont HL. 2014. Real-time polymerase chain reaction detection of asymptomatic Clostridium difficile colonization and rising C. difficile-associated disease rates. Infect Control Hosp Epidemiol 35:667–673. doi: 10.1086/676433. [DOI] [PubMed] [Google Scholar]
- 17.Leibowitz J, Soma VL, Rosen L, Ginocchio CC, Rubin LG. 2015. Similar proportions of stool specimens from hospitalized children with and without diarrhea test positive for Clostridium difficile. Pediatr Infect Dis J 34:261–266. doi: 10.1097/INF.0000000000000556. [DOI] [PubMed] [Google Scholar]
- 18.Kelly CP, LaMont JT. 2008. Clostridium difficile–more difficult than ever. N Engl J Med 359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 19.Kelly CP, Pothoulakis C, Vavva F, Castagliuolo I, Bostwick EF, O'Keane JC, Keates S, LaMont JT. 1996. Anti-Clostridium difficile bovine immunoglobulin concentrate inhibits cytotoxicity and enterotoxicity of C. difficile toxins. Antimicrob Agents Chemother 40:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meridian Bioscience, Inc. 2009. Premier toxins A & B package insert. Meridian Bioscience, Inc., Nice, France: http://www.meridianbioscience.com/Content/Assets/Files/2.1%20%20C.%20difficile%20Products/Package-Insert-Premier-Toxins-A-and-B.pdf. [Google Scholar]
- 21.TechLab. 2008. C. difficile Tox A/B II package insert. TechLab, Inc., Blacksburg, VA: http://www.techlab.com/wp-content/uploads/2013/06/t5015insert_rev_0308.pdf. [Google Scholar]
- 22.Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect 15:1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 23.Eastwood K, Else P, Charlett A, Wilcox M. 2009. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol 47:3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akerlund T, Svenungsson B, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J Clin Microbiol 44:353–358. doi: 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryder AB, Huang Y, Li H, Zheng M, Wang X, Stratton CW, Xu X, Tang YW. 2010. Assessment of Clostridium difficile infections by quantitative detection of tcdB toxin by use of a real-time cell analysis system. J Clin Microbiol 48:4129–4134. doi: 10.1128/JCM.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. 2010. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol 28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rissin DM, Fournier DR, Piech T, Kan CW, Campbell TG, Song L, Chang L, Rivnak AJ, Patel PP, Provuncher GK, Ferrell EP, Howes SC, Pink BA, Minnehan KA, Wilson DH, Duffy DC. 2011. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal Chem 83:2279–2285. doi: 10.1021/ac103161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson DH, Hanlon DW, Provuncher GK, Chang L, Song L, Patel PP, Ferrell EP, Lepor H, Partin AW, Chan DW, Sokoll LJ, Cheli CD, Thiel RP, Fournier DR, Duffy DC. 2011. Fifth-generation digital immunoassay for prostate-specific antigen by single molecule array technology. Clin Chem 57:1712–1721. doi: 10.1373/clinchem.2011.169540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song L, Hanlon DW, Chang L, Provuncher GK, Kan CW, Campbell TG, Fournier DR, Ferrell EP, Rivnak AJ, Pink BA, Minnehan KA, Patel PP, Wilson DH, Till MA, Faubion WA, Duffy DC. 2011. Single molecule measurements of tumor necrosis factor alpha and interleukin-6 in the plasma of patients with Crohn's disease. J Immunol Methods 372:177–186. doi: 10.1016/j.jim.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Cepheid, Inc. 2012. GeneXpert C. difficile package insert. Cepheid, Inc. [Google Scholar]
- 31.Meridian Bioscience, Inc. 2011. illumigene C. difficile package insert. Meridian Bioscience, Inc., Nice, France: http://www.meridianbioscience.com/Content/Assets/PackInsert/8_5_x_11_CLEAN_SN11176_280050_illumigene_C_difficile_REV_08-13_.pdf. [Google Scholar]
- 32.Deshpande A, Pasupuleti V, Rolston DD, Jain A, Deshpande N, Pant C, Hernandez AV. 2011. Diagnostic accuracy of real-time polymerase chain reaction in detection of Clostridium difficile in the stool samples of patients with suspected Clostridium difficile Infection: a meta-analysis. Clin Infect Dis 53:e81–90. doi: 10.1093/cid/cir505. [DOI] [PubMed] [Google Scholar]
- 33.Kyne L, Warny M, Qamar A, Kelly CP. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397. [DOI] [PubMed] [Google Scholar]
- 34.Kan CW, Rivnak AJ, Campbell TG, Piech T, Rissin DM, Mösl M, Peterça A, Niederberger HP, Minnehan KA, Patel PP, Ferrell EP, Meyer RE, Chang L, Wilson DH, Fournier DR, Duffy DC. 2012. Isolation and detection of single molecules on paramagnetic beads using sequential fluid flows in microfabricated polymer array assemblies. Lab Chip 12:977–985. doi: 10.1039/C2LC20744C. [DOI] [PubMed] [Google Scholar]
- 35.Tenover FC, Akerlund T, Gerding DN, Goering RV, Bostrom T, Jonsson AM, Wong E, Wortman AT, Persing DH. 2011. Comparison of strain typing results for Clostridium difficile isolates from North America. J Clin Microbiol 49:1831–1837. doi: 10.1128/JCM.02446-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol 15:443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clabots CR, Gerding SJ, Olson MM, Peterson LR, Gerding DN. 1989. Detection of asymptomatic Clostridium difficile carriage by an alcohol shock procedure. J Clin Microbiol 27:2386–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clabots CR, Johnson S, Bettin KM, Mathie PA, Mulligan ME, Schaberg DR, Peterson LR, Gerding DN. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J Clin Microbiol 31:1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrella LA, Sambol SP, Cheknis A, Nagaro K, Kean Y, Sears PS, Babakhani F, Johnson S, Gerding DN. 2012. Decreased cure and increased recurrence rates for Clostridium difficile infection caused by the epidemic C. difficile BI strain. Clin Infect Dis 55:351–357. doi: 10.1093/cid/cis430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.bioMérieux. 2011. Vidas C. difficile Toxin A & B package insert, bioMérieux, Marcy l'Etoile, France. [Google Scholar]
- 41.Meridian Bioscience, Inc. 2009. ImmunoCard toxins A & B, package insert. Meridian Bioscience, Inc., Nice, France: http://www.meridianbioscience.com/Content/Assets/Files/2.1%20%20C.%20difficile%20Products/Package-Insert-ImmunoCard-Toxins-A-and-B.pdf. [Google Scholar]
- 42.Polage CR, Chin DL, Leslie JL, Tang J, Cohen SH, Solnick JV. 2012. Outcomes in patients tested for Clostridium difficile toxins. Diagn Microbiol Infect Dis 74:369–373. doi: 10.1016/j.diagmicrobio.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker I, Leeming JP, Reynolds R, Ibrahim I, Darley E. 2013. Clinical relevance of a positive molecular test in the diagnosis of Clostridium difficile infection. J Hosp Infect 84:311–315. doi: 10.1016/j.jhin.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. 2015. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6(2):e02569-14. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong E, Marlowe EM, Whitmore J, Persing DH. 2010. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol 48:3719–3724. doi: 10.1128/JCM.00427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antunes A, Dupuy B. 2010. Molecular methods to study transcriptional regulation of Clostridium difficile toxin genes. Methods Mol Biol 646:93–115. doi: 10.1007/978-1-60327-365-7_7. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson S, Burman LG, Akerlund T. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- 48.Kader HA, Piccoli DA, Jawad AF, McGowan KL, Maller ES. 1998. Single toxin detection is inadequate to diagnose Clostridium difficile diarrhea in pediatric patients. Gastroenterology 115:1329–1334. doi: 10.1016/S0016-5085(98)70009-5. [DOI] [PubMed] [Google Scholar]
- 49.Riegler M, Sedivy R, Pothoulakis C, Hamilton G, Zacherl J, Bischof G, Cosentini E, Feil W, Schiessel R, LaMont JT. 1995. Clostridium difficile toxin B is more potent than toxin A in damaging human colonic epithelium in vitro. J Clin Invest 95:2004–2011. doi: 10.1172/JCI117885. [DOI] [PMC free article] [PubMed] [Google Scholar]